Abstract
Hepatocellular carcinoma (HCC) is one of the most common malignancies, with a poor prognosis and high recurrence rate. In the present study, we identified CD133, one of the markers of cancer stem cells, as a novel molecular target of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). In four human HCC cell lines established from primary HCC tumors, we found that CD133-high human liver cancer stem-like cells (CD133hi) derived from the SNU-475 cell line were highly susceptible to TRAIL compared to other HCC cell lines with a small population of CD133. CD133hi SNU-475 cells showed upregulation of TRAIL receptor DR5 and stemness-related genes such as c-Myc and ABC transporters compared to their CD133-low (CD133lo) cells. Hypersensitivity of CD133hi cells to TRAIL was associated with c-Myc-mediated upregulation of DR5 and downregulation of c-FLIPL in the cells. Knockdown of CD133 expression in CD133hi cells resulted in the downregulation of c-Myc, and depletion of c-Myc caused a decrease in the cell surface expression of DR5 and an increase in the expression of c-FLIPL and, consequently, attenuated TRAIL-induced cytotoxicity and apoptosis of CD133hi cells. These results suggest that TRAIL may provide a new strategy for CD133hi CSCs of HCC-targeted therapies and, potentially, for therapies of other CD133-expressing types of cancer.
Key words: TRAIL, Hepatocellular carcinoma (HCC), Cancer stem cells, c-Myc, CD133
INTRODUCTION
Hepatocellular carcinoma (HCC) represents the major histological subtype among liver cancers and is one of the most common malignant human tumors worldwide. Several human cancers contain a small subpopulation of cells called cancer stem-like cells (CSCs) (1), and also functional liver cancer stem cells were found in HCC cell lines (2). CSCs are difficult to treat with conventional methods because of their therapy-resistant phenotype as well as their ability to stimulate angiogenesis and metastasis. The successful eradication of cancer requires anticancer therapy that affects the differentiated cancer cells and the potential CSC population. CSCs showed preferential expression of some stem cell-associated genes and were more resistant to chemotherapeutic agents due to the upregulation of several ATP-binding cassette (ABC) transporters (3). Among all of the protective mechanisms for CSCs, ABC protein overexpression is one of the most reported. Acquisition of drug resistance is the active efflux of chemotherapeutic agents via ABC transporters such as P-glycoprotein (P-gp/ABCB1), breast cancer resistance protein (BCRP/ABCG2), and multidrug resistance proteins (MRP/ABCC1).
CD133, a five-transmembrane cell surface glycoprotein, may play an important role in chemoresistance and recurrence (4) and is widely used as a molecular marker to represent the subset population of CSCs in various types of tumors as well as HCC (5–12). It was reported that CD133+ cells were found in some of the primary human HCC tissue samples and in regenerative liver after massive necrosis but not in normal liver tissues (13), and an elevated CD133 level indicates a poor prognosis for patients with HCC (14). It has been reported that suppression of CD133 enhances the sensitivity of liver CSCs to chemotherapy and radiotherapy (15). Therefore, targeting CD133 as a means to eradicate the therapy-resistant liver CSCs may increase chemotherapeutic efficacy and improve the prognosis for patients with HCC.
The tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) has been suggested as a promising anticancer agent, as it can induce apoptosis in a wide range of cancers without significant cytotoxicity toward normal cells (16). After binding of TRAIL to the death receptors TRAIL-RI (DR4) and/or TRAIL-RII (DR5), the activated receptors recruit an adapter protein called Fas-associated death domain (FADD) and procaspase 8 molecule to form a death-inducing signaling complex (DISC) (17,18). On joining the DISC, caspase 8 is activated and promotes apoptosis by activating downstream effector caspases (19). Recruitment of cellular-FLICE inhibitory protein (c-FLIP) including both FLIPL and FLIPS blocks the processing and activation of procaspase 8 and 10 at the DISC level and inhibits cell death mediated by the DR4 and DR5 (20). As a result, c-FLIP hinders apoptosis by inhibiting the activation of caspase 8, and thus inhibition of c-FLIP enhances TRAIL-induced apoptosis in cancer cells (21).
The deregulation of c-Myc confers a selective advantage on cancer cells by promoting proliferation and cell survival. In CSCs, c-Myc expression is higher than in other cancer cells and plays an important role in CSC proliferation and survival (22). It has been shown that c-Myc can enhance TRAIL susceptibility via direct repression of c-FLIP expression (23) and upregulation of the cell surface level of DR5 (24). In addition, multidrug-resistant (MDR) cancer cells expressing a high level of P-gp and c-Myc were highly susceptible to TRAIL treatment (25). We therefore hypothesize that TRAIL could be used to eliminate liver CSCs or CSC-like cells expressing a high level of c-Myc. This study represents the molecular mechanism of TRAIL susceptibility in liver CSCs with high-level CD133 and suggests TRAIL as a candidate agent for targeting eradication of CD133-enriched CSCs.
MATERIALS AND METHODS
Cell Culture and Reagents
HCC cell lines (SNU-353, SNU-423, SNU-449, and SNU-475) derived from HCC tissues of patients (26) were purchased from the Korean Cell Line Bank. Cells were cultured in RPMI medium (Invitrogen, Carlsbad, CA, USA) containing 10% (v/v) fetal bovine serum (Gibco BRL, Life Technologies, Carlsbad, CA, USA), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Sigma-Aldrich, St. Louis, MO, USA). Cells were maintained in an incubator (5% CO2 and 95% O2 at 37°C) as adherent monolayers. The recombinant human soluble TRAIL was obtained from R&D Systems (Minneapolis, MN, USA). Doxorubicin (DOX) and vinblastine (VBL) were purchased from Sigma-Aldrich.
Cell Proliferation Assay
Cell proliferation was measured by counting viable cells using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye reduction method. Exponentially growing cells (2 × 104 cells/well) were plated in a 96-well plate and incubated in growth medium treated with the indicated doses of TRAIL, DOX, or VBL at 37°C. After 96 h, the medium was removed using centrifugation, and MTT formazan crystals were solubilized in 100 μl of DMSO. The optical density of each sample at 570 nm was measured using ELISA reader. The optical density of the medium was proportional to the number of viable cells. Inhibition of proliferation was evaluated as a percentage of control growth (no drug in the medium). All experiments were repeated in at least two experiments, in triplicate.
Apoptosis Assay
Cells (2 × 105 cells/ml) were treated with or without TRAIL for the indicated time points. Apoptosis was measured by annexin V assay. The cells were centrifuged and resuspended in 500 μl of a staining solution containing annexin V fluorescein (FITC Apoptosis Detection Kit; BD Pharmingen, San Diego, CA, USA) and propidium iodide (PI) in PBS. After incubation at room temperature for 15 min, the cells were analyzed by flow cytometry. Annexin V binds to cells that express phosphatidylserine on the outer layer of their cell membrane, and PI stains the cellular DNA of cells with a compromised cell membrane. This allows for the discrimination of live cells (unstained with either fluorochrome) from apoptotic cells (stained only with annexin V) and necrotic cells (stained with annexin V and PI).
Western Blot Analysis
Cells were washed in ice-cold phosphate buffer, lysed in lysis buffer consisting of 1% (w/v) SDS, 1 mM sodium orthovanadate, and 10 mM Tris (pH 7.4) and sonicated for 5 s. Lysate-containing proteins were quantified using a Bradford Protein Assay Kit (Pierce, Rockford, IL, USA). Protein samples were separated by 8%–12% SDS-PAGE using a mini gel apparatus (Bio-Rad, Hercules, CA, USA) and were transferred onto nitrocellulose membranes (Hybond-ECL; GE Healthcare, Piscataway, NJ, USA). Each membrane was blocked with 5% skim milk in Tris-buffered saline containing 0.05% Tween-20. Protein bands were probed with primary antibody followed by labeling with HRP-conjugated anti-mouse, anti-rabbit secondary antibody (Cell Signaling Technology, Danvers, MA, USA). The antibodies used were poly(ADP-ribose) polymerase (PARP), caspase 3 and β-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA), caspase 8 and c-FLIPL/S (Cell Signaling Technology), DR4 and DR5 (Upstates, Merck-Millipore, Darmstadt, Germany), c-Myc (Abcam, MA, USA), and β-tubulin (Sigma-Aldrich) antibodies. Bands were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ, USA) according to the manufacturer’s instructions.
