Abstract
The ribosomal protein (RP)–p53 pathway has been shown to play a key role in apoptosis and senescence of cancer cells. miR-1908 is a newly found miRNA that was reported to have prognostic potential in melanoma. However, its role and mechanism in the progression of non-small cell lung cancer (NSCLC) are largely unknown. In this study, we found that expression of miR-1908 was significantly downregulated in human NSCLC cell lines, including SK-MES-1, A549, and NCI-H460. Then the role of miR-1908 in NSCLC cell proliferation was explored. The miR-1908 mimic was transfected into NSCLC cell lines, and their proliferation was detected. MTT and Cell Titer-Blue H analyses showed that the cell proliferation was notably reduced by the miR-1908 mimic transfection. Moreover, we found the RP–p53 pathway was activated by miR-1908 mimic. Moreover, the miR-1908 inhibitor transfection had a completely opposite effect on the NSCLC cell proliferation than that of miR-1908 mimic. To explore the underlying mechanism of that, TargetScan bioinformatics server and 3′-UTR luciferase reporter assay were applied to identify the targets of miR-1908. Our results showed that AKT1 substrate 1 (AKT1S1), a newly proven suppressor of the RP–p53 pathway, was a target of miR-1908, suggesting a probable mechanism for miR-191 suppressing NSCLC cell proliferation. Our findings provide a novel molecular target for the regulation of NSCLC cell proliferation.
Key words: Non-small cell lung cancer (NSCLC), miR-1908, AKT1 substrate 1 (AKT1S1), Ribosomal protein (RP)–p53 pathway, Cell proliferation
INTRODUCTION
Lung cancer is one of the leading causes of cancer death in the world, especially in China during the past decade (1,2). Non-small cell lung cancer (NSCLC) accounts for the majority of all lung cancer cases, including squamous cell carcinoma, adenocarcinoma, and large cell carcinoma (3). The growth and division of NSCLC are much quicker than small cell lung cancer, and diffusion and metastasis are so early that the diagnosis of most of NSCLC cases is confirmed in the middle and advanced stages (3,4). Therefore, it is of significance to identify novel genes or factors that regulate NSCLC progression.
The Ser/Thr protein kinase Akt is a key regulator that can inhibit apoptosis and promote survival in multiple types of cells (5). In response to growth signals, Akt phosphorylates a wide array of substrates to activate the mechanistic target of rapamycin (mTOR) through two pathways (6). In the first pathway, Akt phosphorylates and inhibits the GTPase-activating protein TSC2, thus making the GTPase Rheb remain GTP bound and activate mTORC1 (7). The second pathway depends on the proline-rich Akt substrate of 40 kDa (PRAS40, also AKT1S1). On growth signal stimulus, AKT1S1 binds the scaffolding protein 14-3-3 and dissociates from Raptor, thereby allowing mTORC1 access to its downstream effector substrates (8,9). In the above processes, AKT1S1 acted as a negative regulator of mTORC1, so it may be a suppressor of cell growth and proliferation (10). However, many studies indicated that AKT1S1 was upregulated in disease models, and its upregulation was proven to promote cell survival and tumorigenesis (11,12). Moreover, a recent study showed that AKT1S1 negatively regulated the ribosomal protein L11 (RPL11)–p53 nucleolar stress response pathway and suppressed induction of p53-mediated cellular senescence in HeLa cells (13).
MicroRNAs (miRNAs) have been involved in the incidence and progression of nearly all cancers (14). miR-1908 is a newly discovered miRNA that was reported to play a potential role in diverse physiological and pathological processes. Several studies showed that miR-1908 was highly expressed in mature human adipocytes, and its overexpression inhibited adipogenic differentiation and increased cell proliferation (15), suggesting that miR-1908 plays an important role in cell proliferation. Another study revealed that miR-1908 was involved in the regulation of some key regulators in cell proliferation and metastasis in cancerous cells (16–18), such as mitogen-activated protein kinase (MAPK), low-density lipoprotein receptor-related protein (LRP1)/LRP8, and so on. However, the role of miR-1908 in the progression of NSCLC is still elusive.
