Abstract
Raltitrexed (RTX) is an antimetabolite drug used as a chemotherapeutic agent for treating colorectal cancer, malignant mesothelioma, and gastric cancer. The antitumor capacity of RTX is attributed to its inhibitory activity on thymidylate synthase (TS), a key enzyme in the synthesis of DNA precursors. The current study is aimed at investigating the potential antitumor effects of RTX in liver cancer. Using the HepG2 cell line as an in vitro model of liver cancer, we evaluated the effects of RTX on cell proliferation employing both a WST-8 assay and a clone formation efficiency assay. In addition, we monitored the ultrastructure changes of HepG2 cells in response to RTX with transmission electric microscopy. To investigate the mechanism underlying the regulation of cell proliferation by RTX, we analyzed cell cycle using cell flow cytometry. Moreover, real-time PCR and Western blot analyses were conducted to examine expression levels of cell cycle regulatory proteins cyclin A and cyclin-dependent kinase 2 (CDK2), as well as their mediators tumor suppressor genes p53 and p16. Our results demonstrate that RTX inhibits HepG2 proliferation by arresting the cell cycle at G0/G1. This cell cycle arrest function was mediated via downregulation of cyclin A and CDK2. The observed elevated expression of p53 and p16 by RTX may contribute to the reduction of cyclin A/CDK2. Our study indicates that RTX could serve as a potential chemotherapeutic agent in the treatment of hepatocellular carcinoma.
Key words: Raltitrexed (RTX), Hepatocellular carcinoma (HCC), Cell cycle arrest, Cyclin A, Cyclin-dependent kinase 2 (CDK2), p16, p53
INTRODUCTION
Liver cancer is the second highest cause of mortality of any type of cancer (1). Hepatocellular carcinoma (HCC) accounts for up to 90% of all primary liver cancers worldwide and causes up to 600,000 deaths per year globally (1,2). Due to rapid growth and early vascular invasion, most HCC patients present with advanced symptomatic tumors that are not amenable to surgical resection (3). Even after surgical resection, the recurrence rate is very high (4). The poor prognosis of patients with nonresectable or recurrent HCC supports a rationale for the use of chemotherapy. Presently used agents include doxorubicin or, in a combination with cisplatin, interferon, sorafenib (5) and 5-fluorouracil (5-FU) (6). However, chemotherapy has not been used consistently in the treatment of advanced HCC due to the highly refractory nature of HCC tumors to conventional cytotoxic chemotherapy, the presence of significant underlying poor hepatic reserve, and a lack of data from comprehensive clinical trials (7). Continuing research into improved and emerging targeted agents is vital to developing efficacious and safe treatments that combat HCC.
Raltitrexed (RTX; ZD1694, Tomudex), a quinazoline analogue of folinic acid, has been extensively employed in the management of colorectal cancer patients (8,9). RTX alone or in combination with oxaliplatin, epirubicin, or cisplatin has also been used in the treatment and clinical trials of metastatic pancreatic cancer patients (10), advanced pancreatic and biliary carcinomas patients (11), and recurrent or refractory leukemia patients (12). RTX is a robust, long-lasting inhibitor of the enzyme thymidylate synthase (TS), and this is possibly the source of its potent anticancer activity (13). TS is a key enzyme involved in the de novo synthesis of thymidine nucleotides of DNA. Several lines of preclinical data have demonstrated oncogene-like activity for TS, suggesting a link between TS-regulated DNA synthesis and neoplastic transformation (14). A statistically significant association exists between the TYMS3′UTR1494del6 polymorphism of the TS gene and reduced risk of HCC (15). Indeed, one transcription factor of TS, late SV40 factor, has been suggested as novel oncogene for HCC (16). A recent study suggested that the TS inhibitor suberoylanilide hydroxamic acid can increase the chemosensitivity of liver cancer cells to 5-FU when both drugs are combined, synergistically inhibiting cell growth and tumorgenicity in HCC (17). While RTX has been tested alone or as part of a combination therapy in several clinical trials involving HCC management (18,19), details on the molecular mechanism and biological effects are not clearly known. By investigating the biological and physiological functions of RTX, it may reveal insights that make progress on safe and effective treatment strategy for HCC (20,21).
In the current study, we selected HepG2 cells as an in vitro model to evaluate the effects of RTX. HepG2 represents a pure cell line of human liver carcinoma, often used as a HCC model due to the absence of viral infection (22). To assess the potential anticancer activity of RTX in liver cancer, we analyzed cell proliferation, ultrastructure and cell cycle in HepG2 cells. Our study demonstrated that RTX effectively inhibited HepG2 cell proliferation via G0/G1 cell cycle arrest, which could be attributed to changes in expression level of cell cycle regulatory proteins.
MATERIALS AND METHODS
Reagents
HepG2 cells were obtained from Harbin Medical University Cancer Institute. Cells were maintained in RPMI-1640 (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone) and penicillin (100 U/ml)/streptomycin (100 mg/ml) at 37°C in an atmosphere containing 5% CO2. Cells were passaged every 2–3 days, and cells in logarithmic phase were used in experiments.
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless stated otherwise. RTX was purchased from Chia Tai Tianqing (Nanjing, China), and 5-FU was from Kingyork (Tianjin, China). RNaseA was from TianGen (Beijing, China). Giemsa dye was obtained from Amresco (Solon, OH, USA). Cell Counting Kit was purchased from ZomanBio (Beijing, China). Cell lysis buffer was from Beyotime (Shanghai, China). Bradford assay kit was purchased from Jiancheng Bioengineering (Nanjing, China). Antibodies against TS, CDK2, p53, and p16 were from Origene (Rockville, MD, USA). Anti-cyclin A was purchased from Bioworld Technology (Atlanta, GA, USA). Anti-GAPDH was from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-β-actin, goat anti-rabbit IgG, and goat anti-mouse IgG were from ZSGB-Bio (Beijing, China). Transcriptor First Strand cDNA Synthesis Kit and FastStart Universal SYBR Green Master (ROX) were purchased from Roche (Germany). PCR primers were synthesized by Bioneer (Alameda, CA, USA).
Cell Culture and Treatment
HepG2 cells (4 × 104/ml) were seeded in a 96-well plate at 100 µl/well. After cells settled down, RTX (0, 16, 64, 256 nM) or 5-FU (0, 1.25, 2.5, 5 µM), a TS inhibitor chemotherapy agent serving as positive control, were added. HepG2 cells were incubated for an additional 24 h or 48 h as designated. Cell morphology was analyzed by an inverted fluorescence microscopy TE2000-U (Nikon, Japan).
Cell Proliferation Assay
Effect of RTX on HepG2 proliferation was evaluated by Cell Counting Kit that employed a method similar to MTT and used WST-8 reagent instead. HepG2 cells were seeded and treated as described above. After treatment, WST-8 reagent was applied according to the manual and cells were incubated for another 2 h. When the formazan crystals were dissolved completely, the level of optical density (OD) was measured at 450 nm by Microplate Reader (BioTek Instruments, Winooski, VT, USA). IC50 was calculated: percentage of growth (growth %) = ODTreatment/ODControl; percentage of inhibition (inhibition %) = 1 − growth %.
