Abstract
Ovarian cancer is highly malignant with a gradually increasing incidence and a high mortality rate. Immunosuppression is induced in ovarian cancer, although the mechanism detail is not clear. It has been indicated that HVEM (herpesvirus entry mediator) B- and T-lymphocyte attenuator (BTLA) negatively regulates the immune responses of T lymphocytes. Here, HVEM mRNA was found to be elevated in ovarian cancer tissue samples and primary ovarian cancer cells in comparison with benign tissue samples. We then knocked down HVEM expression in an ovarian cancer cell line, OVCAR3, by lentivirus-based small hairpin RNA (shRNA). Cell Counting Kit-8 (CCK-8) assay and flow cytometry analysis showed that HVEM-shRNA had no effect on the proliferation, early apoptosis, or cell cycle distribution of OVCAR3. We then isolated activated T cells and performed coculture experiments in Transwell. Remarkably, HVEM-silenced ovarian cancer cells (primary ovarian cancer cells and OVCAR3) increased the number of T cells and the secretion of tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ), while activated T cells promoted the apoptosis of HVEM-silenced ovarian cancer cells. The current study partially explains the immune escape mechanism of ovarian cancer cells and provides a possible target for immunotherapy.
Key words: Herpesvirus entry mediator (HVEM), Ovarian cancer, T cells, Immunosuppression
INTRODUCTION
Ovarian cancer is highly malignant and has had a gradually increasing incidence over the last 40 years. The mortality rate of ovarian cancer is the highest in gynecological malignant tumors and threatens the life and health of women (1). Traditional treatment, including surgery, chemotherapy, and radiotherapy, can alleviate this disease, but the recurrence rate is still high, and the quality of life for patients is seriously altered by these treatments (2). Cancer immunotherapy attempts to enhance the reactivity of the immune system to the tumor and the immunogenicity of the tumor. With the rapid development of tumor immunology, a series of tumor-associated antigens has been identified, and the immunotherapy targeting these antigens has been used to clear minimal residual disease. Immunotherapy, as a useful supplement to the general treatment of malignant tumors, has a good prospect of application (3).
The immune system plays a vital role in the development of ovarian cancer (4). Several mechanisms on how immunosuppression is induced in ovarian cancer have been proposed, including immune tolerance of dendritic cells and macrophages in the local microenvironment of the pelvic cavity (5), decreased ratio of CD4 to CD8, and impaired lymphocyte response (6), as well as the increased proportion of regulatory T cells (Tregs; CD4+CD25+Foxp3+) (5,7). The key to the progress of ovarian cancer is that the anergic lymphocytes cannot destroy cancer cells.
B- and T-lymphocyte attenuator (BTLA), an immunoglobulin (Ig) domain superfamily protein, is not expressed by naive T cells but is induced during T-cell activation. BTLA is expressed in T helper 1 (Th1) cells but not in Th2 cells. Herpesvirus entry mediator (HVEM), the ligand of BTLA, is a member of the tumor necrosis factor superfamily. Accumulating evidence has indicated that HVEM-BTLA negatively regulates the immune responses of T lymphocytes (8,9). We hypothesized that HVEM-BTLA is involved in the anergy in T lymphocytes, leading to the development of ovarian cancer.
In the present study, knockdown of HVEM expression in ovarian cancer cells had no effect on the proliferation, early apoptosis, or cell cycle distribution. Coculture experiments showed that HVEM-silenced ovarian cancer cells increased the number of T cells as well as the secretion of tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ), and activated T cells promoted the apoptosis of HVEM-silenced ovarian cancer cells. Our study will open a possibility for novel therapeutic strategies in ovarian cancer.
MATERIALS AND METHODS
Isolation of Primary Ovarian Cancer Cells and Cell Culture
The use of patient samples was approved by Shanghai First Maternity and Infant Hospital Institutional Review Board. Ovarian cancer samples were minced to 2-mm3 pieces, incubated with 800 U/ml collagenase type II (Sigma-Aldrich) at 37°C for 30 min, and further disaggregated by mechanical disruption. Following filtration through a 100-µm cell strainer, single cells were washed and then incubated with anti-CD31- and anti-CD45-conjugated microbeads (Miltenyi Biotec, Auburn, CA, USA) at 4°C with rotation for 30 min to remove endothelial and hematopoietic cells. The unbound epithelial cancer cells were collected and cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) and antibiotics in a 5% CO2 incubator at 37°C.
