Abstract
RNA interference (RNAi) has been used for cancer gene therapy in recent years. However, the application of RNAi is hindered in the absence of safe and efficient gene delivery. In this article, a novel vehicle of graphene oxide functionalized with polyethylenimine and polyethylene glycol (GO-PEI-PEG) was successfully synthetized and then used to deliver plasmid-based Stat3 siRNA. The carrier can readily bind plasmid with high transfection efficiency. Moreover, molecular biology studies reveal that Stat3-related gene and protein expressions were significantly inhibited, suggesting that the formation of GO-PEI-PEG complexes could be utilized as a promising gene delivery in cancer therapy.
Key words: Graphene oxide (GO), Stat3, Cancer
INTRODUCTION
Cancer is a leading cause and the most common of the deadliest diseases, severely threatening human health. Gene therapy is an attractive approach for treating diseases caused by cancer. As one of the most promising techniques, the RNA interference (RNAi) method plays an important role in gene therapy (1). The process of RNAi involves specific silencing of targeted protein (2). Signal transducer and activator of transcription 3 (Stat3) has been recognized as an oncogene (3), which is found to be overexpressed in a great number of solid tumors, such as malignant melanoma and prostate cancer (1,4). Constitutive activation of Stat3 has been confirmed to promote the survival of cancer and to be associated with immunologic escape in cancer (5–8). Hence, knockdown of the Stat3 protein expression using the RNAi technique would effectively suppress cell proliferation and induce apoptosis, leading to inhibition of cancer growth (9–11). However, free siRNA is quickly degraded by nuclease in vivo, resulting in uptake inefficiency (12–14). To overcome these challenges, various nanomaterials have been widely used to be gene vectors. Nanomaterials have several favorable features, including excellent repeatability, better biocompatibility, and stability. Moreover, nanomaterial is highly accumulated in tumor in vivo (15), owing to its enhanced permeation and retention effect.
Graphene oxide (GO), a novel carbon nanomaterial, has been studied broadly owing to potential biomedical applications in recent years. For example, GO has been applied to drug and gene delivery and photothermal therapy (16,17). GO has several advantages over other nonviral vehicles. First, GO possesses lots of hydroxyl, carboxylic acid, and other reactive groups, which are easily conjugated to other modifications, making GO tailored for biomedical and other fields. Second, GO has rich hydrophilic groups, hence holding a fine biocompatibility. Finally, the preparation of GO can be developed easily and inexpensively. Cationic polymers polyethylenimine (PEI) are considered to be a gold standard of gene transfections and are widely used for gene delivery (18). PEI can improve DNA binding and condensation and transfection efficiency. Polyethylenimine functionalized graphene oxide (GO-PEI) has been successfully used to bind with plasmid EGFP, which leads to expression of the green fluorescent protein in HeLa cells in vitro (16). In order to improve solubility and stability in physiological solutions, polyethylene glycol (PEG) was introduced to GO-PEI complexes. PEG also has distinctive biomedical features, such as reduced reticuloendothelial system (RES) accumulation and notably improved tumor passive targeting effect (19).
Therefore, GO-PEI functionalized with polyethylene glycol (PEG) was studied as a remedial plasmid carrier in this work. GO-PEI-PEG was used to mediate a plasmid-based Stat3-siRNA for effective gene silencing of malignant melanoma B16 cells, hoping to construct GO-based gene delivery system for intensively future studies in vivo in cancer therapy in clinic.
MATERIALS AND METHODS
Plasmids, Cell Lines, and Animals
Stat3-specific siRNA plasmids were constructed in our laboratory. si-Stat3 (sequence: GCAGCAGCTGAACAACAT, spanning nucleotides 2,144–2,162; GenBank accession No. NM 003150) was used in the present study. A negative control si-Scramble sequence was purchased from Ambion (Austin, TX, USA). The mouse malignant melanoma B16 cells were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). Female C57BL/6 mice weighing 16–18 g, were purchased from the Beijing Institute for Experimental Animals (Beijing, China, Permit No. SCXK2009-0004). All animals were raised in pathogen-free conditions and in conformity with protocols approved by the National Institute of Health Guide for care.
