Abstract
Poly(ADP-ribose) polymerase 1 (PARP-1) is reported to be involved in DNA repair and is now recognized as a key regulator in carcinogenesis. However, the potential role and the molecular mechanism underlying the effect of PARP-1 on osteosarcoma (OS) cells have not been elucidated. In this study, the results showed that knockdown of PARP-1 resulted in decreased cell proliferation, increased cell apoptosis, and G0/G1 phase arrest in U2OS cells. In addition, increased expression of active caspase 3 and Bax, but reduced Bcl-2, cyclin D1, and phosphorylated extracellular signal regulated kinase 1/2 (pERK1/2) were observed in PARP-1 knockdown in U2OS cells. Moreover, knockdown of PARP-1 correlated with elevated chemosensitivity of U2OS cells to cisplatin through inactivation of the ERK1/2 signaling pathway. In conclusion, our findings demonstrated that PARP-1 plays an important role in regulating OS growth, combining PARP-1 gene therapy with traditional chemotherapy, and may serve as a promising approach to OS therapy.
Key words: PARP-1, Osteosarcoma, Cisplatin, Chemoresistance, ERK1/2
INTRODUCTION
Osteosarcoma (OS) is the most common primary bone malignancy, with a high incidence in adolescents and young adults (1). Nowadays, high-dose chemotherapy plus surgical treatment is used in the treatment of OS and dramatically improves the outcome (1). However, a significant number of patients will still relapse due to the resistance of tumor cells to chemotherapeutic agents. Therefore, identification of novel therapeutic targets for overcoming chemoresistance is essential to improve treatment outcomes.
Poly(ADP-ribose) polymerase 1 (PARP-1) [also known as NAD+ ADP-ribosyltransferase 1 or poly(ADP-ribose) synthase 1] is an abundant nuclear enzyme that can be activated by DNA damage and has been implicated in a variety of physiological processes, including DNA repair, genomic stability, and cell apoptosis (2). PARP-1 is a 116-kDa protein consisting of three main domains: the N-terminal DNA-binding domain containing two zinc fingers, the automodification domain, and the C-terminal catalytic domain (3). PARP-1 can function as a DNA damage sensor. Upon binding to damaged DNA, PARP-1 forms homodimers and catalyzes the cleavage of NAD+ into nicotinamide and ADP-ribose to synthesize long branching poly(ADP-ribose) polymers covalently attached to nuclear acceptor proteins (3,4).
Studies have demonstrated that PARP-1 is highly expressed in various types of human cancers, including breast cancer, lung cancer, hepatocellular carcinoma, gastric cancer, and melanomas (5). In addition, several studies have documented that the PARP-1 expression level is a prognostic indicator and is associated with a poor survival prognosis (6–8). In certain types of tumors, PARP-1 inhibition was shown to be an effective means of enhancing tumor sensitivity to radiation and chemotherapy. In OS, early studies have demonstrated that PARP-1 was decreased in human OS cells undergoing spontaneous apoptosis (9). Recently, PARP-1 inhibitor 3-aminobenzamide was shown to suppress cell growth and enhance the suppressive effects of cisplatin in OS cells (10). However, the molecular mechanism underlying the effect of PARP-1 on OS cell chemosensitivity is still unknown.
Extracellular signal regulated kinase (ERK) is the main transducer of growth stimuli. Activation of ERK1/2 mediates cell proliferation, apoptosis, and migration, and is involved in a number of downstream targets, which is important in cancer invasion and metastasis (11). Furthermore, it has been shown that activation of PARP is associated with ERK in several cancer cells (12). Therefore, we hypothesize that ERK1/2 signaling pathway might be involved in cisplatin resistance occurring in OS chemotherapy.
In this study, we utilized small interfering RNA (siRNA) to silence PARP-1 and investigated the action of PARP-1 knockdown in regulating cell proliferation, apoptosis, and chemoresistance to cisplatin in OS cells, and we also determined the correlation between cisplatin resistance and the ERK1/2 signaling pathway in OS cells. Our data suggest that PARP-1 contributes to the growth of OS and may serve as a potential target for OS therapy.
