Abstract
Most of the gastrointestinal stromal tumors (GISTs) have gain-of-function mutations in the KIT gene, which can be used as a prognostic marker for the biological behavior of tumors, predictive marker for the response of tyrosine kinase inhibitors, and diagnostic marker. Researchers have focused on PDGFRA mutations because of both their prognostic and predictive potential and DOG1 positivity for diagnosis on GISTs. The aim of this study is to investigate the effect DOG1, PDGFRA, and KIT mutations on the prediction of the outcome for GIST management. Polymerase chain reaction was performed for KIT gene exons 9, 11, 13, and 17 and PDGFRA gene exons 12 and 18 with the genomic DNA of 46 GIST patients, and amplicons were sequenced in both directions. Immunocytochemical stainings were done by using primary antibodies. Molecular analysis revealed that the KIT mutation was observed in 63% of all cases, while the PDGFRA mutation was observed in 23.9% of cases. Significant relationships were found between age and KIT mutation, tumor location and KIT mutations, and tumor location and PDGFRA mutations (p ≤ 0.05). DOG1 positivity was detected in 65.2% of all GISTs and DOG1-positive cells had a higher KIT mutation ratio than DOG1-negative cells (p ≤ 0.05). KIT gene exon 11 mutations in DOG1-positive cells was higher than DOG1-negative cells (p ≤ 0.05). Conversely, KIT gene exon 13 mutations were higher in DOG1-negative cells than DOG1-positive cells (p ≤ 0.05). In this study, KIT mutation frequency was found similar with the European population; conversely, PDGFRA mutation frequency was similar with an Asian-Chinese-based study. KIT/PDGFRA mutations and tumor location can be used for the prediction of tumor behavior and the management of disease in GISTs. DOG1 positivity might be a candidate marker to support KIT and PDGFRA mutations, due to the higher DOG1 positivity in KIT exon 11 mutant and stomach- and small intestine-localized GISTs.
Key words: Gastrointestinal stromal tumors (GISTs), KIT gene, PDGFRA gene, Mutations, DOG1
INTRODUCTION
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors of the gastrointestinal tract. They originate from the interstitial cells of Cajal (1). Immunocytochemical staining with an antibody against KIT (CD117) led to the discovery that the KIT protein is characteristically expressed in most GISTs (2). Nearly all GISTs have gain-of-function mutations in the KIT gene (approximately 90%), and they are the major cause of GISTs. The KIT gene encodes a transmembrane receptor, the KIT protein, for the cytokine known as stem cell factor. The intracytoplasmic part of the KIT protein functions as a tyrosine kinase. Therefore, mutations in the KIT gene cause constant activation of the receptor tyrosine kinase and increased proliferation and survival due to constant receptor activation (3). GISTs were previously thought to be resistant to cancer chemotherapy, and they were associated with poor prognosis due to the lack of effective therapeutic options, until imatinib mesylate, a selective inhibitor of tyrosine kinases, including KIT, platelet-derived growth factor receptors (PDGFRs), and BCR-ABL, was found to be effective against chemotherapy-resistant GISTs. Researchers characterized platelet-derived growth factor receptor-α (PDGFRA) mutations in a small group of GISTs with the wild-type KIT gene, and this alternative oncogenic mechanism over PDGFRA has been confirmed by other researchers. Imatinib can bind and inhibit PDGFRA (4).
Previously, it was declared that both KIT and PDGFRA mutations have prognostic and predictive potential. KIT mutations were associated with aggressive tumor behavior and poor clinical outcome in GISTs; on the other hand, PDGFRA mutations were identified with a clinically benign outcome, but prognostic results have not been integrated into a risk classification scheme (5). Furthermore, both KIT and PDGFRA mutations have significance for the prediction of response to imatinib (6).
According to generally accepted experience, immunohistochemical staining and gene analysis are considered useful for diagnosis, because nearly 95% of GISTs express CD117 (KIT protein, which stains positively for KIT in immunohistochemistry) and often harbor mutations of a gene that encodes a type III receptor tyrosine kinase (either KIT, approximately 90%, or PDGFRA, approximately 5%). Although CD117 positivity on immunohistochemistry has been considered the gold standard for GIST diagnosis, recent studies have shown that some of these tumors can be negative for CD117 and other markers, such as CD34, S-100, and smooth muscle actin (SMA); therefore, certain diagnosis is often challenging (7). If the tumor is negative for CD117 but is positive for CD34, a histological diagnosis is possible. However, if the tumor is negative for CD117, CD34, S-100, and SMA, making a definitive diagnosis is often challenging. Discovery of GIST-1 (DOG1) has received considerable attention as a useful molecule for the diagnosis of GIST, even in KIT-negative GISTs. DOG1, a membrane channel protein, is known to be overexpressed in GIST. Several publications suggest that DOG1 is more specific and sensitive for the diagnosis of GIST than CD117. Espinosa et al. (8) reported that DOG1-positive staining yielded in 87% of all scorable GIST, whereas CD117 was positive in 74%. Other studies showed that DOG1 positivity was found in 97.8% of scorable GISTs (9,10). In recent years, new systems have been investigated, such as “recurrence risk scoring,” which shows the targeted agents that are useful in patients. Several studies have explored the ability of Ki-67 to predict the malignant potential of GISTs (11). Some authors believe that mitotic index reflects the M phase of mitosis only, but Ki-67 also defines the proliferation of cells in the G1, S, and G2 phases and therefore can be used as an objective criterion in the evaluation of GIST malignancy (11,12).
With molecular studies, GISTs patients carrying KIT gene exon 9 are more likely to show resistance, whereas those with tumors carrying mutations of exon 11 are more likely to show a good response. Similar clinical benefits appear in patients with exon 13 mutations as those with exon 11 mutations, and in vitro studies suggest that exon 17 mutations are resistant to both imatinib and sunitinib (13). The results of these studies provide information about the prognostic factors and their importance in GIST management. The aim of this study is to investigate new and more practical markers, such as DOG1 and PDGFRA mutations and KIT mutations, to predict the outcome in GIST patients and observe importance in the management of the disease.
MATERIALS AND METHODS
Patients and Clinical Evaluation
A total of 46 formalin-fixed and paraffin-embedded (FFPE) GIST samples were used for this retrospective study, which was conducted with the ethical approval of the Clinical Research Ethics Committee of Dokuz Eylul University School of Medicine. Cases with sufficient tumor tissue for molecular analysis were chosen from archive of the Pathology Department of Dokuz Eylul University Hospital. All samples were diagnosed as GIST with immunohistochemical positivity for CD117. Sections (5 µm) were cut from each FFPE sample containing at least 75% tumor tissue.
Immunohistochemistry
In the study group of patients, sections were prepared from paraffin blocks, and histological sections (5 µm) of paraffin-embedded materials were used for immunohistochemistry. The primary antibodies used were as follows: CD117, CD34, SMA, S-100, desmin, Ki-67, and DOG1 (Table 1). Sections were deparaffinized in xylene and hydrated in a graded series of alcohol. Staining was performed using automatic immunohistochemistry staining equipment (Lab Vision Autostainer; Thermo Scientific) and evaluated by pathologists (14). Diffuse- or focal-stained specimens were accepted as positive.
Table 1.