Preparation and Characterization of the Nanosheets
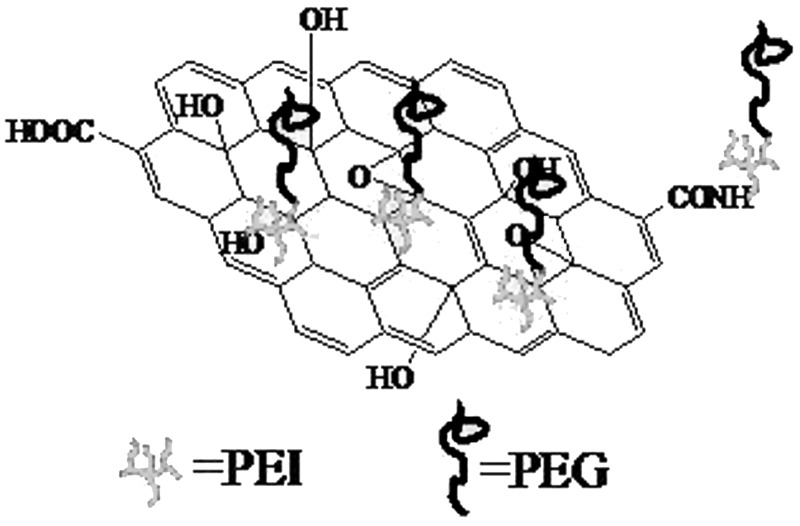
GO was prepared by a modified Hummer’s method (20). Detailed experimental procedures and conditions for the preparation of GO-PEI-PEG were performed following our previous protocol (21). The structure of GO-PEI-PEG is shown in Figure 1.
Figure 1.
Structure of GO-PEI-PEG.
Cell Culture
B16 cells were maintained in culture with Dulbecco’s modified Eagle’s medium (DMEM; Hyclone), supplemented with 10% fetal bovine serum (FBS; Hyclone), penicillin (100 µg/ml; Gibco) and streptomycin (100 µg/ml; Gibco), and cultured in a humidified atmosphere at 37°C under 5% CO2.
Transfection
B16 cells were added onto six-well plates at a density of 8 × 105 cells per well 24 h before transfection. Cells should achieve 60–80% confluence in DMEM without antibiotics at transfections. Four micrograms of si-Stat3 and si-Scramble plasmid were, respectively, diluted in 250 µl FBS-free DMEM, and then the designated amount of GO-PEG-PEI was added to the medium (weight ratio GO-PEI-PEG/plasmid = 15:1). After being gently mixed, the mixtures were kept at room temperature for 60 min before transfection. After 6 h of incubation under 37°C in 2 ml of FBS-free medium, cells were washed with PBS and recultured in fresh DMEM. The gene expression was monitored at 48 h posttransfection. For transfection efficiency examination, laser confocal imaging (A1R-A1; Nikon) and flow cytometry assays (FACScan; Becton Dickinson, USA) were performed.
Semiquantitative Reverse Transcription-PCR (RT-PCR) and Western Blot (WB) Assay
Total RNA and protein were extracted from B16 cells at 48 h posttransfection. Detailed experimental procedures for the detection of the expression of various genes and protein change were carried out as previously described (22).
Cell Cycle Detection
B16 cells (8 × 105/ml) were collected from every group after centrifugation (900 rpm, 5 min). Then the cells were washed with cold PBS twice and added at a concentration of 70% cold ethanol (1 ml). After that, the cells were put at 4°C for 12 h. The next day, the cells were rewashed with cold PBS twice, stained with 500 µl PI, and monitored by flow cytometry assays.