MATERIALS AND METHODS
Cell Culture
Human OS cell line U2OS was obtained from the American Tissue Culture Collection (ATCC, Rockville, MD, USA) and was cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), penicillin (100 U/ml), and streptomycin (100 µg/ml) at 37°C in a humidified incubator supplemented with 5% CO2 and used in experiments at logarithmic growth phase.
siRNA Transfection
Two pairs of siRNA sequences targeting PARP-1 gene sequence with NM_001618 in GeneBank and a negative control siRNA (siCtrl) were designed on the basis of the principles for siRNA design and synthesized by Shanghai Genechem Co. Ltd. These sequences were siPARP-1#1, 5′-CGGATAAGCTCTATCGAGTCGAGTA-3′; siPARP-1#2, 5′-CGAGAAATCTCTTACCTCAAGAAAT-3′; and the siCtrl sequence, 5′-GGCTCCGATCGTCTCACAT-3′. U2OS cells were incubated in culture without antibiotics for 24 h prior to transfection, resulting in 30% to 50% confluence. Then they were transfected with PARP-l siRNA (siPARP-1#1 and siPARP-1#2) or siCtrl in Opti-MEM (Invitrogen) using the Lipofectamine 2000 transfection reagent (Invitrogen) in accordance with the manufacturer’s instructions. Silencing of PARP-1 was assayed for mRNA and protein expression level at 48 h after transfection. Untransfected cells were considered as normal control, and cells transfected with siCtrl were considered as negative control.
Quantitative Real-Time PCR
Total RNA were extracted from cells using TRIzol reagent (Invitrogen). cDNA was synthesized using a reverse transcription kit according to the instructions provided by the manufacturer (Takara, Dalian, China). The following gene-specific primers were used in this study: PARP-1, 5′-CGGAGTCTTCGGATAAGCTCT-3′ (forward) and 5′-TTTCCATCAAACATGGGCGAC-3′ (reverse); GAPDH, 5′-CATGTTCGTCATGGGTGTGAACCA-3′ (forward) and 5′-AGTGATGGCATGGACTGTGGTCAT-3′ (reverse). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed in 20 µl of reaction system consisting of 10 µl of SYBR Green Mix, 4 µl of each forward and reverse primer (10 µmol/L), and 1 µl of cDNA in a Roche LightCycler 480 instrument at conditions of 5 min at 95°C and 40 cycles of 5 min at 95°C, 10 s at 60°C, and 10 s at 72°C. The relative expression level of PARP-1 was normalized to that of GAPDH using the comparative delta CT (2−ΔΔCt) method. Each sample was analyzed in triplicate, and the mean expression level was calculated.
Cell Viability Assay
Cell viability was determined using the Cell Counting Kit-8 (CCK-8) in accordance with the manufacturer’s protocol (Sigma-Aldrich, St. Louis, MO, USA). Briefly, control and transfected cells were seeded at 3 × 103 per well in 96-well plates and treated with different concentrations of cisplatin (Sigma-Aldrich) or ERK inhibitor U0126 (Cell Signaling Technology, Beverly, MA, USA) for 48 h; 10 µl of CCK-8 was added to each well and incubated for an additional 4 h at 37°C. The optical density (OD) was measured using a microplate reader (Bio-Tek Company, Winooski, VT, USA) at 450-nm wavelength. Each time point was repeated in three wells, and the experiment was independently performed three times.
Colony Formation Assay
Control and transfected cells were seeded at 5 × 102 per well in 24-well plates and cultured in an environment with 5% CO2 at 37°C for 10–14 days to allow colonies to form. The plates were stained for the formation of cell colonies with crystal violet in 70% ethanol and counted under the microscope. The experiment was independently performed three times.
Cell Apoptosis and Cell Cycle Assay
Control and transfected cells were seeded at 5 × 104 per well in 96-well plates and cultured for 48 h. For cell cycle analysis, cells were detached by trypsin digestion, fixed with ice-cold 70% ethanol for 4 h, and stained with 50 µg/ml propidium iodide (PI) in the presence of RNase A at 37°C for 30 min. Intracellular DNA content was analyzed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). Cell apoptosis was evaluated by flow cytometry using an Annexin-V-FITC Apoptosis Detection Kit (KeyGen Biotech Co. Roche, Nanjing, China) according to the manufacturer’s instructions. Briefly, the cells were harvested and washed twice in phosphate-buffered saline (PBS) and resuspended in 500 µl of binding buffer. A volume of 5 µl of Annexin-V-FITC and 5 µl of PI was added and mixed gently, and the cells were stained in the dark for 15 min at room temperature. The cells were analyzed immediately by flow cytometry and analyzed using the FlowJo software (FlowJo, Ashland, OR, USA). The experiment was repeated three times.
