Abstract
The cyclic-AMP response element-binding protein (CREB), a well-known nuclear transcription factor, has been shown to play an essential role in many cellular processes, including differentiation, cell survival, and cell proliferation, by regulating the expression of downstream genes. Recently, increased expression of CREB was frequently found in various tumors, indicating that CREB is implicated in the process of tumorigenesis. However, the effects of CREB on Hodgkin lymphoma (HL) remain unknown. To clarify the role of CREB in HL, we performed knockdown experiments in HL. We found that downregulation of CREB by short hairpin RNA (shRNA) resulted in enhancement of cell proliferation and promotion of G1/S phase transition, and these effects can be rescued by expression of shRNA-resistant CREB. Meanwhile, the expression level of cell cycle-related proteins, such as cyclin D1, cyclin E1, cyclin-dependent kinase 2 (CDK2), and CDK4, was elevated in response to depletion of CREB. Furthermore, we performed chromatin immunoprecipitation (ChIP) assay and confirmed that CREB directly bound to the promoter regions of these genes, which consequently contributed to the regulation of cell cycle. Consistent with our results, a clinical database showed that high expression of CREB correlates with favorable prognosis in B-cell lymphoma patients, which is totally different from the function of CREB in other cancers such as colorectal cancer, acute myeloid leukemia, and some endocrine cancers. Taken together, all of these features of CREB in HL strongly support its role as a tumor suppressor gene that can decelerate cell proliferation by inhibiting the expression of several cell cycle-related genes. Our results provide new evidence for prognosis prediction of HL and a promising therapeutic strategy for HL patients.
Key words: Cyclic-AMP response element-binding protein (CREB), Hodgkin lymphoma (HL), Proliferation, Cell cycle
INTRODUCTION
Hodgkin lymphoma (HL) is a malignant B-cell lymphoma characterized by the presence of the mononuclear Hodgkin and multinuclear Reed-Sternberg (HRS) cells (1). Recent studies have identified abnormal B-cell development as a major factor in the pathogenesis of HL (2,3). By multiagent chemotherapy, localized radiation, or hematopoietic stem cell transplantation therapy, HL is one of the most curable human cancers with high survival rates (4). However, because of the adverse effects of treatment that can lead to increased risk of cardiovascular events, secondary malignant conditions, and severe endocrine long-term effects specific for childhood cases, a lot of cured patients do not live up to their expected life span or are subjected to a poor quality of life. Accordingly, new targets for treatment and novel mechanisms of tumorigenesis in HL still need to be explored in depth.
The cyclic-AMP response element-binding protein (CREB) is a leucine zipper (bZIP) transcription factor, which is involved in cell proliferation, survival, and differentiation in a number of cell types (5). CREB has also been shown to be important in memory and hippocampal development (6–8). In neural stem cells, CREB plays a critical role in their proliferation (9). The activated CREB turns on the transcription of many genes, including proto-oncogenes, cell cycle regulatory genes, and other genes related to cell growth, survival, differentiation, and neuronal activities. Recently, overexpressed CREB has been found in various cancers and significantly associated with decreased event-free survival (10), which therefore defined CREB as an oncogene for promoting cancers. However, in HL, the function of CREB is still unknown.
To understand the role of CREB in HL, we performed knockdown and rescue experiments in HL cell lines. Unlike previous reports in other cancers, our results showed that the downregulation of CREB in HL cell lines induced cell proliferation and promoted G1/S phase transition. These phenomena can be rescued by the overexpression of short hairpin RNA (shRNA)-resistant CREB. Furthermore, we found that CREB regulates the expression of several cell cycle-related proteins by directly binding to their promoters. The functions identified for CREB in cell proliferation and cell cycle regulation may suggest a link between NeuroD1 and the progression of HL. Therefore, we wondered whether the findings obtained were relevant to the clinical features of human HL. To this end, we checked the R2 genomics analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Indeed, the clinical data show that high expression of CREB correlates with a good prognosis in B-cell lymphoma, indicating that CREB acts as a tumor suppressor gene in human HL, which is totally different than its roles reported in other cancers. All in all, our results shed light on the special mechanism of CREB-related tumorigenesis in HL and provide new evidence for the complicated roles of CREB in cancer.
MATERIAL AND METHODS
Plasmids
Lentivirus-based nontargeting shRNA and specific shRNA for human CREB were obtained from Sigma-Aldrich (St. Louis, MO, USA). A human CREB complementary DNA (cDNA) was amplified from human brain cDNA and was inserted into CSII-CMV-IRES-VENUS (RIKEN BioResource Center, Tsukuba, Ibaraki, Japan), which is a lentivirus-based cDNA overexpression plasmid. The targeting sequence of CREB shRNA-1 localizes in the untranslated regions of CREB gene. Nevertheless, the CREB overexpression vector we constructed was amplified according to coding sequences of CREB gene, so we used our CREB overexpression vector as a shRNA-resistant CREB expression vector.