Antibodies Used for Immunohistochemistry
| Antigen | Antibody | Dilution |
|---|---|---|
| CD117 | Polyclonal (Dako, Denmark) | 1:400 |
| CD34 | Monoclonal (Clone QBend/10, Neomarkers, USA) | 1:200 |
| SMA | Monoclonal (Clone 1A4, Dako, Denmark) | 1:400 |
| S-100 | Polyclonal (Spring Bioscience Corp., USA) | 1:200 |
| Desmin | Polyclonal (Spring Bioscience Corp., USA) | 1:200 |
| Ki-67 | Monoclonal (Clone SP6, Neomarkers, USA) | 1:200 |
| DOG1 | Monoclonal (Clone K9, Novocastra, UK) | 1:100 |
Molecular Analysis
DNA Extraction
Genomic DNA was extracted from FFPE tumor sections from each sample using a QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Tumor tissue sections were deparaffinized by serial treatment with xylole and a graded series of ethanol and digested with proteinase K.
The Amplification of Target Exons and DNA Sequencing
For the screening of known and unknown exons, exons 9, 11, 13, and 17 of KIT and exons 12 and 18 of PDGFRA were amplified using polymerase chain reaction (PCR; Tag DNA Polymerase-dNTPack; Roche Diagnostics GmbH Mannheim, Germany) with specifically designed primers (Table 2). PCR amplicons were purified using a specific purification kit (High Pure PCR Product Purification Kit; Roche Diagnostics) and sequenced in both the forward and reverse directions (DTCS-Quick Start Kit; Beckman Coulter, USA) with the same primer sets and an automatic genetic analysis system (GenomeLab™ GeXP Genetic Analysis System; Beckman Coulter). The generated DNA sequences were analyzed with GenomeLab Software, version 5.1 (Beckman Coulter) and specific bioinformatics tools. DNA sequences were aligned using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The numbering of specific mutations was referenced from the Ensemble Database (http://www.ensembl.org). The reference sequences used to describe the mutations were ENST00000288135 and ENST00000257290 for the KIT and PDGFRA genes, respectively.
Table 2.
Primers Used for Target Exon Amplification of KIT and PDGFRA Genes
| Target Gene/Exon | Primers (5′–3′) | Melting Point (T m) | Amplicon (bp) |
|---|---|---|---|
| KIT | |||
| Exon 9 | F: GACATTTTCTGTTGATTATGAACCTC | 55.1 | 405 |
| R: CATGGTCAATGTTGGAATGAAC | 54.6 | ||
| Exon 11 | F: CCAGAGTGCTCTAATGACTGAGA | 54.2 | 281 |
| R: AAACAAAGGAAGCCACTGGA | 56.6 | ||
| Exon 13 | F: TACTGCATGCGCTTGACATC | 56.4 | 263 |
| R: TAATCTAGCATTGCCAAAATCA | 52.6 | ||
| Exon 17 | F: CATCATTCAAGGCGTACTTTTG | 55.6 | 327 |
| R: TGCAGGACTGTCAAGCAGAG | 56.6 | ||
| PDGFRA | |||
| Exon 12 | F: TCCAGTCACTGTGCTGCTTC | 55 | 272 |
| R: AAGACTCCCTTTTCCCTTGC | 55 | ||
| Exon 18 | F: ACCATGGATCAGCCAGTCTT | 55 | 251 |
| R: GGTCAGGCTCATCCTCCTTCA | 55 | ||
Statistical Analysis
SPSS (Version 21.0; SPSS, Inc., Chicago, IL, USA) software was used for statistical analysis. The following parameters were analyzed: patient age and gender, tumor type (primary, metastatic), tumor location (gastric, small and large intestine, esophagus, omentum-periton, others), histological cell type (spindle, epitheloid, mixed), tumor size, and mitotic index in 50 HPF, CD117, CD34, Ki-67, SMA, S-100, desmin, DOG1, KIT, and PDGFRA gene molecular status. Chi-square and Fisher tests as a univariate analysis were used to analyze associations between variables. A value of p < 0.05 was considered to be statistically significant.
RESULTS
Clinicopathological Features of GIST Cases
In this study, we analyzed 46 tumor tissues from GIST patients. Demographic and clinicopathological data of patients are shown in Table 3. All tumors presented pathological features consistent with GIST, and all of them expressed CD117. The median age was 64.5 (range: 25–84). It was determined that a greater number of GIST cases were observed in females (56.5%) in comparison with males (43.5%). The same trend toward increased frequency of GIST was noted in older patients (>50 years, 87%) in comparison with younger patients (≤50 years, 13%). The majority of the tumor samples were primary (84.8%) in origin, while only few cases were metastatic (15.2%). Most of the tumor samples were located on the stomach (43.5%), followed by the small intestine (34.8%), large intestine (4.3%), esophagus (4.3%), omentum-periton (4.3%), and other locations (8.7%). Histological examination under a microscope revealed that most of the cases were of the spindle cell type (82.6%), followed by mixed type (10.9%) and epithelioid (6.5%). The mitotic index in 50 high-power fields revealed that a larger number of cases had <5 mitosis/50 HPF (60.9%), while the remaining cases showed ≥5 mitosis/50 HPF (9.1%) (Table 3).
Table 3.
Frequency of KIT and PDGFRA Mutations in the Current Group and Population-Based Studies
| Target Gene/Exon | Current Study [n = 46; % (n)] | Population-Based Studies [% (n)] | ||||
|---|---|---|---|---|---|---|
| Poland (n = 427) (13) | Iceland (n = 56) (14) | France (n = 492) (15) | Italy (n = 54) (16) | Portuguese (n = 78) (17) | ||
| KIT gene | ||||||
| Total KIT mutant | 63 (29) | 69.3 (296) | 87.5 (49) | 70.7 (347) | 74 (40) | 56 (44) |
| Exon 9 | 10.9 (5) | 7.3 (31) | 10.7 (6) | 5.5 (27) | 19 (10) | 5 (4) |
| Insertion | 6.5 (3) | 0 | 0 | 0 | ||
| Substitution | 4.3 (2) | 0 | 0 | 0 | 1.3 (1) | |
| Duplication | 0 | 7.3 (31) | 5.5 (27) | 19 (10) | 3.8 (3) | |
| Exon 11 | 45.7 (21) | 61.1 (261) | 76.8 (43) | 63.2 (311) | 52 (28) | 51 (40) |
| Deletion | 30.4 (14) | 34 (145) | 46.4 (26) | 30.9 (152) | 20.4 (11) | 30.7 (24) |
| Substitution | 10.9 (5) | 15.5 (66) | 28.6 (16) | 21.5 (106) | 26 (14) | 19.2 (15) |
| Deletion-substitution | 2.2 (1) | 0 | 0 | 0 | 0 | |
| Insertion | 2.2 (1) | 0 | 0 | 0 | 1.2 (1) | |
| Duplication | 0 | 7 (30) | 1.8 (1) | 3.7 (18) | 5.6 (3) | 0 |
| Complex | 0 | 4.7 (30) | 0 | 7.1 (35) | 0 | |
| Exon 13 | 15.2 (7) | 0.5 (2) | 0 | 1.4 (6) | 4 (2) | 0 |
| Substitution | 15.2 (7) | 0.5 (2) | 4 (2) | 0 | ||
| Exon 17 | 19.6 (9) | 0.5 (2) | 0 | 0.6 (3) | 0 | 0 |
| Substitution | 19.6 (9) | 0.5 (2) | 0 | 0 | ||
| PDGFRA gene | ||||||
| Total PDGFRA mutant | 23.9 (11) | 12.9 (55) | 5.4 (3) | 15 (73) | 13 (7) | 6.4 (5) |
| Exon 12 | 15.2 (7) | 0.2 (1) | 1.8 (1) | 2 (11) | 0 | 2.6 (2) |
| Substitution | 15.2 (7) | 0 | 1.3 (1) | |||
| Deletion | 0 | 0 | 1.3 (1) | |||
| Exon 18 | 17.4 (8) | 11.9 (51) | 3.6 (2) | 12 (60) | 13 (7) | 3.8 (3) |
| Substitution | 17.4 (8) | 7.4 (4) | 3.8 (3) | |||
| Deletion | 0 | 5.6 (3) | 0 | |||
| No mutation detected | 13.1 (6) | 17.8 (76) | 7.1 (4) | 14.2 (72) | 13 (7) | 37 (29) |
In the population-based studies, reference numbers for each country are shown in parentheses.