OVCAR3 cells were obtained from Shanghai Institutes for Biological Sciences cell bank and cultured in RPMI-1640 medium.
Isolation of Total RNA and Real-Time PCR Analysis of HVEM
Analysis of HVEM expression by real-time polymerase chain reaction (PCR) was performed as described previously (10). Briefly, total RNA was isolated from tissue samples and cancer cells with TRIzol reagent (Life Technologies, Grand Island, NY, USA) and then reverse transcribed into cDNA. Primers used for real-time PCR were synthesized by Generay (Shanghai, China) as follows: HVEM, 5′-CTGCTCCAGGACAGAGAAC-3′ and 5′-CGGAGACGATCACCTTGAC-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-CACCCACTCCTCCACCTTTG-3′ and 5′-CCACCACCCTGTTGCTGTAG-3′. Real-time PCR was performed on 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). HVEM expression was normalized to GAPDH.
Lentiviral Vector Construction and Transfection
To knock down the expression of HVEM, two small hairpin RNA (shRNA) targeting sequence of human HVEM gene and a scramble shRNA (NC) were synthesized (Generay), annealed, and ligated into PGCSIL vector with AgeI/EcoRI sites. The constructed shRNA plasmids were cotransfected with lentiviral packaging vectors into HEK293T cells according to the instruction of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). At 72 h posttransfection, lentivirus particles were collected from cultured medium and used to infect primary ovarian cancer cells and OVCAR3 cells.
Cell Counting Kit-8 (CCK-8) Assay
OVCAR3 cells were plated at a density of 30,000 cells per well in a 96-well plate. After 12 h of culture, OVCAR3 cells were infected with indicated HVEM-shRNA or control virus. At 0, 24, 48, and 72 h after viral infection, 10% CCK-8 solution (v/v) was added to each well. After incubation for 1 h, the optical density at 450 nm (OD450) was measured on a microplate reader (Labsystems, Helsinki, Finland).
Cell Cycle and Cell Apoptosis Analyses
OVCAR3 cells cultured in six-well plates were infected with HVEM-shRNA or control virus. At 48 h after infection, cells were harvested and washed with ice-cold phosphate-buffered saline (PBS). For cell cycle analysis, cells were fixed with cold 70% ethanol at −20°C overnight, washed twice with ice-cold PBS, and stained with propidium iodide (PI; 20 µg/ml; Sigma-Aldrich) and RNase (100 µg/ml; Sigma-Aldrich) in the dark for 20 min. For cell apoptosis analysis, cells were stained with annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kits (KeyGEN Biotech, Nanjing, China). Cell cycle distribution and cell apoptosis were analyzed using a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
T-Cell Isolation and Activation
Human peripheral blood mononuclear cells (PBMCs) were isolated from heparinized whole blood from volunteer donors (Shanghai First Maternity and Infant Hospital) by centrifugation at 300 × g for 30 min on mononuclear/polynuclear cell-resolving medium (Flow Laboratories, Rockville, MD, USA).
Isolation and activation of T cells from PBMCs were performed with T Cell Activation/Expansion Kit (Miltenyi Biotec) according to the manufacturer’s instruction. Briefly, anti-biotin MACSiBead particles were loaded with biotinylated antibodies against human CD2, CD3, and CD28, added to resuspended PBMCs (bead-to-cell ratio 1:2), and mixed well. The mixture was cultured at 37°C in a humidified air atmosphere containing 5% CO2 for 3 days. To validate the activation of T cells, the cells were collected, fluorescently stained using anti-CD4-FITC and anti-CD25-PE or anti-CD4-FITC and anti-CD69-PE, and analyzed in a flow cytometer (BD Biosciences).