Cell Culture and Transfection
Human HL cell lines KMH2, L1236, L428, HDLM2, and L540 were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM) supplied with 10% heat-inactivated fetal bovine serum (FBS). Transfection experiment was performed with lentivirus packaged with HEK293T cells.
Cell Proliferation Assay
Cells were infected with lentivirus-based CREB shRNA with or without CREB-overexpressing lentivirus. The next day, cells (1 × 104) were reseeded in 96-well cell culture plates and allowed to adhere overnight. At day 0, culture medium was changed to DMEM supplied with 2% heat-inactivated FBS. Cell number was counted at day 4.
Real-Time Polymerase Chain Reaction
Total RNA extracted from cultured cells and real-time polymerase chain reaction (PCR) were carried out as previously described (11). Primer information is shown in Table 1.
Table 1.
Primer Sequences for PCR Experiments
| Primer Name | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) |
|---|---|---|
| Human CREB | GATGCAGCTGTAACAGAAGC | ATGACTCCATGGACTTGAACT |
| Human CCNA1 | GAGGTCCCGATGCTTGTCAG | GTTAGCAGCCCTAGCACTGTC |
| Human CCNB1 | AATAAGGCGAAGATCAACATGGC | TTTGTTACCAATGTCCCCAAGAG |
| Human CCND1 | GCTGCGAAGTGGAAACCATC | CCTCCTTCTGCACACATTTGAA |
| Human CCNE1 | GCCAGCCTTGGGACAATAATG | CTTGCACGTTGAGTTTGGGT |
| Human CDK2 | CCAGGAGTTACTTCTATGCCTGA | TTCATCCAGGGGAGGTACAAC |
| Human CDK4 | CTGGTGTTTGAGCATGTAGACC | GATCCTTGATCGTTTCGGCTG |
| Human CDK6 | TCTTCATTCACACCGAGTAGTGC | TGAGGTTAGAGCCATCTGGAAA |
| Human GAPDH | ATCATCCCTGCCTCTACTGG | CCCTCCGACGCCTGCTTCAC |
| CCND1 for ChIP | -1053 TCACGCTCACGAATTCAGTCC -1032 | -927 ATTGCTGGAAATATTAGTCCC -948 |
| CCNE1 for ChIP | -1150 TACAAGTCTTGAGGGAGAAAG -1139 | -1053 AAATCCACACTCCGTGTCCTA -1032 |
| CDK2 for ChIP | -65 AAGAACCAAACTTGCCTGGGG -44 | 69 TGAAACAATGTTGCCGCCTCC 48 |
| CDK4 for ChIP | -106 TGGAAGCAAGCACTCAGTAAA -85 | -340 TACCAGAGGTGATGCTGTCAC -361 |
Western Blot Analysis
For Western blot analysis, anti-CREB (1:1,000; Upstate, Darmstadt, Germany), anti-cyclin A (1:400; Abcam, Cambridge, MA, USA), anti-cyclin E (1:400; Santa Cruz Biotechnology, Dallas, TX, USA), anti-cyclin D1 (1:400; Santa Cruz Biotechnology), anti-cyclin-dependent kinase 2 (CDK2) (1:400; Santa Cruz Biotechnology), anti-CDK4 (1:400; Santa Cruz Biotechnology), and anti-CDK6 (1:400; Santa Cruz Biotechnology) antibodies were used.
Flow Cytometric Analysis of Cell Cycle
Cell cycle analysis was performed as described by Eo et al. (12) and Zhou et al. (13). Briefly, KMH2 cells (1 × 106) were harvested and washed with cold phosphate-buffered saline (PBS). The cells were fixed with 70% ethanol and stored at −20°C overnight. The next day, cells were centrifuged and resuspended with PBS. The cells were treated with 100 μg/ml of RNase, and incubated at 37°C for 30 min. The cells were then incubated with 50 μg/ml of propidium iodide (PI) at 4°C for 30 min. Results obtained from a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) were analyzed with the CellQuest software (BD Biosciences, San Jose, CA, USA).
Chromatin Immunoprecipitation Assay
Chromatin Immunoprecipitation (ChIP) assay with KMH2 cells was carried out as previously described (11), using an anti-CREB antibody. Primer information is listed in Table 2.
Table 2.