Mutational Status and Its Correlation With Clinicopathologic Characteristics
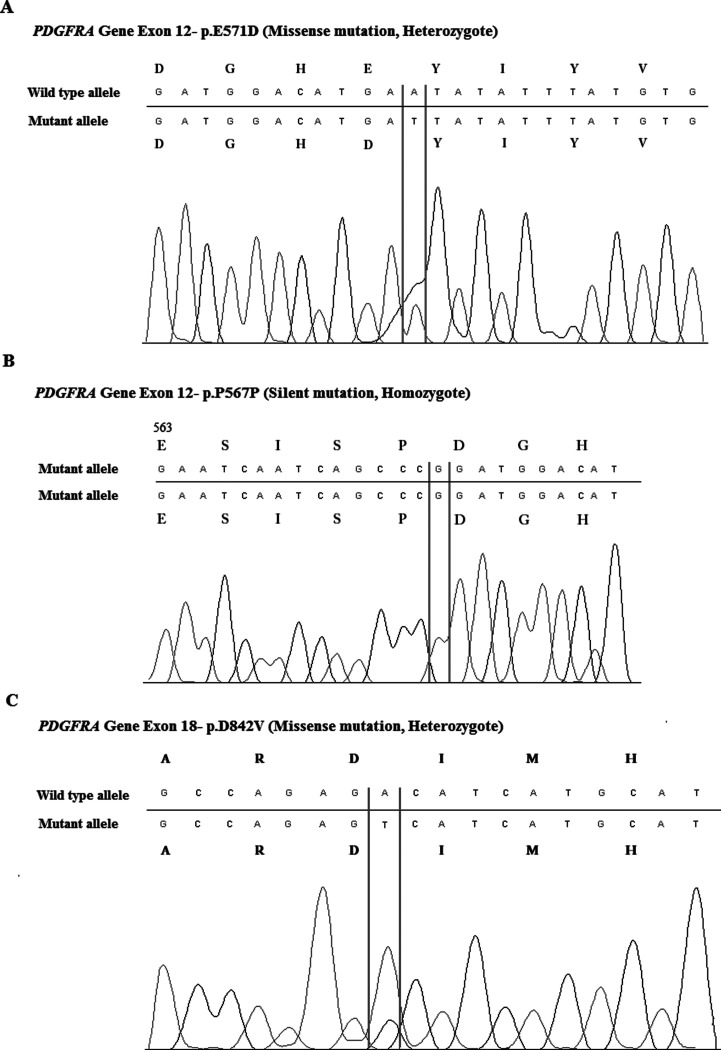
Molecular analysis revealed that the KIT mutation was observed in 63% of all cases, while the PDGFRA mutation was observed in 23.9% of them (Table 4); 13.1% of cases were wild type (WT) for both the KIT and PDGFRA genes. Among cases with the KIT mutation, 10.9% of them had exon 9 mutations, 45.7% of them had exon 11 mutations, 15.2% of them had exon 13 mutations, and 19.6% of them had exon 17 mutations. The number of exon 11 mutations was remarkably higher than those of the other KIT mutations. Deletions were the most common type of mutation in exon 11 (30.4%). Substitutions (10.9%), deletion-substitutions (2.2%), and insertions (2.2%) were also observed in exon 11. Among cases with PDGFRA mutations, 15.2% of them had exon 12 mutations (Fig. 1A, B), and 17.4% of them had exon 18 mutations (Fig. 1C). Only substitutions were observed in the exon 12 and exon 18 regions of PDGFRA.
Table 4.
Clinicopathological Characteristics of GIST Patients According to the KIT and PDGFRA Mutation Status
| Clinicopathologic Parameters (Total Samples n = 46) | % (n) | KIT | PDGFRA | ||||
|---|---|---|---|---|---|---|---|
| Wild Type [% (n)] | Mutation [% (n)] | p | Wild Type [% (n)] | Mutation [% (n)] | p | ||
| Gender | 0.29 | 0.57 | |||||
| Female | 56.5 (26) | 42.3 (11) | 57.7 (15) | 76.9 (20) | 23.1 (6) | ||
| Male | 43.5 (20) | 30 (6) | 70 (14) | 75 (15) | 25 (5) | ||
| Age | 0.02 | 0.13 | |||||
| >50 | 87 (40) | 30 (12) | 70 (28) | 80 (32) | 20 (8) | ||
| ≤50 | 13 (6) | 83.3 (5) | 16.7 (1) | 50 (3) | 50 (3) | ||
| Tumor type | 0.22 | 0.51 | |||||
| Primary | 84.8 (39) | 33.3 (13) | 66.7 (26) | 76.9 (30) | 23.1 (9) | ||
| Metastatic | 15.2 (7) | 57.1 (4) | 42.9 (3) | 71.4 (5) | 28.6 (2) | ||
| Tumor location | 0.07 | 0.08 | |||||
| Stomach | 43.5 (20) | 30 (6) | 70 (14) | 85 (17) | 15 (3) | ||
| Small intestine | 34.8 (16) | 25 (4) | 75 (12) | 81.3 (13) | 18.8 (3) | ||
| Large intestine | 4.3 (2) | 100 (2) | 0 | 0 | 100 (2) | ||
| Esophagus | 4.3 (2) | 100 (2) | 0 | 50 (1) | 50 (1) | ||
| Omentum-periton | 4.3 (2) | 50 (1) | 50 (1) | 0 | 100 (2) | ||
| Others | 8.7 (4) | 50 (2) | 50 (1) | 100 (4) | 0 | ||
| Histological type | 0.67 | 0.91 | |||||
| Spindle | 82.6 (38) | 39.5 (15) | 60.5 (23) | 76.3 (29) | 23.7 (9) | ||
| Mixed type | 10.9 (5) | 20 (1) | 80 (4) | 80 (4) | 20 (1) | ||
| Epitheloid | 6.5 (3) | 33.7 (1) | 66.7 (2) | 66.7 (2) | 33.3 (1) | ||
| Tumor size | 0.11 | 0.24 | |||||
| <5.25 cm | 50 (23) | 26.1 (6) | 73.9 (17) | 82.6 (19) | 17.4 (4) | ||
| ≥5.25 cm | 50 (23) | 47.8 (11) | 52.2 (12) | 69.6 (16) | 30.4 (7) | ||
| Mitosis/50 HPF | 0.23 | 0.44 | |||||
| <5 | 60.9 (28) | 42.9 (12) | 57.1 (16) | 78.6 (22) | 21.4 (6) | ||
| ≥5 | 39.1 (18) | 27.8 (5) | 72.2 (13) | 72.2 (13) | 27.8 (5) | ||
| Risk group | 0.51 | 0.48 | |||||
| High | 67.4 (31) | 35.5 (11) | 64.5 (20) | 74.2 (23) | 25.8 (8) | ||
| Low | 32.6 (15) | 40 (6) | 60 (9) | 80 (12) | 20 (3) | ||
| CD117 | |||||||
| Negative | 0 | 0 | 0 | 0 | 0 | ||
| Positive | 100 (46) | 37 (17) | 63 (29) | 76.1 (35) | 23.9 (11) | ||
| CD34 | 0.23 | 0.02 | |||||
| Negative | 26.1 (12) | 50 (6) | 50 (6) | 50 (6) | 50 (6) | ||
| Positive | 73.9 (34) | 32.4 (11) | 67.6 (23) | 85.3 (29) | 14.7 (5) | ||
| SMA | 0.17 | 0.49 | |||||
| Negative | 41.3 (19) | 26.3 (5) | 73.7 (14) | 78.9 (15) | 21.1 (4) | ||
| Positive | 58.7 (27) | 44.4 (12) | 55.6 (15) | 74.1 (20) | 25.9 (7) | ||
| S-100 | 0.62 | 0.23 | |||||
| Negative | 58.7 (27) | 37 (10) | 63 (17) | 70.4 (19) | 29.6 (8) | ||
| Positive | 41.3 (19) | 36.8 (7) | 63.2 (12) | 84.2 (16) | 15.8 (3) | ||
| Desmin | 0.63 | 0.37 | |||||
| Negative | 82.6 (38) | 36.8 (14) | 63.2 (24) | 73.7 (28) | 26.3 (10) | ||
| Positive | 17.4 (8) | 37.5 (3) | 62.5 (5) | 87.5 (7) | 12.5 (1) | ||
| Ki-67 | 0.59 | 0.44 | |||||
| Negative | 69.6 (32) | 37.5 (12) | 62.5 (20) | 78.1 (25) | 21.9 (7) | ||
| Positive | 30.4 (14) | 35.7 (5) | 64.3 (9) | 71.4 (10) | 28.6 (4) | ||
Tumors with <10% of positive cells were considered as negative for all markers except DOG1. Diffuse- or focal-stained specimens were accepted as positive.