Colony Formation Efficiency Assay
Colony formation efficiency assay was carried out as described previously (23) with minor modifications. Briefly, HepG2 cells were passed through a cell strainer to obtain single cells before being counted and seeded in six-well plates at 500 cells/well, followed by gently shaking for the optimal separation. RTX (0, 16, 64, 256 nM) or 5-FU (0, 1.25, 2.5, 5 μM) were added into the culture medium. Cells were incubated for 24 h, and then supernatant was replaced with 3 ml regular RPMI-1640 containing 10% FBS. HepG2 cells were cultured for an additional 2–3 weeks until visible cell clones formed. HepG2 cells were then fixed by 4% paraformaldehyde (PFA) and stained by 10% Giemsa. Cell colonies were counted in a double-blind manner using a stereomicroscope: colony formation rate % = (colonies counted/cells inoculated) × 100. In the present study, cells inoculated = 500.
Transmission Electron Microscopy
HepG2 cells were treated with RTX (0, 16, 256 nM) or 5-FU (0, 1.25, 2.5, 5 µM) as described previously and incubated for 24 h before harvest. Cells were fixed with 2.5% glutaraldehyde for 2 h. After being rinsed by PBS, cell samples were dehydrated by gradient ethanol and subsequently washed by 100% tert-butanol and embedded in resin. The cell blocks were examined with H-7650 transmission electron microscope (Hitachi, Japan) at an accelerating voltage of 40–120 kV.
Cell Flow Cytometry Analysis
We used fluorescence activated cell sorter (FACS) analysis to examine the effects of RTX on the cell cycle. HepG2 cells were treated by RTX (0, 16, 64, 256 nM) as described. After a 24-h incubation, HepG2 cells were washed three times and resuspended in 500 µl PBS with 20 µl RNaseA and PI. After a 30-min incubation, cells (10,000 events) were acquired using a FACS cytometer FACSCalibur (BD, USA). Data acquisition and cell cycle distribution calculations were carried out using FlowJo software (Tree Star, USA). The number of gated cells in G1, G/M, or S phase were presented as a percentage of the total. Each experiment was performed in triplicate.
Real-Time PCR
Total mRNA was extracted from HepG2 cells using TRIzol reagent. Total cDNAs were synthesized using olig(dT)18 primer. Real-time PCR (RT-PCR) was performed using an ABI Prism7500 Sequence Detection System. The total reaction volume was 10 µl: 1 µl cDNA, 5 µl FastStart Universal, 0.15 µl of each primer and 3.7 µl diethyl pyrocarbonate (DEPC)-treated water. The PCR cycling conditions were as follows: activation at 95°C for 10 min, 45 cycles of denaturation at 95°C for 15 s, and then annealing and extension at 60°C for 30 s. Gene expression levels were carried out using the absolute curve method, and the expression level of β-actin gene was used for normalization. Sequences of primers used are listed in Table 1. We conducted three replicates for the assays and had four replicates of the individual genes in each PCR.
Table 1.
Sequences of Primers Used
| Target | Primer Sequence (5′–3′) |
|---|---|
| TS | F: CCT GGG GCA GAT CCA ACA CA |
| R: TGG ATC CCT TGA TAA ACC ACA GC | |
| Cyclin A | F: TCC ATG TCA GTG CTG AGA GGA |
| R: GAA GGT CCA TGA GAC AAG GC | |
| CDK2 | F: GCT AGC AGA CTT TGG ACT AGC CAG |
| R: AGC TCG GTA CCA CAG GGT CA | |
| p53 | F: GGC CCA CTT CAC CGT ACT AA |
| R: GTG GTT TCA AGG CCA GAT GT | |
| p16 | F: GAG CAG CAT GGA GCC TTC |
| R: TCC TCA TTC CTC TTC CTT GG | |
| β-Actin | F: CTC TTC CAG CCT TCC TTC CT |
| R: AGC ACT GTG TTG GCG TAC AG |
Western Blot Assay
HepG2 were seeded into six-well plates and treated with different stimulants as designated. Cells were then harvested by centrifugation and lysed using cell lysis buffer to obtain total protein. Protein concentration was determined by Bradford assay kit. Proteins (30 µg) were loaded into each lane of a 10% SDS-polyacrylamide gel and transferred onto PVDF membranes (Roche, Germany). Membranes were blocked for 1 h in 5% (w/v) nonfat milk and then incubated with primary antibody at room temperature for 1 h. After being washed three times in TBST, the membranes were incubated with a horseradish peroxidase-coupled goat anti-rabbit IgG (diluted 1:5000) for 1 h at room temperature and then washed three times. Finally, expression levels of protein were detected using an enhanced chemiluminescence system ECL (Roche). Immunoblotted bands were quantified by FluorChemFC2 imaging system (Alpha Innotech, San Leandro, CA, USA), and the protein of interest was normalized to β-actin.
Statistics
All values are presented as means ± standard error of the mean (SEM). Differences between experimental groups were analyzed for statistical significance using one-way analysis of variance (ANOVA). Values of p < 0.05 were considered statistically significant.
RESULTS
RTX Treatment Changed HepG2 Cell Morphology
The morphology of HepG2 cells was investigated using light microscopy with and without RTX treatment. Control group cells, treated with an equivalent amount of PBS, showed clear cell body shape and grew well with significant proliferation (Fig. 1A). In contrast, RTX-treated cells showed delayed growth. Nuclear anomalies (binucleated cells, fragmentation of nuclei, etc.) were observed in some RTX-treated cells (Fig. 1B). Similarly, in the 5-FU-treated group, cells did not grow well and exhibited apoptotic appearance. Cell bodies were reduced in size with disrupted intercellular contacts and damaged cytoplasm (Fig. 1C).
Figure 1.
The morphological changes of HepG2. (A) Control. (B, C) HepG2 cells in response to 24-h treatment with 64 nM RTX (B) or 2.5 μM 5-FU (C). Cells were observed using phase-contrast microscopy. Scale bar: 40 μm. Control, treated with an equivalent volume of PBS; RTX, raltitrexed; 5-FU, 5-fluorouracil.
