Abstract
Cullin 4B (CUL4B), a scaffold protein that assembles CRL4B ubiquitin ligase complexes, was found to be overexpressed in many types of tumors. However, the expression pattern and role of CUL4B in non-small cell lung cancer (NSCLC) remain largely unknown. Therefore, in the present study, we investigated the role of CUL4B in NSCLC, and the underlying mechanism was also explored. Our results showed that CUL4B was highly expressed in NSCLC cell lines. Silencing CUL4B obviously inhibited proliferation and migration/invasion of NSCLC cells, and it also suppressed the epithelial–mesenchymal transition (EMT) progress in NSCLC cells. Furthermore, knockdown of CUL4B significantly inhibited the expression of β-catenin, cyclin D1, and c-Myc in NSCLC cells. Taken together, these results suggest that knockdown of CUL4B inhibited the proliferation and invasion through suppressing the Wnt/β-catenin signaling pathway in NSCLC cells. Therefore, CUL4B may represent a novel therapeutic target for the treatment of NSCLC.
Key words: Cullin 4B (CUL4B), Non-small cell lung cancer (NSCLC), Invasion, Wnt/β-catenin pathway
INTRODUCTION
Lung cancer is one of the leading causes of cancer deaths in the world. More than 80% of cases are classified as non-small cell lung cancer (NSCLC) (1). In recent years, despite great progress in clinical diagnosis and treatment, patients with NSCLC have a 5-year survival rate of less than 15% (2–5). Thus, understanding the molecular mechanisms underlying NSCLC development and progression is of significance in the development of therapeutic strategies for NSCLC patients.
Cullin–RING ligases (CRLs) are multisubunit complexes composed of a cullin, a RING protein, and a substrate recognition subunit and play an important role in regulating diverse cellular processes such as cell cycle progression, transcription, and signal transduction (6–9). Cullin 4B (CUL4B), a scaffold protein in the cullin 4B–RING E3 ligase complex (CRL4B), was found to be overexpressed in most human cancers, including colorectal, cervical, and hepatocellular carcinomas (10–12). Several years ago, Jiang et al. reported that high CUL4B expression was significantly associated with the depth of colon cancer invasion, lymph node metastasis, distant metastasis, histological differentiation, vascular invasion, and advanced tumor stage (13). Moreover, knockdown of CUL4B reduced the proliferation, colony formation, and invasiveness of hepatocellular carcinoma cells in vitro and inhibited tumor growth in vivo (14). However, the biological function of CUL4B in NSCLC has not yet been well documented. In this study, we investigate the role of CUL4B in NSCLC. Our data showed that expression of CUL4B was significantly reduced in NSCLC cell lines, and knockdown of CUL4B markedly suppressed the proliferation and migration/invasion in NSCLC cells.
MATERIALS AND METHODS
Cell Culture
NSCLC cell lines including A549, H157, H1299, and H1650 as well as the human lung bronchus epithelial cell line BEAS-2B were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). They were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA), 100 U/ml penicillin, and 100 µg/ml streptomycin in a 5% CO2 humidified atmosphere at 37°C.
RNA Interference and Transfection
The specific small interfering RNA (siRNA) against human CUL4B and its negative control were synthesized by Invitrogen. For transfection, NSCLC cells were seeded in 24-well plates at a density of 5 × 104 per well and incubated at 37°C in a humidified incubator containing 5% CO2. The next day, when these cells were about 80% confluent, cells were transfected with siRNA-CUL4B or scramble-CUL4B using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from cells using TRIzol (Invitrogen) according to the manufacturer’s instructions. Approximately 5 µg of total RNA for each sample was reverse transcribed into first-strand cDNA for RT-polymerase chain reaction (PCR) analysis using a High-Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City, CA, USA). The primer sequences used for PCR amplification were as follows: CUL4B, 5′-CCTGGAGTTTGTAGGGTTTGAT-3′ (forward) and 5′-GAGACGGTGGTAGAAGATTTGG-3′ (reverse); β-actin, 5′-TCATGAAGTGTGACGTTGACATCCGT-3′ (forward) and 5′-CCTAGAAGCATTTGCGGTGCACGATG-3′ (reverse). Subsequently, the differential expression of these genes was analyzed using the 2−ΔΔCt method.