Cell Ultrastructure
Animal models were prepared in a similar fashion as detailed in our previous report (21). Briefly, the animals were housed in plastic cages (with five or six mice/cage) under controlled conditions of humidity (44 ± 5%), light (12-h light/dark cycle), and temperature (22 ± 2°C) in the animal laboratory of Jilin University. The mice were randomly separated into five groups (n = 8) that were subcutaneously planted with B16 cells. The research procedures were described in detail previously (21). After 9 days, the mice were injected intratumorally with different experimental substances. The mice of the mock group were injected with 100 µl PBS, and experimental groups were treated with different recombinant plasmids by GO vehicle. The mice were inspected for body weight and general state (including food and drink, hair gloss level, and activity of the four limbs) of each mouse daily. The mice were executed on day 30. Before killing the animals, they were anesthetized with ketamine 100 mg/kg body weight, IP, under aseptic conditions. For conventional histology, tissues were collected immediately, prefixed with 2.5% glutaraldehyde, postfixed in 1% osmiumtetroxide, dehydrated in a graded alcohol series, embedded in epoxy resin, and cut with an ultramicrotome. Thin sections poststained with uranyl acetate and lead citrate were inspected with transmission electron microscope (TEM).
Data Analysis
The data are shown as mean ± standard deviation (SD). Statistical significance of the comparisons among samples was determined using one-way analysis of variance (ANOVA). A value of p < 0.05 was recognized as statistically significant.
RESULTS
Transfection Efficiency
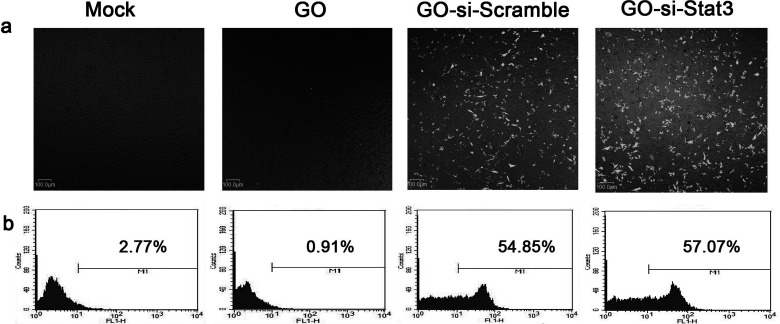
Transfection was used to evaluate the potential ability of GO-PEI-PEG to carry plasmids into B16 cells. The si-Stat3 and si-Scramble plasmids were both labeled with a green fluorescent protein (GFP) for visualization. Confocal microscopy images showed that GFP gene was expressed in GO-si-Stat3 and GO-si-Scramble (Fig. 2a). The cells transfection with mock and GO-PEI-PEG served as a negative control. The above results demonstrated that the plasmids were successfully delivered into the cells by conjugating with GO-PEI-PEG.
Figure 2.
(a) Confocal microscopy images of B16 cells treated with mock, GO-PEI-PEG, GFP-si-Scramble plasmid carried by GO-PEI-PEG and GFP-si-Stat3 plasmid carried by GO-PEI-PEG. (b) Flow cytometry analysis for transfection efficiency of B16 cells in each group.
Flow cytometry analysis was used to further quantify the delivery efficiency of plasmid by GO-PEI-PEG. The results were in accordance with the confocal imaging analysis. B16 cells treated with GO-PEI-PEG/si-Stat3-GFP and GO-PEI-PEG/si-Scramble-GFP showed strong fluorescent signals (Fig. 2b). The fraction of cells with strong fluorescent signals in the GO–PEI–PEG/si-Stat3-GFP group was counted to be almost 60%, which were significantly higher than the mock and GO groups. The results were assessed directly proportional to the delivery efficiency of plasmid Stat3 siRNA by GO-PEI-PEG, indicating that GO-PEI-PEG could be an encouraging candidate of gene delivery nanocarriers.
Ultrastructure Investigation
In order to explore the uptake of GO and the changes of ultrastructure, the ultrasection of B16 cells was observed by TEM. As shown in Figure 3, GO was indeed internalized by cells and located inside the cytoplasm around the nucleus. There were some GO nanomatrials inside the cytoplasm in different sizes. GO appeared as black dots and showed high electron density. Moreover, GO sheets were encapsulated by lysosomes. GO sheets were not observed inside the nucleus.
Figure 3.
Transmission electron microscope image of malignant melanoma cells treated with GO-si-Stat3. The arrows indicate GO nanomaterials.