Figure 1.
PDGFRA gene mutations in exon 12 and 18. (A) The missense mutation of p.E571D (heterozygote) is one of the most common mutations in exon 12. (B) The silent mutation of p.P567P (homozygote). (C) The missense mutation of p.D842V (heterozygote) is one of the most common mutations in exon 18.
According to the statistical analysis between gender/age/tumor type/tumor location/histologic type/tumor size/mitosis/risk group and mutational status, significant relationships were found between age and KIT mutation, tumor location and KIT mutations, and tumor location and PDGFRA mutations (Table 3). The KIT mutation rate was significantly higher in the old patient group (>50) than in the young patient group (≤50) (p = 0.02). The KIT mutation rate in the stomach (70%) and small intestine-localized (75%) tumors was significantly higher than those of other tumors (p = 0.07). The PDGFRA mutation rate was significantly higher in the same tumor groups [stomach (85%), small intestine (81.3%)] (p = 0.08).
Mutational Status and Its Correlation With the Expression of Immunohistochemical Parameters, Including DOG1
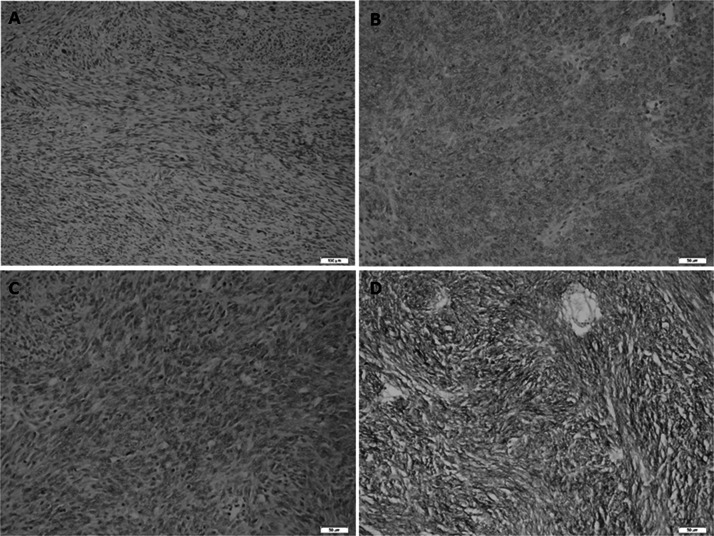
All cases were immunopositive for CD117. There were no statistically significant relationships between CD117, SMA, S-100, desmin, and Ki-67 immunoreactivity and mutation status (Table 5). CD34 staining was detected in 73.9% of all cases. The mutational status had the following distribution: KIT mutations were detected in 67.6% of all cases, and PDGFRA mutations were detected in 14.7% of all patients. CD34-positive cells presented a greater wild-type character for the PDGFRA gene than CD34-negative cells (p ≤ 0.05) (Table 5). DOG1 staining was detected in 65.2% of all cases (Fig. 2). Diffuse and strong staining was observed in most of the cases; however, focal staining was also observed in some cases. The mutational status had the following distribution for DOG1-positive cells: KIT mutations were detected in 73.3% of all cases, and PDGFRA mutations were detected in 23.3% of them. According to the statistical analysis, it was observed that DOG1-positive cells had a higher KIT mutation ratio than DOG1-negative cells (75.9%; p = 0.04) (Table 6). Additionally, the number of exon 11 mutations in DOG1-positive cells was higher than in DOG1-negative cells (81%; p = 0.04). Conversely, DOG1-positive cells were wild type at exon 13 (71.8%; p = 0.04).
Table 5.
DOG1 Expression Status and Correlation With Mutational Status of KIT and PDFGRA Genes
| Target Gene/Exon | % (n) | DOG1 Negative [% (n)] | DOG1 Positive [% (n)] | p |
|---|---|---|---|---|
| Total | 100 (46) | 34.8 (16) | 65.2 (30) | |
| KIT gene | ||||
| Overall | ||||
| Wild type | 37 (17) | 52.9 (9) | 47.1 (8) | 0.04 |
| Mutant | 63 (29) | 24.1 (7) | 75.9 (22) | |
| Exon 9 | ||||
| Wild type | 89.1 (41) | 39 (16) | 61 (25) | 0.1 |
| Mutant | 10.9 (5) | 0 | 100 (5) | |
| Exon 11 | ||||
| Wild type | 54.3 (25) | 48 (12) | 52 (13) | 0.04 |
| Mutant | 45.7 (21) | 19 (4) | 81 (17) | |
| Exon 13 | ||||
| Wild type | 84.8 (39) | 28.2 (11) | 71.8 (28) | 0.04 |
| Mutant | 15.2 (7) | 71.4 (5) | 28.6 (2) | |
| Exon 17 | ||||
| Wild type | 80.4 (37) | 37.8 (14) | 62.2 (23) | 0.32 |
| Mutant | 19.6 (9) | 22.2 (2) | 77.8 (7) | |
| PDGFRA gene | ||||
| Overall | ||||
| Wild type | 76.1 (35) | 34.3 (12) | 65.7 (23) | 0.58 |
| Mutant | 23.9 (11) | 36.4 (4) | 63.6 (7) | |
| Exon 12 | ||||
| Wild type | 84.8 (39) | 33.3 (13) | 66.7 (26) | 0.46 |
| Mutant | 15.2 (7) | 42.9 (3) | 57.1 (4) | |
| Exon 18 | ||||
| Wild type | 82.6 (38) | 36.8 (14) | 63.2 (24) | 0.42 |
| Mutant | 17.4 (8) | 25 (2) | 75 (6) | |
Figure 2.
Spectrum of DOG1 immunoreactivity in gastrointestinal stromal tumors. (A) Negative (original magnification 100×), (B) weak–cytoplasmic (original magnification 200×), (C) moderate–cytoplasmic (original magnification 200×), (D) strong–cytoplasmic and membranous (original magnification 200×).
Table 6.