Western Blot Analysis
Cells were homogenized and lysed using radioimmunoprecipitation assay lysis buffer (100 mM NaCl; 50 mM Tris-HCl, pH 7.5; 1% Triton X-100; 1 mM EDTA; 10 mM β-glycerophosphate; 2 mM sodium vanadate; and protease inhibitor). Protein concentrations were detected by using the Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions. Forty micrograms of protein was separated by 10% SDS-PAGE and transferred onto a nitrocellulose membrane using a commercial semidry blotting apparatus (Bio-Rad, Richmond, CA, USA). The immunoblot was incubated for 1 h with blocking solution (5% skim milk) at room temperature. Next, the membranes were incubated overnight at 4°C with the following primary antibodies: anti-CUL4B (1:500 dilution), anti-E-cadherin (1:500 dilution), anti-N-cadherin (1:1,000 dilution), anti-vimentin (1:500 dilution), anti-β-catenin (1:1,000 dilution), anti-c-Myc (1:500 dilution), and anti-cyclin D1 (1:200 dilution) (all Santa Cruz Biotechnology, Santa Cruz, CA, USA). After washing with TBS-T, the membranes were incubated at room temperature for 1 h with a 1:2,000 dilution of horseradish peroxidase-conjugated immunoglobulin G (IgG) (Santa Cruz Biotechnology). Target protein was detected by an enhanced chemiluminescence substrate kit (Pierce Biotechnology, Rockford, IL, USA).
Cell Proliferation Assay
Cell proliferation was detected using a CCK-8 assay according to the manufacturer’s instructions. In brief, infected cells were harvested and reseeded in 96-well plates (1 × 104 per well) and cultured for 1–3 days, respectively. The absorbance was subsequently measured on a Bio-Rad Microplate Reader (Bio-Rad, Hercules, CA, USA) using a test wavelength of 490 nm.
In Vitro Migration and Invasion Assay
To measure changes in cell migration, we performed the Transwell migration assay using a 24-well Boyden chamber (BD Biosciences). In brief, 1 × 104 cells were seeded into the upper well in DMEM (without serum), while the lower compartment was filled with 500 µl of RPMI-1640 supplemented with 10% FBS. After incubation at 37°C for 24 h, the cells on the lower surface of the membrane were fixed, stained, and then counted under a light microscope. The average number of migrated cells from five random optical fields (100× magnification) and triplicate filters was determined. The invasion assay was done by the same procedure, except that the inserts of the chambers were coated with Matrigel.