The Effect of Stat3 Knockdown on B16 Cells In Vitro
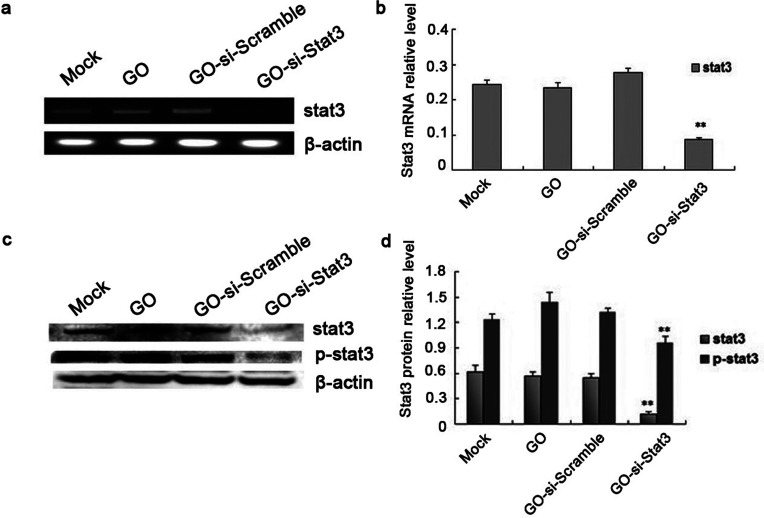
WB and RT-PCR analysis were used to detect Stat3 protein and gene expression, aiming at studying the ability of these plasmids to silence Stat3 expression in B16 cells in vitro. As shown in Figure 4, the mRNA and protein expressions of Stat3 were decreased sharply in B16 cells transfected with GO-si-Stat3 compared with the other three groups (p < 0.01). The results indicated that GO-based nanovectors for delivery of the si-Stat3 plasmid specifically knocked down the expression of Stat3 in the B16 cells effectively.
Figure 4.
(a) Assay of relative Stat3 mRNA level using RT-PCR. (b) Calculation of relative Stat3 mRNA level in each group. (c) Stat3 and p-Stat3 protein expression was determined using Western blot. (d) Calculation of relative Stat3 and p-Stat3 protein expression in each group. **p < 0.01 versus mock, GO, and GO-si-Scramble.
The Antitumor Mechanisms of Stat3 Knockdown on B16 Cells
To study the potential antitumor mechanism of GO-si-Stat3 in B16 cells in vitro, PI-stained assay was examined to measure the tumor cell cycle. B16 cells treated with GO-si-Stat3 in the G1 phase (70.69 ± 0.01%) significantly increased (Table 1). Therefore, B16 cells treated with GO-si-Stat3 in the S phase (8.53 ± 0.57%) proportionally decreased, comparable with mock, GO, and GO-si-Scramble groups. The results demonstrated that B16 cells transfected with Stat3 siRNA delivered by GO-PEI-PEG were blocked in the S phase of the tumor; thus DNA of tumor cells was significantly inhibited.
Table 1.
Induction of Cell Cycle Analysis by GO-PEI-PEG/si-Stat3 in B16 Cells (n = 3)
| G0–G1 Phase | S Phase | |
|---|---|---|
| Mock | 50.83 ± 2.78 | 41.02 ± 7.35 |
| GO | 52.94 ± 0.34 | 34.71 ± 0.07 |
| GO-si-Scramble | 54.91 ± 3.32 | 30.49 ± 1.02 |
| GO-si-Stat3 | 70.69 ± 0.01* | 8.53 ± 0.57* |
p < 0.01 versus Mock, GO, and GO-si-Scramble.