Flow Cytometric Analysis of TRAIL Receptors
The treated cells (3 × 105 cells/well) were centrifuged at 500 × g and resuspended in 500 μl PBS. The cells were then incubated with 5 μl of mouse IgG, anti-DR4, or anti-DR5 monoclonal mouse antibody (1:100; R&D Systems) for 30 min. After washing with PBS, FITC-conjugated rabbit anti-mouse IgG (1:200) was added to the cell suspension and incubated for 30 min on ice followed by washing with PBS. After rinsing, the samples were analyzed by flow cytometry using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA). The data were analyzed using the CellQuest program.
Conventional Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted from cells using the RNeasy Mini Kit (Qiagen, Hilden, Germany), and the RNA concentration was determined from the absorbance at 260 nm. First-strand DNA was reverse transcribed from 1 μg of total RNA in a final volume of 10 μl. The DNA was added to a 20-μl PCR reaction mixture with each set of gene-specific primers: the 5′-forward and 3′-reverse complement PCR primers for amplification of each gene were as follows: β-catenin, 5′-TGGGGTAAGGGTGTAGGGAG-3′ (forward) and 5′-GACCCCTGATCTTCTGGCAT-3′ (reverse); DR4, 5′-AGGGATGGTCAAGGTCAAGG-3′ (forward) and 5′-ATGAGCTGGTCCCAGG AGTC-3′ (reverse); DR5, 5′-CGAGATGCCTCTGTCCACAC-3′ (forward) and 5′-CACAAACGGAATGATCCAG-3′ (reverse); ABCG2, 5′-GACAGTTTCCAATGACCTGA-3′ (forward) and 5′-TTACATTTGA AATTGGCAGG-3′ (reverse); ABCC1, 5′-GGTGCCTTGTTTTTACCTCT-3′ (forward) and 5′-AAGACTGAACTCCCTTCCTC-3′ (reverse); ABCB1, 5′-GGAAGCCAATGCCTATGACTTTA-3′ (forward) and 5′-GAACCACTGCTTCGCTTTCTG-3′ (reverse); c-Myc, 5′-GCAGCCGTATTTCTACTGCG-3′ (forward) and 5′-GTCGTTGAGAGGGTAGGGGA-3′ (reverse); CD133, 5′-AGTCGGAAACTGGCAGATAGC-3′ (forward) and 5′-GGTAGTGTTGTAGGGGCCAAT-3′ (reverse); CD44, 5′-AAATGCACCATTTCCTGAGA-3′ (forward) and 5′-AGAAGGTGTGGGCAGAAGAA-3′ (reverse); Oct4, 5′-CCTGAAGCAGAAGAGGATCA-3′ (forward) and 5′-CCGCAGCTTACACATGTTCT-3′ (reverse); FLIP L, 5′-TTCCAGGCTTTCGGTTTCTT-3′ (forward) and 5′-GTCCGAAACAAGGTGAGGGT-3′ (reverse); FLIP S, 5′-ACCCTCACCTTGTTTCGGAC-3′ (forward) and 5′-CTTTTGGATTGCTGCTTGGA-3′ (reverse); β-actin, 5′-ACCAACTGGGACGACATGGA-3′ (forward) and 5′-GTGAGGATCTTCATGAGGTA-3′ (reverse). The thermal cycling conditions used consisted of initial denaturation at 95°C for 2 min, followed by 30 cycles of 95°C for 20 s, 60°C for 40 s, and 72°C for 60 s, and a final extension for 5 min at 72°C. The final PCR products were electrophoresed on 1.2% agarose gel and photographed under UV illumination. In all experiments, β-actin was used as the endogenous control.
Statistical Analysis
The results obtained were expressed as the mean ± SE of at least three independent experiments. The statistical significance of differences was assessed using the Student’s t-test. Values of p < 0.05, p < 0.01, and p < 0.001 were considered statistically significant in all experiments.
RESULTS
Comparison of TRAIL Sensitivity Among Different Human HCC Cell Lines Established From Primary HCC Tumors and CD133 as a Novel Molecular Target of TRAIL
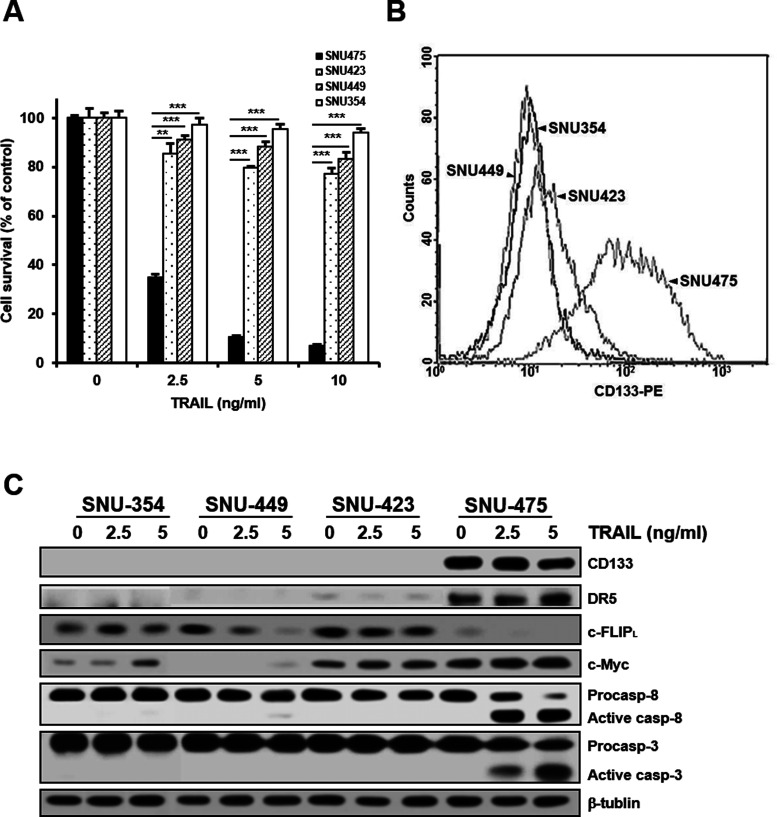
CD133 has attracted increasing attention as an important liver CSC marker (27) since elevated CD133 expression levels in patients with HCC correlated with increased tumor grade, advanced disease stage, and higher recurrence rates compared to patients who had low CD133 expression (14). We examined whether CD133 targeting with TRAIL could be an efficient treatment for human HCC. To compare TRAIL-induced cytotoxicity in well-characterized HCC cell lines from human primary liver cancers, four different HCC cell lines were treated with increasing concentrations of TRAIL, and TRAIL-induced cytotoxicity in these cells was determined by MTT assay. SNU-475 cells were hypersensitive to TRAIL, whereas the other three HCC cell lines (SNU-423, SNU-449, and SNU-354) were less sensitive or resistant to TRAIL (Fig. 1A). Therefore, we next compared the expression of CD133 in four HCC cell lines using fluorescence-activated cell sorting (FACS) analysis. The level of CD133 in SNU-475 cells was significantly higher than that in the other three cell lines, suggesting that the expression of CD133 could be able to confer susceptibility to TRAIL (Fig. 1B). To better understand the relationship between CD133 expression and TRAIL sensitivity, SNU-475 and other cell lines treated with TRAIL were subjected to Western blot analysis to determine the expression of endogenous proteins involving CD133 and caspase activities. The level of CD133 in SNU-475 cells was significantly higher than that in the other three cell lines. In addition, SNU-475 cells showed that the levels of c-Myc and DR5 were increased, and the level of c-FLIPL was decreased, when compared with other HCC cell lines with a low level of CD133. Moreover, the activation of caspase 8 and 3 by TRAIL in SNU-475 cells was more prominent in other HCC cell lines (Fig. 1C). These results strongly suggest that CD133 could serve as a target for TRAIL treatment in HCC.
Figure 1.