Western Blot
Total proteins were extracted using radioimmuno precipitation assay (RIPA)-phenylmethylsulfonyl fluoride (PMSF) solution and were quantified by the BCA assay (Beyotime, China). A total of 20 µg of proteins from each sample was subjected to 8% polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were then blocked with 5% skim milk at room temperature for 1 h and incubated overnight with the primary antibodies directly against PARP-1 (1:500 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA), ERK1/2, phosphorylated ERK1/2 (pERK1/2) (1:500 dilution), active caspase 3 (1:500 dilution), Bcl-2 (1:500 dilution), Bax (1:500 dilution), and β-actin (1:2,000 dilution) overnight at 4°C (all from Santa Cruz Biotechnology). After being washed with TBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody (Beyotime) for 1 h at 37°C, and visualized using ECL chemiluminiscence detection reagent.
Statistical Analysis
Data were presented as means ± SD of three independent determinations. Differences between experimental groups were analyzed using an unpaired Student’s t-test. All data were analyzed with GraphPad Prism version 5.0 (GraphPad Software Inc., La Jolla, CA, USA). A value of p < 0.05 was considered a significant difference.
RESULTS
Knockdown of PARP-1 Inhibits Cell Growth of U2OS Cells In Vitro
To explore the biological functions of PARP-1, we identified and validated two independent and nonoverlapping siRNA sequences to deplete endogenous PARP-1 expression in U2OS cells. We used siPARP-1#1 and siPARP-1#2 to transfect U2OS cells, siCtrl as the negative control, and untransfected OS cells as the normal control. The knockdown efficacy of siRNA was confirmed by RT-PCR and Western blot at 48 h posttransfection. As shown in Figure 1A and B, PARP-1 in cells transfected with specific siRNA for PARP-1 was significantly reduced by ∼42% with siPARP-1#1 and ∼79% with siPARP-1#2 for mRNA levels, and ∼26% with siPARP-1#1 and ∼70% with siPARP-1#2 for protein levels, compared with siCtrl transfected cells. Thus, siPARP-1#2 was accordingly chosen as the most efficient and specific sequence to silence the expression of PARP-1 in the U2OS cells and was used in the subsequent experiments.
Figure 1.
Inhibition of poly(ADP-ribose) polymerase 1 (PARP-1) expression in U2OS cells by siRNA. (A) Relative PARP-1 mRNA expression in U2OS cells measured by quantitative real-time polymerase chain reaction (qRT-PCR). (B) PARP-1 protein expression measured by Western blot. Data are shown as means ± SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Next we evaluated the effect of knockdown of PARP-1 on U2OS cell growth by colony formation assay. As shown in Figure 2A, cells transfected with siPARP-1#2 formed fewer and smaller colonies as compared with cells transfected with siCtrl. In addition, the effect of PARP-1 knockdown on cell proliferation was also assessed by the CCK-8 assay. As shown in Figure 2B, PARP-1 knockdown resulted in a significant decrease in the proliferation rate of U2OS cells at 48, 72, and 96 h. Taken together, our results indicate that knockdown of PARP-1 evoked a marked inhibitive effect on cell proliferation and colony formation capacity in U2OS cells in vitro.
Figure 2.