Sequences of shRNAs
| shRNA Clone | shRNA Sequence (5′–3′) |
|---|---|
| Human CREB shRNA-1 | CCGGGCTCGATAAATCTAACAGTTACTCGAGTAACTGTTAGATTTAATCGAGCTTTTT |
| Human CREB shRNA-2 | CCGGCGTCTAATGAAGAACAGGGAACTCGAGTTCCCTGTTCTTCATTAGACGTTTTT |
Statistical Analysis
All experiments were performed in triplicates and found to be reproducible. The p value was calculated with Student’s t-test, and results are presented as mean ± standard deviation.
RESULTS
Knockdown of CREB Promotes Cell Proliferation in HL Cell Lines
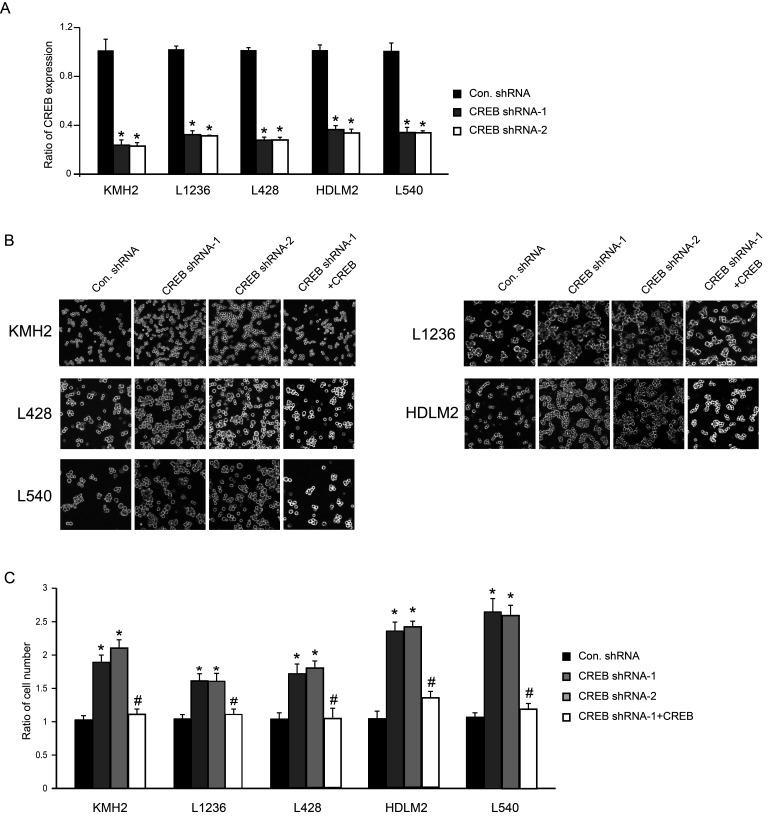
CREB has been reported as an oncogene in several kinds of cancers, but there is almost no report in terms of CREB function in HL. In order to clarify the role of CREB in HL, we performed knockdown experiments with two different shRNA targeting CREB. The expression level of CREB was successfully knocked down by both shRNAs in five HL cell lines: KMH2, L1236, L428, HDLM2, and L540 cells (Fig. 1A). Compared to the control group, the cell number was significantly increased after infection with shRNA-expressing lentivirus (Fig. 1B, C). Furthermore, the rescued experiment was performed by using shRNA-resistant CREB (Fig. 1B, C). As a consequence, the excessive growth of CREB shRNA-promoted cells was decelerated, suggesting that the proliferation of the cells was truly induced by knockdown of CREB expression in HL cell line (Fig. 1B, C).
Figure 1.
Downregulation of CREB promotes cell proliferation of KMH2, L1236, L428, HDLM2, and L540 cells. (A) A quantitative PCR showed significant decrease in CREB expression in KMH2, L1236, L428, HDLM2, and L540 cells after CREB shRNA treatment. (B) Phase-contrast images of cells at day 4 after infection of shRNA-expressing lentivirus with or without shRNA-resistant CREB-expressing lentivirus. (C) Cell number was counted at day 4. *p < 0.001 versus control shRNA. #p < 0.001 versus CREB shRNA-1.