RTX Treatment Inhibited HepG2 Cell Proliferation
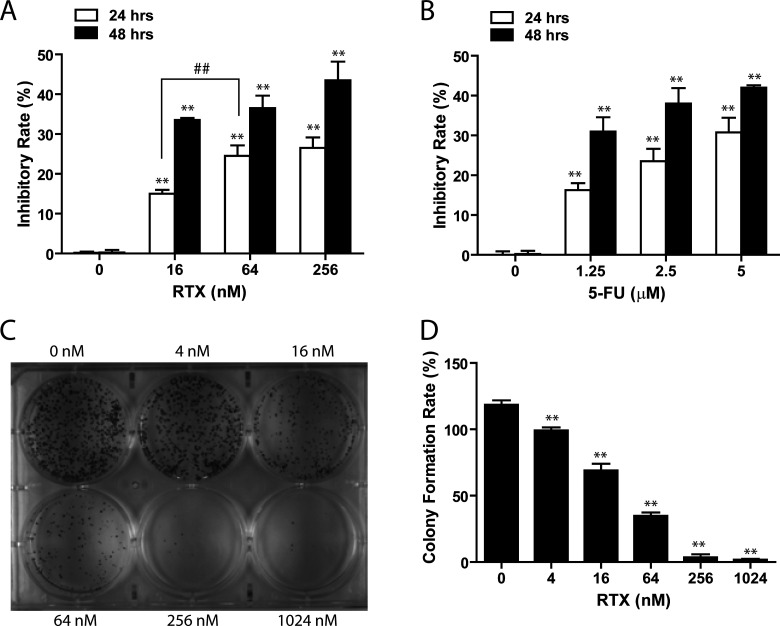
The effect of RTX on HepG2 cell proliferation was evaluated by WST-8 assay as described in Materials and Methods. This cell proliferation analysis revealed rapid cell growth of HepG2 cells in regular culture medium that was significantly reduced when the cells were first treated by RTX or 5-FU for 24 h. Figure 2A demonstrates that all doses of RTX (16, 64, and 256 nM) significantly inhibited HepG2 proliferation compared to control (p < 0.05), and the magnitude of this inhibition was dose dependent. After 24-h treatment, the growth inhibition percentages exerted by RTX (16, 64, and 256 nM) were 16.3 ± 0.5%, 22.1 ± 0.2%, and 24.5 ± 0.2%, respectively (Fig. 2A). The cell proliferation inhibition rate of RTX was similar to that of 5-FU. The growth inhibition rate of 5-FU at 1.25 μM was 18.2 ± 0.3%, and at 2.5 μM was 21.1 ± 0.9% (Fig. 2B). Using a 24-h treatment, the IC50 for RTX was determined to be 78.919 nM, and the IC50 for 5-FU was determined to be 3.245 µM. This inhibitory effect was also time dependent, as increasing the RTX treatment duration to 48 h showed an intensified inhibitory capacity: RTX (16, 64, and 256 nM) for 48 h led to 33.2 ± 0.3%, 34.3 ± 0.9%, and 36.1 ± 0.4% inhibition in HepG2 cells growth.
Figure 2.
Inhibitory effect of RTX on HepG2 proliferation. (A) WST-8 assay showed that RTX inhibited HepG2 cell proliferation in a time- and dose-dependent manner. (B) 5-FU, a positive control, induced similar inhibition in HepG2 proliferation. (C) Representative image of a six-well plate from the colony formation efficiency assay. (D) Quantitative analysis shows that the colony formation rate in cells treated with RTX was lower than that in untreated cells. Control, treated with an equivalent volume of PBS; RTX, raltitrexed; 5-FU, 5-fluorouracil. **p < 0.01 versus control; ##p < 0.01 versus different concentration.
To validate the inhibitory effects of RTX on HepG2 proliferation, we conducted colony formation efficiency assay. The results showed that colony formation was significantly suppressed with increased concentration of RTX (Fig. 2C, D). Specifically, the number of cell colonies was reduced by 40% in cells treated with 16 nM RTX compared to control cells (p < 0.05). A concentration-related viability decrease was also observed, corroborating the results of the WST-8 assay.
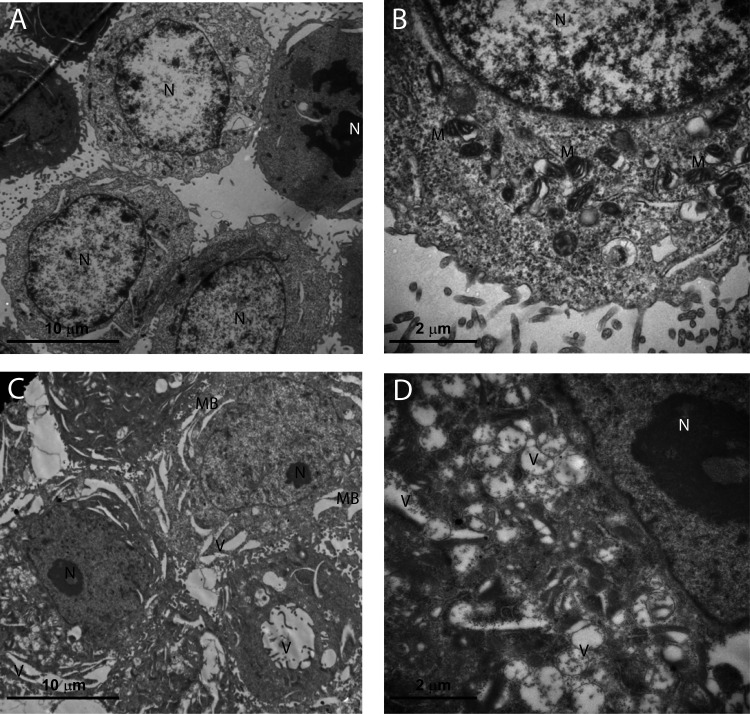
RTX Treatment Altered Ultrastructure of HepG2 Cell
To investigate if RTX treatment led to ultrastructure changes in proliferation-inhibited HepG2 cells, we applied transmission electron microscopy, a useful tool for observing morphologic changes. Alterations in ultrastructure by TEM are shown in Figure 3. The control group presented regular HepG2 morphology characterized by fine-textured nuclear chromatin, intact nuclear membranes and cytoplasmic membranes, plenty of mitochondria that showed parallel and intact cristae. A number of mitotic cells were observed (Fig. 3A, B). In contrast, the RTX group exhibited severe plasma membrane interruption, dilated nuclear membranes, nuclear chromatin condensation and margination, reduced cellular organelles, as well as lytic changes such as foamy-vacuolated cytoplasm and numerous, presumably autophagic, vacuoles. Cells in early apoptotic stage were observed but no cells in mitotic phase were detected (Fig. 3C, D).
Figure 3.
Transmission electron microscope views of RTX-treated HepG2 cells at 24 h. (A) The control group at a 5,000× magnification (HV = 80.0 kV). (B) The control group at a 20,000× magnification (HV = 80.0 kV). Untreated cells demonstrate a structure and morphology typical of HepG2 cells. The nucleus contains evenly distributed chromatin, and large and numerous mitochondria exist. (C) The RTX (256 nM)-treated group at a 5,000× magnification (HV = 80.0 kV). (D) The RTX (256 nM)-treated group at a 20,000× magnification (HV = 80.0 kV). Cells treated with 256 nM RTX for 24 h show membrane blebbing and reduced number of subcellular organelles and vacuolization. Some cells exhibited formation of apoptotic micronuclei. RTX, raltitrexed; N, nucleus; M, mitochondria; MB, membrane blebbing; V, vacuolization.