Clinical and Pathological Characteristics of GIST Patients According to the DOG1 Positivity
| Clinicopathologic Parameters (Total Samples, n = 46) | % (n) | DOG1 Negative [% (n)] | DOG1 Positive [% (n)] | p |
|---|---|---|---|---|
| Total | 100 (46) | 34.8 (16) | 65.2 (30) | |
| Gender | 0.35 | |||
| Female | 56.5 (26) | 42.3 (11) | 57.7 (15) | |
| Male | 43.5 (20 | 25 (5) | 75 (15) | |
| Age | 0.16 | |||
| >50 | 87 (40) | 30 (12) | 70 (28) | |
| ≤50 | 13 (6) | 66.7 (4) | 33.3 (2) | |
| Tumor type | 0.68 | |||
| Primary | 84.8 (39) | 33.3 (13) | 66.7 (26) | |
| Metastatic | 15.2 (7) | 42.9 (3) | 57.1 (4) | |
| Tumor location | 0.022 | |||
| Stomach | 43.5 (20) | 35 (7) | 65 (13) | |
| Small intestine | 34.8 (16) | 12.5 (2) | 87.5 (14) | |
| Large intestine | 4.3 (2) | 100 (2) | 0 | |
| Esophagus | 4.3 (2) | 100 (2) | 0 | |
| Omentum-periton | 4.3 (2) | 50 (1) | 50 (1) | |
| Others | 8.7 (4) | 50 (2) | 50 (1) | |
| Histological type | 0.061 | |||
| Spindle | 82.6 (38) | 36.8 (14) | 63.2 (24) | |
| Mixed type | 10.9 (5) | 0 | 100 (5) | |
| Epitheloid | 6.5 (3) | 66.7 (2) | 33.3 (1) | |
| Tumor size | 0.75 | |||
| <5.25 mm | 50 (23) | 30.4 (7) | 69.6 (16) | |
| ≥5.25 mm | 50 (23) | 39.1 (9) | 60.9 (14) | |
| Mitosis/50HPF | 0.53 | |||
| <5 | 60.9 (28) | 39.3 (11) | 60.7 (17) | |
| ≥5 | 39.1 (18) | 27.8 (5) | 72.2 (13) | |
| Risk group | 0.52 | |||
| High | 67.4 (31) | 38.7 (12) | 61.3 (19) | 0.52 |
| Low | 32.6 (15) | 26.7 (4) | 73.3 (11) | |
| CD117 | ||||
| Negative | 0 | 0 | 0 | |
| Positive | 100 (46) | 34.8 (16) | 65.2 (30) | |
| CD34 | 0.06 | |||
| Negative | 26.1 (12) | 58.3 (7) | 41.7 (5) | |
| Positive | 73.9 (34) | 26.5 (9) | 73.5 (25) | |
| SMA | 0.76 | |||
| Negative | 41.3 (19) | 31.6 (6) | 68.4 (13) | |
| Positive | 58.7 (27) | 37 (10) | 63 (17) | |
| S100 | 0.52 | |||
| Negative | 58.7 (27) | 33.3 (9) | 66.7 (18) | |
| Positive | 41.3 (19) | 36.8 (7) | 63.2 (12) | |
| Desmin | 0.42 | |||
| Negative | 82.6 (38) | 31.6 (12) | 68.4 (26) | |
| Positive | 17.4 (8) | 50 (4) | 50 (4) | |
| Ki-67 | 0.59 | |||
| Negative | 69.6 (32) | 34.4 (11) | 65.6 (21) | |
| Positive | 30.4 (14) | 35.7 (5) | 64.3 (9) |
DISCUSSION
In recent years, the inhibition of tyrosine kinases has played an important role in the pathogenesis of GIST. Observations and research conducted in recent years have indicated changes in the prognosis and the response of tyrosine kinase inhibitors due to KIT and PDGFRA gene mutations; in this context, KIT and PDGFRA mutations and their role in GIST pathogenesis and management have been the most important targets (15,16).
Mutations in KIT and PDGFRA genes cause the activation of tyrosine kinase and uncontrolled tyrosine kinase activation; therefore, proliferation of cells and survival are increased due to constant receptor activation. Both KIT and PDGFRA mutations have a prognostic role for the biological behavior of tumors and a predictive role for the response of tyrosine kinase inhibitors (3).
In this study, we retrospectively collected and reviewed data from 46 GIST patients according to the sufficiency of archival tumor tissue. We collected information regarding patients’ clinicopathological and demographic characteristics and focused on the mutation data of KIT and PDGFRA genes. Therefore, we have evaluated the spectrum, frequency, and prognostic effect of KIT and PDGFRA mutations in GIST in terms of clinicopathological parameters, including DOG1, which is a promising marker for GIST diagnosis.
In the literature, KIT and PDGFRA mutations are variable. Generally, the frequency of the KIT gene mutation was reported to be between 38.5% and 87.5%, and KIT mutations were associated with aggressive tumor behavior and poor clinical outcome in GISTs (15–17). In our study, the overall mutation frequency for the KIT gene was 63%, which is comparable to frequencies observed in a population study from Poland (69.3%) and France (70.7%) (16,18) (Table 4).
KIT gene exon 9 and 11 mutations are very important due to their effect on tumor behavior and response of imatinib. Exon 11 mutations were found to be more common in the aggressive type, and a correlation was observed between KIT gene exon 11 mutations and poor clinical results compared with WT GISTs (19).
In terms of imatinib response, patients with exon 11 mutations have higher partial response the other patients who are treated with 400 mg/day. Patients with exon 9 mutations had significantly longer PFS when treated with imatinib 800 mg/day than others. Therefore, KIT gene exon 9 mutations are the only predictive marker for the 800 mg/day high-dose imatinib therapy (20).
In this study, the mutation frequency of KIT exon 9 and 11 mutants accounted for 10.9% and 45.7% of GISTs, respectively. The frequency of the exon 9 mutation is in agreement with Mediterranean population-based studies, specifically Italian (11%) and Portuguese (9%) (21,22). However, the frequency of the exon 9 mutation is generally higher than in other European countries, such as Poland (7.3%) and France (5.5%) (16,18). The reported frequencies of the mutations in exon 11 are relatively variable in the literature, and our results are slightly lower than those reported in some of the previous studies (Italian, 67%; Portuguese, 91%; Polish, 61.1%; French, 63.2%) (16,18,21,22). These discrepancies may mostly reflect the methodological (pathological or molecular methods) and material differences (fresh or formalin-fixed paraffin-embedded tissue) but, at the same time, can reflect the population-based variations due to ethnicity. Because there has been no multinational study on the relationship between mutations in the KIT gene and ethnicity, it is difficult to evaluate the effects of the ethnicity on these mutations. However, GISTs patients with carrying KIT gene exon 9 are more likely to show resistance, whereas those with tumors carrying mutations of exon 11 are more likely to show a good response. For possible resistance and survival, it was an advantage to have higher levels of the exon 11 mutation in our group.
Regardless of the type of mutation, these alterations were clustered in a small region between codon 552 and codon 586, as previously described. Mutation type in exon 11 is also important for outcomes. Deletions on exon 11 of the KIT gene have been reported to be more aggressive and metastatic than substitutions. These deletions may mostly effect survival (23–25). Patients with KIT gene exon 11 deletion/insertion have shorter recurrence-free survival (RFS) than patients with tumors missense mutations, which are more frequent in favorable outcome, low-risk GIST (26). In our group, 30.4% of the patients had deletions, and 10.9% of them had substitutions. The higher frequency of deletions may be the main reason for the shorter overall survival in our group. Most of the deletions were found on the W557 and K558 codons. Several studies noted that this deletion represents a significant adverse factor for patients’ outcome (27).