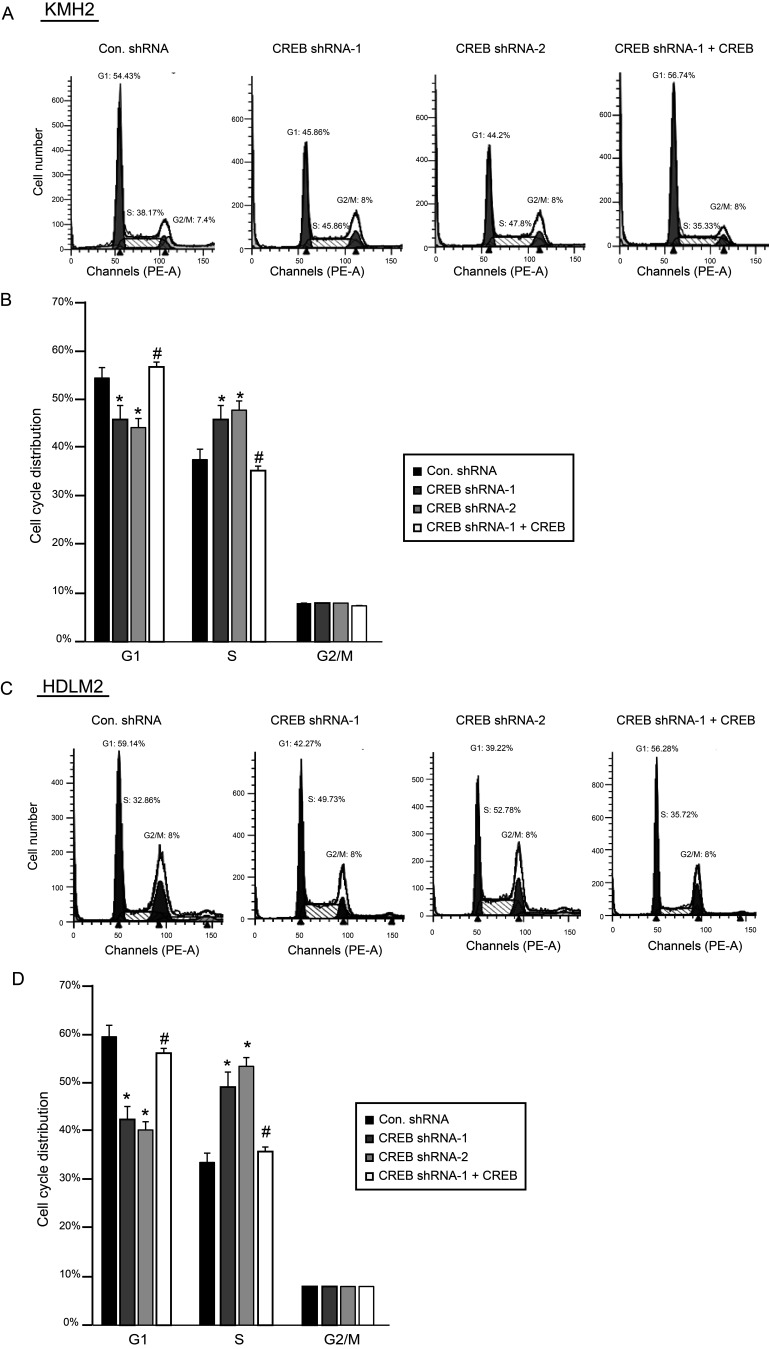
CREB Regulates G1/S Phase Transition in KMH2 and HDLM2 Cells
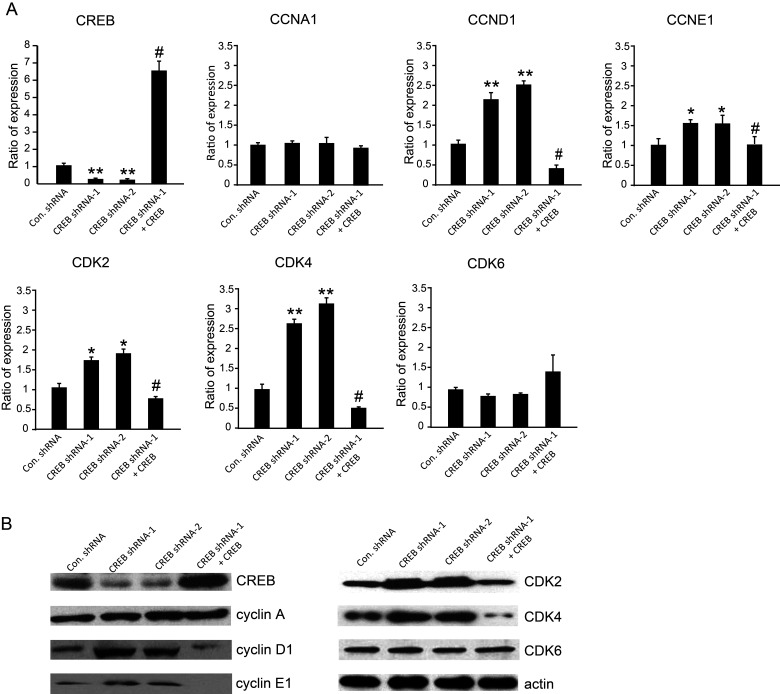
The role of CREB in cell cycle regulation has been reported in several researches. Therefore, we wondered whether the disturbance of cell proliferation described in Figure 1B and C was attributed to regulation of cell cycle by CREB in the HL cell line. We analyzed cell cycle with flow cytometry by staining cells with PI. Consequently, we found that knockdown of CREB by CREB shRNA affected the G1/S phase in KMH2 and HDLM2 cells. The cell number in the G1 phase decreased, but increased almost 10% in the S phase with downregulation of CREB (Fig. 2A–D), while the G2/M phase was not obviously changed. The results also show that the shRNA-resistant CREB expression can rescue the phenomena described above. In order to clarify the molecular mechanism underlying CREB-regulated cell cycle in HL, cell cycle-related genes involved in the G1 phase, G1/S phase transition, and S phase were investigated. The expression of cyclin D1, cyclin E1, CDK2, and CDK4 determined by real-time PCR was significantly elevated with downregulation of CREB. However, there was no effect on the expression of cyclin A and CDK6 (Fig. 3A). Real-time PCR results also showed that shRNA-resistant CREB-expressing lentivirus can successfully restore the expression of CREB. Similarly, the expression of these cell cycle-related genes was reversed by overexpression of shRNA-resistant CREB. Western blot analysis (Fig. 3B) confirmed the results in Figure 3A.