Knockdown of PARP-1 inhibits cell growth and colony formation of U2OS cells in vitro. (A) Control and transfected U2OS cells were seeded at 5 × 102 per well in 24-well plates and cultured for 10–14 days to allow colonies to form. Positive colonies were counted under the microscope. (B) Control and transfected U2OS cells were seeded at 3 × 103 per well in 96-well plates for indicated time points; cell viability was determined using a Cell Counting Kit-8 (CCK-8) assay. Data are shown as means ± SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Knockdown of PARP-1 Induces Cell Apoptosis and G0/G1 Arrest in U2OS Cells In Vitro
To explore the potential mechanism by which PARP-1 suppresses OS cell growth, we evaluated the cell cycle distribution in siPARP-1#2 transfected cells and siCtrl cells using flow cytometry. The results showed that knockdown of PARP-1 in U2OS cells elicited an accumulation of cells in the G0/G1 phase and a decrease in the S phase and G2/M phase as compared with siCtrl-transfected cells (Fig. 3A), and a significant decreased of cyclin D1 was detected in siPARP-1#2-transfected cells (Fig. 3B). Additionally, results from annexin V/PI analysis showed that U2OS cells transfected with siPARP-1#2 (12.52 ± 4.21%) underwent obvious apoptosis when compared to siCtrl-transfected cells (5.02 ± 1.24%, p < 0.01) (Fig. 3C). Furthermore, we examined the levels of several apoptosis-associated proteins by Western blot. As shown in Figure 3D, the protein levels of active caspase 3 and Bax were significantly increased, whereas the expression level of Bcl-2 was remarkably reduced in siPARP-1#2-transfected U2OS cells. Collectively, our data suggest that suppression of cell growth by PARP-1 knockdown is partially attributable to increased cell apoptosis and G0/G1 phase arrest in vitro.
Figure 3.
Knockdown of PARP-1 induces cell apoptosis and G0/G1 arrest in U2OS cells in vitro. Control and transfected cells were seeded at 5 × 104 per well in 96-well plates. (A) Cells were harvested at 48 h and stained with propidium iodide (PI), and then quantified for the cycle distribution by flow cytometry. (B) Western blot analysis of cyclin D1 protein expression in transfected cells. (C) Flow cytometry analysis with annexin V/PI staining was performed to evaluate the percentage of apoptotic cells. (D) Western blot analysis of caspase 3, Bax, and Bcl-2 protein expressions in transfected cells. Data are shown as means ± SD of three independent experiments. *p < 0.05, **p < 0.01.
PARP-1 Expression Positively Correlates With pERK1/2 in U2OS Cells
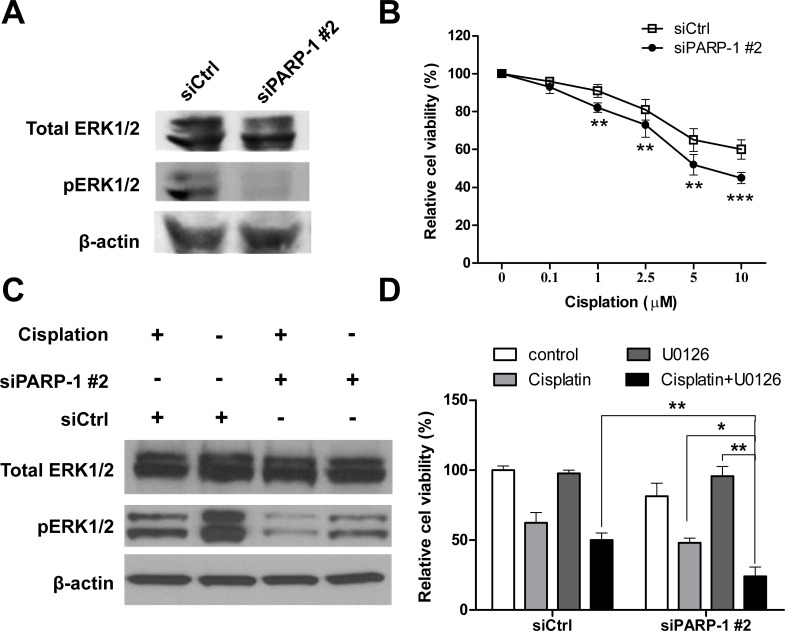
It has been reported that the phosphorylation of ERK1/2 is involved in multiple biological behaviors, such as cell proliferation and cell apoptosis, as well as malignant transformation and drug resistance (11). Previous studies have demonstrated that activation of ERKl/2 has been shown to enhance tumor progression in OS (13,14). We hypothesized that PARP-1 might affect the ERK1/2 pathway in OS cells. Thus, we first investigated the effect of PARP-1 knockdown on the ERK1/2 pathway in U2OS cells. As shown in Figure 4A, 48 h after transfection, knockdown of PARP-1 exhibited significantly decreased pERK1/2 expression in U2OS cells, while total ERK1/2 expression was unchanged. The correlation between PARP-1 and pERK1/2 expression in OS cell lines suggested that PARP-1 might be involved in the regulation of the ERK1/2 signaling pathway.