Statistical Analysis
All statistical analyses were performed using the SPSS13.0 software. Data were expressed as mean ± SD from at least three independent experiments. Statistical comparisons between groups were performed using a one-way analysis of variance followed by a Student’s t-test. A value of p < 0.05 was considered statistically significant.
RESULTS
CUL4B Is Overexpressed in Human NSCLC Lines
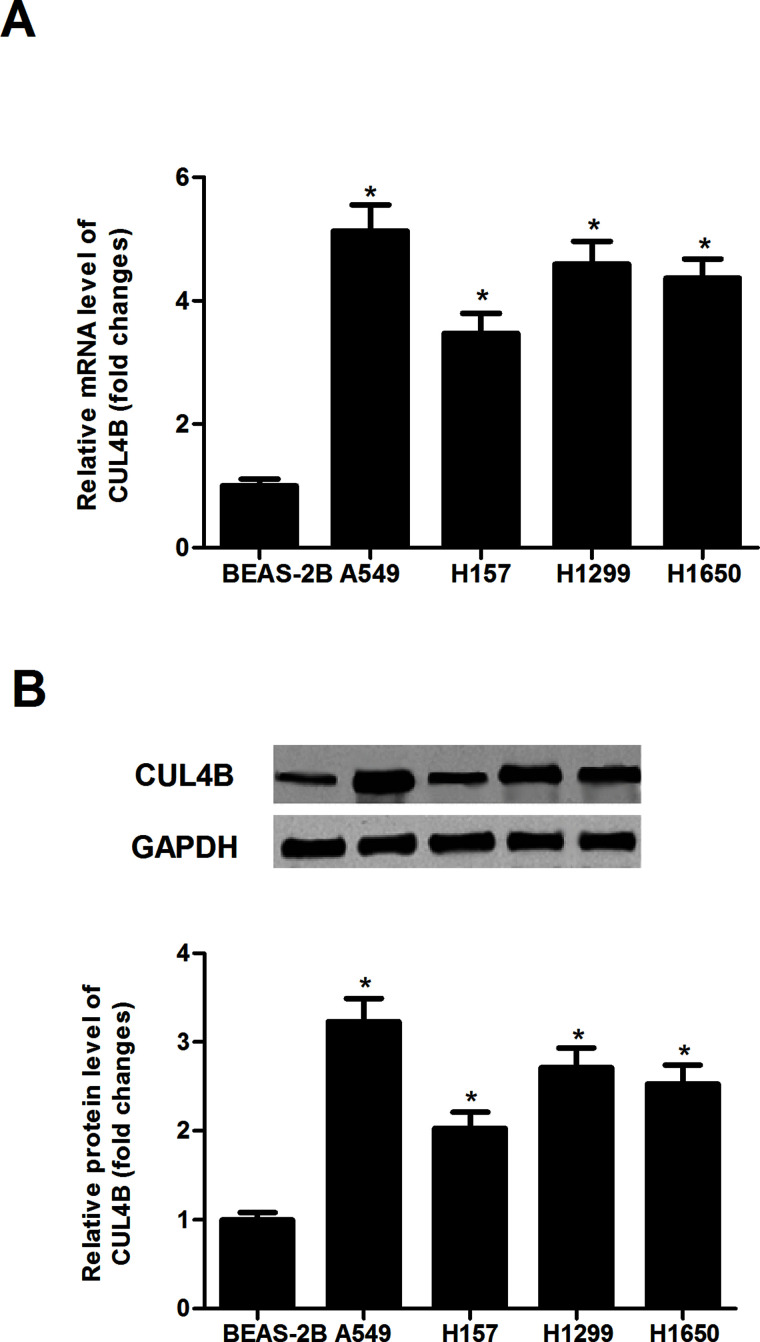
To determine the possible role of CUL4B in human NSCLC, we detected the expression of CUL4B in four different NSCLC cell lines using qRT-PCR. We observed that the mRNA expression of CUL4B in NSCLC cell lines was much higher in comparison with that in the normal lung cell line (Fig. 1A). We further investigated the protein expression of CUL4B in these cells by Western blotting, and the results showed that the protein level of CUL4B was also higher in the NSCLC cell lines, compared with those of the normal lung cell line (Fig. 1B).
Figure 1.
CUL4B is overexpressed in human NSCLC lines. The CUL4B mRNA and protein levels were detected by quantitative real-time polymerase chain reaction (qRT-PCR) (A) and Western blot assays (B) in four different NSCLC cell lines and the normal lung cell line. Data are mean ± SD values from three experiments, each performed in triplicate. *p < 0.05 versus BEAS-2B cells.
Knockdown of CUL4B Inhibits NSCLC Cell Proliferation
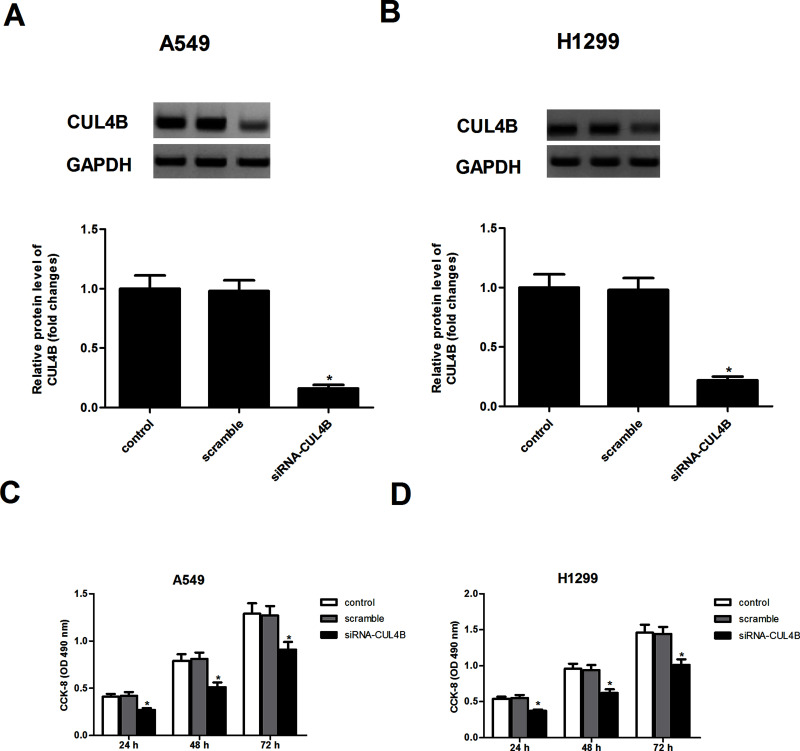
To explore the role of CUL4B in NSCLC, A549 and H1299 cells stably expressing siRNA-CUL4B were generated, respectively. Western blot analysis confirmed the low expression of CUL4B in A549 (Fig. 2A) and H1299 (Fig. 2B) cells, respectively, as compared to controls. CCK-8 proliferation assay showed that knockdown of CUL4B obviously decreased the proliferation of A549 (Fig. 2C) and H1299 (Fig. 2D) cells, respectively, when compared with respective controls.