Figure 2.
Knockdown of CREB promotes G1/S phase transition in KMH2 and HDLM2 cells. (A, C) Cell distribution was detected by flow cytometry. (B, D) Cell cycle distribution was calculated and shown as graphs. *p < 0.001 versus control shRNA. #p < 0.05 versus CREB shRNA-1.
Figure 3.
CREB regulates expression of several cell cycle-related genes in KMH2 cells. (A) The mRNA levels of CREB and several cell cycle-related genes were examined by real-time PCR. *p < 0.01, **p < 0.001 versus control shRNA. #p < 0.001 versus CREB shRNA-1. (B) Protein levels were detected by Western blot.
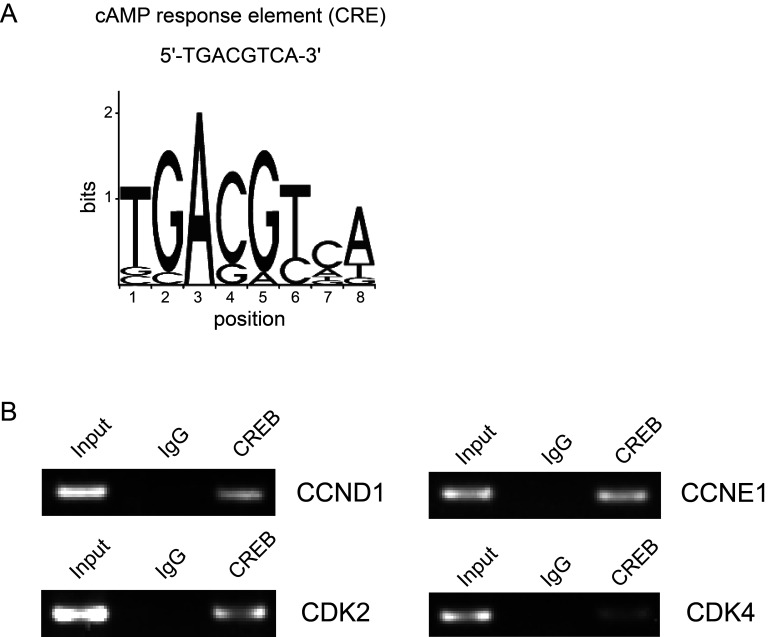
CREB Regulates Cell Cycle by Directly Binding to the Promoter Regions of Several Cell Cycle-Related Genes
It is already known that CREB is a transcription factor that binds to the cAMP response element (CRE) in the regulatory region of target genes. To investigate whether CREB directly regulates these cell cycle-related genes by binding to their promoter regions, we utilized the JASPAR database (http://jaspar.genereg.net/) to predict the putative CRE-like binding sites. According to the predicted sites, we designed primers and performed ChIP assay. The results showed that CREB can directly bind to the promoter region of cyclin D1, cyclin E1, CDK2, and CDK4 (Fig. 4).
Figure 4.
CREB directly binds to the promoter regions of several cell cycle-related genes. (A) The sequence of classic CRE is shown. (B) ChIP assay was performed by using anti-CREB antibody with KMH2 cells. Rabbit immunoglobulin G (IgG) was used as a control.