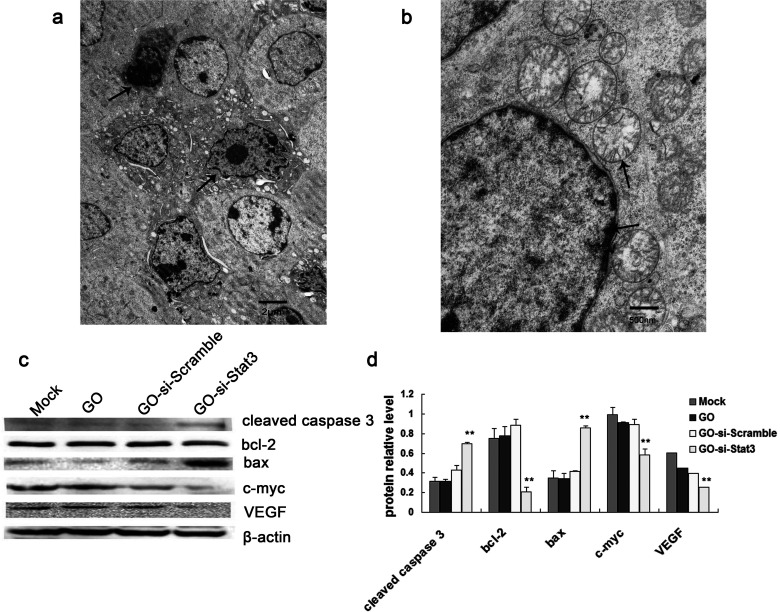
The possible mechanism to silence Stat3 was determined next using Western blot analyses. It is observed that the Stat3-related protein expressions of cleaved caspase 3 and Bax protein levels were significantly increased in GO-si-Stat3, compared with control groups. In contrast, the expressions of c-Myc, Bcl-2, and VEGF were markedly decreased in GO-si-Stat3, compared with controls (Fig. 5c, d).
Figure 5.
(a) Transmission electron microscope image of B16 cells treated with GO-si-Stat3. The arrows indicate typical apoptotic cells. (b) TEM image of B16 cells treated with GO-si-Stat3. The upper arrow indicates fractured mitochondrial cristaes. The lower arrow indicates chromatin margination. (c) Western blot method of investigation of Stat3-related proteins in B16 cells. (d) Calculation of relative Stat3-related protein expression in each group. **p < 0.01 versus mock, GO, and GO-si-Scramble.
The images of TEM (Fig. 5a, b) showed that B16 cells treated with GO-si-Stat3 had numerous apoptotic cells, characteristic of small cells in size, shrinkage of cell nucleus, high chromatin condensation, and chromatin margination. The mitochondria in the cytoplasm became swollen, and the mitochondrial cristaes were fractured.
DISCUSSION
To testify to the efficiency of the GO-PEI-PEG in delivering plasmids, B16 cells were simultaneously selected as cell models. The plasmids labeled with GFP in the transfected cells were visualized, expressing GFP by confocal microscopy. According to a previous report, PEI has been widely used as a gene delivery (16). PEI possesses the proton sponge effect, thus effectively protecting plasmids from nuclease degradation (23) and successfully transporting them to the cells by penetrating the cell membrane. PEI demonstrated enhanced cellular uptake and transfection efficiency when it was linked to carbon materials (24). The cells transfected with GO-PEI had rather good transfection efficiency in accordance with a previous literature report (16). PEG was coupled to enhance the stability of the GO-PEI complex in a physicochemical environment (25). The fluorescent signals from the laser confocal images (Fig. 2) indicated that plasmid Stat3-siRNA was successfully delivered into the cells by coupling with GO-PEI-PEG. Flow cytometry analysis was consistent with the results of laser confocal analysis. B16 cells transfected with si-Stat3-GFP plasmid and si-Scramble-GFP plasmid revealed strong fluorescent signals, which indicated the abundant accumulation of plasmid in the cells. The results suggested that the GO-PEI-PEG could be used as efficient gene vectors.
To observe the uptake of GO nanomaterials loaded with plasmids for B16 cells clearly, TEM analysis was performed. These experimental results indicated that GO entered the cytoplasm around the nucleus. The uptake mechanism of GO inside the cell may be due to cell endocytosis. Plasmids carried by GO may be delivered into the nucleus, whereas GO nanomaterials are mainly located inside the cytoplasm such as lysosomes, mitochondrion, and endoplasm. GO was encapsulated by lysosomes under TEM. GO nanomaterials in different sizes were seen, mainly due to ultrasound methods in preparation of GO, resulting in GO nanosheets in diverse sizes (Fig. 3).