Figure 2.
Knockdown of CUL4B inhibits NSCLC cell proliferation. (A, B) A549 and H1299 cells were transfected with siRNA-CUL4B or scramble for 24 h, respectively. The transfection efficiency was determined by Western blot assay. (C, D) Cell proliferation was detected by the CCK-8 assay. Data are mean ± SD values from three experiments, each performed in triplicate. *p < 0.05 versus scramble.
Knockdown of CUL4B Inhibits NSCLC Cell Migration and Invasion
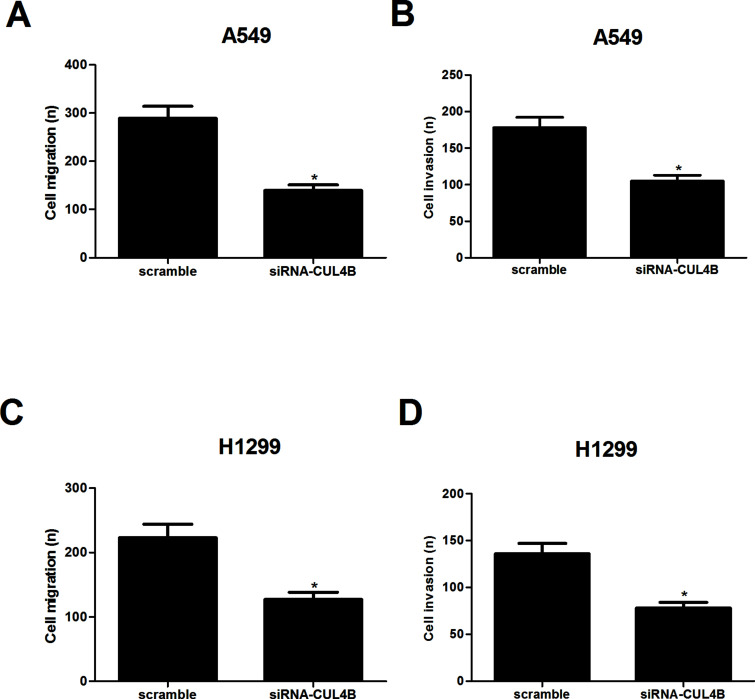
We then investigated the effects of CUL4B on NSCLC cell migration using the Boyden chamber Transwell without Matrigel. The results demonstrated that A549 cells derived from siRNA-CUL4B transfectants displayed a lower ability for migration, compared with those derived from scramble-CUL4B-transfected cells (Fig. 3A). In addition, we examined the role of CUL4B in NSCLC cell invasion using Transwell chamber with Matrigel. As illustrated in Figure 3B, knockdown of CUL4B obviously suppressed the number of A549 cells that invaded through Matrigel, as compared with the scramble group. Similar results were observed in H1299 cells (Fig. 3C and D).
Figure 3.
Knockdown of CUL4B inhibits NSCLC cell migration and invasion. A549 and H1299 cells were transfected with siRNA-CUL4B or scramble for 24 h, respectively. (A, C) Cell migration was evaluated by a Transwell assay. (B, D) Cell invasion was evaluated by a Matrigel-coated Transwell assay. Data are mean ± SD values from three experiments, each performed in triplicate. *p < 0.05 versus scramble.