Figure 4.
Knockdown of PARP-1 enhances sensitivity of U2OS cells to cisplatin through modulating extracellular signal regulated kinase 1/2 (ERK1/2) signaling. (A) U2OS cells were transfected with siPARP-1#2 or siCtrl, and 48 h later proteins were extracted and subjected to Western blot analysis using antibodies against ERK1/2 and pERK1/2. (B) Control and transfected cells were seeded at 3 × 103 per well in 96-well plates and treated with different concentrations of cisplatin for 48 h; cell viability was determined using a CCK-8 assay. (C) Western blot analysis of ERK1/2 and pERK1/2 in U2OS cells transfected with siPARP-1#2 or siCtrl after treatment of 10 µM for 48 h. (D) U2OS cells transfected with siPARP-1#2 or siCtrl were treated with 10 µM U0126 for 12 h prior to being treated with l0 µM cisplatin; cell viability was determined using a CCK-8 assay. Data are shown as means ± SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Knockdown of PARP-1 Enhances Sensitivity of U2OS Cells to Cisplatin Through Modulating ERK1/2 Signaling
It has been reported that PARP inhibitors have increased activity in platinum-sensitive tumors, including OS (5,10). We further investigated the effect of PARP-1 knockdown on the sensitivity of U20S cells to cisplatin. To determine the sensitivity of cells to cisplatin, 48 h after transfection with siPARP-1#2 or siCtrl, U2OS cells were exposed to different concentrations of cisplatin ranging from 0 to 10 µM for 48 h. Cell viability was examined by CCK-8 assay. The cell survival rate appeared to show a dose-dependent manner in response to cisplatin treatment, and knockdown of PARP-1 resulted in enhanced sensitivity to cisplatin treatment in U2OS cells (Fig. 4B). We further explored whether PARP-1 knockdown-enhanced cisplatin sensitivity of OS cells was due to the inactivation of ERK1/2 signaling pathway. U2OS cells transfected with siPARP-1#2 or siCtrl were exposed to 10 µM cisplatin, and the level of pERK1/2 was analyzed by Western blot. As shown in Figure 4C, cisplatin suppressed phosphorylation of ERK in U2OS cells. However, knockdown of PARP-1 induced a lower reduction of pERK1/2 expression, while the total ERK1/2 was unaffected. Then we used the specific MEK inhibitor U0126 to attenuate ERK1/2 activity in U2OS cells. Upon treatment with 10 µM U0126 for 12 h prior to being treated with l0 µM cisplatin, the results showed that U0126 effectively enhanced siPARP-1#2-mediated cisplatin sensitivity of U2OS cells (Fig. 4D). Collectively, our findings indicate that knockdown of PARP-1 expression could enhance the chemosensitivity of OS cells to cisplatin by suppression of ERK1/2 activity.
DISCUSSION
PARPs are defined as cell signaling enzymes that catalyze the transfer of ADP-ribose units from NAD+ to a number of acceptor proteins. PARP-1, the best characterized member of the PARP family, is essential to the repair of DNA single-strand breaks via the base excision repair pathway. It is clearly shown that PARP-1 is upregulated in breast cancer (8,15), prostate cancer (16,17), pancreatic cancer (18,19), and ovarian cancer (20), and conducive for cells to resist genotoxic stress and to enhance DNA damage repair (5). However, the molecular mechanism regulating PARP-1 expression in OS remains unknown.
To better understand the potential connection between PARP-1 expression and the biological features of OS cells, we employed a vector-based RNAi strategy to silence endogenous PARP-1 expression in U2OS OS cells. qRT-PCR and Western blot showed that PARP-1 expression was significantly suppressed by PARP-1 siRNA. Further experiments demonstrated that knockdown of PARP-1 in U2OS cells significantly inhibited cell proliferation and colony formation in vitro. In addition, cell cycle analysis showed that knockdown of PARP-1 increased the number of cells in the G0/G1 phase while reducing the number of cells in the S phase and G2/M phase. These data are consistent with the previous findings on other tumor cells (10,18,21). Furthermore, it is also reported that knockdown of PARP-1 can finally induce cell apoptosis (10,18). Our present study also showed that knockdown of PARP-1 significantly induced cell apoptosis in U2OS cells. In accordance with these results, knockdown of PARP-1 resulted in the elevation of active caspase 3 and Bax, as well as reduced Bcl-2 and cyclin D1 in U2OS cells. Taken together, these findings generally support the mechanisms to explain how silencing PARP-1 leads to inhibition of proliferation in OS cells. However, the precise mechanisms by which knockdown of PARP-1 causes tumor growth inhibition need further studies.