DISCUSSION
Aberrant expression of nuclear transcription factors has frequently been found in many cancers. As an important transcription factor, highly expressed CREB was also presented in several kinds of cancer and correlated with a high risk of tumorigenesis and a low survival rate. CREB regulates the expression of many downstream genes and subsequently controls cellular transformation, cell proliferation, and cell survival in a variety of cancers, including breast cancer (14), colorectal cancer (15), acute lymphoblastic leukemia (16–20), glioma (10), and some endocrine tumors (21). In previous articles, the results indicated that CREB serves as an oncogene to promote tumorigenesis through various cancer signaling pathways.
Unexpectedly, we discovered that CREB exhibits tumor inhibitory activity, which prevents tumorigenesis in HL. In HL cell lines, the proliferation of cells was promoted by knockdown of CREB in low serum condition, and this shRNA-induced phenomena could be reversed by overexpression of shRNA-resistant CREB. Meanwhile, the G1/S phase transition was accelerated in response to depletion of CREB. We supposed that CREB may act as a tumor suppressor in HL disease. Additionally, to clarify whether our findings are consistent with the clinical features of human HL, we utilized the R2 genomics analysis and visualization platform. Nevertheless, there is no clinical database specific for HL. Since HL is also a type of B-cell lymphoma, we further checked the dataset (Tumor B-cell Lymphoma - Xiao - 420 - MAS5.0 - u133p2), which includes specimens gained from B-cell lymphoma patients who had been diagnosed with diffuse large B-cell lymphoma. Analyzing the overall survival probabilities with regard to the expression level of CREB, the results show that the high expression level of CREB is associated with favorable survival, whereas the low expression level of CREB is related to poor prognosis, which are consistent with our findings in the HL cell line. Although no analytical results were achieved from HL, to some extent, we thought it could offer important clues that are able to benefit us in pursuit of discovering the cure for HL. Moreover, the report by Fang et al. found that loss of the CREB-regulated transcription coactivator (CRTC2), as well as CREB and its coactivator CREB-binding protein (CBP), results in a deficiency in DNA mismatch repair (MMR) and a resultant increased mutation frequency, indicating that the CREB pathway could be involved in protecting cells from proceeding toward cancer (22). They described an unanticipated role for CREB as a tumor suppressor in lymphoma, which is similar to our standpoint.
Cell cycle is an essential event that leads to cell division and duplication, and is crucial for cell survival and prevention of uncontrolled cell division (23,24). Increased expression of cell cycle-related genes, such as cyclin D1 (25), cyclin E1 (26,27), and CDKs (28), has been found in a variety of tumor tissues. Therefore, targeting cell cycle has emerged as an attractive strategy for the treatment of cancer. In HL, high expression of cell cycle-related proteins that account for accelerating cell cycle progression has been described (29–32). Ideally, cell cycle could be a potential target for HL treatment (33). In previous studies, one important mechanism of CREB was its ability to regulate cell proliferation through management of the cell cycle, either in cancer or in noncancer cell types. In several kinds of cancers, CREB could directly bind to the promoter region of cell cycle-related genes to regulate their expression (34). Our results demonstrated that CREB serves as a tumor suppressor gene, and downregulation of CREB expression promotes G1/S phase transition in the HL cell line, which is distinct from some reports in which CREB was described as an oncogene. This indicated that gene expression regulated by CREB closely depends on cell type, cell environment, or condition of cofactors. Furthermore, our ChIP assay result showed that CREB directly binds to promoter regions of cyclin D1, cyclin E1, CDK2, and CDK4. The similar case reported by Melnikova et al. concludes that CREB is a major suppressor for AP-2α by directly binding to its promoter region in melanoma (35). Accordingly, we hypothesize that CREB would regulate cell cycle-related protein expression in HL via directly binding to their promoter region and repressing their expression, as shown in Figures 3 and 4. Further work should be done to confirm this hypothesis in the future. Overall, our study indicates that CREB is a tumor suppression gene in HL by inhibiting G1/S phase transition.