Knockdown of CUL4B Inhibits the EMT Progress in NSCLC Cells
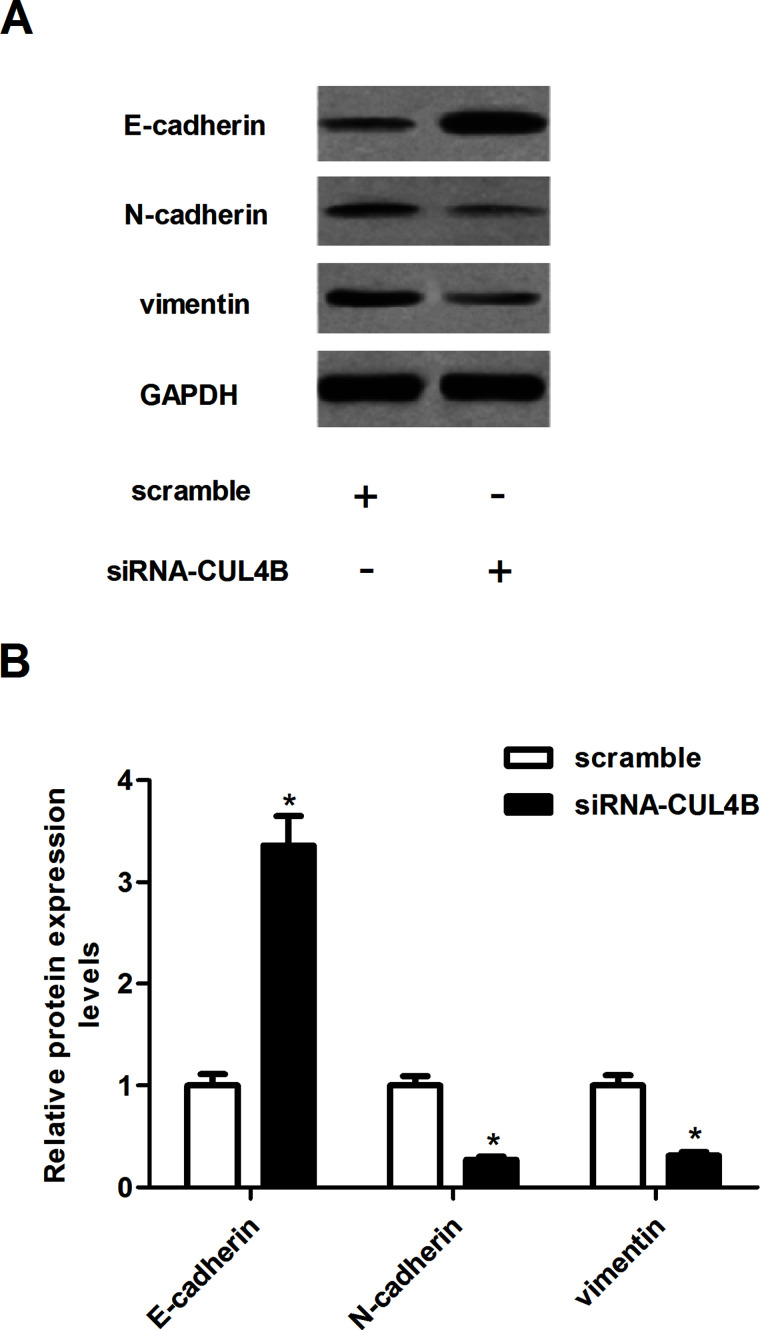
We next asked whether siRNA-CUL4B could influence the EMT process in NSCLC cells. As shown in Figure 4, the epithelial marker E-cadherin was significantly increased in A549 cells transfected with siRNA-CUL4B. Meanwhile, knockdown of CUL4B obviously reduced N-cadherin and vimentin protein levels, compared to the scramble group (Fig. 4A). Quantification analysis of E-cadherin, N-cadherin, and vimentin is shown in Figure 4B.
Figure 4.
Knockdown of CUL4B inhibits the epithelial–mesenchymal transition (EMT) progress in NSCLC cells. A549 cells were transfected with siRNA-CUL4B or scramble for 24 h. (A) The protein expression of E-cadherin, N-cadherin, and vimentin was analyzed using Western blotting. (B) Quantification analysis was performed using Gel-Pro Analyzer version 4.0 software. Data are mean ± SD values from three experiments, each performed in triplicate. *p < 0.05 versus scramble.