Studies have shown that ERK signaling mediates tumor cell proliferation, apoptosis, and migration, and it has shown previously that PARP-1 is closely related to ERK signaling pathway in acute myeloid leukemia (22). We next studied whether PARP-1 affects the ERK1/2 pathway in OS cells. Western blot analysis showed that the expression of pERK1/2 was markedly decreased upon PARP-1 downregulation, but the level of total ERK1/2 was not changed. Furthermore, we explored the association between PARP-1 expression and cisplatin sensitivity in U2OS cells. Our results demonstrated that knockdown of PARP-1 sensitized U2OS cells to cisplatin in vitro in U2OS cells. So targeting PARP-1 as a means to chemosensitize clinically established drugs could be a potent strategy to overcome drug resistance in OS treatment. We next studied whether PARP-1 regulates the chemosensitivity to cisplatin by modulating ERK1/2 in OS cells. The results showed that specific ERK inhibitor U0126 effectively enhanced PARP-1 siRNA-mediated cisplatin sensitivity. These findings indicated that PARP-1 may regulate chemosensitivity to cisplatin through a pERK1/2 signaling pathway.
In summary, our data demonstrate that knockdown of PARP-1 inhibits cell growth and increases sensitivity of U2OS cells to cisplatin, with the increasing drug sensitivity possibly associated with the inhibition of ERK signals. Our findings suggest that combining PARP-1 gene therapy with traditional chemotherapy may be more effectively used in treating OS.
REFERENCES
- 1. Isakoff M. S.; Bielack S. S.; Meltzer P.; Gorlick R. Osteosarcoma: Current treatment and a collaborative pathway to success. J. Clin. Oncol. 33(27):3029–3035; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aguilar-Quesada R.; Munoz-Gamez J. A.; Martin-Oliva D.; Peralta-Leal A.; Quiles-Perez R.; Rodriguez-Vargas J. M.; Ruiz de Almodovar M.; Conde C.; Ruiz-Extremera A.; Oliver F. J. Modulation of transcription by PARP-1: Consequences in carcinogenesis and inflammation. Curr. Med. Chem. 14(11):1179–1187; 2007. [DOI] [PubMed] [Google Scholar]
- 3. Kraus W. L.; Hottiger M. O. PARP-1 and gene regulation: Progress and puzzles. Mol. Aspects Med. 34(6):1109–1123; 2013. [DOI] [PubMed] [Google Scholar]
- 4. Langelier M. F.; Pascal J. M. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol. 23(1):134–143; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vyas S.; Chang P. New PARP targets for cancer therapy. Nat. Rev. Cancer 14(7):502–509; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Z.; Lv T.; Liu Y.; Huang X.; Qiu Z.; Li J. PARP1 is a novel independent prognostic factor for the poor prognosis of chordoma. Cancer Biomark. 16(4):633–639; 2016. [DOI] [PubMed] [Google Scholar]
- 7. Xu X.; Liu Z.; Wang J.; Xie H.; Li J.; Cao J.; Zhou L.; Zheng S. Global proteomic profiling in multistep hepatocarcinogenesis and identification of PARP1 as a novel molecular marker in hepatocellular carcinoma. Oncotarget 7(12):13730–13741; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mazzotta A.; Partipilo G.; De Summa S.; Giotta F.; Simone G.; Mangia A. Nuclear PARP1 expression and its prognostic significance in breast cancer patients. Tumour Biol. 37(5):6143–6153; 2016. [DOI] [PubMed] [Google Scholar]
- 9. Simbulan-Rosenthal C. M.; Rosenthal D. S.; Iyer S.; Boulares H.; Smulson M. E. Involvement of PARP and poly(ADP-ribosyl)ation in the early stages of apoptosis and DNA replication. Mol. Cell. Biochem. 193(1–2):137–148; 1999. [PubMed] [Google Scholar]