Transwell Experiments
Transwell experiments were performed in 24-well plates (0.4-µm pore size; Corning Inc., New York, NY, USA). Activated T cells (6 × 106) were placed in the upper chamber, and OVCAR3 or primary cancer cells (2 × 104) were placed in the lower chamber for 72 h. At the end of the culture, T cells were harvested for cell number counting with a flow cytometer, the cultured media were collected for enzyme-linked immunosorbent assay (ELISA) analysis and lactate dehydrogenase (LDH) assay, and the cancer cells were harvested for CCK-8 assay and Western blotting analysis.
T-Cell Number Counting
T cells in the upper chamber were harvested, spun at 3,000 rpm for 10 min, and resuspended in 600 µl of PBS. A total of 100 µl of cell suspension was subjected to flow cytometry analysis for cell number counting.
ELISA for TNF-α and IFN-γ in Cell Culture Supernatants
Cell culture supernatants were collected. The concentrations of TNF-α and IFN-γ were measured using the human TNF-α and IFN-γ ELISA kit (Bio-Swap, Life Science, Shanghai, China), respectively. Assays were carried out on microtiter plates following the instructions of the manufacturer.
LDH Measurement
After treatment, LDH released into the culture medium was determined by using LDH-Cytotoxicity Colorimetric Assay Kit II (BioVision, Mountain View, CA, USA) following the manufacturer’s protocol on a microplate reader.
Western Blot Analysis
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer and separated on 15% SDS-polyacrylamide gel electrophoresis (PAGE) in denaturing conditions at 25 µg of protein per lane. Proteins were electrotransferred onto nitrocellulose membrane. After blocking with 5% skim milk, the membranes were probed with anti-Bax (Sc-493, 1:400; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Bcl-2 (Sc-492, 1:400; Santa Cruz Biotechnology), or anti-GAPDH (#5174, 1:1,500; Cell Signaling Technology, Danvers, MA, USA) followed by horseradish peroxidase (HRP)-conjugated secondary antibody (Beyotime, Shanghai, China). Signals were detected by enhanced chemiluminescence system (Bio-Rad, Richmond, CA, USA).
Statistics
All results are presented as mean ± standard deviation (SD). Comparisons between groups were made using Student’s t-test. A value of p < 0.05 was considered as statistically significant. Calculations were performed with GraphPad Prism 6.0 (GraphPad Software Inc., La Jolla, CA, USA).
RESULTS
Expression of HVEM mRNA
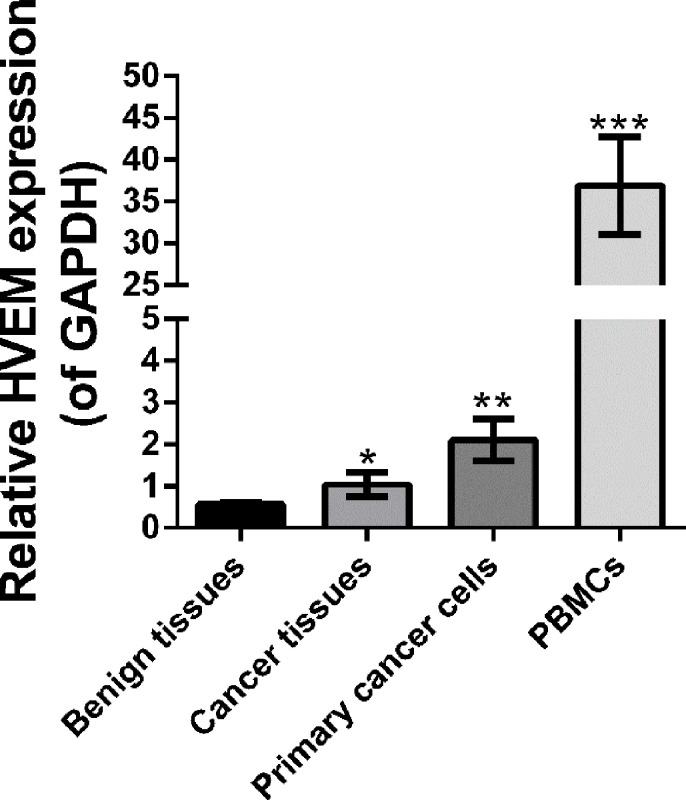
We detected the mRNA expression level of HVEM mRNA in benign tissues, ovarian cancer tissues, primary ovarian cancer cells, and PBMCs by using real-time PCR. PBMCs served as a positive control. Figure 1 showed that HVEM mRNA was significantly higher in ovarian cancer tissues and primary ovarian cancer cells than in benign tissues. These data suggest HVEM may be associated with ovarian cancer.