In this study, we found that expression of miR-1908 was significantly reduced in NSCLC cell lines including SK-MES-1 human lung squamous cell, A549 human lung adenocarcinoma, and NCI-H460 human lung large-cell carcinoma cells. Overexpression of miR-1908 markedly suppressed the proliferation of these three cell lines. Moreover, we explored the underlying mechanism of that. Our findings reveal a novel p53-dependent tumor suppressor in NSCLC cells.
MATERIALS AND METHODS
Cell Culture and Mild Hypoxia Stimulation
HBE normal lung, SK-MES-1 human lung squamous cell, A549 human lung adenocarcinoma, and NCI-H460 human lung large-cell carcinoma cells were purchased from ATCC (Manassas, VA, USA). The cells were taken from liquid nitrogen and thawed in a 37°C water bath. The cells were centrifuged at 1,000 × g for 7 min, and then the cells were suspended by DMEM containing 4.5 g/L glucose, 4 mmol/L l-glutamine (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Invitrogen). The NCI-H460 cells were 1640 containing 4.5 g/L glucose, 4 mmol/L l-glutamine supplemented with 10% FBS. The cells were incubated in a humidified incubator with an atmosphere of 95% air/5% CO2 at 37°C. For mild hypoxia stimulation, the cells were incubated in a humidified incubator with an atmosphere of 80% N2/15% O2/5% CO2 at 37°C.
Transfection
Single-strand miRNA mimic negative control (NC) and miR-1908 mimic were designed, synthesized, and confirmed effective by Ribobio Company (Guangzhou, China). On reaching 70% confluence, 40 pmol NC or 40 pmol miR-1908 mimic was transfected into the cell lines with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. The medium was changed every 3 days.
Cell Proliferation Analysis
Cell proliferation was evaluated using the MTT method (Sigma-Aldrich, St. Louis, MO, USA) and Cell Titer-Blue H® Cell Viability Assay Kit (Promega). After treatment, the cells were incubated for 0, 24, 48, and 72 h before adding the MTT reagent to each well at a final concentration of 0.5 mg/ml and incubated at 37°C for 4 h. After medium removal, 500 µl of dimethyl sulfoxide was added to each well. Viable cells were measured by absorbance at a 550 nm wavelength using a microplate reader (Bio-Rad, Hercules, CA, USA). Cell Titer-Blue H® Cell Viability Assay was carried out according to the manufacturer’s instructions.
Real-Time Quantitative PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. The RNA concentration was quantified using a spectrophotometer measuring OD260/280 ratio (1.80–1.95). The integrity of RNA was checked by electrophoresis on 1.0% agarose gel with ethidium bromide staining. Real-time qPCR reactions were carried out in a final volume of 25 µl, using SYBR Premix Ex Taq (TaKaRa), 0.4 mM of each primer, and 200 ng of cDNA template. Mature miR-191 stem–loop primer and quantitative primers, as well as U6 RNA primers, were designed and produced by Ribobio Company. Each individual sample was run in triplicate wells. PCR amplification cycles were performed using iQ™5 Multicolor Real-Time PCR Detection System (Bio-Rad) and SYBR Premix Ex Taq II kit (Invitrogen). The reactions were initially denatured at 95°C for 3 min followed by 35 cycles of 95°C for 15 s, and 60°C for 60 s. The data were calculated using the 2−ΔΔCt method.
3′-UTR Luciferase Reporter Assay
The cDNA fragment corresponding to the 3′-UTR of AKT1S1 mRNA contains the binding site of miR-1908 with XhoI and NotI cutting sites. The constructs were confirmed by sequencing. The fragments were cloned into psiCHECK™-2 Vectors (Promega, Madison, WI, USA) at the 3′ end of the Renilla gene. The mutant psiCHECK™-3′-UTR vector (with a three-consecutive-base mutation at the miR-1908 seed targeting site) was produced with Hieff Mut™ Site-Directed Mutagenesis Kit (Yeasen Biological Technology Co., Ltd., Shanghai, China) according to the manufacturer’s instructions. HEK293T cells (2 × 104) were seeded in 96-well plates, and 24 h later transfections were performed using 1.0 µl Lipofectamine 3000 transfection reagent (Invitrogen), 100 ng of vector constructs, and either 40 nM of miR-1908 mimic or NC per well. Cells were harvested at 48 h after transfection. Luciferase activity was measured using the DualGlo Luciferase Assay System (Promega). Renilla luciferase activity was measured and normalized to corresponding firefly luciferase activity.
