Abstract
Hyperhomocysteinemia (HHcy) is considered as a risk factor for several complications, including cardiovascular and neurological disorders. A high methionine low folate (HMLF) diet chronically causes HHcy by accumulating homocysteine in the systemic circulation. Elevated Hcy level is also associated with the incidence of diabetes mellitus. However, very few studies focus on the impact of HMLF diet on glucose homeostasis, and that on gut microbiome profile. HHcy was induced by feeding C57BL/6 mice a HMLF diet for 8 weeks. The HMLF diet feeding resulted in a progressive body weight loss, and development of slight glucose intolerance and insulin resistance in HHcy mice. Notably, the HMLF diet alters the gut microbiome profile and increases the relative abundance of porphyromonadaceae family of bacteria in HHcy mice. These findings provide new insights into the roles of dysregulated glucose homeostasis and gut flora in the pathogenesis of HHcy-related complications.
Keywords: HMLF diet, Hyperhomocysteinemia, Glucose homeostasis, Gut microbiome, 16S rRNA sequencing, Porphyromonadaceae
Abbreviations: Hcy, homocysteine; HDL, high-density lipoprotein; HHcy, hyperhomocysteinemia; HMLF, high methionine low folate; LEfSe, linear discriminant analysis effect size; NAFLD, non-alcoholic fatty liver disease; NMDS, non-metric multi-dimensional scaling; OTU, operational taxonomic unit; PCA, principal component analysis; SCFA, short-chain fatty acids; TC, total cholesterol; TG, triglyceride
Highlights
-
•
A high methionine low folate (HMLF) diet induces hyperhomocysteinemia in mice.
-
•
HMLF diet slightly but significantly induces glucose intolerance and insulin resistance in mice.
-
•
HMLF diet alters gut microbiome profile in mice.
-
•
HMLF diet increases the abundance of porphyromonadaceae family of bacteria.
1. Introduction
Homocysteine (Hcy) is a non-proteinogenic sulfur-containing homologue of cysteine. Hcy is synthesized through elimination of a terminal methyl group from methionine [1]. In normal physiological conditions, Hcy is converted to either methionine or cysteine in the presence of B-vitamins, which serve as critical co-factors for normal function of methionine synthase and cystathionine synthase [2]. Dietary consumption of excessive methionine and dietary deficiency of B-vitamins (pyridoxine (B6), folate (B9) and B12) cause Hcy accumulation in the systemic circulation [3]. A diet of high methionine and low folate (HMLF) content has been therefore used to induce hyperhomocysteinemia (HHcy) in rodents [4]. An elevated level of plasma Hcy (200–300 μmol L−1) is regarded as HHcy [5], a risk factor for cardiovascular complications (e.g. coronary heart disease, hypertension) and neurological disorders (e.g. the Alzheimer's disease, the Parkinson's disease) [6]. Furthermore, increased level of serum Hcy has been reported in patients with type 2 diabetes mellitus [7]. Previous studies also showed that an elevated Hcy level correlates with higher risk of vascular complications and mortality in type 2 diabetic patients [8].
Non-enzymatic factors, particularly insulin, can affect the circulatory level of Hcy. In brief, higher plasma level of insulin down-regulates the hepatic expression of cystathionine beta-synthase, a crucial enzyme involved in the biosynthesis of cysteine from Hcy [9], implying that hyperinsulinemia may result in elevated Hcy level. Most often, hyperinsulinemia results from insulin resistance. When the cells in liver, fat and muscles fail to respond to insulin due to insulin resistance, the pancreas compensatively secretes more insulin, a condition called hyperinsulinemia [10]. Insulin resistance was shown to be associated with HHcy in a clinical trial of 2214 individuals [11]. Another study suggested that high glucose level can facilitate Hcy clearance by enhancing remethylation of Hcy, and hence methionine synthesis [12]. However, whether HHcy negatively impacts on glucose metabolism and insulin sensitivity remains inconclusive and contradictory. A previous clinical study reported a correlation between HHcy and insulin resistance in patients with hypothyroidism [13], whereas another Mendelian randomization study suggested no such causal relationship among levels of Hcy, fasting glucose and fasting insulin in European individuals [14].
In addition, the effect of HMLF diet and HMLF diet-induced HHcy on glucose homeostasis are scarcely investigated. Before gut absorption and hepatic metabolism of Hcy, the dietary HMLF content might have exerted an effect on gut microbiome, which emerges as a new organ system in the host [15]. Whether HMLF content affects gut microbiome, in terms of relative abundance and biodiversity of gut bacteria, remains to be determined. Therefore, in the present study, we aim to provide novel insights into the predisposing effects of HMLF diet on glucose homeostasis and gut microbiome in HHcy-related complications.