Differential TRAIL sensitivity and expression pattern of CD133 and TRAIL-induced caspase activation among four HCC cell lines of primary HCC tumors. (A) The indicated cell lines were treated with various concentrations of TRAIL for 96 h. Percentage of cell survival was determined after 96 h of incubation using MTT assay. Each bar represents the mean ± SD of triplicate experiments. **p < 0.01 and ***p < 0.001. (B) The cell surface expression of CD133 after labeling each cell line with anti-CD133 antibody (1:10) was quantified by flow cytometry. (C) For the indicated cell lines treated with TRAIL (2.5 or 5 ng/ml for 6 h), the levels of CD133, DR5, c-FLIPL, and c-Myc and activation of caspase (casp) were determined by Western blot analysis. β-Tubulin (tubulin) was used as a loading control.
Isolation of CSC-Enriched CD133-High Cell Population From SNU-475 HCC Cells and Its Expression of Stemness-Related Genes and Differential Chemosensitivity
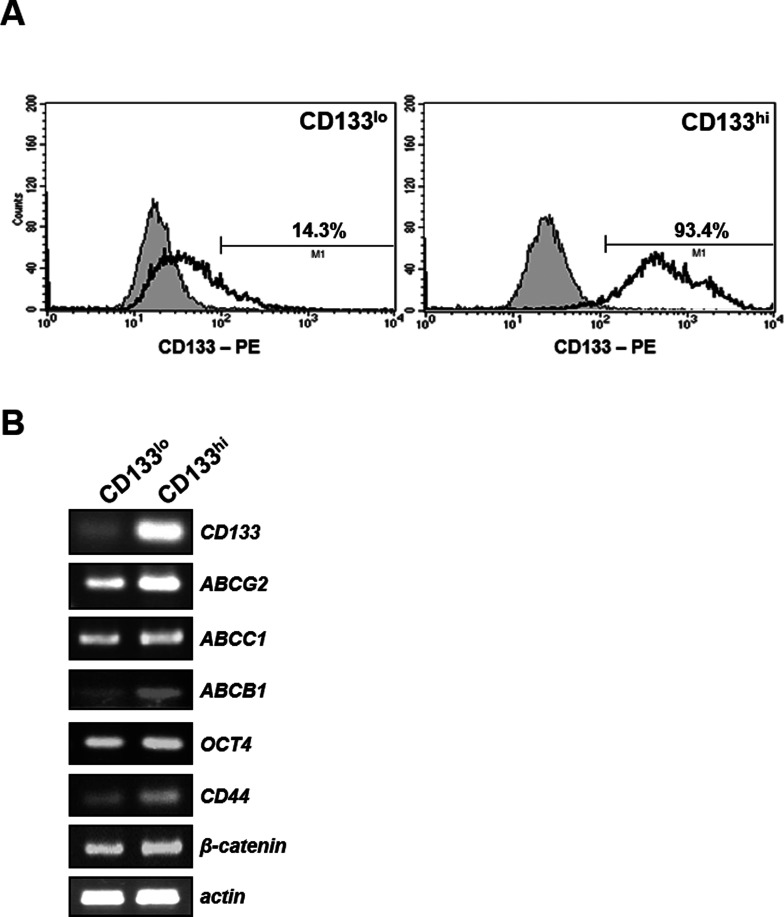
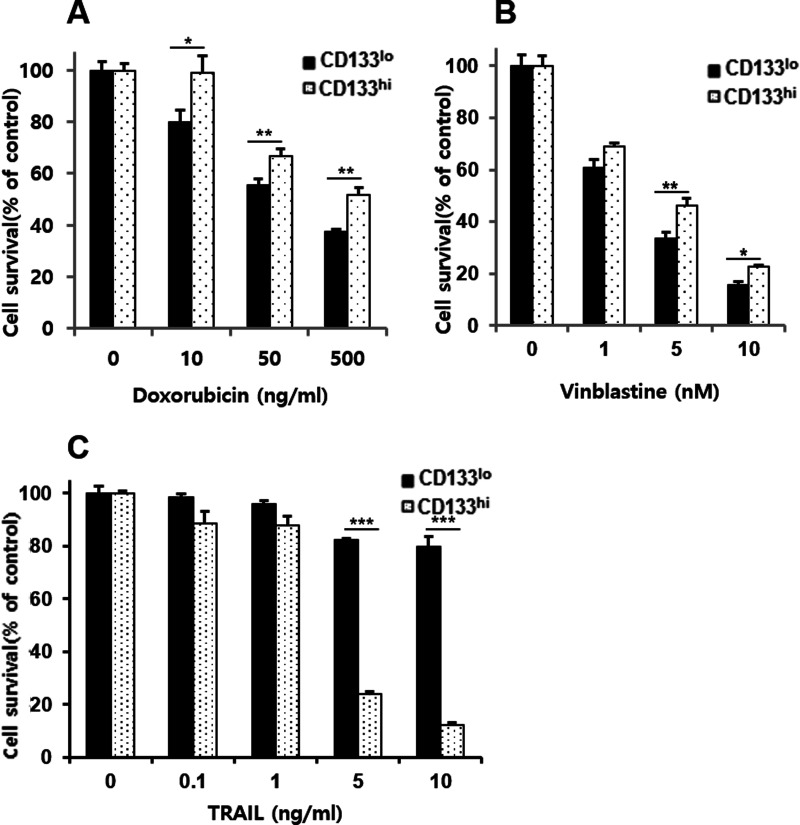
In order to address the mechanism by which SNU-475 cells confer hypersensitivity to TRAIL, CD133-enriched fraction was isolated from SNU-475 cells by FACS (Fig. 2A). The parental SNU-475 cells showed approximately 40% positive for CD133, and, after sorting, CD133-high (CD133hi) and CD133-low (CD133lo) fractions contained approximately 93% and 14% of CD133+ cells, respectively. One characteristic of CSCs is drug resistance, possibly due to enrichment of ABC drug transporter proteins in CSCs and resultant rapid and effective efflux of drugs out of the cells (28). To understand the distinction between the CD133-enriched and -low fraction of SNU-475 cells, we therefore determined the mRNA expression of the drug transporter genes (ABCB1, ABCC1, and ABCG2) and stemness-related genes (Oct4, CD44, and β-catenin) in both CD133hi and CD133lo SNU-475 cells using RT-PCR assay (Fig. 2B). Higher mRNA levels of ABC drug transporter and stemness-related genes were observed in CD133hi cells compared to their CD133lo counterparts, suggesting that CD133hi SNU-475 cells might be a CSCs phenotype of HCC. Next, we compared the viability of CD133hi and CD133low SNU-475 cells treated with increasing doses of DOX and VBL as MDR-related drugs, and TRAIL as MDR-unrelated drug. CD133hi SNU-475 cells showed differential susceptibility against MDR-related drugs and TRAIL. The CD133hi cells were more resistant to MDR-related drugs (DOX and VBL) than their CD133lo counterparts (Fig. 3A and B), suggesting that the CD133hi cells are enriched in CSCs with MDR phenotype resulting from overexpression of ABC drug transporter genes. In contrast with resistance to MDR-related drugs, the CD133hi cells were more sensitive to TRAIL than their CD133lo counterparts (Fig. 3C), indicating that expression of CD133 may cause hypersensitivity to TRAIL of SNU-475 cells.
Figure 2.
Identification of CD133hi SNU-475 cells using the cell surface marker CD133 and expression of stemness-related molecules. (A) After labeling SNU-475 cells with anti-CD133 antibody (1:10), the cell surface expression of CD133 was quantified by flow cytometry, and CD133lo (left) and CD133hi (right) cells were isolated from SNU-475 cells. Histograms showed fluorescence intensity of isotype control (shaded peak) and CD133-PE-stained cells (unshaded peak). (B) RT-PCR was performed to compare the mRNA levels of drug transporter and stemness-related genes in both CD133lo and CD133hi SNU-475 cells. β-Actin (actin) was used as a loading control of both cells.
Figure 3.
Differential sensitivity of CD133hi SNU-475 cells to chemotherapeutic drugs and TRAIL. CD133lo and CD133hi SNU-475 cells were treated with the indicated concentrations of doxorubicin (DOX) (A), vinblastine (VBL) (B), and TRAIL (C) for 96 h. Percentage of cell survival was determined after 96 h of incubation using MTT assay. Each bar represents the mean ± SD of triplicate experiments. *p < 0.05, **p < 0.01, and ***p < 0.001.