Western Blotting
Twenty micrograms of protein from each sample was separated by 12% SDS-PAGE and electrotransferred to PVDF membrane (Millipore) for immunoblotting analysis. The following primary antibodies were used: anti-AKT1S1 (1:200; Abcam), anti-p53 (1:300; Abcam), anti-p21 (1:200; Abcam), anti-CDK4 (1:200; Abcam), anti-RPL11 (1:200; Abcam), and anti-GAPDH, which was used as the internal reference. After incubation with the appropriate HRP-conjugate secondary antibody, proteins were detected using a ChemiDoc XRS imaging system and analysis software Quantity One (Bio-Rad).
Website References
The targeting relationship between AKT1S1 and miR-1908 was predicted by online server TargetScan (http://www.targetscan.org/cgi-bin/targetscan/vert_61/view_gene.cgi?taxid=9606&rs=NM_001098632&members=miR-663/663a/1908&showcnc=1&shownc=1&showncf=1).
Statistical Analysis
All data were obtained from at least three independent experiments. Values were expressed as means ± SEM. Statistics were calculated with SPSS 19.0. Multiple comparisons were assessed by one-way ANOVA followed by Dunnett’s tests. The difference between groups was considered statistically significant with a value of p < 0.05.
RESULTS
Expression of miR-1908 Was Significantly Downregulated in NSCLC Cells
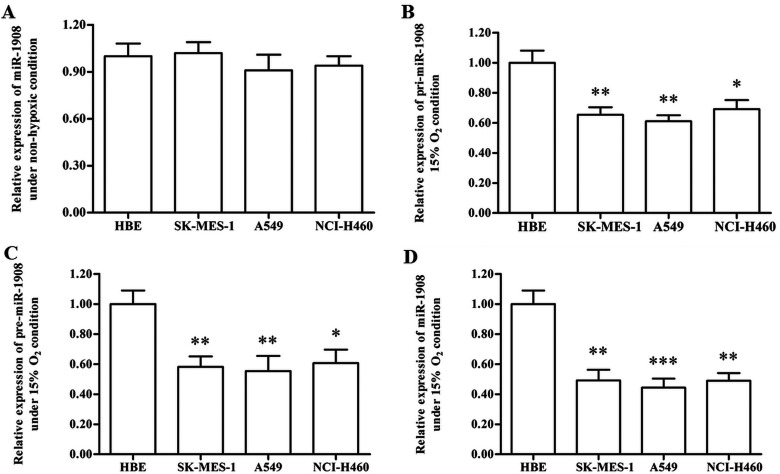
To explore the potential role of miR-1908 in the progression of NSCLC, the expression of miR-1908 was detected with real-time qPCR. The results showed that miR-1908 expression levels did not change between HBE normal lung epithelial cells and NSCLC cell lines (Fig. 1A). Then the expression levels of pri-, pre-, and mature miR-1908 in mild hypoxic NSCLC cell lines and HBE cells were detected. The results showed that all these transcripts were significantly downregulated in the NSCLC cell lines (Fig. 1B–D), suggesting that miR-1908 might play a role in the progression of NSCLC.
Figure 1.
miR-1908 is downregulated in NSCLC cell lines under mild hypoxia condition. (A) There is no significant difference in the miR-1908 expression levels between normal lung cells and NSCLC cell lines. (B) Primary miR-1908 transcript (pri-miR-1908) was downregulated in NSCLC cell lines under mild hypoxia condition. (C) Precursor miR-1908 transcript (pre-miR-1908) was downregulated in NSCLC cell lines under mild hypoxia condition. (D) Mature miR-1908 was downregulated in NSCLC cell lines under mild hypoxia condition. After adhesion, normal lung cells HBE, and NSCLC cell lines SK-MES-1, A549, and NCI-H460 were incubated in normal condition (nonhypoxia) or a mild hypoxia atmosphere with 15% O2. On reaching confluence, the levels of pri-, pre-, and mature miR-1908 were detected with real-time qPCR. *p < 0.05, **p < 0.01.