2. Materials and methods
2.1. Animal protocol
The current study was conducted under the approval of the Animal Experimentation Ethics Committee, the Chinese University of Hong Kong (CUHK). All animal experiments complied with the ARRIVE guidelines [16], and the guide for the care and use of laboratory animals established by the National Institutes of Health. Male C57BL/6 mice (8 weeks old, 22–25 g) were supplied by the Laboratory Animal Services Center, CUHK. Randomization was undertaken to lower the risk of bias in this study. All C57BL/6 mice (n = 10) were randomized before treatment. Before changing the diet, the male C57BL/6 mice were orally administrated 200 μL antibiotic cocktail (1 g L-1 ampicillin, 1 g L-1 metronidazole, 0.5 g L-1 neomycin and 0.5 g L-1 vancomycin) for 3 consecutive days [17]. The mice were subsequently fed on either normal chow (n = 5) or HMLF diet (n = 5) (Table 1; Envigo Teklad, Madison, WI, USA) for 8 weeks. Thereafter, fecal samples were collected for subsequent bacterial 16S rRNA sequencing (Fig. 1A). The mice were kept in individual and well-ventilated caging systems under a 12 h light: 12 h dark cycle. The mice were guaranteed free access to laboratory diets and water, at a constant temperature (22 ± 1 °C) and humidity (55% ± 5%). Body weights of mice were monitored weekly.
Table 1.
The compositions of normal chow and HMLF diet.
| Formula (g kg−1) | Normal chow | HMLF diet |
|---|---|---|
| Casein | 200.0 | 200.0 |
| l-Methionine | 3.0 | 20.0 |
| Sucrose | 524.747 | 506.7475 |
| Maltodextrin | 100.0 | 100.0 |
| Soybean oil | 30.0 | 30.0 |
| Anhydrous milkfat | 25.0 | 25.0 |
| Cellulose | 80.0 | 80.0 |
| Mineral Mix | 35.0 | 35.0 |
| Succinylsulfathiazole | 0 | 1.0 |
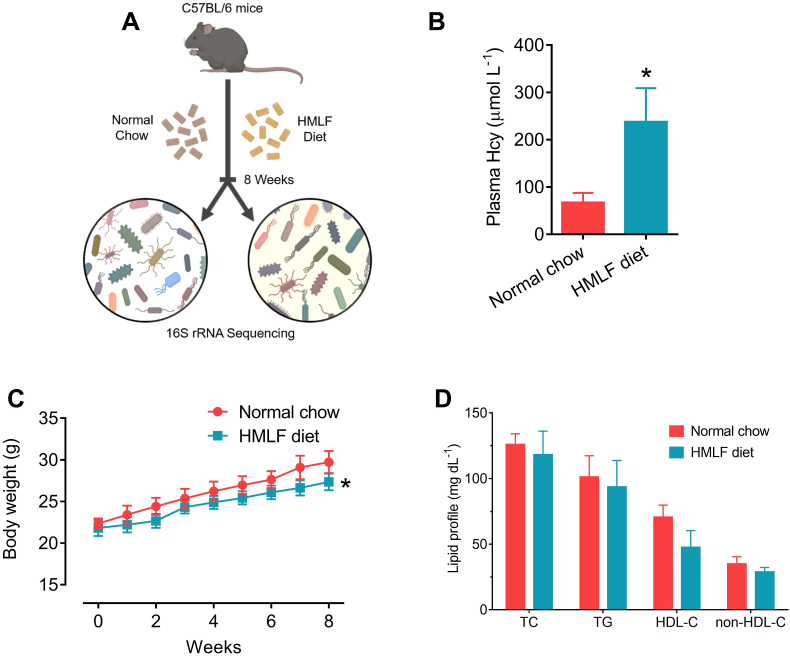
Fig. 1.
HMLF diet induced HHcy in C57BL/6 mice. (A) Schematic representation of the present study. (B) Plasma Hcy concentration of C57BL/6 mice fed with either normal chow or HMLF diet for 8 weeks. (C) Changes in body weights of mice during 8 weeks of feeding. (D) Lipid profile of two groups of mice (i.e. normal chow and HMLF diet groups). HDL-C: high-density lipoprotein cholesterol; HMLF: high methionine low folate; non-HDL-C: non-HDL cholesterol; TC: total cholesterol; TG: triglyceride. *P < 0.05 vs normal chow group. n = 5 per group. All data are expressed as mean ± SD.
2.2. Plasma Hcy measurement by ELISA
In the presence of heparin, mouse blood was collected by cardiac puncture. The blood was centrifuged for 10 min at 1000×g at 4 °C for plasma collection. The plasma Hcy level was evaluated by a commercially available ELISA kit (Cell Biolabs, San Diego, CA, USA) following the manufacturer's protocol [18].