High Susceptibility of CD133hi SNU-475 Cells to TRAIL Through c-Myc-Mediated Upregulation of DR5 and Downregulation of c-FLIPL
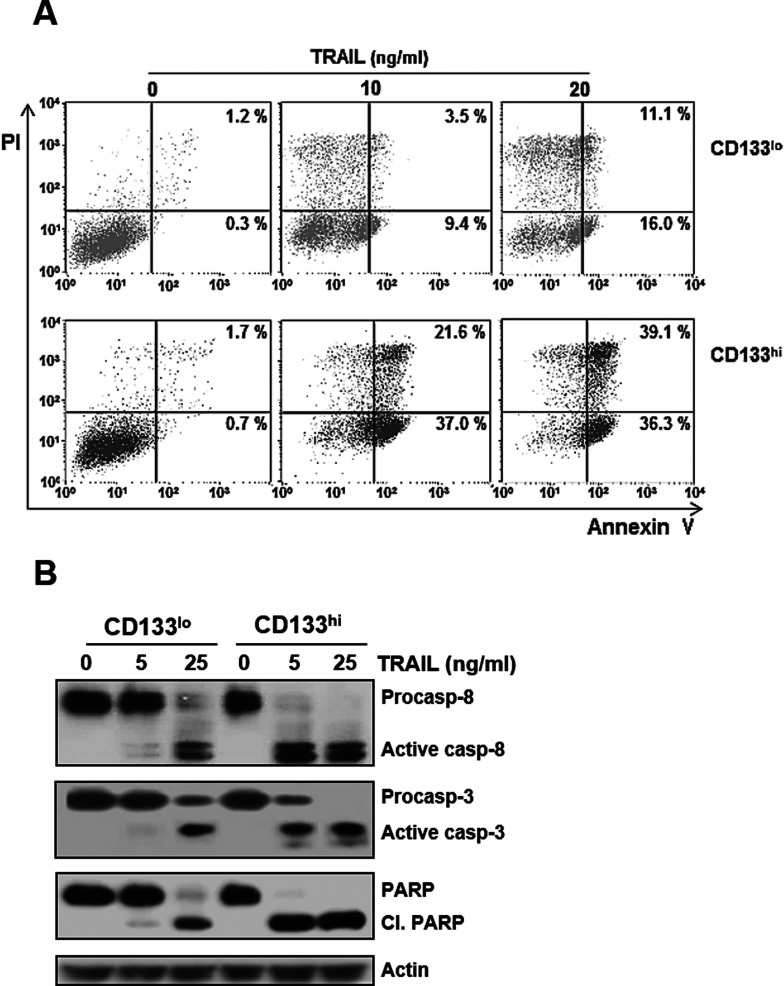
Since TRAIL acts through the TRAIL receptors (DR4 and DR5) to induce apoptosis, we next determined differences in cell surface expression of DR4 and DR5 between CD133hi and CD133lo SNU-475 cells. In the flow cytometric analysis, cell surface expression of DR5, but not DR4, in CD133hi cells was significantly increased when compared to that in their CD133lo counterparts (Fig. 4A). It has been reported that c-Myc plays a key role in regulating proliferation and growth of the CD133+ cancer stem cells (22,29) and in modulating TRAIL sensitivity (23,24). Therefore, we evaluated changes in mRNA and protein levels of c-Myc, as both positive regulator of DR5 and negative regulator of c-FLIP, between CD133hi and CD133lo SNU-475 cells (Fig. 4B). The mRNA and protein levels of c-Myc in the CD133hi cells were significantly increased compared to those in their CD133lo counterparts. Moreover, the mRNA and protein levels of DR5 in the CD133hi cells were higher than those in their CD133low counterparts, whereas DR4 mRNA and protein levels were similar in both cells. We also checked the differential expression level of c-FLIP, a negative regulator of TRAIL signal, in both cells. The mRNA and protein levels of c-FLIPL in the CD133hi cells were decreased compared with those in their CD133lo counterparts. However, the c-FLIPS mRNA and protein levels were similar in both cells. Therefore, these results indicate that hypersensitivity of CD133hi SNU-475 cells to TRAIL is closely associated with c-Myc-mediated upregulation of DR5 and downregulation of c-FLIPL. Since the CD133hi cells showed upregulation of DR5 and downregulation of c-FLIPL, we next examined whether the CD133hi cells were more susceptible to TRAIL-induced apoptosis than their CD133lo counterparts (Fig. 5A). The degree of apoptotic cell death was approximately 59% in the CD133hi cells after treatment with 10 ng/ml TRAIL, while that in their CD133lo counterparts was approximately 27%, even after treatment with 20 ng/ml TRAIL, suggesting that CD133hi SNU-475 cells might be more susceptible to TRAIL-induced apoptosis than their CD133lo counterparts caused by both upregulation of DR5 and downregulation of c-FLIPL. Consistent with the increased susceptibility of the CD133hi SNU-475 cells to TRAIL, activation of procaspase 8, procaspase 3, and PARP in the CD133hi cells was more prominent than in their CD133lo counterparts (Fig. 5B). As cleavage of caspases is a common indicator of activation, procaspase 8 and 3 were cleaved, and thus, these caspases were activated. Activation of caspase 3 subsequently leads to apoptotic cell death through cleavage of a broad spectrum of cellular target protein, including PARP. The increased caspase 3 activity in the CD133hi cells was accompanied by cleavage of PARP, suggesting that TRAIL-induced apoptosis of CD133hi SNU-475 cells occurred via caspase-dependent pathway.
Figure 4.
Upregulation of c-Myc and DR5 and downregulation of c-FLIPL in CD133hi SNU-475 cells. (A) CD133lo and CD133hi SNU-475 cells were stained with control IgG or anti-DR4 (or anti-DR5) antibody (1:100), and subsequently labeled with FITC-conjugated secondary antibodies (1:100) to determine the surface expression of DR4 and DR5. The cell surface expression was measured by a flow cytometer. Shaded and unshaded peaks correspond to control and specific stainings, respectively. (B) The mRNA and protein levels of c-Myc, DR4/DR5, and c-FLIPL/S were determined by RT-PCR and Western blot, respectively.
Figure 5.
High susceptibility of CD133hi SNU-475 cells to TRAIL-induced apoptosis and caspase activation. (A) After treatment of CD133lo and CD133hi SNU-475 cells with the indicated concentrations of TRAIL for 6 h, percentage of apoptotic cells in both cells was determined with flow cytometry using annexin V and PI double staining. The bottom right quadrant of each dot plot represents early apoptotic cells (annexin V positive/PI negative). The upper right quadrant represents late apoptotic cells (annexin V positive/PI positive). (B) For both cells treated with TRAIL (5 or 25 ng/ml for 6 h), the activation of caspases and cleavage of PARP (cl. PARP) were determined by Western blot analysis.