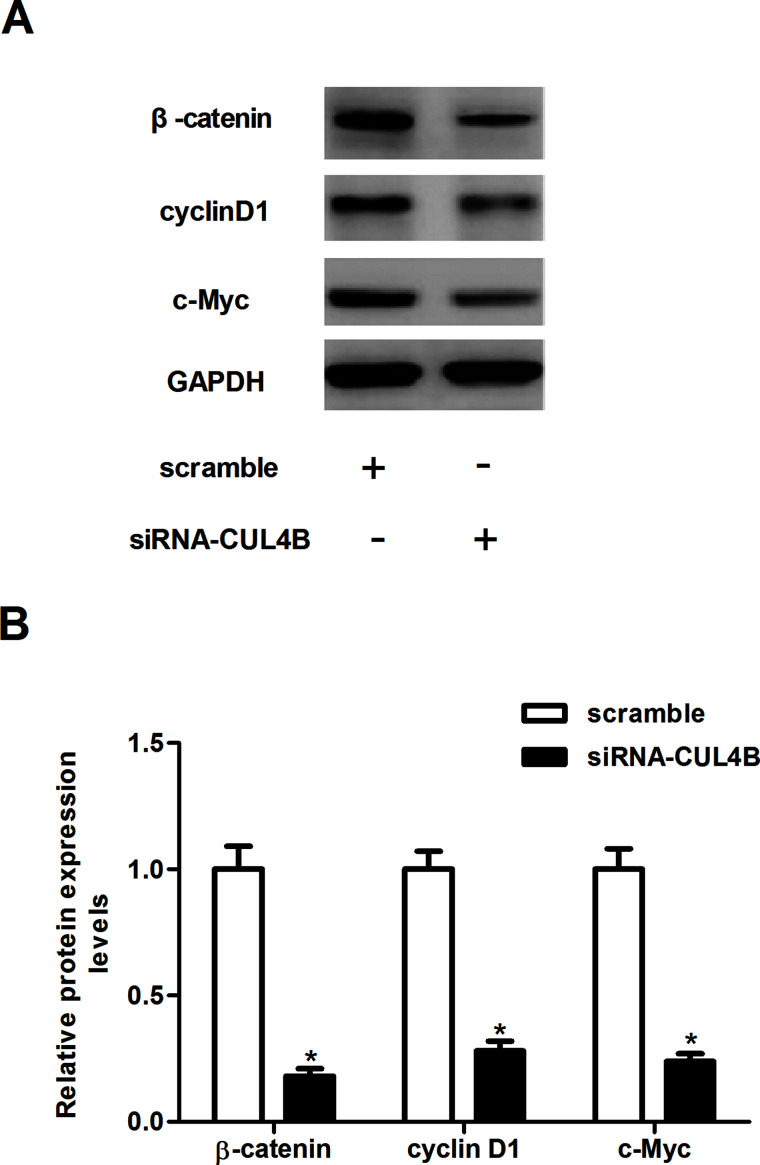
Knockdown of CUL4B Inhibits the Activation of Wnt/β-Catenin Pathway in NSCLC Cells
The Wnt/β-catenin pathway plays a critical role in tumor initiation, progression, and metastasis. Thus, we examined the effects of siRNA-CUL4B on the key components of the Wnt signaling pathway in NSCLC cells. As shown in the Western blot analysis in Figure 5A, knockdown of CUL4B significantly suppressed the protein levels of β-catenin, cyclin D1, and c-Myc in A549 cells, as compared with the scramble group. Quantification analysis of β-catenin, cyclin D1, and c-Myc is shown in Figure 5B.
Figure 5.
Knockdown of CUL4B inhibits the activation of Wnt/β-catenin pathway in NSCLC cells. A549 cells were transfected with siRNA-CUL4B or scramble for 24 h. (A) The protein expression of β-catenin, cyclin D1, and c-Myc was analyzed using Western blotting. (B) Quantification analysis was performed using Gel-Pro Analyzer version 4.0 software. Data are mean ± SD values from three experiments, each performed in triplicate. *p < 0.05 versus scramble.
DISCUSSION
In this study we found that CUL4B expression was highly expressed in human NSCLC cells. In addition, knockdown of CUL4B inhibited proliferation and invasion, as well as suppressed the EMT process in NSCLC cells. Furthermore, knockdown of CUL4B inhibited the expression of β-catenin, cyclin D1, and c-Myc in NSCLC cells.