REFERENCES
- 1. Janz M.; Hummel M.; Truss M.; Wollert-Wulf B.; Mathas S.; Johrens K. Classical Hodgkin lymphoma is characterized by high constitutive expression of activating transcription factor 3 (ATF3), which promotes viability of Hodgkin/Reed-Sternberg cells. Blood 107:2536–2539; 2006. [DOI] [PubMed] [Google Scholar]
- 2. Kuppers R. The biology of Hodgkin’s lymphoma. Nat. Rev. Cancer. 9:15–27; 2009. [DOI] [PubMed] [Google Scholar]
- 3. Schwering I.; Brauninger A.; Klein U.; Jungnickel B.; Tinguely M.; Diehl V.; Hansmann M. L.; Dalla-Favera R.; Rajewsky K.; Kuppers R. Loss of the B-lineage-specific gene expression program in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 101:1505–1512; 2003. [DOI] [PubMed] [Google Scholar]
- 4. Mauz-Korholz C.; Metzger M. L.; Kelly K. M.; Schwartz C. L.; Castellanos M. E.; Dieckmann K.; Kluge R.; Korholz D. Pediatric Hodgkin lymphoma. J. Clin. Oncol. 33:2975–2985; 2015. [DOI] [PubMed] [Google Scholar]
- 5. Siu Y. T.; Jin D. Y. CREB—a real culprit in oncogenesis. FEBS J. 274:3224–3232; 2007. [DOI] [PubMed] [Google Scholar]
- 6. Bito H.; Deisseroth K.; Tsien R. W. CREB phosphorylation and dephosphorylation: A Ca2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell 87:1203–1214; 1996. [DOI] [PubMed] [Google Scholar]
- 7. Deisseroth K.; Bito H.; Tsien R. W. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron 16:89–101; 1996. [DOI] [PubMed] [Google Scholar]
- 8. Ortega-Martinez S. A new perspective on the role of the CREB family of transcription factors in memory consolidation via adult hippocampal neurogenesis. Front. Mol. Neurosci. 8:46; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dworkin S.; Heath J. K.; DeJong-Curtain T. A.; Hogan B. M.; Lieschke G. J.; Malaterre J.; Ramsay R. G.; Mantamadiotis T. CREB activity modulates neural cell proliferation, midbrain-hindbrain organization and patterning in zebrafish. Dev. Biol. 307:127–141; 2007. [DOI] [PubMed] [Google Scholar]
- 10. Daniel P.; Filiz G.; Brown D. V.; Hollande F.; Gonzales M.; D’Abaco G.; Papalexis N.; Phillips W. A.; Malaterre J.; Ramsay R. G.; Mantamadiotis T. Selective CREB-dependent cyclin expression mediated by the PI3K and MAPK pathways supports glioma cell proliferation. Oncogenesis 3:10; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu F. J.; Kishida S.; Mu P.; Huang P.; Cao D. L.; Tsubota S.; Kadomatsu K. NeuroD1 promotes neuroblastoma cell growth by inducing the expression of ALK. Cancer Sci. 106:390–396; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eo S. H.; Kim J. H.; Kim S. J. Induction of G2/M arrest by berberine via activation of PI3K/Akt and p38 in human chondrosarcoma cell line. Oncol. Res. 22:147–157; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou C.; Zhang P.; Xu G. C.; Wu D. M.; Liu R. Y.; Zeng Q.; Wang C. T. RNA Interference of Biot2 induces G1 phase arrest and apoptosis in mouse colorectal cancer cell line. Oncol. Res. 22:93–103; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nguyen T. T. P.; Lim S. C.; Kim Y. M.; Kang K. W. Aromatase induction in tamoxifen-resistant breast cancer: role of phosphoinositide 3-kinase-dependent CREB activation. Cancer Lett. 351:91–99; 2014. [DOI] [PubMed] [Google Scholar]
- 15. Steven A.; Heiduk M.; Recktenwald C. V.; Hiebl B.; Wickenhauser C.; Massa C.; Seliger B. Colorectal carcinogenesis: connecting K-RAS-induced transformation and CREB activity in vitro and in vivo. Mol. Cancer Res. 13:1248–1262; 2015. [DOI] [PubMed] [Google Scholar]
- 16. Cheng J. C.; Kinjo K.; Judelson D. R.; Chang J.; Wu W. S.; Schmid I.; Shankar D. B.; Kasahara N.; Stripecke R.; Bhatia R.; Landaw E. M.; Sakamoto K. M. CREB is a critical regulator of normal hematopoiesis and leukemogenesis. Blood 111:1182–1192; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cho E.-C.; Mitton B.; Sakamoto K. M. CREB and leukemogenesis. Crit. Rev. Oncog. 16:37–46; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pellegrini M.; Cheng J. C.; Voutila J.; Judelson D.; Taylor J.; Nelson S. F.; Sakamoto K. M. Expression profile of CREB knockdown in myeloid leukemia cells. BMC Cancer 8:264; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sandoval S.; Sakamoto K.; Deepa S. Acceleration of leukemogenesis in CREB transgenic mice by retroviral insertional mutagenesis. Blood 108:310B–310B; 2006. [Google Scholar]