Reduced Susceptibility of CD133hi SNU-475 Cells to TRAIL by Depletion of CD133
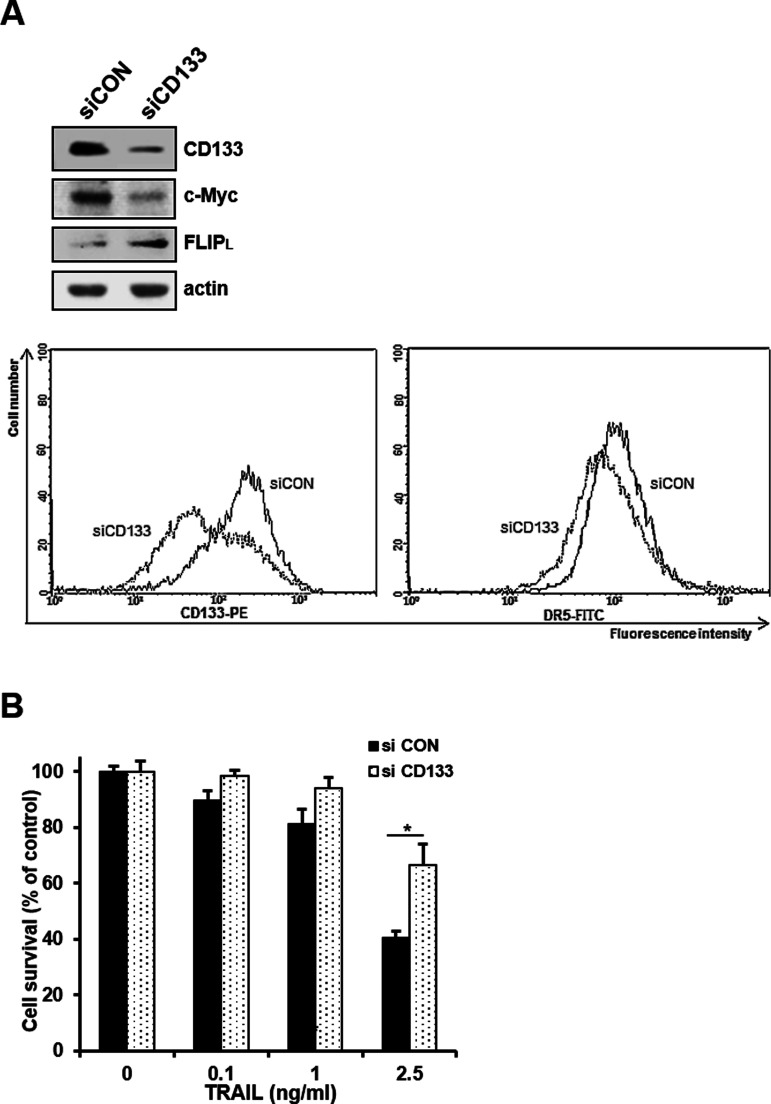
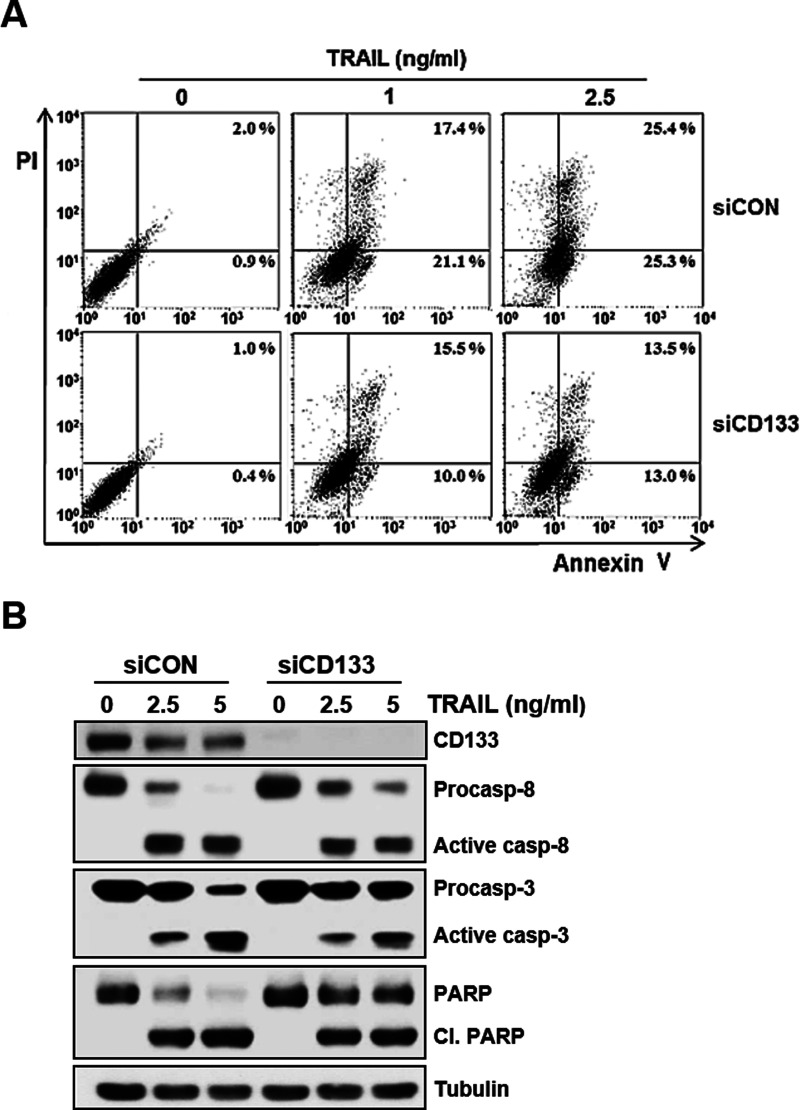
Therefore, we next determined whether the c-Myc-induced susceptibility of CD133hi SNU-475 cells to TRAIL was caused by the expression of CD133. We used siRNA to knock down CD133 expression and examined the changed level of c-Myc and TRAIL susceptibility in CD133-depleted cells. When the expression of c-Myc in CD133hi SNU-475 cells was significantly reduced after CD133 knockdown, an increase in c-FLIPL expression and a decrease in cell surface expression of DR5 occurred in the CD133-depleted cells (Fig. 6A). In addition, TRAIL-induced cytotoxicity of the CD133-depleted cells was significantly reduced (Fig. 6B). To further confirm these results, we also checked the modulation of TRAIL-induced apoptosis in the CD133-depleted cells. As expected, TRAIL-induced apoptosis was attenuated in CD133hi SNU-475 cells after CD133 knockdown (Fig. 7A). We next determined whether the activation of caspases was changed by the depletion of CD133. TRAIL-induced activation of procaspase 8 and 3 and cleavage of PARP was significantly attenuated in the CD133-depleted cells (Fig. 7B). Taken together, our results strongly suggest that the hypersensitivity to TRAIL of CD133hi SNU-475 cells might be associated with CD133-mediated upregulation of c-Myc.
Figure 6.
Effects of CD133 depletion on the expression of c-Myc/FLIPL/DR5 and TRAIL-induced cytotoxicity. (A) CD133hi SNU-475 cells were transfected with 50 nM CD133 siRNA (siCD133) or siCON for 24 h and then subjected to Western blot analysis to monitor the levels of CD133, c-Myc, and c-FLIPL (upper), and the cells transfected with siCON or siCD133 were stained with anti-CD133 antibody (1:10) or anti-DR5 antibody (1:100) and subsequently labeled with FITC-conjugated secondary antibody (1:100) to determine the surface expression of DR5 (lower). The cell surface expression of DR5 was measured by a flow cytometer. (B) These transfectants were treated with TRAIL (0.1, 1, or 2.5 ng/ml for 96 h), and percentage of cell survival in these transfectants was determined by MTT assay. *p < 0.05.
Figure 7.
Attenuation of TRAIL-induced apoptosis and caspase activity by CD133 depletion. (A) CD133hi SNU-475 cells transfected with 50 nM siCD133 or siCON for 24 h were treated with TRAIL (1 or 2.5 ng/ml for 6 h), and percentage of apoptotic cells in these transfectants was determined with flow cytometry using annexin V and PI double staining. (B) For these transfectants treated with TRAIL (2.5 or 5 ng/ml for 6 h), the activation of caspase (casp) and cleavage of PARP (cl. PARP) were determined by Western blot analysis. β-Tubulin (tubulin) was used as a loading control.