2.3. Lipid profile determination
The levels of total cholesterol (TC), triglyceride (TG) and high-density lipoprotein (HDL) cholesterol in mouse plasma were measured by commercial assay kits (Stanbio, Boerne, TX), according to the manufacturer's instructions. In addition, the level of non-HDL cholesterol was determined by using the following formula: non-HDL cholesterol = TC - (TG/5) – HDL [19].
2.4. Blood glucose determination
After 4 and 8 weeks of HMLF diet, glucose tolerance test (GTT) and insulin tolerance test (ITT) were performed in C57/BL6 mice 6 h post-fasting. By intraperitoneal injection of glucose (1 g kg−1) or insulin (1 unit kg−1), the glucose levels were evaluated in venous blood of mouse tail at different time points (0, 15, 30, 60, 90 and 120 min).
2.5. DNA extraction from fecal samples
After 8 weeks, fecal pellets were collected from mice fed on normal chow and HMLF diet and stored at −80 °C for later processing. Bacterial DNA was isolated from the stored fecal samples by using the NucleoSpin® DNA Stool kit (Macherey-Nagel, Düren, Germany). In brief, 180–220 mg of fecal samples were homogenized in bead beating tubes by vortex at a maximal speed for 10 min. Total genomic DNA was subsequently captured on the silica membrane and NucleoSpin® Inhibitor Removal Column was applied to remove impurities that interfere with downstream procedures. At last, purified DNA was eluted by 80 μL Elution Buffer SE [20]. DNA concentrations were determined by maximum absorbance at 260 nm.
2.6. 16S rRNA sequencing: library preparation and sequencing
High-throughput sequencing of bacterial 16S rRNA gene was conducted by Novogene Technology Co. (Beijing, China). Briefly, after DNA quality was determined by agarose gel electrophoresis, the verified DNA was amplified by the 515F forward (GTGCCAGCMGCCGCGGTAA) and 806R reverse (GGACTACVSGGGTATCTAAT) primers, which aim at the V4 hypervariable region of bacterial 16S rRNA gene. The PCR products were purified by gel extraction kit (Qiagen, Germany). Sequencing libraries were constructed by NEBNext® Ultra™ DNA Library Pre Kit for Illumina (New England Biolabs), according to manufacturer's instructions. In the final step, the library was sequenced via an Illumina sequencing platform to generate 250 bp paired-end reads [21].
2.7. Data analysis for bacterial 16S rRNA sequencing
Bacterial 16S rRNA sequencing analysis was performed by using QIIME (v1.7.0; http://qiime.org/index.html) [22]. Quality filtering on successfully merged reads were conducted based on the default settings of QIIME. Sequences were assigned to operational taxonomic units (OTUs) according to the similarity to annotated bacterial sequences by using Uparse (v7.0.1001; http://drive5.com/uparse/; 97% sequence similarity cut-off) [23]. The most abundant sequence was selected to represent each OTU and classified to a taxonomic group by RDP classifier (v2.2; http://sourceforge.net/projects/rdp-classifier/) [24]. Chimeric OTUs were discarded based on the analysis against the sequence in SILVA database (http://www.mothur.org/wiki/Silva-reference-files) [25]. OTUs were intended for beta diversity analysis where unweighted Unifrac distances were computed by QIIME. Consequently, these distances were displayed by both principal component analysis (PCA) and non-metric multi-dimensional scaling (NMDS). Both PCA and NMDS are intended to obtain principal coordinates for subsequent visualization from the complex and multidimensional data [26]. In addition, the difference in microbiota composition among groups was analyzed by both MetaStat and linear discriminant analysis effect size (LEfSe) [27], in which the results of LEfSe were displayed in a cladogram.
2.8. Statistical analysis
All data were expressed as mean ± SD of 5 animals. Differences between the 2 experimental groups (normal chow and HMLF diet groups) were assessed by GraphPad Prism 6 software by two-tailed non-parametric Mann-Whitney test. A P-value ≤ 0.05 indicates statistical significance. Statistical difference in bacterial family between groups based on 16S rRNA sequencing data was determined by T-test, using P-value ≤ 0.05 as a cut-off.
3. Results
3.1. A HMLF diet induced HHcy in C57BL/6 mice
To evaluate the effect of HMLF diet on glucose homeostasis and gut microbiome, C57BL/6 mice were fed either normal chow or HMLF diet for 8 weeks (Fig. 1A). The 8-week feeding of HMLF diet elevated the plasma Hcy concentration in C57BL/6 mice to a level higher than 200 μmol L−1, approximately 3.5 folds higher than that in normal chow-fed mice (Fig. 1B). Moreover, the HMLF diet slightly lowered the body weight gains of C57BL/6 mice when compared to normal chow (Fig. 1C). However, the HMLF diet did not alter the lipid profiles (i.e. TC, TG, HDL-C and non-HDL-C) in C57BL/6 mice (Fig. 1D).