- 20. van der Sligte N. E.; Kampen K. R.; ter Elst A.; Scherpen F. J. G.; Meeuwsen-de Boer T. G. J.; Guryev V.; van Leeuwen F. N.; Kornblau S. M.; de Bont E. Essential role for cyclic-AMP responsive element binding protein 1 (CREB) in the survival of acute lymphoblastic leukemia. Oncotarget 6:14970–14981; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drozdov I.; Svejda B.; Gustafsson B. I.; Mane S.; Pfragner R.; Kidd M.; Modlin I. M. Gene network inference and biochemical assessment delineates GPCR pathways and CREB targets in small intestinal neuroendocrine neoplasia. PLoS One 6:10; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fang M. G.; Pak M. L.; Chamberlain L.; Xing W.; Yu H. B.; Green M. R. The CREB coactivator CRTC2 is a lymphoma tumor suppressor that preserves genome integrity through transcription of DNA mismatch repair genes. Cell Rep. 11:1350–1357; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vermeulen K.; Van Bockstaele D. R.; Berneman Z. N. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 36:131–149; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McDonald E. R.; El-Deiry W. S. Cell cycle control as a basis for cancer drug development (review). Int. J. Oncol. 16:871–886; 2000. [PubMed] [Google Scholar]
- 25. Loden M.; Stighall M.; Nielsen H. N.; Roos G.; Emdin S. O.; Ostlund H.; Landberg G. The cyclin D1 high and cyclin E high subgroups of breast cancer: separate pathways in tumorogenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene 21:4680–4690; 2002. [DOI] [PubMed] [Google Scholar]
- 26. Wu Z.; Cho H.; Hampton G. M.; Theodorescu D. Cdc6 and cyclin E2 are PTEN-regulated genes associated with human prostate cancer metastasis. Neoplasia 11:66–76; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao S. P.; Yi M.; Yuan Y. Y.; Zhuang W. X.; Zhang D. C.; Yu X. Y.; Chen X. X.; Teng B. G.; Guan Z. Y.; Zhang Y. X. Expression of AKAP95, Cx43, cyclinE1 and cyclinD1 in esophageal cancer and their association with the clinical and pathological parameters. Int. J. Clin. Exp. Med. 8:7324–7332; 2015. [PMC free article] [PubMed] [Google Scholar]
- 28. Malinkova V.; Vylicil J.; Krystof V. Cyclin-dependent kinase inhibitors for cancer therapy: a patent review (2009–2014). Expert Opin. Ther. Pat. 25:953–970; 2015. [DOI] [PubMed] [Google Scholar]
- 29. Bai M.; Tsanou E.; Agnantis N. J.; Kamina S.; Grepi C.; Stefanaki K.; Rontogianni D.; Galani V.; Kanavaros P. Proliferation profile of classical Hodgkin’s lymphomas. Increased expression of the protein cyclin D2 in Hodgkin’s and Reed-Sternberg cells. Mod. Pathol. 17:1338–1345; 2004. [DOI] [PubMed] [Google Scholar]
- 30. Tzankov A.; Zimpfer A.; Went P.; Maurer R.; Pileri S. A.; Geley S.; Dirnhofer S. Aberrant expression of cell cycle regulators in Hodgkin and Reed-Sternberg cells of classical Hodgkin’s lymphoma. Mod. Pathol. 18:90–96; 2005. [DOI] [PubMed] [Google Scholar]
- 31. Bai M.; Papoudou-Bai A.; Kitsoulis P.; Horianopoulos N.; Kamina S.; Agnantis N. J.; Kanavaros P. Cell cycle and apoptosis deregulation in classical Hodgkin lymphomas. In Vivo 19:439–453; 2005. [PubMed] [Google Scholar]
- 32. Chang K. C.; Chang Y.; Jones D.; Su I. J. Aberrant expression of cyclin A correlates with morphogenesis of Reed-Sternberg cells in Hodgkin lymphoma. Am. J. Clin. Pathol. 132:50–59; 2009. [DOI] [PubMed] [Google Scholar]
- 33. Foell J. L.; Max D.; Giersberg C.; Korholz D.; Staege M. S. Sensitivity of Hodgkin’s lymphoma cell lines to the cell cycle inhibitor roscovitine. Anticancer Res. 28:887–894; 2008. [PubMed] [Google Scholar]
- 34. Wang P.; Huang S. S.; Wang F.; Ren Y.; Hehir M.; Wang X.; Cai J. Cyclic AMP-response element regulated cell cycle arrests in cancer cells. PLoS One 8:7; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Melnikova V. O.; Dobroff A. S.; Zigler M.; Villares G. J.; Braeuer R. R.; Wang H.; Huang L.; Bar-Eli M. CREB inhibits AP-2 alpha expression to regulate the malignant phenotype of melanoma. PLoS One 5:11; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]