Effect of c-Myc Depletion on TRAIL-Induced Apoptosis in CD133hi SNU-475 Cells
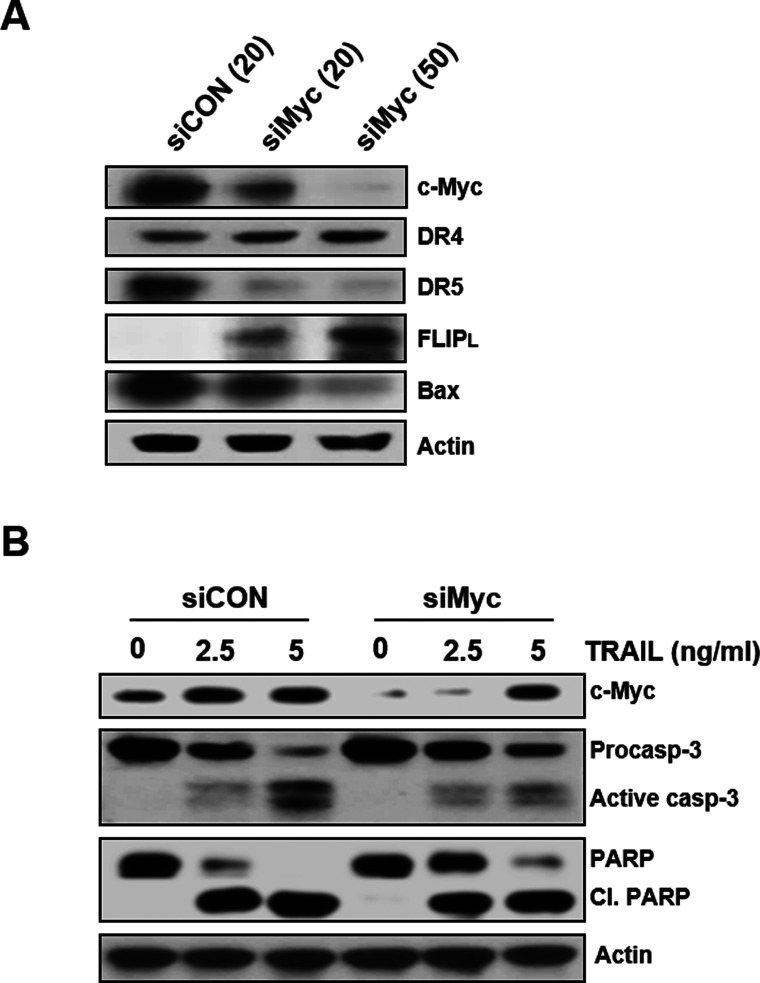
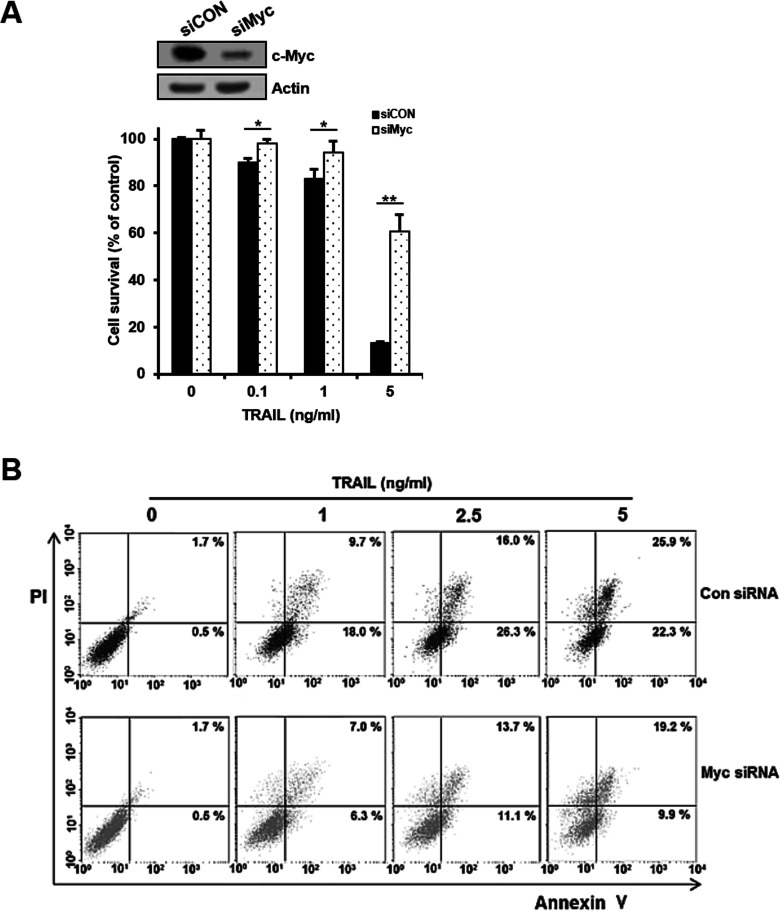
c-Myc is highly expressed in cancer stem cells relative to non-stem cancer cells (22) and mandates cell fate by inducing stemness and blocking cellular senescence and differentiation (30). To investigate the direct role of c-Myc in regulating the susceptibility of CD133hi SNU-475 cells with a high level of c-Myc to TRAIL, we used siRNA to knock down c-Myc expression and determined whether depletion of c-Myc could affect the expression of death receptors and c-FLIP in the cells (Fig. 8A). After transfection of CD133hi SNU-475 cells with increasing doses of siRNA against c-Myc (siMyc) or scrambled control siRNA (siCON), the modulation of DR5/DR4 and c-FLIPL expressions in these transfectants was evaluated. Depletion of c-Myc in the CD133hi cells resulted in the downregulation of DR5 but not DR4 and the upregulation of c-FLIPL in a concentration-dependent manner. Since c-Myc transcriptionally regulates Bax, and Bax activation is c-Myc dependent, we also examined whether the expression of Bax in the CD133hi cells could be modulated by depletion of c-Myc. As expected, depletion of c-Myc in the CD133hi cells led to downregulation of Bax. These results indicate that the upregulation of c-Myc in CD133-enriched cells of HCC might be potentially susceptible to TRAIL. Moreover, treatment of siCON-transfected CD133hi SNU-475 cells with TRAIL resulted in accelerated cleavage of procaspase 3 and PARP, whereas siMyc-transfected cells treated with TRAIL showed attenuated activation of caspase 3 and PARP cleavage (Fig. 8B), suggesting that the expression of c-Myc might be an important role in the susceptibility of CSCs overexpressing CD133 to TRAIL. Therefore, we next determined the effect of c-Myc depletion on regulating the susceptibility of CD133hi SNU-475 cells to TRAIL. The cytotoxic and apoptotic effects of TRAIL on CD133hi SNU-475 cells were evaluated after transfecting the cells with either siMyc or siCON by MTT assay and flow cytometric analysis using annexin V-PI staining, respectively (Fig. 9). When CD133hi SNU-475 cells were treated with the indicated doses of TRAIL after c-Myc knockdown, the susceptibility of siMyc-transfected cells to TRAIL was significantly attenuated compared to that of siCON-transfected cells treated with TRAIL. As shown in Figure 9A, TRAIL-induced cytotoxicity of CD133hi SNU-475 cells was significantly reduced by transfection of siMyc when compared to that of siCON-transfected cells treated with TRAIL. To further validate apoptotic cooperation between TRAIL and c-Myc, CD133hi SNU-475 cells were treated with TRAIL after transfecting the cells with siMyc or siCON, and then the degree of apoptotic cell death was determined by flow cytometry. TRAIL-induced apoptosis was significantly attenuated in the siMyc-transfected cells when compared to siCON-transfected CD133hi SNU-475 cells (Fig. 9B). These results suggest that depletion of c-Myc attenuates the cytotoxic and apoptotic activities of TRAIL in CD133hi SNU-475 cells.
Figure 8.
Effects of c-Myc depletion on the expression of TRAIL-signaling molecules and TRAIL-induced caspase activity in CD133hi SNU-475 cells. (A) Cells were transfected with 20 or 50 nM c-Myc siRNA (siMyc) or 20 nM nontargeted control siRNA (siCON) for 24 h and then subjected to Western blot analysis to monitor the levels of c-Myc, DR4/DR5, c-FLIPL, and Bax. (B) Cells were transfected with 50 nM siMyc or siCON for 24 h, and these transfectants were treated with TRAIL (2.5 or 5 ng/ml for 6 h) and the activation of caspase (casp) and cleavage of PARP were determined by Western blot analysis.
Figure 9.
Attenuation of TRAIL-induced cytotoxicity and apoptosis of CD133hi SNU-475 cells by Myc depletion. (A) Cells were transfected with 50 nM siCON or siMyc. After 24 h, these transfectants were subjected to Western blot analysis to monitor the c-Myc level and treated with various concentrations of TRAIL for 96 h. Cell survival was determined using MTT assay. *p < 0.05 and **p < 0.01. (B) Percentage of apoptotic cells in these transfectants treated with the indicated concentrations of TRAIL for 6 h was determined with flow cytometry using annexin V and PI double staining.
DISCUSSION
HCC is one of the most common malignancies, with a poor prognosis and high recurrence rate. CD133+ cells in HCC display cancer stem-like properties and are thought to be responsible for chemoradioresistance (15). In the present study, we evaluated the differential expression pattern of CD133 in various HCC cell lines that were established from primary HCC tumors to aim targeting CD133 as a novel therapeutic molecule of TRAIL. We found that the SNU-475 cell line exhibited the highest level of CD133 and susceptibility to TRAIL among the four human HCC cell lines.