Mutations in tyrosine kinase domains on exon 13 and exon 17 were found at higher frequencies, 15.2% and 19.6%, respectively; 15.2% of cases exhibited p.K642E mutations in exon 13, and 19.6% of cases exhibited p.D820A mutations in exon 17, which play a role in the resistance of the tyrosine kinase inhibitor imatinib. Higher mutation frequency on exon 13 and 17 is presenting the reason of drug resistance and resistance-related shorter survival. After analyzing the relationship between mutational status and clinicopathological/demographic parameters, the association between age and KIT gene mutations was observed to have statistical significance (p = 0.02), as expected. It is well known that the accumulation of age-related mutations can be observed in cancerous tumors due to general problems in DNA repair mechanisms. However, patients older than 50 present wild-type characteristics for the PDGFRA gene. This may explain why PDGFRA mutations are mostly found in cases with the wild-type KIT gene. In the literature, some papers, such as that of Wozniak and colleagues (28), suggested that stomach and intestinal tumors have significantly higher frequencies of the KIT and PDGFRA mutations. In our group, stomach- and small intestine-localized tumors exhibited a higher mutation frequency for the KIT gene and lower mutation frequency for the PDGFRA gene. The association was also reported between KIT gene exon 9 mutations and small intestine location and patients with exon 9 mutations have poorer outcomes compared with other patients (29).
PDGFRA gene mutations were reported to be between 5.4% and 20% in the literature (16,17,30). PDGFRA mutations have prognostic and predictive value. PDGFRA exon 18 mutant tumors have a lower chance of metastasis when compared with KIT exon 9 and 11 mutant and PDGFRA exon 12 mutant tumors. They have epitheloid morphology and low mitotic count (31). In the present study, PDGFRA was mutated in 23.9% of cases. Among PDGFRA mutants, changes in exon 12 were identified in 15.2% of cases, including all cases with substitutions. The determined substitutions were as follows: p.K552R, p.E556K, p.E557G, p.W559*, p.E571D, p.E571X, p.I573 M, p.Y574D, and p.Q579L. Moreover, we observed silent mutations, such as a base substitution in exon 12 (CCA > CCG) at codon 567 (P567P) in 87% of cases (Fig. 1A, B). Due to the same protein sequence, this substitution did not affect the mutation frequency. Some researchers have also reported this silent substitution in their GIST patient groups (32,33). It is still interesting to observe this change in nearly all cases. It may be characteristic of our population. On exon 18, 17.4% of the cases had mutations. All cases showed substitutions; 2.2% of cases had p.R822H mutation, and 15.2% of them had the p.D842V mutation (Fig. 1C). There was a large variation between the frequencies of the PDGFRA mutation (5.4–20%) in different studies, which can be at least partially explained by the material, methodology, or ethnicity. In our group, PDGFRA mutation frequency is comparable with Zheng et al.’s study, the PDGFRA mutation frequency was found as 20% in their Chinese GIST patient-based group (30). Miettinen et al. also established the frequency to be as high as 22.6% in a gastric series of GISTs (23). In this study, overall PDGFRA mutation frequency was determined as 23%. Stomach-localized GIST tumors were 43.5% over all cases, but interestingly, only three of them had PDGFRA mutation. Further studies should be done with larger patient groups.
In the literature, it was reported that approximately 12–15% of GIST were WT for either kinases KIT and PDGFRA (6). In our group, 13.1% of cases were WT for both the KIT and PDGFRA genes. WT-GISTs should be analyzed for further gene mutations such as succinate dehydrogenase (SDH). Loss of SDH results with the accumulation of succinate, inhibitors of DNA demethylation enzymes and causes turning on of oncogenic fenotypes with the hypermethylation phenotype and loss of genes (34).
A clear diagnosis was established by immunostaining tissue specimens for CD117, CD34, SMA, desmin, S-100, and Ki-67. Approximately 80–95% of GISTs show positive staining for CD117, while the other 5–20% exhibit negative staining. If the tumor is negative for CD117 but positive for CD34, a histological diagnosis is possible (3). CD34 is also an important adhesion molecule, and cells expressing CD34 (CD34-positive cells) are normally found in mesenchymal cells, endothelial cells, endothelial progenitor cells, and cells in soft tissue tumors, such as GISTs. In our group, CD34-positive cells had the wild-type PDGFRA gene (p = 0.02). However, if the tumors are negative for CD117, CD34, S-100, and SMA, similar to our patient, a definitive diagnosis is often challenging. However, recently DOG1 has received considerable attention as a useful molecule for the diagnosis of GIST. DOG1, a membrane channel protein, is known to be overexpressed in GIST. Because the sensitivity and specificity of DOG1 are higher than those of CD117, positive staining for DOG1 has been reported, even in CD117-negative GIST (12,35). In our group, 65.2% of all cases were DOG1 positive, and 34.8% of them were negative. DOG1 exhibited +1 positivity in 13% of all cases, +2 in 37% of cases, and +3 in 15.2% of cases. West et al. (9) showed that the frequency of DOG1 positivity was 97.8% and CD117 positivity, and Miettinen et al. (36) showed that DOG1 positivity was 94.8% and CD117 positivity was 94.9%. With these results, the frequency of DOG1 positivity observed in our study was lower than all of these groups (DOG1 positivity 65.2%, CD117 positivity 100%).
Approximately half of the patients with DOG1 negativity are CD117 positive in different reports, whereas 36% to 46% of KIT-negative cases are DOG1 positive. Therefore, DOG1 can identify a significant part of GIST with KIT and PDGFR mutations. Nearly 66% of CD117-positive samples showed a strong DOG1 expression in the literature. This study showed that 65.2% of CD117-positive samples showed a strong DOG1 expression. Under this information, our DOG1 results were not different from the literature. But it should be kept in mind in this evaluation the variability of DOG1 expression can be mostly caused by different monoclonal antibodies used for immunohistochemistry techniques and different characteristics of the tumor specimens (8,37).
According to our results, it is clear that CD117 is still the most sensitive marker for the diagnosis of GIST. A relationship could not be found between CD117 and DOG1, which was diagnosed via immunohistochemical methods because all cases showed CD117 immunopositivity. However, an association was found between the existence of the KIT mutation and DOG1 positivity (p = 0.04). DOG1 positivity was higher in KIT mutant cases. The same relationship was observed between mutation of exon 11 of KIT and DOG1 (p = 0.04). DOG1 positivity can be used as an alternative immunohistochemical marker for the analysis of KIT gene exon 11 mutations. Stomach- and small intestine-localized tumors have also higher DOG1 positivity than other localized tumors (p = 0.022). The overall KIT mutation and the existence of KIT exon 11 mutation were also higher for these stomach- and small intestine-localized tumors; therefore, the association between KIT mutations and DOG1 positivity may originate from the location. Moreover, KIT exon 13 mutant cases presented DOG1 negativity (p = 0.04). Exon 13 mutations are indicated as the reason for resistance to tyrosine kinase inhibitors. DOG1 negativity may be an indicator for the prediction of the possible resistance to tyrosine kinase inhibitors.