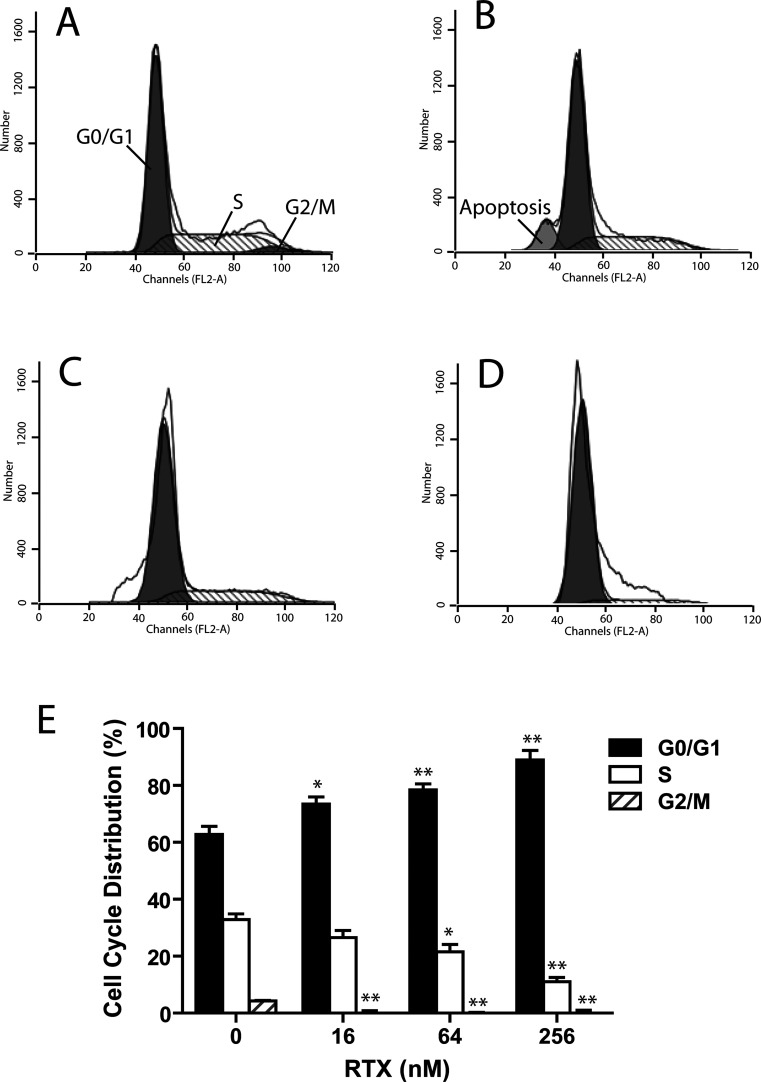
RTX Induced HepG2 Cell Cycle Arrest in G0/G1 Phase
To gain insight into the mechanism underlying the cell proliferation inhibition triggered by RTX, we tested if RTX treatment changed cell cycle distribution in HepG2 cells using cell flow cytometry. As shown in Figure 4A through D, concomitant with the inhibitory effect on HepG2 cell proliferation, RTX induced a strong G1 phase arrest after 24-h treatment. Cell populations in the G0/G1, S, and G2/M phases were 60.5%, 35.2%, and 4.3% of the total population, respectively, in control HepG2 cells (Fig. 4E). After incubation with RTX (16, 64, and 256 nM), the S phase population was significantly reduced to 28.9%, 23.9%, and 13.4%, respectively (p < 0.05 vs. control). RTX-induced S phase population decrease was accompanied with an increase in G0/G1 phase population to 71.5%, 76.1%, and 86.7% of the total population, respectively in response to increased concentrations.
Figure 4.
Cell cycle distribution of HepG2 was analyzed by flow cytometry. (A) Cell cycle distribution in control group. (B–D) Arrests of cell cycle at G0/G1 in HepG2 in response to increased dosages of RTX 24 h after treatment. (B) RTX = 16 nM, (C) RTX = 64 nM, (D) RTX = 256 nM. (E) The distribution of the cell cycle of HepG2 cells was assessed by flow cytometry after staining with propidium iodide (PI). RTX significantly inhibited the proliferation of HepG2 cells by triggering cell cycle arrest at G0/G1 phase in a dose-dependent manner. RTX, raltitrexed. *p < 0.05 versus control; **p < 0.01 versus control.
RTX-Inhibited HepG2 Cells Enter S Phase via Downregulating Cyclin A/CDK2 Complex
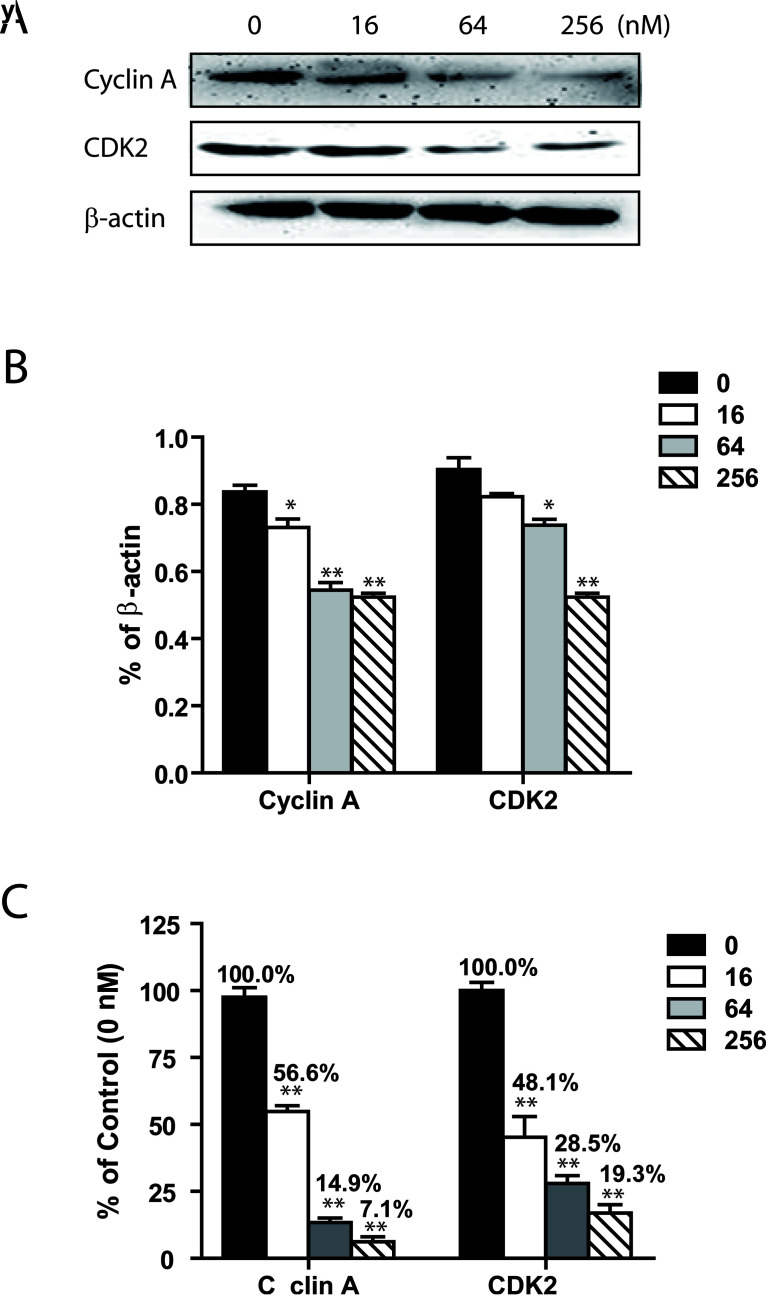
Cell cycle progression is tightly controlled by the regulation of expression and activity of cyclin/cyclin-dependent kinase complexes. To elucidate signaling pathways involved in RTX-induced G0/G1 phase cell cycle arrest in HepG2 cells, we measured the expression levels of cyclin A/CDK2 complex. Activation of this complex enables cells to pass from G1 to S phase (24–26).