Direct CSC killing by targeting CSC-specific markers has been reported in experimental cancer stem cell models (31). CD133+ cells have stem cell properties and are responsible for multidrug resistance in cell lines used as an HCC model (32), and CD133-enriched liver CSCs appeared more frequently in advanced stages and is correlated with early recurrence and poor prognosis (33). Moreover, we showed that CD133hi SNU-475 cells enriched for the CD133+ subpopulation exhibited a higher expression of stemness-related genes such as c-Myc, CD44, Oct4, and β-catenin as well as ABC transporter genes such as ABCB1, ABCG2, and ABCC1 than their CD133lo counterparts. CD133hi SNU-475 cells were more resistant to MDR-related drugs such as DOX and VBL, but paradoxically were more sensitive to TRAIL when compared to their CD133lo counterparts, indicating that TRAIL could be a candidate agent for targeting CD133-expressing liver CSCs of HCC.
c-Myc has been recently recognized as an important regulator of stem cell biology. The activity of c-Myc is required for cellular proliferation and growth and/or survival of the cancer stem cells (22) and may serve as a link connecting malignancy and stemness (30). c-Myc expression is required for self-renewal of CD133+ glioma cancer stem cells, and knockdown of c-Myc inhibits the tumorigenic potential of the cells (22). Deregulated c-Myc is found in diverse human tumors and is often associated with advanced malignancy and poor prognosis (34). c-Myc and β-catenin positively regulate ABCB1/MDR1 gene promoter and increase P-gp expression (35,36), which might be associated with resistance to DOX and VBL in CD133hi SNU-475 cells. The role of c-Myc in regulating apoptosis remains controversial. Previous reports have demonstrated that c-Myc promotes apoptosis through the extrinsic apoptotic pathway, such as death receptor pathways, at multiple junctions and also participates in the intrinsic pathways and, conversely, downregulation of c-Myc leads to apoptosis under certain circumstances (36,37).
TRAIL has been demonstrated to induce apoptosis in a wide range of cancer types both in vitro and in various mouse models of human cancers (38). Indeed, overexpression of c-Myc dramatically sensitizes cells to the apoptotic action of the death ligands such as CD95 ligand and TRAIL (36). The role of TRAIL signaling pathway in the regulation of CSCs is emerging. The chemotherapy-resistant side population (SP) of SW480 human colon tumor cells has a higher sensitivity to TRAIL than the non-SP, and TRAIL-sensitive SP cells have significantly increased levels of c-Myc (9). Expression of DR5 has been detected with a high frequency in tumor cell lines and clinical tumor specimens (39). Indeed, MDR cells overexpressing both P-gp and c-Myc were hypersensitive to TRAIL due to upregulation of DR5 and downregulation of c-FLIP (25). Moreover, DR5 is enriched in pancreatic CSCs (40) and colon CSCs (41) compared with the non-stem cell bulk tumor populations and contributed to TRAIL-induced apoptosis rather than DR4 in cancer cells that expressed both death receptors (42).
It has been reported that c-Myc expression could be associated with upregulation of DR5 and, conversely, downregulation of c-FLIP (23,24), and c-Myc knockdown reduces the responsiveness to TRAIL and the expression of DR5 (43). In this study, CD133hi SNU-475 cells with a high level of c-Myc showed a marked increase in DR5 mRNA and protein levels and decrease in c-FLIPL mRNA and protein levels when compared to their CD133lo counterparts with a low level of c-Myc. Therefore, high susceptibility of CD133hi SNU-475 cells to TRAIL might be associated with enhanced level of c-Myc in the cells. It has been reported that the level of c-FLIP in CD133+ fraction within glioblastoma was higher than that in the CD133− population (44). In contrast, our data showed that the expression of c-FLIPL in CD133hi HCC cells was rather decreased compared to that in their CD133lo counterparts. Moreover, the expression of c-Myc was downregulated in CD133hi SNU-475 cells by depletion of CD133 using CD133-inhibiting siRNA, which resulted in the downregulation of DR5, upregulation of c-FLIPL, and subsequent attenuation of caspase 8 and 3 activities, and TRAIL-induced cytotoxicity and apoptosis were attenuated in CD133hi SNU-475 cells after CD133 knockdown. Therefore, differential expression of c-FLIP in CD133+ cancer stem cells might be responsible for cell type-specific regulation of CD133 expression. Knockdown of c-Myc in CD133hi SNU-475 cells resulted in the downregulation of DR5 and upregulation of c-FLIPL, which is associated with attenuation of TRAIL susceptibility, indicating susceptibility of CD133hi SNU-475 cells to TRAIL might be associated with CD133-mediated upregulation of c-Myc.
In conclusion, our data demonstrated that CD133hi CSCs of HCC were resistant to MDR-related drugs but rather were hypersensitive to TRAIL, suggesting that despite the refractory nature of CSCs to conventional therapies, TRAIL may be useful to eradicate CD133-expressing CSCs of HCC. Our study suggests that the stem-targeted therapy via CD133 silencing using TRAIL could provide a novel strategy for the treatment of HCC and for the therapies of other CD133-expressing types of cancer.
ACKNOWLEDGMENTS
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2014R1A1A2053321) and the National R&D Program for Cancer Control, Ministry for Health, Welfare, and Family Affairs (No. 0920050), Korea.
REFERENCES
- 1. Pardal R.; Clarke M. F.; Morrison S. J. Applying the principles of stem-cell biology to cancer. Nat. Rev. Cancer 3(12):895–902; 2003. [DOI] [PubMed] [Google Scholar]
- 2. Chiba T.; Kamiya A.; Yokosuka O.; Iwama A. Cancer stem cells in hepatocellular carcinoma: Recent progress and perspective. Cancer Lett. 286(2):145–153; 2009. [DOI] [PubMed] [Google Scholar]
- 3. Yamashita T.; Wang X. W. Cancer stem cells in the development of liver cancer. J. Clin. Invest. 123(5):1911–1918; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Warrier S.; Pavanram P.; Raina D.; Arvind M. Study of chemoresistant CD133+ cancer stem cells from human glioblastoma cell line U138MG using multiple assays. Cell Biol. Int. 36(12):1137–1143; 2012. [DOI] [PubMed] [Google Scholar]
- 5. Tang K. H.; Ma S.; Lee T. K.; Chan Y. P.; Kwan P. S.; Tong C. M.; Ng I. O.; Man K.; To K. F.; Lai P. B. and others. CD133(+) liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology 55(3):807–820; 2012. [DOI] [PubMed] [Google Scholar]
- 6. Collins A. T.; Berry P. A.; Hyde C.; Stower M. J.; Maitland N. J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 65(23):10946–10951; 2005. [DOI] [PubMed] [Google Scholar]
- 7. Olempska M.; Eisenach P. A.; Ammerpohl O.; Ungefroren H.; Fandrich F.; Kalthoff H. Detection of tumor stem cell markers in pancreatic carcinoma cell lines. Hepatobiliary Pancreat. Dis. Int. 6(1):92–97; 2007. [PubMed] [Google Scholar]
- 8. O’Brien C. A.; Pollett A.; Gallinger S.; Dick J. E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445(7123):106–110; 2007. [DOI] [PubMed] [Google Scholar]
- 9. Sussman R. T.; Ricci M. S.; Hart L. S.; Sun S. Y.; El-Deiry W. S. Chemotherapy-resistant side-population of colon cancer cells has a higher sensitivity to TRAIL than the non-SP, a higher expression of c-Myc and TRAIL-receptor DR4. Cancer Biol. Ther. 6(9):1490–1495; 2007. [DOI] [PubMed] [Google Scholar]
- 10. Wilson R. J.; Thomas C. D.; Fox R.; Roy D. B.; Kunin W. E. Spatial patterns in species distributions reveal biodiversity change. Nature 432(7015):393–396; 2004. [DOI] [PubMed] [Google Scholar]