Figure 1.
The expression level of HVEM was determined by real-time PCR (RT-PCR) in benign tissues (n = 3), ovarian cancer tissues (n = 3), primary ovarian cancer cells (n = 3), and PBMCs (n = 6). GAPDH served as an internal control. *p < 0.05, **p < 0.01, and ***p < 0.001 as compared with benign tissues.
Silencing of HVEM Had no Effect on the Proliferation, Early Apoptosis, or Cell Cycle Distribution of OVCAR3 Cells
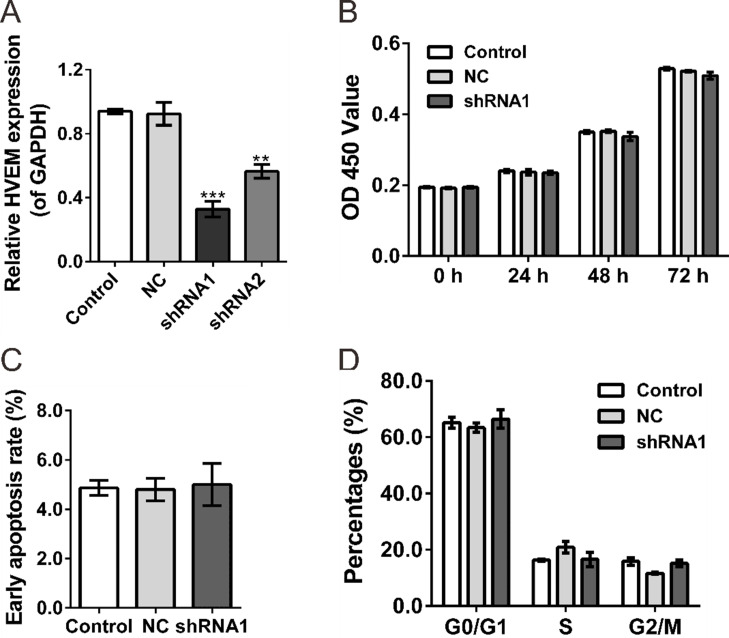
To investigate the functional role of HVEM in ovarian cancer cells, HVEM-shRNA lentivirus (shRNAi1 and shRNA2) and control shRNA lentivirus (NC) were produced and infected in a human ovarian cancer cell line, OVCAR3. Real-time PCR assays revealed that HVEM expression was significantly reduced in shRNAi lentivirus-infected cells compared to those of control and NC-infected cells (NC) (Fig. 2A). shRNAi1 had better efficiency in knocking down HVEM expression and was chosen for the following experiments.
Figure 2.
Effects of HVEM-shRNA on the proliferation of OVCAR3 cells. (A) Knockdown efficiency was determined by quantitative RT-PCR (qRT-PCR) in OVCAR3 cells. (B) Knockdown of HVEM in OVCAR3 cells did not affect the proliferative capacities as determined by the CCK-8 assay. (C, D) Knockdown of HVEM in OVCAR3 cells did not change the early apoptosis rate (C) and cell cycle distribution (D) as determined by flow cytometry analysis. Control, cells without infection; NC, cells infected with control shRNA virus; shRNA1, cells infected with HVEM-shRNA1 virus. **p < 0.01 and ***p < 0.001 compared with NC cells.
The CCK-8 assay revealed that knockdown of HVEM expression in OVCAR3 cells did not change cell proliferation compared with the control and NC cells (Fig. 2B). Next, similar early apoptosis rate (Fig. 2C) and cell cycle distribution (Fig. 2D) were observed in control, NC, and shRNAi1 cells as determined by flow cytometry analysis.