We first analyzed protein and mRNA expression levels of TS in response to RTX treatment. Our result indicated that RTX did not significantly alter the expressions of TS in HepG2 cell. We then assessed expression levels of CDK2. Both a significantly reduced mRNA and significantly reduced protein expression level of CDK2 was observed after 24 h of RTX incubation (Fig. 5A, B). At a dosage of 256 nM, RTX treatment resulted in a 50% reduction in the protein expression levels of CDK2 compared to control. In addition, the mRNA levels of CDK2 markedly dropped to 19.0% of the control with the use of 256 nM RTX. In mammalian cells, cyclin A binds to CDK2 to create an active complex that is critical for CDK2 activation (24). We found that both mRNA and protein expression levels of cyclin A in HepG2 were significantly reduced after 24-h incubation with RTX (Fig. 5C).
Figure 5.
RTX triggered downregulation of CDK2 and cyclin A in HepG2 cells 24 h after treatment. (A) Representative images of Western blot analysis of CDK2 and cyclin A protein expressions in HepG2 cells. CDK2 and cyclin A protein expressions in HepG2 were inhibited by RTX in a dose-dependent manner. (B) Relative protein levels of CDK2 and cyclin A. All data are presented as mean ± SEM (n = 3). All message levels were normalized to that of β-actin. (C) Relative mRNA message levels of TS, CDK2, and cyclin A in HepG2 cells in response to RTX treatment. All data are presented as mean ± SEM (n = 4). All message levels were normalized to that of control (0 nM RTX). Average message RNA level of target gene is labeled above each bar. RTX, raltitrexed; CDK2, cyclin-dependent kinase 2. *p < 0.05 versus control; **p < 0.01 versus control.
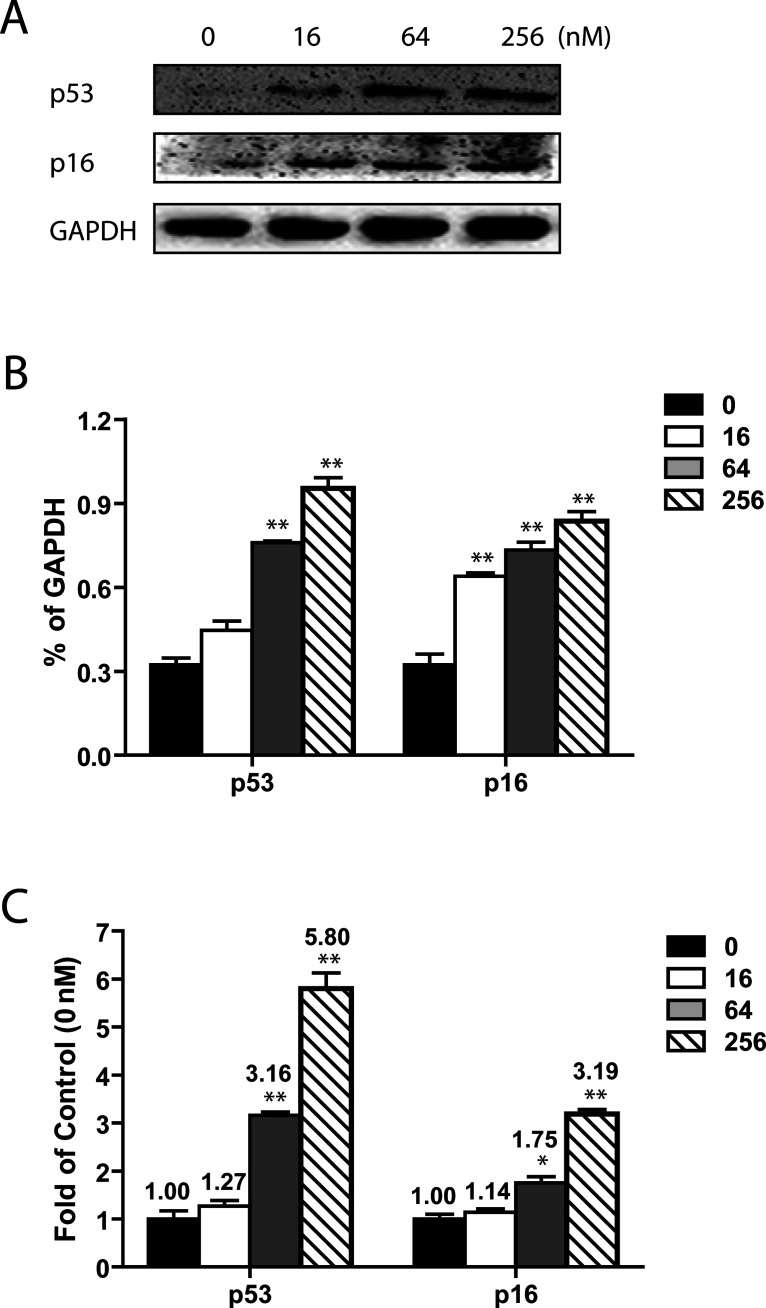
RTX Upregulated Tumor Suppressor Genes p53 and p16
CDKs are regulated by two families of inhibitors: the INK4 inhibitors (including p16) and the Cip/Kip inhibitors (including p53) (26). To further investigate the mechanism underlying RTX-mediated cell cycle arrest, we analyzed the well-known cellular tumor suppressor, p53. p53 is involved in cell cycle regulation as a trans-activator that negatively regulates cell division by controlling expression of a set of genes required for the process. Protein levels of p53 were also significantly upregulated when assessed by Western blot (Fig. 6A, B). Consistent with that, real-time PCR data showed that RTX significantly enhanced mRNA expression of p53. In the HepG2 cells, a 1.3- up to 5.8-fold increase in p53 mRNA expression was observed compared to the control group (Fig. 6C).
Figure 6.
RTX induced upregulation of p53 and p16 in HepG2 cells 24 h after treatment. (A) Representative images of Western blot analysis of p53 and p16 protein expressions in HepG2 cells. Protein expressions of p53 and p16 in HepG2 were increased by RTX in a dose-dependent manner. (B) Relative protein levels of p53 and p16. All data are presented as mean ± SEM (n = 3). All mRNA message levels were normalized to that of β-actin. (C) Relative message levels of p53 and p16 in HepG2 cells in response to RTX treatment. All data are presented as mean ± SEM (n = 4). All message levels were normalized to that of control (0 nM RTX). Average message RNA level of target gene is labeled above each bar. RTX, raltitrexed. *p < 0.05 versus control; **p < 0.01 versus control.