- 10. Zheng Y. D.; Xu X. Q.; Peng F.; Yu J. Z.; Wu H. The poly(ADP-ribose) polymerase-1 inhibitor 3-aminobenzamide suppresses cell growth and migration, enhancing suppressive effects of cisplatin in osteosarcoma cells. Oncol. Rep. 25(5):1399–1405; 2011. [DOI] [PubMed] [Google Scholar]
- 11. McCubrey J. A.; Steelman L. S.; Chappell W. H.; Abrams S. L.; Wong E. W.; Chang F.; Lehmann B.; Terrian D. M.; Milella M.; Tafuri A.; Stivala F.; Libra M.; Basecke J.; Evangelisti C.; Martelli A. M.; Franklin R. A. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 1773(8):1263–1284; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohen-Armon M. PARP-1 activation in the ERK signaling pathway. Trends Pharmacol. Sci. 28(11):556–560; 2007. [DOI] [PubMed] [Google Scholar]
- 13. Si H.; Peng C.; Li J.; Wang X.; Zhai L.; Li X.; Li J. RNAi-mediated knockdown of ERK1/2 inhibits cell proliferation and invasion and increases chemosensitivity to cisplatin in human osteosarcoma U2-OS cells in vitro. Int. J. Oncol. 40(4):1291–1297; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Na K. Y.; Kim Y. W.; Park Y. K. Mitogen-activated protein kinase pathway in osteosarcoma. Pathology 44(6):540–546; 2012. [DOI] [PubMed] [Google Scholar]
- 15. Park S. H.; Noh S. J.; Kim K. M.; Bae J. S.; Kwon K. S.; Jung S. H.; Kim J. R.; Lee H.; Chung M. J.; Moon W. S. Expression of DNA damage response molecules PARP1, gammaH2AX, BRCA1, and BRCA2 predicts poor survival of breast carcinoma patients. Transl. Oncol. 8(4):239–249; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barboro P.; Ferrari N.; Capaia M.; Petretto A.; Salvi S.; Boccardo S.; Balbi C. Expression of nuclear matrix proteins binding matrix attachment regions in prostate cancer. PARP-1: New player in tumor progression. Int. J. Cancer 137(7):1574–1586; 2015. [DOI] [PubMed] [Google Scholar]
- 17. Li L.; Chang W.; Yang G.; Ren C.; Park S.; Karantanos T.; Karanika S.; Wang J.; Yin J.; Shah P. K. and others. Targeting poly(ADP-ribose) polymerase and the c-Myb-regulated DNA damage response pathway in castration-resistant prostate cancer. Sci. Signal. 7(326):ra47; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuan K.; Sun Y.; Zhou T.; McDonald J.; Chen Y. PARP-1 regulates resistance of pancreatic cancer to TRAIL therapy. Clin. Cancer Res. 19(17):4750–4759; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klauschen F.; von Winterfeld M.; Stenzinger A.; Sinn B. V.; Budczies J.; Kamphues C.; Bahra M.; Wittschieber D.; Weichert W. Striefler High nuclear poly-(ADP-ribose)-polymerase expression is prognostic of improved survival in pancreatic cancer. Histopathology 61(3):409–416; 2012. [DOI] [PubMed] [Google Scholar]
- 20. Ganzinelli M.; Mariani P.; Cattaneo D.; Fossati R.; Fruscio R.; Corso S.; Ricci F.; Broggini M.; Damia G. Expression of DNA repair genes in ovarian cancer samples: Biological and clinical considerations. Eur. J. Cancer 47(7):1086–1094; 2011. [DOI] [PubMed] [Google Scholar]
- 21. Park S. H.; Jang K. Y.; Kim M. J.; Yoon S.; Jo Y.; Kwon S. M.; Kim K. M.; Kwon K. S.; Kim C. Y.; Woo H. G. Tumor suppressive effect of PARP1 and FOXO3A in gastric cancers and its clinical implications. Oncotarget 6(42):44819–44831; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang L.; Cai W.; Zhang W.; Chen X.; Dong W.; Tang D.; Zhang Y.; Ji C.; Zhang M. Inhibition of poly(ADP-ribose) polymerase 1 protects against acute myeloid leukemia by suppressing the myeloproliferative leukemia virus oncogene. Oncotarget 6(29):27490–27504; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]