Effects of HVEM-Silenced Ovarian Cancer Cells on Activated T Cells
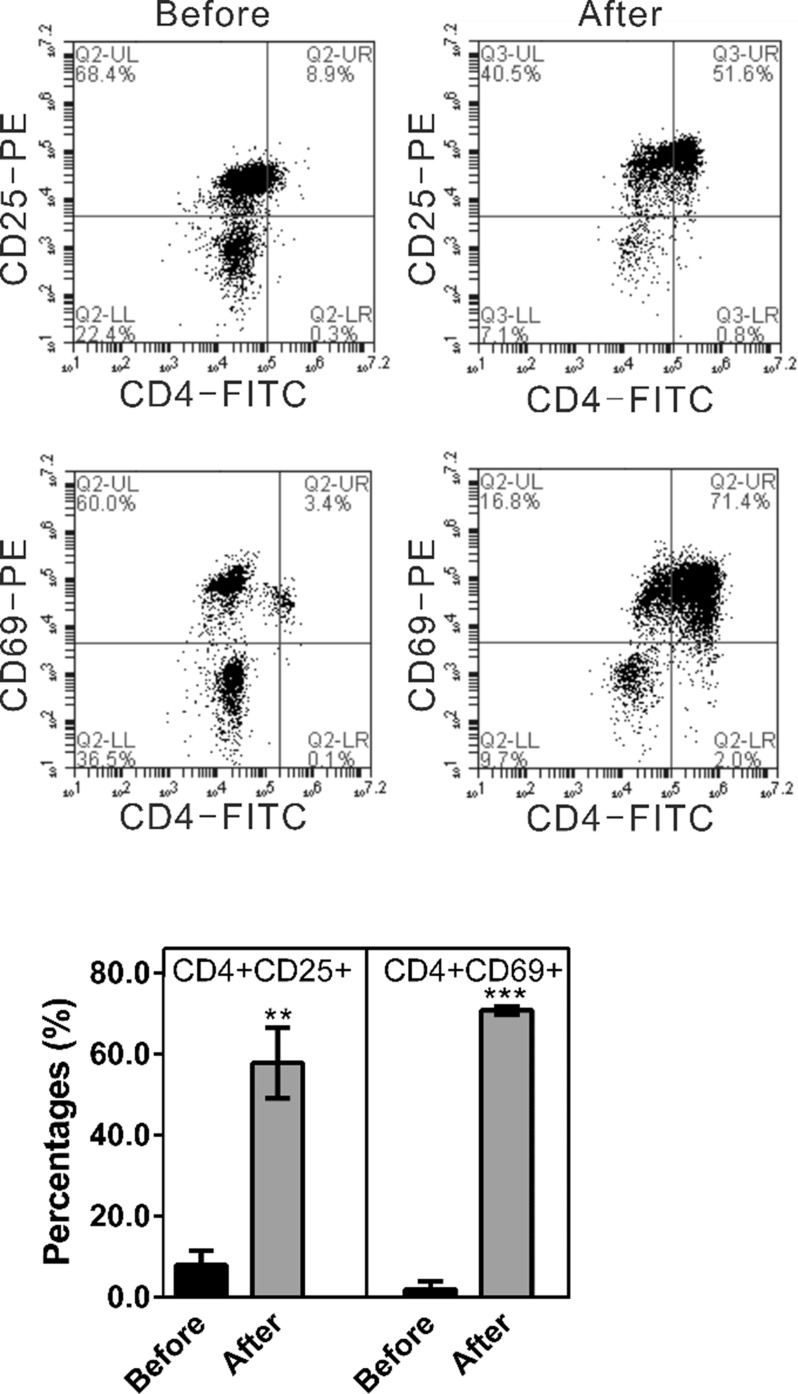
BTLA, the receptor of HVEM, is induced during T-cell activation (8,9). In order to explore the effects of HVEM-BTLA on T cells, we cocultured activated T cells with HVEM-silenced ovarian cancer cells. PBMCs were first isolated from volunteer donors (Before), and then T cells were isolated and activated from the PBMCs with the T Cell Activation/Expansion Kit (After). CD25 and CD69 are activation markers for T cells (11). As shown in Figure 3, after activation, the ratio of CD4+/CD25+ cells was increased from 7.9 ± 3.5% to 57.8 ± 8.7%, while the ratio of CD4+/CD69+ cells went from 2.9 ± 0.8% to 70.7 ± 1.1%. These data suggest that the percentages of CD4+ cells bearing activation markers, CD25 and CD69, were high and suitable for the subsequent coculture experiments.