CONCLUSION
Most of the GISTs harbor activating mutations either in the KIT or PDGFRA genes, whereas a small group of GISTs is WT. The role of KIT and PDGFRA mutations in GIST pathogenesis and management have revealed them as the most important targets for the management of disease and diagnosis. Mutation status can predict the response to treatment with tyrosine kinase inhibitors and give insights as a prognostic factor for the nature of tumors. Therefore, to know the differences on the mutation status of KIT and PDGFRA genes is crucial for effective disease management. Investigation of DOG1, KIT, and PDGFRA mutations together for the prediction of outcome in GIST patients is rare in the literature. This study brings three markers together and observes their importance in GIST. This study is also the first report on the molecular analysis of KIT and PDGFRA genes together from Turkish patients. In this study, KIT mutation frequency was found to be similar with European population-based literature. However, PDGFRA mutation frequency (23.9%) was higher than the indicated in the literature (5.4–20%); the mutation results were close to a Chinese population-based study (PDGFRA mutation frequency was 20%) (30). Further studies should be done with larger patient groups. In our group, stomach- and small intestine-localized tumors exhibited a higher mutation frequency for the KIT gene and lower mutation frequency for the PDGFRA gene; therefore, KIT and PDGFRA mutations and tumor location can be used for the prediction of tumor behavior and the management of disease in GISTs.
DOG1 is used as a marker for differential diagnosis in many studies, regardless of oncogenic mutations of either kinases KIT and PDGFRA, but to avoid the variability of DOG1 results in GIST diagnosis and management processes it should be kept in mind DOG1, CD117, KIT, and PDGFRA mutations should be evaluated together (8,37). However, the prognostic role of DOG1 is still unclear. In contrast with limited previous studies (8,36,38), our data showed that DOG1 expression has significant relation with overall existence of KIT gene mutation, KIT gene exon 11 mutation, and stomach/small intestine location of tumor, although a significant correlation was demonstrated with DOG1 negativity and existence of KIT gene exon 13 mutation. DOG1 positivity might be a candidate marker to support KIT mutations.
ACKNOWLEDGMENTS
This study was supported by Dokuz Eylul University Scientific Research Foundation (DEU-BAP 2010.KB.SAG.006 and 2010.KB.SAG.034).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Corless C. L. Gastrointestinal stromal tumors: What do we know now. Mod. Pathol. 27(Suppl. 1):S1–16; 2014. [DOI] [PubMed] [Google Scholar]
- 2. Corless C. L.; Fletcher J. A.; Heinrich M. C. Biology of gastrointestinal stromal tumors. J. Clin. Oncol. 22:3813–3825; 2014. [DOI] [PubMed] [Google Scholar]
- 3. Fletcher C. D.; Berman J. J.; Corless C.; Gorstein F.; Lasota J.; Longley B. J.; Miettinen M.; O’Leary T. J.; Remotti H.; Rubin B. P.; Shmookler B.; Sobin L. H.; Weiss S. W. Diagnosis of gastrointestinal stromal tumors: A consensus approach. Hum. Pathol. 33(5):459–465; 2002. [DOI] [PubMed] [Google Scholar]
- 4. Demoulin J. B.; Essaghir A. PDGF receptor signaling networks in normal and cancer cells. Cytokine Growth Factor Rev. 25(3):273–283; 2014. [DOI] [PubMed] [Google Scholar]
- 5. Zong L.; Chen P. Prognostic value of KIT/PDGFRA mutations in gastrointestinal stromal tumors: A meta-analysis. World J. Surg. Oncol. 12:71; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen P.; Zong L.; Zhao W.; Shi L. Efficacy evaluation of imatinib treatment in patients with gastrointestinal stromal tumors: A meta-analysis. World J. Gastroenterol. 16(33):4227–4232; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Joensuu H.; Rutkowski P.; Nishida T.; Steigen S. E.; Brabec P.; Plank L.; Nilsson B.; Braconi C.; Bordoni A.; Magnusson M. K.; Sufliarsky J.; Federico M.; Jonasson J. G.; Hostein I.; Bringuier P. P.; Emile J. F. KIT and PDGFRA mutations and the risk of GI stromal tumor recurrence. J. Clin. Oncol. 33(6):634–642; 2015. [DOI] [PubMed] [Google Scholar]
- 8. Espinosa I.; Lee C. H.; Kim M. K.; Rouse B. T.; Subramanian S.; Montgomery K.; Varma S.; Corless C. L.; Heinrich M. C.; Smith K. S.; Wang Z.; Rubin B.; Nielsen T. O.; Seitz R. S.; Ross D. T.; West R. B.; Cleary M. L.; van de Rijn M. A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am. J. Surg. Pathol. 32:210–218; 2008. [DOI] [PubMed] [Google Scholar]
- 9. West R. B.; Corless C. L.; Chen X.; Rubin B. P.; Subramanian S.; Montgomery K.; Zhu S.; Ball C. A.; Nielsen T. O.; Patel R.; Goldblum J. R.; Brown P. O.; Heinrich M. C.; van de Rijn M. The novel marker, DOG1 is expressed ubiquitously in gastrointestinal stromal tumors irrespective of KIT or PDGFRA mutation status. Am. J. Pathol. 165:107–113; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sozutek D.; Yanik S.; Akkoca A. N.; Sozutek A.; Ozdemir Z. T.; Avsar C. U.; Gunald M.; Sahin B.; Doron F. Diagnostic and prognostic roles of DOG1 and Ki-67, in GIST patients with localized or advanced/metastatic disease. Int. J. Clin. Exp. Med. 7(7):1914–1922; 2014. [PMC free article] [PubMed] [Google Scholar]
- 11. Nakamura N.; Yamamoto H.; Yao T.; Oda Y.; Nishiyama K.; Imamura M.; Yamada T.; Nawata H.; Tsuneyoshi M. Prognostic significance of expressions of cell-cycle regulatory proteins in gastrointestinal stromal tumor and the relevance of the risk grade. Hum. Pathol. 36(7):828–837; 2005. [DOI] [PubMed] [Google Scholar]
- 12. Jones R. L. Practical aspects of risk assessment in gastrointestinal stromal tumors. J. Gastrointest. Cancer 45(3):262–267; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barnett C. M.; Corless C. L.; Heinrich M. C. Gastrointestinal stromal tumors: Molecular markers and genetic subtypes. Hematol. Oncol. Clin. North Am. 27(5):871–888; 2013. [DOI] [PubMed] [Google Scholar]
- 14. Turner M. S.; Goldsmith J. D. Best practices in diagnostic immunohistochemistry: Spindle cell neoplasms of the gastrointestinal tract. Arch. Pathol. Lab. Med. 133(9):1370–1374; 2009. [DOI] [PubMed] [Google Scholar]
- 15. Ahmad F.; Lad P.; Bhatia S.; Das B. R. Molecular spectrum of c-KIT and PDGFRA gene mutations in gastro intestinal stromal tumor: Determination of frequency, distribution pattern and identification of novel mutations in Indian patients. Med. Oncol. 32(1):424; 2015. [DOI] [PubMed] [Google Scholar]