3.2. HMLF diet impaired glucose homeostasis in diet-induced HHcy mice
We performed GTT and ITT to determine glucose absorption by metabolic organs and insulin sensitivity, respectively, at weeks 4 and 8 of diet feeding. At week 4, no significant glucose intolerance (Fig. 2A and E) and insulin resistance (Fig. 2C and F) were observed in diet-induced HHcy mice when compared to normal chow-fed mice. However, diet-induced HHcy mice were slightly but significantly glucose intolerant (Fig. 2B and E) and insulin resistant (Fig. 2D and F) when comparted to normal chow-fed mice at week 8.
Fig. 2.
HMLF diet impaired glucose homeostasis in C57BL/6 mice. (A) OTT of C57BL/6 mice at (A) week 4 and (B) week 8 of HMLF diet feeding. (C) ITT of C57BL/6 mice at (C) week 4 and (D) week 8 of HMLF diet feeding. AUCs of (E) OTT and (F) ITT at weeks 4 and 8 of HMLF diet feeding. HMLF: high methionine low folate. *P < 0.05 vs normal chow group. n = 5 per group. All data are expressed as mean ± SD.
3.3. HMLF diet altered the gut microbiome profile in diet-induced HHcy mice
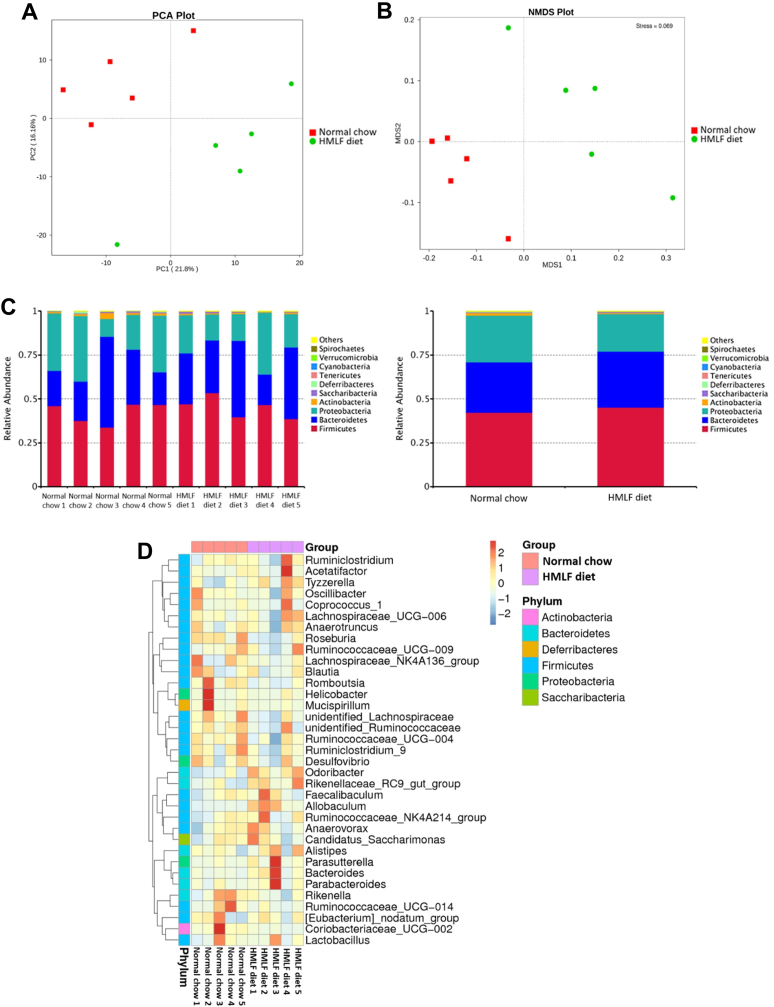
To compare the gut microbiota compositions between normal chow and HMLF diet groups, 16S rRNA sequencing of V4 hypervariable region was performed. Beta diversity analysis was displayed on PCA and NMDS plots, which were based on the calculations of Bray-Curtis distance. Both PCA and NMDS plots revealed that the community compositions of intestinal microbiota in HMLF diet group was distinct from that of normal chow group (Fig. 3A and B). Furthermore, the relative abundance of predominant taxa in the two groups were compared. At the phylum level, both Bacteroidetes and Firmicutes were dominant phyla, contributing to about 70% of relative abundance (Fig. 3C). Of note, the relative abundance of Bacteroidetes was higher (P = 0.0340), whereas that of Proteobacteria was lower in HMLF diet group (Fig. 3C, P = 0.0204). The species abundance heatmap was constructed to compare the predominant genera of two groups. Notably, the HMLF diet-fed mice were associated with slight enrichment in Tyzzerella (P = 0.0647), Odoribacter (P = 0.0512) and Allobaculum (P = 0.0684) (Fig. 3D). Lower abundance of Roseburia (P = 0.0361) and Romboutsia (P = 0.0491) were observed in HMLF diet-fed mice (Fig. 3D).