Overexpression of miR-1908 Suppressed the Proliferation of NSCLC Cells
To investigate the effect of miR-1908 on the proliferation of NSCLC cells, miR-1908 mimic or a negative control mimic (NC) were transfected into human NSCLC cell lines SK-MES-1, A549, and NCI-H460 under mild hypoxia conditions, and then their proliferation was detected. The results of MTT analysis showed that the proliferation of these three cell lines was markedly reduced by the miR-1908 mimic transfection (Fig. 2A–C). Data on Cell Titer-Blue H analysis also revealed that miR-1908 mimic transfection caused a significant decrease in the proliferation of these cells (Fig. 2D–F).
Figure 2.
Overexpression of miR-1908 suppresses NSCLC cell proliferation under mild hypoxia condition. (A–C) The effect of miR-1908 mimic on the proliferation of SK-MES-1, A549, and NCI-H460 cells was determined by MTT under mild hypoxia condition. (D–F) The effect of miR-1908 mimic on the proliferation of SK-MES-1, A549, and NCI-H460 cells was determined by Cell Titer-Blue H analysis under mild hypoxia condition. *p < 0.05.
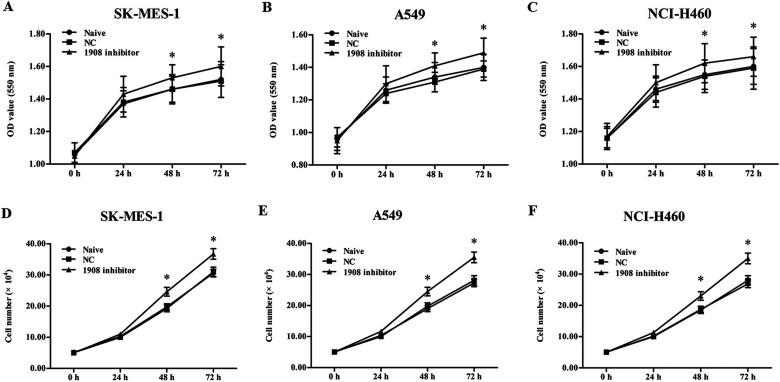
Silencing miR-1908 Promoted the Proliferation of NSCLC Cell Lines
Subsequently, to further explore the role of miR-1908 in the progression of NSCLC cell lines, oligo miR-1908 inhibitor was transfected into the three cell lines under mild hypoxia conditions. After incubation, the proliferation of the cell lines was detected with MTT and Cell Titer-Blue H assays. The results indicated that the proliferation of these cells was notably increased by the miR-1908 inhibitor transfection (Fig. 3A–F). These results, together with those of miR-1908 mimic transfection, demonstrated that miR-1908 plays a negative role in the regulation of NSCLC cell proliferation.
Figure 3.
Silencing miR-1908 increases NSCLC cell proliferation under mild hypoxia condition. (A–C) The effect of miR-1908 inhibitor on the proliferation of SK-MES-1, A549, and NCI-H460 cells was determined by MTT under mild hypoxia condition. (D–F) The effect of miR-1908 inhibitor on the proliferation of SK-MES-1, A549, and NCI-H460 cells was determined by Cell Titer-Blue H analysis under mild hypoxia condition. *p < 0.05.
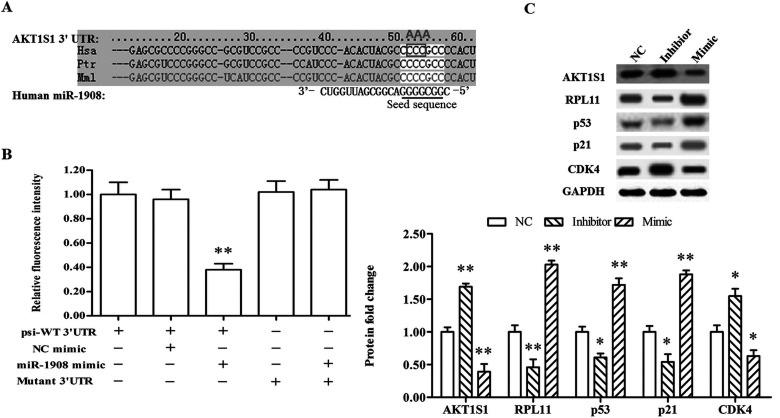
miR-1908 Targeted AKT1S1 and Upregulated the RP-p53-p21 Tumor-Suppressing Pathway
Finally, to explore the potential mechanism of miR-1908 suppressing the proliferation of NSCLC cell lines, the online server TargetScan was applied to screening its potential target genes. The output showed that the AKT1S1 mRNA was completely matched by the miR-1908 seed sequence at its 3′-UTR region (Fig. 4A). The 3′-UTR luciferase reporter assay was applied to verify their targeting relationship. The results revealed that miR-1908 sharply reduced the relative fluorescence intensity of psi-WT 3′-UTR, but had no effect on that of mutant 3′-UTR (Fig. 4B). Subsequently, the miR-1908 mimic or inhibitor was transfected into A549 cells. Western blotting analysis showed that expression AKT1S1 protein was sharply reduced by miR-1908 mimic transfection and increased by the inhibitor (Fig. 4C). Moreover, several marker genes in the downstream tumor-suppressing pathway were upregulated by miR-1908 mimic and suppressed by the inhibitor, including RPL11, p53, and p21 (Fig. 4C). CDK4 expression was altered in response to the alternation of p21 level caused by the mimic and inhibitor (Fig. 4C).