- 11. Bao S.; Wu Q.; McLendon R. E.; Hao Y.; Shi Q.; Hjelmeland A. B.; Dewhirst M. W.; Bigner D. D.; Rich J. N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444(7120):756–760; 2006. [DOI] [PubMed] [Google Scholar]
- 12. Bruno S.; Bussolati B.; Grange C.; Collino F.; Graziano M. E.; Ferrando U.; Camussi G. CD133+ renal progenitor cells contribute to tumor angiogenesis. Am. J. Pathol. 169(6):2223–2235; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Craig C. E.; Quaglia A.; Selden C.; Lowdell M.; Hodgson H.; Dhillon A. P. The histopathology of regeneration in massive hepatic necrosis. Semin. Liver Dis. 24(1):49–64; 2004. [DOI] [PubMed] [Google Scholar]
- 14. Song W.; Li H.; Tao K.; Li R.; Song Z.; Zhao Q.; Zhang F.; Dou K. Expression and clinical significance of the stem cell marker CD133 in hepatocellular carcinoma. Int. J. Clin. Pract. 62(8):1212–1218; 2008. [DOI] [PubMed] [Google Scholar]
- 15. Lan X.; Wu Y. Z.; Wang Y.; Wu F. R.; Zang C. B.; Tang C.; Cao S.; Li S. L. CD133 silencing inhibits stemness properties and enhances chemoradiosensitivity in CD133-positive liver cancer stem cells. Int. J. Mol. Med. 31(2):315–324; 2013. [DOI] [PubMed] [Google Scholar]
- 16. Merino D.; Lalaoui N.; Morizot A.; Solary E.; Micheau O. TRAIL in cancer therapy: Present and future challenges. Expert Opin. Ther. Targets 11(10):1299–1314; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. LeBlanc H. N.; Ashkenazi A. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 10(1):66–75; 2003. [DOI] [PubMed] [Google Scholar]
- 18. Ashkenazi A. Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat. Rev. Drug Discov. 7(12):1001–1012; 2008. [DOI] [PubMed] [Google Scholar]
- 19. Bodmer J. L.; Holler N.; Reynard S.; Vinciguerra P.; Schneider P.; Juo P.; Blenis J.; Tschopp J. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat. Cell Biol. 2(4):241–243; 2000. [DOI] [PubMed] [Google Scholar]
- 20. Majkut J.; Sgobba M.; Holohan C.; Crawford N.; Logan A. E.; Kerr E.; Higgins C. A.; Redmond K. L.; Riley J. S.; Stasik I. and others. Differential affinity of FLIP and procaspase 8 for FADD’s DED binding surfaces regulates DISC assembly. Nat. Commun. 5:3350; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thome M.; Schneider P.; Hofmann K.; Fickenscher H.; Meinl E.; Neipel F.; Mattmann C.; Burns K.; Bodmer J. L.; Schroter M.; Scaffidi C.; Krammer P. H.; Peter M. E.; Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386(6624):517–521; 1997. [DOI] [PubMed] [Google Scholar]
- 22. Wang J.; Wang H.; Li Z.; Wu Q.; Lathia J. D.; McLendon R. E.; Hjelmeland A. B.; Rich J. N. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One 3(11):e3769; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ricci M. S.; Jin Z.; Dews M.; Yu D.; Thomas-Tikhonenko A.; Dicker D. T.; El-Deiry W. S. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol. Cell Biol. 24(19):8541–8555; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y.; Engels I. H.; Knee D. A.; Nasoff M.; Deveraux Q. L.; Quon K. C. Synthetic lethal targeting of MYC by activation of the DR5 death receptor pathway. Cancer Cell 5(5):501–512; 2004. [DOI] [PubMed] [Google Scholar]
- 25. Kim D. Y.; Kim M. J.; Kim H. B.; Lee J. W.; Bae J. H.; Kim D. W.; Kang C. D.; Kim S. H. Suppression of multidrug resistance by treatment with TRAIL in human ovarian and breast cancer cells with high level of c-Myc. Biochim. Biophys. Acta 1812(7):796–805; 2011. [DOI] [PubMed] [Google Scholar]
- 26. Ku J. L.; Park J. G. Biology of SNU cell lines. Cancer Res. Treat. 37(1):1–19; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu Z.; Hao X.; Yan M.; Yao M.; Ge C.; Gu J.; Li J. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int. J. Cancer 126(9):2067–2078; 2010. [DOI] [PubMed] [Google Scholar]
- 28. Moitra K.; Lou H.; Dean M. Multidrug efflux pumps and cancer stem cells: Insights into multidrug resistance and therapeutic development. Clin. Pharmacol. Ther. 89(4):491–502; 2011. [DOI] [PubMed] [Google Scholar]
- 29. Feng B. H.; Liu A. G.; Gu W. G.; Deng L.; Cheng X. G.; Tong T. J.; Zhang H. Z. CD133+ subpopulation of the HT1080 human fibrosarcoma cell line exhibits cancer stem-like characteristics. Oncol. Rep. 30(2):815–823; 2013. [DOI] [PubMed] [Google Scholar]
- 30. Murphy M. J.; Wilson A.; Trumpp A. More than just proliferation: Myc function in stem cells. Trends Cell Biol. 15(3):128–137; 2005. [DOI] [PubMed] [Google Scholar]
- 31. Yang Z. F.; Ho D. W.; Ng M. N.; Lau C. K.; Yu W. C.; Ngai P.; Chu P. W.; Lam C. T.; Poon R. T.; Fan S. T. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 13(2):153–166; 2008. [DOI] [PubMed] [Google Scholar]
- 32. Ma S.; Lee T. K.; Zheng B. J.; Chan K. W.; Guan X. Y. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene 27(12):1749–1758; 2008. [DOI] [PubMed] [Google Scholar]
- 33. Ma S.; Tang K. H.; Chan Y. P.; Lee T. K.; Kwan P. S.; Castilho A.; Ng I.; Man K.; Wong N.; To K. F.; Zheng B. J.; Lai P. B.; Lo C. M.; Chan K. W.; Guan X. Y. miR-130b promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 7(6):694–707; 2010. [DOI] [PubMed] [Google Scholar]
- 34. Vita M.; Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin. Cancer Biol. 16(4):318–330; 2006. [DOI] [PubMed] [Google Scholar]
- 35. Blanc E.; Goldschneider D.; Ferrandis E.; Barrois M.; Le Roux G.; Leonce S.; Douc-Rasy S.; Benard J.; Raguenez G. MYCN enhances P-gp/MDR1 gene expression in the human metastatic neuroblastoma IGR-N-91 model. Am. J. Pathol. 163(1):321–331; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoffman B.; Liebermann D. A. Apoptotic signaling by c-MYC. Oncogene 27(50):6462–6472; 2008. [DOI] [PubMed] [Google Scholar]
- 37. Prendergast G. C. Mechanisms of apoptosis by c-Myc. Oncogene 18(19):2967-2987; 1999. [DOI] [PubMed] [Google Scholar]
- 38. Ashkenazi A.; Herbst R. S. To kill a tumor cell: The potential of proapoptotic receptor agonists. J. Clin. Invest. 118(6):1979–1990; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Daniels R. A.; Turley H.; Kimberley F. C.; Liu X. S.; Mongkolsapaya J.; Ch’En P.; Xu X. N.; Jin B. Q.; Pezzella F.; Screaton G. R. Expression of TRAIL and TRAIL receptors in normal and malignant tissues. Cell Res. 15(6):430–438; 2005. [DOI] [PubMed] [Google Scholar]
- 40. Rajeshkumar N. V.; Rasheed Z. A.; Garcia-Garcia E.; Lopez-Rios F.; Fujiwara K.; Matsui W. H.; Hidalgo M. A combination of DR5 agonistic monoclonal antibody with gemcitabine targets pancreatic cancer stem cells and results in long-term disease control in human pancreatic cancer model. Mol. Cancer Ther. 9(9):2582–2592; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee S. H.; Kim M. J.; Kim D. W.; Kang C. D.; Kim S. H. Amurensin G enhances the susceptibility to tumor necrosis factor-related apoptosis-inducing ligand-mediated cytotoxicity of cancer stem-like cells of HCT-15 cells. Cancer Sci. 104(12):1632–1639; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat. Rev. Cancer 2(6):420–430; 2002. [DOI] [PubMed] [Google Scholar]
- 43. Kim H. B.; Kim M. J.; Kim D. Y.; Lee J. W.; Bae J. H.; Kim D. W.; Kang C. D.; Kim S. H. High susceptibility of metastatic cells derived from human prostate and colon cancer cells to TRAIL and sensitization of TRAIL-insensitive primary cells to TRAIL by 4,5-dimethoxy-2-nitrobenzaldehyde. Mol. Cancer 10:46; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu G.; Yuan X.; Zeng Z.; Tunici P.; Ng H.; Abdulkadir I. R.; Lu L.; Irvin D.; Black K. L.; Yu J. S. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 5:67; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]