Fig. 3.
HMLF diet caused compositional alterations in gut microbiota of C57BL/6 mice. Beta diversity of gut microbiome, as represented by (A) PCA and (B) NMDS plots. In these two plots, samples of two groups are denoted by data points of different colors and shapes. The further the distance between points, the more distinct in species composition. (C) Microbiota composition of fecal samples at phylum level. Distinct species are denoted by different colors. Proportion of each species is represented by the length of columns. (D) Species abundance heatmap of the fecal samples of the two groups at genus level. HMLF: high methionine low folate; NMDS: non-metric multi-dimensional scaling; PCA: principal component analysis. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. HMLF diet increased the abundance of porphyromonadaceae family of bacteria
T-test analysis revealed significant variation between two groups, where the HMLF diet group was characterized by a higher abundance of Porphyromonadaceae family (P < 0.05) (Fig. 4A). MetaStat method was used to analyze the microbial species abundance data for fecal samples of normal chow and HMLF diet groups. Based on the q value at family level, a significant variation among the species (Q < 0.05) was observed in the Porphyromonadaceae family between two groups (Fig. 4B). LEfSe analysis was performed to study the enrichment of bacterial taxa upon different dietary contents. Cladogram generated from LEfSe analysis demonstrated that both Porphyromonadaceae and Erysipelotrichaceae families of bacteria were enriched in the HMLF diet group (Fig. 4C).
Fig. 4.
HMLF diet increased the abundance of porphyromonadaceae family of bacteria. (A) Between-group T-test analysis and (B) MetaStat of fecal samples of normal chow and HMLF diet groups. *P < 0.05 or *Q < 0.05 vs normal chow group. n = 5 per group. (C) Cladogram diagram showing the microbial species with significant differences. Circles radiating from inside to outside represent the taxonomic rank: phylum, class, order, family, and genus. Red nodes in the phylogenetic tree denote microbial species that play crucial role in the HMLF diet group. Yellow nodes stand for those microbial species with no significant difference. HMLF: high methionine low folate. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
A HMLF diet causes the accumulation of non-proteinogenic amino acid Hcy in the circulation [28]. In the present investigation, we demonstrated that a dietary pattern of HMLF content brought about multiple effects in vivo. In addition to the induction of HHcy, which is considered as an independent risk factor for several cardiovascular diseases [29], we provided novel evidence that the HMLF diet altered the glucose homeostasis and gut microbiome profile in C57BL/6 mice.
An 8-week feeding of HMLF diet successfully induced HHcy in C57BL/6 mice by increasing the concentration of plasma Hcy to approximately 200 μmol L−1, a level comparable to human patients with severe HHcy (>100 μmol L−1) [30]. Body weight gains of HMLF diet-fed mice were observed to be lower than that of normal chow-fed mice. This is consistent with a previous clinical finding that an elevated Hcy level is associated with body weight loss in humans [31]. A lower body weight gain can be partly attributed to appetite loss caused by folate deficiency in diet [32]. In addition, HMLF diet did not elevate levels of plasma cholesterol and triglycerides in C57BL/6 mice, implying that HHcy represents a risk factor different from hypercholesterolemia and obesity.
A gradual impairment of glucose homeostasis was observed, as revealed by GTT and ITT at weeks 4 and 8 of HMLF diet feeding. A previous study suggested that high insulin level raises Hcy level in rats [33]. Human body generally produces more insulin, i.e. hyperinsulinemia, in response to insulin resistance [9]. Under diabetic conditions, elevated Hcy level is often observed [34]. The present study provides novel findings that HMLF diet can gradually induce insulin resistance and hyperglycemia in non-diabetic C57BL/6 mice, implying that HMLF diet could alter glucose homeostasis.
Both PCA and NMDS plots indicate the compositional difference in gut microbiome between the HMLF diet and normal chow groups. Bacteroidetes represent the bacterial phylum responsible for protein synthesis and polysaccharide absorption. In brief, Bacteroidetes decompose complex polysaccharides into short-chain fatty acids (SCFAs), which mediate intestinal permeability and inflammation [35]. Higher Bacteroidetes abundance in HMLF diet group implies a higher risk for various diseases, such as inflammatory bowel disease, alcoholic liver disease and steatohepatitis, due to leaky gut and low-grade inflammation [36]. Meanwhile, the phylum Proteobacteria consists of bacteria contributing to carbohydrate and protein metabolism, and oxygen homeostasis inside the intestinal tract [37]. Often overrepresented in obese individuals, the Proteobacteria phylum is a marker of dysbiosis and hence dysbiosis-related diseases such as colitis and metabolic disorders [38]. The sequencing results indicates a lower risk of Proteobacteria-related dysbiosis in the HMLF diet group. Weight loss is often associated with reduced Proteobacteria abundance [38]. Notably, the lower abundance of Proteobacteria correlates with the lower body weight gain in HMLF diet group. HMLF diet induced gut microbiome alterations distinct from obesity.