Since GO was indeed internalized by B16 cells, can si-Stat3 plasmid be delivered by GO inside the cells to act as a biomedical function? In order to investigate the RNAi effect of plasmid-based si-Stat3, WB and RT-PCR analyses were carried out. These experimental results revealed that the Stat3 mRNA and protein expression decreased significantly in the transfection with GO-si-Stat3 compared with the other three groups (p < 0.01). Stat3 has been recognized as an oncogene. Previous data showed that constitutive activation of Stat3 was implicated in various tumor cells, including malignant melanoma (26–28). Stat3 plays an important part in promoting proliferation, differentiation, antiapoptosis, and cell cycle progression. For these reasons, the Stat3 signaling pathway can be applied as a potential target in antitumor therapy (29). Therefore, inhibition of Stat3 expression accompanies tumor growth suppression and induction of tumor cell apoptosis. In this article, plasmid-based Stat3 siRNA carried by GO may be released into the nucleus and retain normal biologic activity. Hence, the expression levels of Stat3 were markedly decreased in GO-si-Stat3.
The potential antitumor mechanism of GO-si-Stat3 in B16 cells was evaluated using Western blot. As shown in Figure 5c and d, the Stat3-related protein expression levels of Bcl-2, c-Myc, and VEGF in the B16 cells transfected with GO-si-Stat3 were markedly decreased, whereas expressions of Bax and cleaved caspase 3 were notably elevated in GO-si-Stat3 compared with controls. A possible explanation for this phenomenon is that knockdown Stat3 gene downregulated antiapoptotic factors such as Bcl-2 and VEGF and also downregulated cell cycle-related factors such as c-Myc (Table 1). B16 cells transfected with Stat3 siRNA delivered by GO-PEI-PEG were blocked in the S phase of the tumor; thus the DNA of the tumor cells was significantly inhibited.
The results demonstrated that knockdown Stat3 upregulated the expression of several proapoptotic proteins, including Bax and cleaved caspase 3, which are key components of mitochondrial apoptotic pathways (30). As shown in Figure 5a, lots of apoptotic cells were seen at different stages by TEM. In the meantime, the morphology of mitochondria in the cytoplasm became swollen, and the mitochondrial cristaes were fractured (Fig. 5b).
In summary, GO-PEI-PEG is able to effectively transfer plasmid-based si-Stat3 into B16 cells. Moreover, Stat3 siRNA carried by GO retains normal biologic activity. Based on the molecular biology, Stat3 protein and related protein expressions were significantly inhibited. From morphology, there were symptoms of apoptotic cells, indicating that the formation of GO-PEI-PEG complexes could be utilized as an effective gene delivery strategy. These plasmid-based Stat3 siRNA delivered by functional graphene oxide complexes are expected to be applied in cancer therapy in clinic in the future.
ACKNOWLEDGMENTS
This work was funded by the National Natural Science Foundation of China (Nos. 81201188, 81472344, 81370240), Nature Science Fund of Guangdong Province (No. 2014A030310159), and Guangzhou Medical University Foundation of China (No. 2013C07).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Alshamsan A.; Hamdy S.; Samuel J.; El-Kadi A. O.; Lavasanifar A.; Uludag H. The induction of tumor apoptosis in B16 melanoma following STAT3 siRNA delivery with a lipid-substituted polyethylenimine. Biomaterials 31:1420–1428; 2010. [DOI] [PubMed] [Google Scholar]