Figure 4.
miR-1908 targets AKT1S1 and negatively regulates RPL11–p53–p21 signaling. (A) The output of TargetScan for targeting relationship between miR-1908 and AKT1S1. The gray box indicates the substituted bases in the mutant AKT1S1 3′-UTR sequence, and the gray letters above the box represent the bases that substitute them. (B) MiR-1908 targeted WT AKT1S1 3′-UTR but not mutant AKT1S1 3′-UTR. (C) Western blotting analysis for protein levels of AKT1S1, RPL11, p53, p21, and CDK4. After the mimic or inhibitor transfection for 72 h under chronic hypoxia condition, the cells were collected and the AKT1S1, RPL11, p53, p21, and CDK4 protein levels were detected. *p < 0.05, **p < 0.01.
DISCUSSION
As a newly discovered miRNA, the role of miR-1908 has been poorly studied. High throughput sequencing revealed that the functions of its target genes were mostly predicted as regulators of the Wnt receptor signaling, glucose/insulin metabolism, cell cycle, or cell apoptosis (15). Studies have indicated that miR-1908 was highly expressed in human mature adipocytes and further proven to play an important role in the regulation of differentiation and proliferation of normal adipocytes and is involved in the regulation of multiple cytokines (15,19). Recently, miR-1908 was found to be involved in the regulation of chordoma and melanoma cell proliferation and metastasis. In this study, we reported that expression of miR-1908 was significantly downregulated in human NSCLC cell lines, including SK-MES-1, A549, and NCI-H460, suggesting it has a suppressive effect on the progression of NSCLC. Then the miR-1908 mimic was transfected into three cell lines, and our data on the cell proliferation detection indicated that miR-1908 had a negative effect on the proliferation of NSCLC cells. What is more, the results show that miR-1908 inhibitor transfection significantly promoted cell proliferation.
Interestingly, most of the existing reports show a promoting effect of miR-1908 on cell proliferation in cancerous and noncancerous cells. However, our study came to an opposite conclusion. Our explanation is that the mild hypoxia atmosphere might change its targeting manner, which we would validate through further study. In fact, although the exact role of miR-1908 in lung cancers was rarely reported, we recognized from a recent report that serum miR-1908 was lowered in patients with primary lung cancer compared with control individuals (20), which also suggested a suppressive role of miR-1908 in lung cancer. Moreover, it is not a singular event that miRNAs played multiple roles in different cancerous cells or in the same type of cancerous cells under different conditions. A typical miRNA that plays multiple roles under hypoxic conditions is miR-210. As a signature of hypoxia and an extensively investigated miRNA in cancer, miR-210 could function as an oncogene or a tumor suppressor and could be a positive or negative prognostic biomarker (21). Another similar miRNA is miR-191, which was proven to be a tumor promotor in many cancers, but not NSCLC (22–24). Overexpression of miR-191 had no effect on any phenotypes of A549 NSCLC cells, including cell cycle distribution, cell proliferation, adherent colony formation, and soft agar colony formation (24).