Correlated to higher risk of cardiovascular disease [39], the spore-forming Tyzzerella and non-spore-forming Allobaculum bacteria are slightly enriched in the HMLF diet group. Notably, the butyrate-producing Roseburia bacteria was shown in lower frequency in the HMLF diet group, consistent with previous case-control studies that lower Roseburia abundance correlates with glucose intolerance and higher risk of type 2 diabetes [40]. Our results therefore imply that the HMLF dietary pattern may pose a higher risk of cardiometabolic diseases by altering the gut microbiome profile.
More importantly, an expansion in the family Porphyromonadaceae was observed in the fecal samples of HMLF diet group. Porphyromonadaceae represents the bacterial family under Bacteroidetes phylum, potentially exerting harmful effects on the host [41]. Enrichment of Porphyromonadaceae family is associated with non-alcoholic fatty liver disease (NAFLD), characterized by hepatic inflammation and risk of nonalcoholic steatohepatitis [42]. Previously, another study correlated the higher frequency of Porphyromonadaceae with the development of GLP-1 resistance, resulting in the impairment on glucose-induced insulin secretion [43]. Our results provide novel hint that the HHcy-inducing HMLF diet might impair glucose homeostasis, as revealed by slight glucose intolerance and insulin resistance, by increasing the Porphyromonadaceae abundance, possibly through the induction of GLP-1 resistance. In short, a HMLF diet might have posed pro-diabetic threats by firstly altering gut microbiome before subsequent absorption and metabolism to induce HHcy. However, detailed mechanism in-between the alterations in gut microbiome and metabolic parameters after a HMLF diet remains elusive. Further studies are needed to confirm which gut-derived metabolites contribute to the altered metabolic parameters after a HMLF diet.
Data availability statement
Data are available from the authors upon reasonable request.
Author Statement
Cheng Chak Kwong: Conceptualization, Methodology, Investigation, Writing – original draft, Visualization, Wang Chenguang: Investigation, Resources, Shang Wenbin: Investigation, Resources, Lau Chi Wai: Investigation, Resources, Luo Jiang-Yun: Writing – review & editing, Wang Li: Writing – review & editing, Huang Yu: Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was financially supported by the Vice-Chancellor's One-off Discretionary Fund [Grant number 4930709] and Hong Kong RGC Senior Research Fellow Scheme 2020/21 [Grant number SRFS2021‐4S04].
References
- 1.Troen A.M., Lutgens E., Smith D.E., Rosenberg I.H., Selhub J. The atherogenic effect of excess methionine intake. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15089–15094. doi: 10.1073/pnas.2436385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar A., Palfrey H.A., Pathak R., Kadowitz P.J., Gettys T.W., Murthy S.N. The metabolism and significance of homocysteine in nutrition and health. Nutr. Metab. 2017;14:78. doi: 10.1186/s12986-017-0233-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fratoni V., Brandi M.L. B vitamins, homocysteine and bone health. Nutrients. 2015;7:2176–2192. doi: 10.3390/nu7042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinohara M., Ji C., Kaplowitz N. Differences in betaine-homocysteine methyltransferase expression, endoplasmic reticulum stress response, and liver injury between alcohol-fed mice and rats. Hepatology. 2010;51:796–805. doi: 10.1002/hep.23391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brattström L., Wilcken D.E. Homocysteine and cardiovascular disease: cause or effect? Am. J. Clin. Nutr. 2000;72:315–323. doi: 10.1093/ajcn/72.2.315. [DOI] [PubMed] [Google Scholar]
- 6.Rozycka A., Jagodzinski P.P., Kozubski W., Lianeri M., Dorszewska J. Homocysteine level and mechanisms of injury in Parkinson's disease as related to MTHFR, MTR, and MTHFD1 genes polymorphisms and L-dopa treatment. Curr. Genom. 2013;14:534–542. doi: 10.2174/1389202914666131210210559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seghieri G., Breschi M.C., Anichini R., DeBellis A., Alviggi L., Maida I., Franconi F. Serum homocysteine levels are increased in women with gestational diabetes mellitus. Metabolism. 2003;52:720–723. doi: 10.1016/s0026-0495(03)00032-5. [DOI] [PubMed] [Google Scholar]