Figure 3.
Percentages of CD4+ cells bearing activation markers, CD4+CD25+ and CD4+CD69, were significantly elevated after T-cell activation (After) in comparison with isolated PBMCs (Before). PBMCs were isolated from volunteer donors (Before), and then T cells were isolated and activated from the PBMCs with the T Cell Activation/Expansion Kit (After). To validate the activation of T cells, the cells were collected, stained using anti-CD4-FITC and anti-CD25-PE or anti-CD4-FITC and anti-CD69-PE, and analyzed in a flow cytometer. **p < 0.01 and ***p < 0.001 compared with Before.
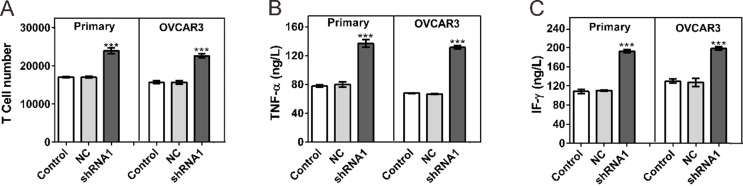
At 72 h following coculture with ovarian cancer cells, the number of T cells was counted. As shown in Figure 4A, coculture with HVEM-silenced primary ovarian cancer cells or OVCAR3 significantly increased the number of T cells.
Figure 4.
Effects of HVEM-silenced ovarian cancer cells on activated T cells. Coculture experiments were performed in Transwell chambers (0.4 µm pore size; Corning Inc.). Activated T cells (6 × 106) were placed in the upper chamber, and OVCAR3 or primary cancer cells (2 × 104) were placed in the lower chamber. (A) T-cell numbers were counted at 72 h after coculture with ovarian cancer cells. (B, C) Secretion of TNF-α and IFN-γ was evaluated at 72 h after coculture with ovarian cancer cells by ELISA assay. Control, cells without infection; NC, cells infected with control shRNA virus; shRNA1, cells infected with HVEM-shRNA1 virus. ***p < 0.001 compared with NC cells.
TNF-α and IFN-γ are known to play an important role in regulating immune responses and repressing tumor cell growth (12). We then assessed the secretion of TNF-α and IFN-γ by ELISA assay. Figure 4B and C shows that coculture with HVEM-silenced cancer cells significantly enhanced the secretion of detected cytokines.
Effect of Activated T Cells on HVEM-Silenced Ovarian Cancer Cells
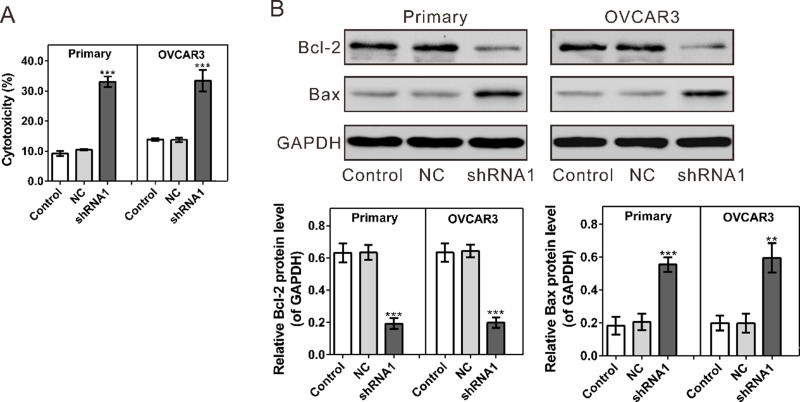
At 72 h following coculture with activated T cells, significantly increased cytotoxicity was observed in both primary cultured ovarian cancer cells and OVCAR3 cells with HVEM silenced as assessed by LDH release measurement (Fig. 5A).