- 16. Wozniak A.; Rutkowski P.; Piskorz A.; Ciwoniuk M.; Osuch C.; Bylina E.; Sygut J.; Chosia M.; Rys J.; Urbanczyk K.; Kruszewski W.; Sowa P.; Siedlecki J.; Debiec-Rychter M.; Limon J.; Polish Clinical GIST Registry. Prognostic value of KIT/PDGFRA mutations in gastrointestinal stromal tumors (GIST): Polish Clinical GIST Registry experience. Ann. Oncol. 23(2):353–360; 2012. [DOI] [PubMed] [Google Scholar]
- 17. Tryggvason G.; Hilmarsdottir B.; Gunnarsson G. H.; Jónsson J. J.; Jónasson J. G.; Magnússon M. K. Tyrosine kinase mutations in gastrointestinal stromal tumors in a nation-wide study in Iceland. APMIS. 118(9):648–656; 2010. [DOI] [PubMed] [Google Scholar]
- 18. Emile J. F.; Brahimi S.; Coindre J. M.; Bringuier P. P.; Monges G.; Samb P.; Doucet L.; Hostein I.; Landi B.; Buisine M. P.; Neuville A.; Bouché O.; Cervera P.; Pretet J. L.; Tisserand J.; Gauthier A.; Le Cesne A.; Sabourin J. C.; Scoazec J. Y.; Bonvalot S.; Corless C. L.; Heinrich M. C.; Blay J. Y.; Aegerter P. Frequencies of KIT and PDGFRA mutations in the MolecGIST prospective population-based study differ from those of advanced GISTs. Med. Oncol. 29(3):1765–1772; 2012. [DOI] [PubMed] [Google Scholar]
- 19. Lasota J.; Jasinski M.; Sarlomo-Rikala M.; Miettinen M. Mutations in exon 11 of c-Kit occur preferentially in malignant versus benign gastrointestinal stromal tumors and do not occur in leiomyomas or leiomyosarcomas. Am. J. Pathol. 154:53–60; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maleddu A.; Pantaleo M. A.; Nannini M.; Biasco G. The role of mutational analysis of KIT and PDGFRA in gastrointestinal stromal tumors in a clinical setting. J. Transl. Med. 9:75; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Braconi C.; Bracci R.; Bearzi I.; Bianchi F.; Costagliola A.; Catalani R.; Mandolesi A.; Ranaldi R.; Galizia E.; Cascinu S.; Rossi G.; Giustini L.; Latini L.; Valeri N.; Cellerino R. KIT and PDGFRalpha mutations in 104 patients with gastrointestinal stromal tumors (GISTs): A population-based study. Ann. Oncol. 19(4):706–710; 2008. [DOI] [PubMed] [Google Scholar]
- 22. Gomes A. L.; Gouveia A.; Capelinha A. F.; de la Cruz D.; Silva P.; Reis R. M.; Pimenta A.; Lopes J. M. Molecular alterations of KIT and PDGFRA in GISTs: Evaluation of a Portuguese series. J. Clin. Pathol. 61(2):203–208; 2008. [DOI] [PubMed] [Google Scholar]
- 23. Miettinen M.; Sobin L. H.; Lasota J. Gastrointestinal stromal tumours of the stomach: A clinicopathologic, immunohistochemical, and molecular genetic study of 1765 cases with long-term follow-up. Am. J. Surg. Pathol. 29:52–68; 2005. [DOI] [PubMed] [Google Scholar]
- 24. Andersson J.; Bümming P.; Meis-Kindblom J. M.; Sihto H.; Nupponen N.; Joensuu H.; Odén A.; Gustavsson B.; Kindblom L. G.; Nilsson B. Gastrointestinal stromal tumours with KIT exon 11 deletions are associated with poor prognosis. Gastroenterology. 130:1573–1581; 2006. [DOI] [PubMed] [Google Scholar]
- 25. Wardelmann E.; Losen I.; Hans V.; Neidt I.; Speidel N.; Bierhoff E.; Heinicke T.; Pietsch T.; Büttner R.; Merkelbach-Bruse S. Deletion of Trp-557 and Lys-558 in the juxtamembrane domain of the c-kit protooncogene is associated with metastatic behaviour of gastrointestinal stromal tumours. Int. J. Cancer 106:887–895; 2003. [DOI] [PubMed] [Google Scholar]
- 26. Singer S.; Rubin B. P.; Lux M. L.; Chen C. J.; Demetri G. D.; Fletcher C. D.; Fletcher J. A. Prognostic value of KIT mutation type, mitotic activity, and histologic subtype in gastrointestinal stromal tumors. J. Clin. Oncol. 20:3898–3905; 2002. [DOI] [PubMed] [Google Scholar]
- 27. Martín J.; Poveda A.; Llombart-Bosch A.; Ramos R.; López-Guerrero J. A.; García del Muro J.; Maurel J.; Calabuig S.; Gutierrez A.; González de Sande J. L.; Martínez J.; De Juan A.; Laínez N.; Losa F; Alija V.; Escudero P.; Casado A.; García P.; Blanco R.; Buesa J. M.; Spanish Group for Sarcoma Research. Deletions affecting codons 557-558 of the c-KIT gene indicate a poor prognosis in patients with completely resected gastrointestinal stromal tumours: A study by the Spanish Group for Sarcoma Research (GEIS). J. Clin. Oncol. 23:6190–6198; 2005. [DOI] [PubMed] [Google Scholar]
- 28. Lasota J.; Wozniak A.; Sarlomo-Rikala M.; Rys J.; Kordek R.; Nassar A.; Sobin L. H.; Miettinen M. Mutations in exons 9 and 13 of KIT gene are rare events in gastrointestinal stromal tumors. A study of 200 cases. Am. J. Pathol. 157:1091–1095; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sakurai S.; Oguni S.; Hironaka M.; Fukayama M.; Morinaga S.; Saito K. Mutations in c-kit gene exons 9 and 13 in gastrointestinal stromal tumors among Japanese. Jpn. J. Cancer Res. 92:494–498; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng S.; Chen L. R.; Wang H. J.; Chen S. Z. Analysis of mutation and expression of c-kit and PDGFR-alpha gene in gastrointestinal stromal tumor. Hepatogastroenterology 54(80):2285–2290; 2007. [PubMed] [Google Scholar]
- 31. Lasota J.; Dansonka-Mieszkowska A.; Leslie S. H.; Miettinen M. A great majority of GISTs with PDGFRA mutations represent gastric tumors of low or no malignant potential. Lab. Invest. 84:874–883; 2004. [DOI] [PubMed] [Google Scholar]
- 32. Sihto H.; Sarlomo-Rikala M.; Tynninen O.; Tanner M.; Andersson L. C.; Franssila K.; Nupponen N. N.; Joensuu H. KIT and platelet-derived growth factor receptor alpha tyrosine kinase gene mutations and KIT amplifications in human solid tumors. J. Clin. Oncol. 23(1):49–57; 2005. [DOI] [PubMed] [Google Scholar]
- 33. Holden J. A.; Willmore-Payne C.; Coppola D.; Garrett C. R.; Layfield L. J. High-resolution melting amplicon analysis as a method to detect c-kit and platelet-derived growth factor receptor alpha activating mutations in gastrointestinal stromal tumors. Am. J. Clin. Pathol. 128(2):230–238; 2007. [DOI] [PubMed] [Google Scholar]
- 34. Rubin B. P.; Heinrich M. C. Genotyping and immunohistochemistry of gastrointestinal stromal tumors: An update. Semin. Diagn. Pathol. 32(5):392–399; 2015. [DOI] [PubMed] [Google Scholar]
- 35. Lee C. H.; Liang C.; Espinos I. The utility of discovered on gastrointestinal stromal tumor 1 (DOG1) antibody in surgical pathology-the GIST of it. Adv. Anat. Pathol. 17:222–232; 2010. [DOI] [PubMed] [Google Scholar]
- 36. Miettinen M.; Wang Z. F.; Lasota J. DOG1 antibody in the differential diagnosis of gastrointestinal stromal tumors. A study of 1840 cases. Am. J. Surg. Pathol. 33:1401–1408; 2009. [DOI] [PubMed] [Google Scholar]
- 37. Joensuu H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum. Pathol. 39:1411–1419; 2008. [DOI] [PubMed] [Google Scholar]
- 38. Sui X. L.; Wang H.; Sun X. W. Expression of DOG1, CD117 and PDGFRA in gastrointestinal stromal tumors and correlations with clinicopathology. Asian Pac. J. Cancer Prev. 13(4):1389–1393; 2012. [DOI] [PubMed] [Google Scholar]