- 8.Huang T., Ren J., Huang J., Li D. Association of homocysteine with type 2 diabetes: a meta-analysis implementing Mendelian randomization approach. BMC Genom. 2013;14:867. doi: 10.1186/1471-2164-14-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarty M.F. Insulin secretion as a potential determinant of homocysteine levels. Med. Hypotheses. 2000;55:454–455. doi: 10.1054/mehy.1999.1008. [DOI] [PubMed] [Google Scholar]
- 10.Kim S.H., Reaven G.M. Insulin resistance and hyperinsulinemia: you can't have one without the other. Diabetes Care. 2008;31:1433–1438. doi: 10.2337/dc08-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meigs J.B., Jacques P.F., Selhub J., Singer D.E., Nathan D.M., Rifai N., D'Agostino R.B., Wilson P.W. Framingham Offspring Study, Fasting plasma homocysteine levels in the insulin resistance syndrome: the Framingham offspring study. Diabetes Care. 2001;24:1403–1410. doi: 10.2337/diacare.24.8.1403. [DOI] [PubMed] [Google Scholar]
- 12.Chiang E.P.I., Wang Y.C., Chen W.W., Tang F.Y. Effects of insulin and glucose on cellular metabolic fluxes in homocysteine transsulfuration, remethylation, S -adenosylmethionine synthesis, and global deoxyribonucleic acid methylation. J. Clin. Endocrinol. Metab. 2009;94:1017–1025. doi: 10.1210/jc.2008-2038. [DOI] [PubMed] [Google Scholar]
- 13.Yang N., Yao Z., Miao L., Liu J., Gao X., Fan H., Hu Y., Zhang H., Xu Y., Qu A., Wang G. Novel clinical evidence of an association between homocysteine and insulin resistance in patients with hypothyroidism or subclinical hypothyroidism. PloS One. 2015;10 doi: 10.1371/journal.pone.0125922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar J., Ingelsson E., Lind L., Fall T. No evidence of a causal relationship between plasma homocysteine and type 2 diabetes: a mendelian randomization study. Front. Cardiovasc. Med. 2015;2:11. doi: 10.3389/fcvm.2015.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rastelli M., Cani P.D., Knauf C. The gut microbiome influences host endocrine functions. Endocr. Rev. 2019;40:1271–1284. doi: 10.1210/er.2018-00280. [DOI] [PubMed] [Google Scholar]
- 16.Kilkenny C., Browne W., Cuthill I.C., Emerson M., Altman D.G. NC3Rs Reporting Guidelines Working Group, Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br. J. Pharmacol. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bárcena C., Valdés-Mas R., Mayoral P., Garabaya C., Durand S., Rodríguez F., Fernández-García M.T., Salazar N., Nogacka A.M., Garatachea N., Bossut N., Aprahamian F., Lucia A., Kroemer G., Freije J.M.P., Quirós P.M., López-Otín C. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat. Med. 2019;25:1234–1242. doi: 10.1038/s41591-019-0504-5. [DOI] [PubMed] [Google Scholar]
- 18.Siervo M., Shannon O., Kandhari N., Prabhakar M., Fostier W., Köchl C., Rogathi J., Temu G., Stephan B.C.M., Gray W.K., Haule I., Paddick S.M., Mmbaga B.T., Walker R. Nitrate-rich beetroot juice reduces blood pressure in Tanzanian adults with elevated blood pressure: a double-blind randomized controlled feasibility trial. J. Nutr. 2020;150:2460–2468. doi: 10.1093/jn/nxaa170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Justice M., Ferrugia A., Beidler J., Penprase J.C., Cintora P., Erwin D., Medrano O., Brasser S.M., Hong M.Y. Effects of moderate ethanol consumption on lipid metabolism and inflammation through regulation of gene expression in rats. Alcohol Alcohol. 2019;54:5–12. doi: 10.1093/alcalc/agy079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiedorová K., Radvanský M., Němcová E., Grombiříková H., Bosák J., Černochová M., Lexa M., Šmajs D., Freiberger T. The impact of DNA extraction methods on Stool bacterial and fungal microbiota community recovery. Front. Microbiol. 2019;10:821. doi: 10.3389/fmicb.2019.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L., Jin L., Xue B., Wang Z., Peng Q. Characterizing the bacterial community across the gastrointestinal tract of goats: composition and potential function. Microbiologyopen. 2019;8 doi: 10.1002/mbo3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P., Gao J., Ke W., Wang J., Li D., Liu R., Jia Y., Wang X., Chen X., Chen F., Hu X. Resveratrol reduces obesity in high-fat diet-fed mice via modulating the composition and metabolic function of the gut microbiota. Free Radic. Biol. Med. 2020;156:83–98. doi: 10.1016/J.FREERADBIOMED.2020.04.013. [DOI] [PubMed] [Google Scholar]