Figure 5.
Effects of activated T cells on HVEM-silenced ovarian cancer cells. (A) Following 72 h of coculture with T cells, cell cytotoxicity was significantly increased in primary ovarian cancer cells (Primary) and OVCAR3 with HVEM silenced as determined by LDH release. (B) Protein levels of Bcl-2 and Bax were evaluated by Western blot. Control, cells without infection; NC, cells infected with control shRNA virus; shRNA1, cells infected with HVEM-shRNA1 virus. **p < 0.01 and ***p < 0.001 compared with NC cells.
Moreover, the protein level of apoptosis-regulating proteins was evaluated by Western blot. At 72 h after coculture with activated T cells, HVEM knockdown led to a remarkable reduction in the antiapoptosis protein Bcl-2 and a significant increase in the apoptosis-promoting protein Bax (Fig. 5B). These results indicate that the activated T cells could promote the apoptosis of ovarian cancer cells with HVEM silenced.
DISCUSSION
The tumor microenvironment (TME) composed of noncancer cells and their stroma can affect the growth and evolution of cancerous cells (13,14). Cancer cells can also affect TME by promoting angiogenesis and inducing immune tolerance. HVEM-BTLA is reported to negatively regulate T lymphocytes (8,9). In the present study, we found that activated T cells induced the apoptosis of HVEM-silenced ovarian cancer cells, and HVEM-silenced ovarian cancer cells increased the number of T cells, as well as the secretion of TNF-α and IFN-γ. Ovarian cancer cells and TME can affect each other.
Immunosuppression is induced in most cancer patients, although they have a number of CD8 and CD4 T cells that are specific for the tumor antigen. Numerous studies have suggested that Tregs, a population of CD4+ T cells with a high expression of CD25, contribute to cancer-related immunosuppression (7,15–18). We isolated and activated T cells from PBMCs of volunteer donors. The proportion of Tregs in the activated T cells used for coculture experiments was more than 50% (Fig. 3) and can be used to mimic the condition in ovarian cancer patients. Coculture experiments showed that HVEM-silenced cancer cells significantly increased the number of T cells and enhanced the secretion of detected cytokines. Our data suggested that the overexpression of HVEM in ovarian cancer cells inhibited the proliferation and the immune functions of T cells (Fig. 4). These results propose a potential mechanism for the immunosuppressive state of ovarian cancer, although further investigation is still needed. Furthermore, HVEM knockdown had no effect on the proliferation, apoptosis, or cell cycle distribution of OVCAR3 cells (Fig. 2). Increased cancer cell apoptosis was observed when HVEM was knocked down in both primary ovarian cancer cells and ovarian cancer cell line (Fig. 5), and cocultured with activated T cells. These results demonstrated the cytotoxicity of activated T cells on HVEM-silenced cancer cells. Our data indicate that HVEM might be a novel target for ovarian cancer treatment.
In summary, the overexpression of HVEM in ovarian cancer cells may suppress the proliferation and immune function of T cells, thus leading to the development of ovarian cancer. The current study partially explains the immune escape mechanism of ovarian cancer cells.
ACKNOWLEDGMENT
This work was supported by grants from Shanghai Health Development and Planning Commission Issues (No. 20124111).