Following this, we examined whether p16 (cyclin-dependent kinase inhibitor 2A, P16INK4A) signaling was involved in RTX-induced G0/G1 phase arrest. In our hands, we identified significant upregulation of p16 in response to RTX. Western blot analysis revealed a marked increase in the protein levels of p16 under RTX treatment (Fig. 6A, B). Similar results were obtained from RT-PCR study; the RTX dose-dependently enhanced mRNA levels of p16 (Fig. 6C).
DISCUSSION
In the work presented here, we demonstrate that RTX significantly inhibited HepG2 proliferation and arrested the cell cycle at the G0/G1 phase. This cell cycle arrest was mediated through downregulation of cyclin A and CDK2, the key regulators responsible for the transition from G1 phase to S phase. Moreover, elevated expressions of p53 and p16 by RTX were identified, which could contribute to the reduction of cyclin A/CDK2 complex. Our study indicates that RTX may serve as a potential chemotherapeutic agent in the treatment of hepatocellular carcinoma patients.
Synthesized as part of a program in rational drug discovery, RTX has attracted great interest in investigating its mechanism and optimal dose/schedule against cancer. A number of studies support that RTX is well tolerated and associated with a relatively low risk of hematological toxicity, diarrhea, and mucositis, demonstrating it has potential as a major player in cancer management (8,27,28). RTX has been suggested to possess a different toxicity profile from that of 5-FU, another TS inhibitor (8). In the current study using HepG2 as in vitro model, we found that RTX effectively inhibited HepG2 cell proliferation in a time- and dose-dependent manner and triggered cell cycle arrest in the G0/G1 phase. This finding was inconsistent with previous reports that RTX exerted growth inhibitory effect in SGC7901 human gastric cancer cells (29), which was accompanied by cell cycle arrest at the G0/G1 phase. The cell cycle arrest function of RTX may be cell-type specific as RTX has been reported to induce a buildup of cells in the S phase in SW620 colon carcinoma cells (30). Interestingly, although both RTX and 5-FU act as TS inhibitors, our result indicates that RTX exhibited different mechanisms in preventing cell proliferation, compared to 5-FU. It has been reported that low-dose 5-FU (1 mg/ml) induced cell cycle G2 arrest and apoptosis in keloid fibroblasts (31). In contrast, in response to 7.5 μg/ml of 5-FU, S phase cell arrest was observed in HepG2 after 48-h treatment (32). Other studies showed that exposure to 100 ng/ml of 5-FU in SW480 and COLO320DM caused G1 arrest after 24 h and G2 arrest after 72–144 h (33). These studies elucidated that cell cycle arrest function of 5-FU was cell-type dependent, dose dependent, and time dependent, suggesting that similar effects may also be found in RTX.
Furthermore, we identified that RTX-induced cell cycle inhibition was associated with downregulation of cyclin A and CDK2. Cyclin A/CDK2 complexes phosphorylate serine residues on pRb, which is crucial for the G1 to S phase transition in proliferating HepG2 cells. Decreased cyclin A expression contributed to the cell cycle arrest at G1 in HepG2 cells (34). In addition, cyclin A accumulates prior to cyclin B in the cell cycle and appears to be involved in control of S phase. Previous study has determined that expression of cyclin A was significantly elevated in HCC (35). Recent study using a conditional genetic knockout mouse model also showed that mice with cyclin A2-deficient hepatocytes had delayed liver tumor formation. In addition, in oncogene-transformed mouse embryo fibroblasts lacking both cyclin A2 and CDK2, tumor formation was strongly suppressed in a manner associated with decreased proliferation (36). These previous studies, together with our data, suggested that RTX could inhibit liver tumor growth by stalling the cell cycle through downregulation of cyclin A/CDK2 expression.
Various signals are involved in regulation of cell cycle proteins. p53 is particularly interesting to us because a previous study found that expression of p53 is critical for sensitivity of colorectal cancer cells to RTX (37). In another study, transfecting colorectal cancer cells with mutant p53 prevented cells from responding to RTX (38). To add importance to the role of p53 in RTX-mediated cyclin A/CDK2 expression, Orlandi et al. confirmed that RTX could not change the expression of CDK2 and cyclin A in the MDA-MB-435 human breast cancer cell line that contains a mutated p53 gene (39). Moreover, another group reported RTX promoted a 1.9-fold increase in the expression of cyclin A and 3.4-fold increase in CDK2 in the same MDA-MB-435 cells. Interestingly, these upregulation effects could be diminished if TS was overexpressed (40). Our result elucidated that increased p53 expression was associated with RTX-induced cyclin A/CDK2 reduction. It will be interesting to examine whether p53 is dispensable to RTX’s inhibitory function in HepG2 cells or other liver cancer cell lines, because p53-independent apoptosis in RTX-induced toxicity was also indicated in human colon carcinoma cell lines as well as a murine model (41,42).
Tumor suppressor gene p16 belongs to the INK4 family of CDK inhibitors, contributing to the regulation of cell cycle progression by inhibiting progression from G to S phase (43). p16 has been recognized as a tumor suppressor gene due to the prevalence of its genetic inactivation in a variety of human cancers (44), including HCC (45,46). p16 is capable of inducing cell cycle arrest in the G1 phase (47). Min et al. reported that upregulation of p16 was critical to the antitumor activity of HepG2 cells treated with fucoidan (48). To date, we are not aware of any investigation into regulation of p16 by RTX. Our preclinical work here represents the first demonstration of an interaction between p16 and RTX. Our study could represent a rationale for further clinical investigation into the mechanisms underlying the inhibition of proliferation by RTX on cancer cells. Furthermore, it will be particularly interesting to conduct future studies investigating if RTX interferes with expression of p14/p19ARF (ARF), a distinct protein encoded by the same INK4A/ARF locus as p16. ARF, at the same time, interestingly, is capable of inducing both G1 and G2 arrest due to its stabilizing effects on p53 (47).
In conclusion, the current study elucidated the effectiveness of RTX in prohibiting HepG2 cells from proliferating. HepG2 cells treated with RTX were arrested in the G0/G1 phase via downregulation of the cyclin A/CDK2 complex. Increased p53 and p16 expressions were found to be associated with RTX’s biological activity. Our study supports the potential antitumor capacity of RTX in liver cancer and provides insight into the mechanism underlying this effect. A deeper and more comprehensive understanding of the mechanisms and molecular events involved in RTX-mediated growth suppression will aid in the diagnosis, prognosis, and treatment of liver cancer.