As a substrate of both Akt1 and mTOR complex 1, AKT1S1 functioned crucially in the Akt/mTOR signaling pathway to regulate multiple biological processes (12,25). For a long time, AKT1S1 was regarded as an inhibitor of Akt/mTOR signaling and a suppressor in cell cycle and proliferation (26,27), until Pallares-Cartes and colleagues showed a contrasting finding regarding the function of AKT1S1 in mTORC1 signaling in Drosophila (28). An increasing number showed that silencing AKT1S1 increased apoptosis, lowered cell viability, and reduced tumor development in several cancers (29,30). Additionally, a recent study demonstrated that AKT1S1 negatively regulates the RPL11-HDM2-p53 nucleolar stress response pathway and suppresses induction of p53-mediated cellular senescence. Therefore, AKT1S1 had a protumorigenic effect and might be a potential target for the regulation of p53-mediated cell growth arrest (13). In this current study, our data on TargetScan bioinformatics server and 3′-UTR luciferase reporter assay showed that AKT1S1 was a target of miR-1908. Moreover, miR-1908 had a positive impact on the expression of RPL11, p53, and p21.
In conclusion, miR-1908 was downregulated in NSCLC cell lines under a mild hypoxia condition. miR-1908 suppressed the proliferation of NSCLC cells through targeting AKT1S1.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Jemal A.; Bray F.; Center M. M.; Ferlay J.; Ward E.; Forman D. Global cancer statistics. CA-Cancer J. Clin. 61:69–90; 2011. [DOI] [PubMed] [Google Scholar]
- 2. Chen W.; Zheng R.; Zhang S.; Zhao P.; Zeng H.; Zou X. Report of cancer incidence and mortality in China, 2010. Ann. Transl. Med. 2:61–90; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldstraw P.; Ball D.; Jett J. R.; Le Chevalier T.; Lim E.; Nicholson A. G.; Shepherd F. A. Non-small-cell lung cancer. Lancet 378:1727–1740; 2011. [DOI] [PubMed] [Google Scholar]
- 4. Ettinger D. S.; Akerley W.; Borghaei H.; Chang A. C.; Cheney R. T.; Chirieac L. R.; D’Amico T. A.; Demmy T. L.; Ganti A. K. P.; Govindan R. Non-small cell lung cancer. J. Natl. Compr. Cancer Netw. 10:1236–1271; 2012. [DOI] [PubMed] [Google Scholar]
- 5. Lawlor M. A.; Alessi D. R. PKB/Akt a key mediator of cell proliferation, survival and insulin responses? J. Cell. Sci. 114:2903–2910; 2001. [DOI] [PubMed] [Google Scholar]
- 6. Song G.; Ouyang G.; Bao S. The activation of Akt/PKB signaling pathway and cell survival. J. Cell. Mol. Med. 9:59–71; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manning B. D.; Cantley L. C. Akt/PKB signaling: Navigating downstream. Cell 129:1261–1274; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oshiro N.; Takahashi R.; Yoshino K. I.; Tanimura K.; Nakashima A.; Eguchi S.; Miyamoto T.; Hara K.; Takehana K.; Avruch J. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J. Biol. Chem. 282:20329–20339; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kovacina K. S.; Park G. Y.; Bae S. S.; Guzzetta A. W.; Schaefer E.; Birnbaum M. J.; Roth R. A. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J. Biol. Chem. 278:10189–10194; 2003. [DOI] [PubMed] [Google Scholar]
- 10. Wang H.; Zhang Q.; Wen Q.; Zheng Y.; Lazarovici P.; Jiang H.; Lin J.; Zheng W. Proline-rich Akt substrate of 40kDa (PRAS40): A novel downstream target of PI3K/Akt signaling pathway. Cell Signal. 24:17–24; 2012. [DOI] [PubMed] [Google Scholar]
- 11. Das F.; Dey N.; Venkatesan B.; Kasinath B. S.; Ghosh-Choudhury N.; Choudhury G. G. High glucose upregulation of early-onset Parkinson’s disease protein DJ-1 integrates the PRAS40/TORC1 axis to mesangial cell hypertrophy. Cell Signal. 23:1311–1319; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wiza C.; Nascimento E. B.; Ouwens D. M. Role of PRAS40 in Akt and mTOR signaling in health and disease. J. Physiol. Endocrinol. Metab. 302:E1453–E1460; 2012. [DOI] [PubMed] [Google Scholar]