REFERENCES
- 1. Siegel R.; Naishadham D.; Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 63:11–30; 2013. [DOI] [PubMed] [Google Scholar]
- 2. Marsden D. E.; Friedlander M.; Hacker N. F. Current management of epithelial ovarian carcinoma: A review. Semin. Surg. Oncol. 19:11–19; 2000. [DOI] [PubMed] [Google Scholar]
- 3. Mellman I.; Coukos G.; Dranoff G. Cancer immunotherapy comes of age. Nature 480:480–489; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Visser K. E.; Eichten A.; Coussens L. M. Paradoxical roles of the immune system during cancer development. Nat. Rev. Cancer 6:24–37; 2006. [DOI] [PubMed] [Google Scholar]
- 5. Yigit R.; Massuger L. F.; Figdor C. G.; Torensma R. Ovarian cancer creates a suppressive microenvironment to escape immune elimination. Gynecol. Oncol. 117:366–372; 2010. [DOI] [PubMed] [Google Scholar]
- 6. Sato E.; Olson S. H.; Ahn J.; Bundy B.; Nishikawa H.; Qian F.; Jungbluth A. A.; Frosina D.; Gnjatic S.; Ambrosone C. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc. Natl. Acad. Sci. USA 102:18538–18543; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wolf D.; Wolf A. M.; Rumpold H.; Fiegl H.; Zeimet A. G.; Muller-Holzner E.; Deibl M.; Gastl G.; Gunsilius E.; Marth C. The expression of the regulatory T cell-specific forkhead box transcription factor FoxP3 is associated with poor prognosis in ovarian cancer. Clin. Cancer Res. 11:8326–8331; 2005. [DOI] [PubMed] [Google Scholar]
- 8. Jun-Fa X. U. BTLA-A novel co-inhibitory molecule of CD28 family. Int. J. Immunol. 2007. [Google Scholar]
- 9. Murphy T. L.; Murphy K. M. Slow down and survive: Enigmatic immunoregulation by BTLA and HVEM. Annu. Rev. Immunol. 28:389–411; 2010. [DOI] [PubMed] [Google Scholar]
- 10. Saha S. K.; Roy S.; Khuda-Bukhsh A. R. Ultra-highly diluted plant extracts of Hydrastis canadensis and Marsdenia condurango induce epigenetic modifications and alter gene expression profiles in HeLa cells in vitro. J. Integr. Med. 13:400–411; 2015. [DOI] [PubMed] [Google Scholar]
- 11. Caruso A.; Licenziati S.; Corulli M.; Canaris A. D.; De Francesco M. A.; Fiorentini S.; Peroni L.; Fallacara F.; Dima F.; Balsari A.; Turano A. Flow cytometric analysis of activation markers on stimulated T cells and their correlation with cell proliferation. Cytometry 27:71–76; 1997. [DOI] [PubMed] [Google Scholar]
- 12. Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer 4:11–22; 2004. [DOI] [PubMed] [Google Scholar]
- 13. Pietras K.; Östman A. Hallmarks of cancer: Interactions with the tumor stroma. Exp. Cell Res. 316:1324–1331; 2010. [DOI] [PubMed] [Google Scholar]
- 14. Whiteside T. The tumor microenvironment and its role in promoting tumor growth. Oncogene 27:5904–5912; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woo E. Y.; Chu C. S.; Goletz T. J.; Schlienger K.; Yeh H.; Coukos G.; Rubin S. C.; Kaiser L. R.; June C. H. Regulatory CD4(+)CD25(+) T cells in tumors from patients with early-stage non-small cell lung cancer and late-stage ovarian cancer. Cancer Res. 61:4766–4772; 2001. [PubMed] [Google Scholar]
- 16. Sasada T.; Kimura M.; Yoshida Y.; Kanai M.; Takabayashi A. CD4+CD25+ regulatory T cells in patients with gastrointestinal malignancies: Possible involvement of regulatory T cells in disease progression. Cancer 98:1089–1099; 2003. [DOI] [PubMed] [Google Scholar]
- 17. Woo E. Y.; Yeh H.; Chu C. S.; Schlienger K.; Carroll R. G.; Riley J. L.; Kaiser L. R.; June C. H. Cutting edge: Regulatory T cells from lung cancer patients directly inhibit autologous T cell proliferation. J. Immunol. 168:4272–4276; 2002. [DOI] [PubMed] [Google Scholar]
- 18. Liyanage U. K.; Moore T. T.; Joo H. G.; Tanaka Y.; Herrmann V.; Doherty G.; Drebin J. A.; Strasberg S. M.; Eberlein T. J.; Goedegebuure P. S.; Linehan D. C. Prevalence of regulatory T cells is increased in peripheral blood and tumor microenvironment of patients with pancreas or breast adenocarcinoma. J. Immunol. 169:2756–2761; 2002. [DOI] [PubMed] [Google Scholar]