REFERENCES
- 1. Llovet J. M.; Villanueva A.; Lachenmayer A.; Finn R. S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat. Rev. Clin. Oncol. 12:436; 2015. [DOI] [PubMed] [Google Scholar]
- 2. Yang J. D.; Roberts L. R. Hepatocellular carcinoma: A global view. Nat. Rev. Gastroenterol. Hepatol. 7:448–458; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pang R. W.; Joh J. W.; Johnson P. J.; Monden M.; Pawlik T. M.; Poon R. T. Biology of hepatocellular carcinoma. Ann. Surg. Oncol. 15:962–971; 2008. [DOI] [PubMed] [Google Scholar]
- 4. Lacaze L.; Scotté M. Surgical treatment of intra hepatic recurrence of hepatocellular carcinoma. World J. Hepatol. 7:1755–1760; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhai J. M.; Yin X. Y.; Lai Y. R.; Hou X.; Cai J. P.; Hao X. Y.; Liang L. J.; Zhang L. J. Sorafenib enhances the chemotherapeutic efficacy of S-1 against hepatocellular carcinoma through downregulation of transcription factor E2F-1. Cancer Chemother. Pharmacol. 71:1255–1264; 2013. [DOI] [PubMed] [Google Scholar]
- 6. Masuzaki R.; Yoshida H.; Tateishi R.; Omata M. Staging systems: Is there a surgical staging and a medical one? Hepatologist’s perspective. J. Hepatobiliary Pancreat. Sci. 17:440–442; 2010. [DOI] [PubMed] [Google Scholar]
- 7. Attwa M. H.; El-Etreby S. A. Guide for diagnosis and treatment of hepatocellular carcinoma. World J. Hepatol. 7:1632–1651; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y.; Wu W.; Hong W.; Sun X.; Wu J.; Huang Q. Raltitrexed-based chemotherapy for advanced colorectal cancer. Clin. Res. Hepatol. Gastroenterol. 38:219–225; 2014. [DOI] [PubMed] [Google Scholar]
- 9. Judson I. R. ‘Tomudex’ (raltitrexed) development: Preclinical, phase I and II studies. Anticancer Drugs 8(Suppl. 2):S5–S9; 1997. [DOI] [PubMed] [Google Scholar]
- 10. Reni M.; Pasetto L.; Aprile G.; Cordio S.; Bonetto E.; Dell’Oro S.; Passoni P.; Piemonti L.; Fugazza C.; Luppi G.; Milandri C.; Nicoletti R.; Zerbi A.; Balzano G.; Di Carlo V.; Brandes A. A. Raltitrexed-eloxatin salvage chemotherapy in gemcitabine-resistant metastatic pancreatic cancer. Br. J. Cancer 94:785–791; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. François E.; Hebbar M.; Bennouna J.; Mayeur D.; Perrier H.; Dorval E.; Martin C., Bourgeois H.; Barthélemy P.; Douillard J. Y. A phase II trial of raltitrexed (Tomudex) in advanced pancreatic and biliary carcinoma. Oncology 68:299–305; 2005. [DOI] [PubMed] [Google Scholar]
- 12. Horton T. M.; Blaney S. M.; Langevin A. M.; Kuhn J.; Kamen B.; Berg S. L.; Bernstein M.; Weitman S. Phase I trial and pharmacokinetic study of raltitrexed in children with recurrent or refractory leukemia: A pediatric oncology group study. Clin. Cancer Res. 11:1884–1889; 2005. [DOI] [PubMed] [Google Scholar]
- 13. Avallone A.; Di Gennaro E.; Silvestro L.; Iaffaioli V. R.; Budillon A. Targeting thymidylate synthase in colorectal cancer: Critical re-evaluation and emerging therapeutic role of raltitrexed. Expert Opin. Drug Saf. 13:113–129; 2014. [DOI] [PubMed] [Google Scholar]
- 14. Voeller D.; Rahman L.; Zajac-Kaye M. Elevated levels of thymidylate synthase linked to neoplastic transformation of mammalian cells. Cell Cycle 3:1005–1007; 2004. [PubMed] [Google Scholar]
- 15. Yuan J. M.; Lu S. C.; Van Den Berg D.; Govindarajan S.; Zhang Z. Q.; Mato J. M.; Yu M. C. Genetic polymorphisms in the methylenetetrahydrofolate reductase and thymidylate synthase genes and risk of hepatocellular carcinoma. Hepatology 46:749–758; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santhekadur P. K.; Rajasekaran D.; Siddiq A.; Gredler R.; Chen D.; Schaus S. E.; Hansen U.; Fisher P. B.; Sarkar D. The transcription factor LSF: A novel oncogene for hepatocellular carcinoma. Am. J. Cancer Res. 2:269–285; 2012. [PMC free article] [PubMed] [Google Scholar]
- 17. Liao B.; Liang H.; Chen J.; Liu Q.; Zhang B.; Chen X. Suberoylanilide hydroxamic acid enhances chemosensitivity to 5-fluorouracil in hepatocellular carcinoma via inhibition of thymidylate synthase. Tumour Biol. 36:9347–56; 2015. [DOI] [PubMed] [Google Scholar]
- 18. Geurs F.; Vandewaeter S.; Ponette S.; Ponette J.; Knape S.; Demetter P. Successful and well-tolerated second line therapy with cetuximab, irinotecan, and raltitrexed in progressive liver disease due to metastatic colon cancer. J. Gastrointest. Cancer 39:26–28; 2008. [DOI] [PubMed] [Google Scholar]
- 19. Rougier P.; Ducreux M.; Kerr D.; Carr B. I.; François E.; Adenis A.; Sevmour L. A phase II study of raltitrexed (‘Tomudex’) in patients with hepatocellular carcinoma. Ann. Oncol. 8:500–502; 1997. [DOI] [PubMed] [Google Scholar]
- 20. Santini D.; Picardi A.; Vincenzi B.; Binetti P.; Massacesi C.; La Cesa A.; Tonini G. Severe liver dysfunction after raltitrexed administration in an HCV-positive colorectal cancer patient. Cancer Invest. 21:162–163; 2003. [DOI] [PubMed] [Google Scholar]
- 21. Raderer M.; Fiebiger W.; Wrba F.; Scheithauer W. Fatal liver failure after the administration of raltitrexed for cancer chemotherapy: A report of two cases. Cancer 89:890–892; 2000. [DOI] [PubMed] [Google Scholar]
- 22. Costantini S.; Di Bernardo G.; Cammarota M.; Castello G.; Colonna G. Gene expression signature of human HepG2 cell line. Gene 518:335–345; 2013. [DOI] [PubMed] [Google Scholar]
- 23. Rafehi H.; Orlowski C.; Georgiadis G. T.; Ververis K.; El-Osta A.; Karagiannis T. C. Clonogenic assay: Adherent cells. J. Vis. Exp. 49:2573; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Collins K.; Jacks T.; Pavletich N. P. The cell cycle and cancer. Proc. Natl. Acad. Sci. USA 94:2776–2778; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malumbres M.; Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 9:153–166; 2009. [DOI] [PubMed] [Google Scholar]