- 2. Wu M. T.; Wu R. H.; Hung C. F.; Cheng T. L.; Tsai W. H.; Chang W. T. Simple and efficient DNA vector-based RNAi systems in mammalian cells. Biochem. Biophys. Res. Commun. 330:53–59; 2005. [DOI] [PubMed] [Google Scholar]
- 3. Bromberg J. F.; Wrzeszczynska M. H.; Devgan G.; Zhao Y.; Pestell R. G.; Albanese C.; Darnell J. E. Jr. Stat3 as an oncogene. Cell 98:295–303; 1999. [DOI] [PubMed] [Google Scholar]
- 4. Gao L.; Zhang L.; Hu J.; Li F.; Shao Y.; Zhao D. Kalvakolanu D. V.; Kopecko D. J.; Zhao X.; Xu D. Q. Down-regulation of signal transducer and activator of transcription 3 expression using vector-based small interfering RNAs suppresses growth of human prostate tumor in vivo. Clin. Cancer Res. 11:6333–6341; 2005. [DOI] [PubMed] [Google Scholar]
- 5. Hung W.; Elliott B. Cooperative effect of c-Src tyrosine kinase and Stat3 in activation of hepatocyte growth factor expression in mammary carcinoma cells. J. Biol. Chem. 276:12395–12403; 2001. [DOI] [PubMed] [Google Scholar]
- 6. Catlett-Falcone R.; Landowski T. H.; Oshiro M. M.; Turkson J.; Levitzki A.; Savino R.; Ciliberto G.; Moscinski L.; Fernandez-Luna J. L.; Nunez G.; Dalton W. S.; Jove R. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 10:105–115; 1999. [DOI] [PubMed] [Google Scholar]
- 7. Vogelstein B.; Lane D.; Levine A. J. Surfing the p53 network. Nature 408:307–310; 2000. [DOI] [PubMed] [Google Scholar]
- 8. Kortylewski M.; Yu H. Role of Stat3 in suppressing anti-tumor immunity. Curr. Opin. Immunol. 20:228–233; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang L.; Gao L.; Li Y.; Lin G.; Shao Y.; Ji K.; Yu H.; Hu J.; Kalvakolanu D. V.; Kopecko D. J.; Zhao X.; Xu D. Q. Effects of plasmid-based Stat3-specific short hairpin RNA and GRIM-19 on PC-3M tumor cell growth. Clin. Cancer Res. 14:559–568; 2008. [DOI] [PubMed] [Google Scholar]
- 10. Zhang L.; Gao L.; Zhao L.; Guo B.; Ji K.; Tian Y.; Wang J.; Yu H.; Hu J.; Kalvakolanu D. V.; Kopecko D. J.; Zhao X.; Xu D. Q. Intratumoral delivery and suppression of prostate tumor growth by attenuated Salmonella enterica serovar typhimurium carrying plasmid-based small interfering RNAs. Cancer Res. 67:5859–5864; 2007. [DOI] [PubMed] [Google Scholar]
- 11. Liang Z. W.; Guo B. F.; Li Y.; Li X. J.; Li X.; Zhao L. J.; Gao L. F.; Yu H.; Zhao X. J.; Zhang L.; Yang B. X. Plasmid-based Stat3 siRNA delivered by hydroxyapatite nanoparticles suppresses mouse prostate tumour growth in vivo. Asian J. Androl. 13:481–486; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Whitehead K. A.; Langer R.; Anderson D. G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 8:129–138; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emeagi P. U.; Maenhout S.; Dang N.; Heirman C.; Thielemans K.; Breckpot K. Downregulation of Stat3 in melanoma: Reprogramming the immune microenvironment as an anticancer therapeutic strategy. Gene Ther. 20:1085–1092; 2013. [DOI] [PubMed] [Google Scholar]
- 14. Manuel E. R.; Blache C. A.; Paquette R.; Kaltcheva T. I.; Ishizaki H.; Ellenhorn J. D.; Hensel M.; Metelitsa L.; Diamond D. J. Enhancement of cancer vaccine therapy by systemic delivery of a tumor-targeting Salmonella-based STAT3 shRNA suppresses the growth of established melanoma tumors. Cancer Res. 71:4183–4191; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tian B.; Wang C.; Zhang S.; Feng L.; Liu Z. Photothermally enhanced photodynamic therapy delivered by nano-graphene oxide. ACS Nano 5:7000–7009; 2011. [DOI] [PubMed] [Google Scholar]