- 13. Havel J.; Li Z.; Cheng D.; Peng J.; Fu H. Nuclear PRAS40 couples the Akt/mTORC1 signaling axis to the RPl11-HDN2-p53 nucleolar stress response pathway. Oncogene 34:1487–1498; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Calin G. A.; Croce C. M. Microrna signatures in human cancers. Nat. Rev. Cancer 6:857–866; 2006. [DOI] [PubMed] [Google Scholar]
- 15. Jiang X.; Yang L.; Pang L.; Chen L.; Guo X.; Ji C.; Shi C.; Ni Y. Expression of obesity-related mir-1908 in human adipocytes is regulated by adipokines, free fatty acids and hormones. Mol. Med. Rep. 10:1164–1169; 2014. [DOI] [PubMed] [Google Scholar]
- 16. Pencheva N.; Tran H.; Buss C.; Huh D.; Drobnjak M.; Busam K.; Tavazoie S. F. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell 151:1068–1082; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Long C.; Jiang L.; Wei F.; Ma C.; Zhou H.; Yang S.; Liu X.; Liu Z. Integrated miRNA-mRNA analysis revealing the potential roles of miRNAs in chordomas. PLoS One 8:e66676; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pencheva N.; Tavazoie S. F. Control of metastatic progression by microrna regulatory networks. Nat. Cell Biol. 15:546–554; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang L.; Shi C. M.; Chen L.; Pang L. X.; Xu G. F.; Gu N.; Zhu L. J.; Guo X. R.; Ni Y. H.; Ji C. B. The biological effects of HSA-miR-1908 in human adipocytes. Mol. Biol. Rep. 42:927–935; 2015. [DOI] [PubMed] [Google Scholar]
- 20. Keller A.; Leidinger P.; Gislefoss R.; Haugen A.; Langseth H.; Staehler P.; Lenhof H. P.; Meese E. Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 8:506–516; 2011. [DOI] [PubMed] [Google Scholar]
- 21. Qin Q.; Furong W.; Baosheng L. Multiple functions of hypoxia-regulated miR-210 in cancer. J. Exp. Clin. Cancer Res. 33:50–84; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang Y. Z.; Zhang J.; Shao H. Y.; Chen J. P.; Zhao H. Y. MicroRNA-191 promotes osteosarcoma cells proliferation by targeting checkpoint kinase 2. Tumor Biol. 36:6095–6101, 2015. [DOI] [PubMed] [Google Scholar]
- 23. Zhang X. F.; Li K. K.; Gao L.; Li S. Z.; Chen K.; Zhang J. B.; Wang D.; Tu R. F.; Zhang J. X.; Tao K. X. Mir-191 promotes tumorigenesis of human colorectal cancer through targeting C/EBPβ. Oncotarget 6:4144–4158; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patnaik S. K.; Kannisto E.; Yendamuri S. Overexpression of microRNA miR-30a or miR-191 in A549 lung cancer or BEAS-2B normal lung cell lines does not alter phenotype. PLoS One 5:e9219–e9225; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zoncu R.; Efeyan A.; Sabatini D. M. Mtor: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12:21–35; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Porta C.; Paglino C.; Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front. Oncol. 4:64–82, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vander Haar E.; Lee S. I.; Bandhakavi S.; Griffin T. J.; Kim D. H. Insulin signalling to mtor mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9:316–323; 2007. [DOI] [PubMed] [Google Scholar]
- 28. Pallares-Cartes C.; Cakan-Akdogan G.; Teleman A. A. Tissue-specific coupling between insulin/igf and torc1 signaling via pras40 in drosophila. Dev. Cell. 22:172–182; 2012. [DOI] [PubMed] [Google Scholar]
- 29. Madhunapantula S. V.; Sharma A.; Robertson G. P. PRAS40 deregulates apoptosis in malignant melanoma. Cancer Res. 67:3626–3636; 2007. [DOI] [PubMed] [Google Scholar]
- 30. Malla R.; Ashby C. R.; Narayanan N. K.; Narayanan B.; Faridi J. S.; Tiwari A. K. Proline-rich Akt substrate of 40-kDa (PRAS40) in the pathophysiology of cancer. Biochem. Biophys. Res. Commun. 463:161–166; 2015. [DOI] [PubMed] [Google Scholar]