- 26. Neganova I.; Lako M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J. Anat. 213:30–44; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Papanastasopoulos P.; Stebbing J. Molecular basis of 5-fluorouracil-related toxicity: Lessons from clinical practice. Anticancer Res. 34:1531–1535; 2014. [PubMed] [Google Scholar]
- 28. Kelly C.; Bhuva N.; Harrison M.; Buckley A.; Saunders M. Use of raltitrexed as an alternative to 5-fluorouracil and capecitabine in cancer patients with cardiac history. Eur. J. Cancer 49:2303–2310; 2013. [DOI] [PubMed] [Google Scholar]
- 29. Xue S.; Chen Y. X.; Qin S. K.; Yang A. Z.; Wang L.; Xu H. J.; Geng H. Y. Raltitrexed induces mitochondrial–mediated apoptosis in SGC7901 human gastric cancer cells. Mol. Med. Rep. 10:1927–1934; 2014. [DOI] [PubMed] [Google Scholar]
- 30. Smith T. A.; Maisey N. R.; Titley J. C.; Jackson L. E.; Leach M. O.; Ronen S. M. Treatment of SW620 cells with Tomudex and oxaliplatin induces changes in 2-deoxy-D-glucose incorporation associated with modifications in glucose transport. J. Nucl. Med. 41:1753–1759; 2000. [PubMed] [Google Scholar]
- 31. Huang L.; Wong Y. P.; Cai Y. J.; Lung I.; Leung C. S.; Burd A. Low-dose 5-fluorouracil induces cell cycle G2 arrest and apoptosis in keloid fibroblasts. Br. J. Dermatol. 163:1181–1185; 2010. [DOI] [PubMed] [Google Scholar]
- 32. Liang S. R.; Hu G. R.; Fang L. J.; Huang S. J.; Li J. S.; Zhao M. Y.; Meng M. J. CpG oligodeoxynucleotides enhance chemosensitivity of 5-fluorouracil in HepG2 human hepatoma cells via downregulation of the antiapoptotic factors survivin and livin. Cancer Cell Int. 13:106; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoshikawa R.; Kusunoki M.; Yanagi H.; Noda M.; Furuyama J. I.; Yamamura T.; Hashimoto-Tamaoki T. Dual antitumor effects of 5-fluorouracil on the cell cycle in colorectal carcinoma cells: A novel target mechanism concept for pharmacokinetic modulating chemotherapy. Cancer Res. 61:1029–1037; 2001. [PubMed] [Google Scholar]
- 34. Tsukada Y.; Tanaka T.; Miyazawa K.; Kitamura N. Involvement of down-regulation of Cdk2 activity in hepatocyte growth factor-induced cell cycle arrest at G1 in the human hepatocellular carcinoma cell line HepG2. J. Biochem. 136:701–709; 2004. [DOI] [PubMed] [Google Scholar]
- 35. Masaki T.; Shiratori Y.; Rengifo W.; Igarashi K.; Yamagata M.; Kurokohchi K.; Uchida N.; Miyauchi Y.; Yoshiji H.; Watanabe S.; Omata M.; Kuriyama S. Cyclins and cyclin-dependent kinases: Comparative study of hepatocellular carcinoma versus cirrhosis. Hepatology 37:534–543; 2003. [DOI] [PubMed] [Google Scholar]
- 36. Gopinathan L.; Tan S. L.; Padmakumar V. C.; Coppola V.; Tessarollo L.; Kaldis P. Loss of Cdk2 and cyclin A2 impairs cell proliferation and tumorigenesis. Cancer Res. 74:3870–3879; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Di Gennaro E.; Bruzzese F.; Pepe S.; Leone A.; Delrio P.; Subbarayan P. R.; Avallone A.; Budillon A. Modulation of thymidilate synthase and p53 expression by HDAC inhibitor vorinostat resulted in synergistic antitumor effect in combination with 5FU or raltitrexed. Cancer Biol. Ther. 8:782–791; 2009. [DOI] [PubMed] [Google Scholar]
- 38. Giovannetti E.; Backus H. H.; Wouters D.; Ferreira C. G.; van Houten V. M.; Brakenhoff R. H.; Poupon M. F.; Azzarello A.; Pinedo H. M.; Peters G. J. Changes in the status of p53 affect drug sensitivity to thymidylate synthase (TS) inhibitors by altering TS levels. Br. J. Cancer 96:769–775; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Orlandi L.; Bearzatto A.; Abolafio G.; De Marco C.; Daidone M. G.; Zaffaroni N. Involvement of bcl-2 and p21waf1 proteins in response of human breast cancer cell clones to Tomudex. Br. J. Cancer 81:252–260; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Longley D. B.; Ferguson P. R.; Boyer J.; Latif T.; Lynch M.; Maxwell P.; Harkin D. P.; Johnston P. G. Characterization of a thymidylate synthase (TS)-inducible cell line: A model system for studying sensitivity to TS- and non-TS-targeted chemotherapies. Clin. Cancer Res. 7:3533–3539; 2001. [PubMed] [Google Scholar]
- 41. Backus H. H.; Wouters D.; van der Wilt C. L.; Kuiper C. M.; van Groeningen C. J.; Pinedo H. M.; Peters G. J. Thymidylate synthase inhibition induces p53 dependent and independent cell death. Adv. Exp. Med. Biol. 486:303–306; 2000. [DOI] [PubMed] [Google Scholar]
- 42. Pritchard D. M.; Bower L.; Potten C. S.; Jackman A. L.; Hickman J. A. The importance of p53-independent apoptosis in the intestinal toxicity induced by raltitrexed (ZD1694, Tomudex): Genetic differences between BALB/c and DBA/2 mice. Clin. Cancer Res. 6:4389–4395; 2000. [PubMed] [Google Scholar]
- 43. Romagosa C.; Simonetti S.; López-Vicente L.; Mazo A.; Lleonart M. E.; Castellvi J.; Ramon y Cajal S. p16(Ink4a) overexpression in cancer: A tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 30:2087–2097; 2011. [DOI] [PubMed] [Google Scholar]
- 44. Akçay N. İ.; Bashirov R.; Tüzmen Ş. Validation of signalling pathways: Case study of the p16-mediated pathway. J. Bioinform. Comput. Biol. 13:1550007; 2015. [DOI] [PubMed] [Google Scholar]
- 45. Kita R.; Nishida N.; Fukuda Y.; Azechi H.; Matsuoka Y.; Komeda T.; Sando T.; Nakao K.; Ishizaki K. Infrequent alterations of the p16INK4A gene in liver cancer. Int. J. Cancer. 67:176–180; 1996. [DOI] [PubMed] [Google Scholar]
- 46. Pulling L. C.; Klinge D. M.; Belinsky S. A. p16INK4a and beta-catenin alterations in rat liver tumors induced by NNK. Carcinogenesis. 22:461–466; 2001. [DOI] [PubMed] [Google Scholar]
- 47. Ivanchuk S. M.; Mondal S.; Dirks P. B.; Rutka J. T. The INK4A/ARF locus: Role in cell cycle control and apoptosis and implications for glioma growth. J. Neurooncol. 51:219–229; 2001. [DOI] [PubMed] [Google Scholar]
- 48. Min E. Y.; Kim I. H.; Lee J.; Kim E. Y.; Choi Y. H.; Nam T. J. The effects of fucodian on senescence are controlled by the p16INK4a-pRb and p14Arf-p53 pathways in hepatocellular carcinoma and hepatic cell lines. Int. J. Oncol. 45:47–56; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]