- 27.yanPei L., shiKe Y., huZhao H., Wang L., Jia C., zhiLiu W., huiFu Q., niShi M., Cui J., chunLi S. Role of colonic microbiota in the pathogenesis of ulcerative colitis. BMC Gastroenterol. 2019;19:1–11. doi: 10.1186/s12876-019-0930-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Škovierová H., Vidomanová E., Mahmood S., Sopková J., Drgová A., Červeňová T., Halašová E., Lehotský J. The molecular and cellular effect of homocysteine metabolism imbalance on human health. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17101733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tinelli C., Pino A.D., Ficulle E. Hyperhomocysteinemia as a Risk Factor and Potential Nutraceutical Target for Certain Pathologies. Front Nutr. 2019;6 doi: 10.3389/fnut.2019.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price B.R., Wilcock D.M., Weekman E.M. Hyperhomocysteinemia as a risk factor for vascular contributions to cognitive impairment and dementia. Front. Aging Neurosci. 2018;10 doi: 10.3389/fnagi.2018.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon J.B., Dixon M.E., O'Brien P.E. Elevated homocysteine levels with weight loss after Lap-Band® surgery: higher folate and vitamin B12 levels required to maintain homocysteine level. Int. J. Obes. 2001;25:219–227. doi: 10.1038/sj.ijo.0801474. [DOI] [PubMed] [Google Scholar]
- 32.Namdari M., Abadi A., Taheri S.M., Rezaei M., Kalantari N., Omidvar N. Effect of folic acid on appetite in children: ordinal logistic and fuzzy logistic regressions. Nutrition. 2014;30:274–278. doi: 10.1016/j.nut.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Gursu M.F., Baydas G., Cikim G., Canatan H. Insulin increases homocysteine levels in a dose-dependent manner in diabetic rats. Arch. Med. Res. 2002;33:305–307. doi: 10.1016/S0188-4409(01)00379-4. [DOI] [PubMed] [Google Scholar]
- 34.Platt D.E., Hariri E., Salameh P., Merhi M., Sabbah N., Helou M., Mouzaya F., Nemer R., Al-Sarraj Y., El-Shanti H., Abchee A.B., Zalloua P.A. Type II diabetes mellitus and hyperhomocysteinemia: a complex interaction. Diabetol. Metab. Syndrome. 2017;9:1–7. doi: 10.1186/s13098-017-0218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu C., Su Z., Li Y., Li Y., Liu K., Chu F., Liu T., Chen R., Ding X. Dysbiosis of gut microbiota is associated with gastric carcinogenesis in rats. Biomed. Pharmacother. 2020;126:110036. doi: 10.1016/j.biopha.2020.110036. [DOI] [PubMed] [Google Scholar]
- 36.Fukui H. Increased intestinal permeability and decreased barrier function: does it really influence the risk of inflammation? Inflamm. Intest. Dis. 2016;1:135–145. doi: 10.1159/000447252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon C.D., Young W., Maclean P.H., Cookson A.L., Bermingham E.N. Metagenomic insights into the roles of Proteobacteria in the gastrointestinal microbiomes of healthy dogs and cats. Microbiologyopen. 2018;7 doi: 10.1002/mbo3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizzatti G., Lopetuso L.R., Gibiino G., Binda C., Gasbarrini A. Proteobacteria: a common factor in human diseases. BioMed Res. Int. 2017;2017 doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ascher S., Reinhardt C. The gut microbiota: an emerging risk factor for cardiovascular and cerebrovascular disease. Eur. J. Immunol. 2018;48:564–575. doi: 10.1002/eji.201646879. [DOI] [PubMed] [Google Scholar]
- 40.Gurung M., Li Z., You H., Rodrigues R., Jump D.B., Morgun A., Shulzhenko N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu L., Qiao N., Li T., Yu R., Zhai Q., Tian F., Zhao J., Zhang H., Chen W. Dietary supplementation with probiotics regulates gut microbiota structure and function in Nile tilapia exposed to aluminum. PeerJ. 2019;2019 doi: 10.7717/peerj.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blacher E., Levy M., Tatirovsky E., Elinav E. Microbiome-modulated metabolites at the interface of host immunity. J. Immunol. 2017;198:572–580. doi: 10.4049/jimmunol.1601247. [DOI] [PubMed] [Google Scholar]
- 43.Grasset E., Puel A., Charpentier J., Collet X., Christensen J.E., Tercé F., Burcelin R. A specific gut microbiota dysbiosis of type 2 diabetic mice induces GLP-1 resistance through an enteric NO-dependent and gut-brain Axis mechanism. Cell Metabol. 2017;25:1075–1090. doi: 10.1016/j.cmet.2017.04.013. e5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from the authors upon reasonable request.