- 16. Feng L.; Zhang S.; Liu Z. Graphene based gene transfection. Nanoscale 3:1252–1257; 2011. [DOI] [PubMed] [Google Scholar]
- 17. Zhang L.; Lu Z.; Zhao Q.; Huang J.; Shen H.; Zhang Z. Enhanced chemotherapy efficacy by sequential delivery of siRNA and anticancer drugs using PEI-grafted graphene oxide. Small 7:460–464; 2011. [DOI] [PubMed] [Google Scholar]
- 18. Neu M.; Fischer D.; Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J. Gene Med. 7:992–1009; 2005. [DOI] [PubMed] [Google Scholar]
- 19. Liu Z.; Cai W.; He L.; Nakayama N.; Chen K.; Sun X.; Chen X.; Dai H. In vivo biodistribution and highly efficient tumour targeting of carbon nanotubes in mice. Nat. Nanotechnol. 2:47–52; 2007. [DOI] [PubMed] [Google Scholar]
- 20. Zhang L.; Xia J.; Zhao Q.; Liu L.; Zhang Z. Functional graphene oxide as a nanocarrier for controlled loading and targeted delivery of mixed anticancer drugs. Small 6:537–544; 2010. [DOI] [PubMed] [Google Scholar]
- 21. Yin D.; Li Y.; Lin H.; Guo B.; Du Y.; Li X.; Jia H.; Zhao X.; Tang J.; Zhang L. Functional graphene oxide as a plasmid-based Stat3 siRNA carrier inhibits mouse malignant melanoma growth in vivo. Nanotechnology 24:105102; 2013. [DOI] [PubMed] [Google Scholar]
- 22. Jia H.; Li Y.; Zhao T.; Li X.; Hu J.; Yin D.; Guo B.; Kopecko D. J.; Zhao X.; Zhang L.; Xu D. Q. Antitumor effects of Stat3-siRNA and endostatin combined therapies, delivered by attenuated Salmonella, on orthotopically implanted hepatocarcinoma. Cancer Immunol. Immunother. 61:1977–1987; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Godbey W. T.; Barry M. A.; Saggau P.; Wu K. K.; Mikos A. G. Poly(ethylenimine)-mediated transfection: A new paradigm for gene delivery. J. Biomed. Mater. Res. 51:321–328; 2000. [DOI] [PubMed] [Google Scholar]
- 24. Nunes A.; Amsharov N.; Guo C.; Van den Bossche J.; Santhosh P.; Karachalios T. K.; Nitodas S. F.; Burghard M.; Kostarelos K.; Al-Jamal K. T. Hybrid polymer-grafted multiwalled carbon nanotubes for in vitro gene delivery. Small 6:2281–2291; 2010. [DOI] [PubMed] [Google Scholar]
- 25. Lee H.; Jeong J. H.; Park T. G. A new gene delivery formulation of polyethylenimine/DNA complexes coated with PEG conjugated fusogenic peptide. J. Control Release 76:183–192; 2001. [DOI] [PubMed] [Google Scholar]
- 26. Lee S. O.; Lou W.; Qureshi K. M.; Mehraein-Ghomi F.; Trump D. L.; Gao A. C. RNA interference targeting Stat3 inhibits growth and induces apoptosis of human prostate cancer cells. Prostate 60:303–309; 2004. [DOI] [PubMed] [Google Scholar]
- 27. Mora L. B.; Buettner R.; Seigne J.; Diaz J.; Ahmad N.; Garcia R.; Bowman T.; Falcone R.; Fairclough R.; Cantor A.; Muro-Cacho C.; Livingston S.; Karras J.; Pow-Sang J.; Jove R. Constitutive activation of Stat3 in human prostate tumors and cell lines: Direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 62:6659–6666; 2002. [PubMed] [Google Scholar]
- 28. Garcia R.; Bowman T. L.; Niu G.; Yu H.; Minton S.; Muro-Cacho C. A.; Cox C. E.; Falcone R.; Fairclough R.; Parsons S.; Laudano A.; Gazit A.; Levitzki A.; Kraker A.; Jove R. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene 20:2499–2513; 2001. [DOI] [PubMed] [Google Scholar]
- 29. Germain D.; Frank D. A. Targeting the cytoplasmic and nuclear functions of signal transducers and activators of transcription 3 for cancer therapy. Clin. Cancer Res. 13:5665–5669; 2007. [DOI] [PubMed] [Google Scholar]
- 30. Al Z. S. K.; Turkson J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 18:254–267; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]