Recent studies have shown that CUL4B is upregulated in various tumors and that this expression is correlated with tumor growth. For example, CUL4B expression is significantly upregulated in glioblastoma cells, and knockdown of CUL4B via shRNA-delivering lentiviruses obviously decreased cell proliferation and colony formation, as well as alleviated in vivo tumorigenesis in glioma xenograft nude mice (15). Another study demonstrated that CUL4B is highly expressed in human osteosarcoma cells, and silencing of CUL4B gene expression can effectively inhibit osteosarcoma cell proliferation (16). Consistent with the previous reports, in this study we found that CUL4B was highly expressed in human NSCLC cells. Moreover, knockdown of CUL4B inhibited the proliferation of NSCLC cells. These data suggest that CUL4B may function as an oncogene involved in the development and progression of NSCLC.
EMT is considered as one of the critical steps in tumor metastasis and invasion (17,18). Generally, increased motility and invasion are positively correlated with EMT, which is characterized by repression of epithelial markers and induction of mesenchymal markers, and reduction or a loss of E-cadherin expression is one of the well-established hallmarks of EMT (19). In this study we observed that knockdown of CUL4B significantly inhibited the migration/invasion and EMT process in NSCLC cells. Overall, our results suggest that CUL4B regulates EMT and consequently affects cell migration and invasion in vitro.
The Wnt/β-catenin pathway is a major pathway involved in the malignant progression of various tumors, and regulates the proliferation, migration, and invasion of cancer cells (20–22). Wnt activation results in the stabilization of β-catenin and the regulation of the transcription of the downstream members of the pathway, including c-Myc, cyclin D1, and MMP7 (23). In NSCLC, aberrant Wnt signaling correlates with tumor recurrence and poor patient survival (24). β-Catenin expression is frequently lost in NSCLC, and the loss of β-catenin expression may be due to hypermethylation of the β-catenin promoter. Loss of β-catenin was reportedly correlated with lymph node metastasis, high TNM (tumor, node, and metastasis) stages in multiple cancers, and poor prognosis in NSCLC (25,26). Recently, Yuan et al. demonstrated that CUL4B and β-catenin were frequently upregulated and positively correlated in hepatocellular carcinoma tissues; CUL4B activated Wnt/β-catenin signaling by protecting β-catenin from GSK3-mediated degradation (14). In this study we found that knockdown of CUL4B inhibited the expression levels of β-catenin, cyclin D1, and c-Myc in NSCLC cells. These data support the notion that knockdown of CUL4B inhibits NSCLC cell proliferation and invasion through suppressing the Wnt/β-catenin signaling pathway.
In conclusion, our findings show that knockdown of CUL4B efficiently inhibited NSCLC cell proliferation and invasion in vitro, perhaps via suppressing the Wnt/β-catenin signaling pathway. Therefore, the results of our study indicate the potential of CUL4B as a new epigenetic target for the treatment of NSCLC.
ACKNOWLEDGMENTS
This study was supported by the Crossing Research Projects of Zhengzhou University (Grant No. 340700532007).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Siegel R.; Naishadham D.; Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 63:11–30; 2013. [DOI] [PubMed] [Google Scholar]
- 2. Yang P. Epidemiology of lung cancer prognosis: Quantity and quality of life. Cancer Epidemiol. 471:469–486; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anglim P. P.; Alonzo T. A.; Laird-Offringa I. A. DNA methylation-based biomarkers for early detection of non-small cell lung cancer: An update. Mol. Cancer 7:81; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer 71:3–10; 2011. [DOI] [PubMed] [Google Scholar]
- 5. Maione P.; Gridelli C.; Troiani T.; Ciardiello F. Combining targeted therapies and drugs with multiple targets in the treatment of NSCLC. Oncologist 11:274–284; 2006. [DOI] [PubMed] [Google Scholar]
- 6. Hotton S. K.; Callis J. Regulation of cullin ring ligases. Annu. Rev. Plant Biol. 59:467–489; 2008. [DOI] [PubMed] [Google Scholar]
- 7. Bosu D. R.; Kipreos E. T. Cullin-ring ubiquitin ligases: Global regulation and activation cycles. Cell Div. 3:7; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duda D. M.; Scott D. C.; Calabrese M. F.; Zimmerman E. S.; Zheng N.; Schulman B. A. Structural regulation of cullin-ring ubiquitin ligase complexes. Curr. Opin. Struc. Biol. 21:257–264; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petroski M. D.; Deshaies R. J. Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 6:9–20; 2005. [DOI] [PubMed] [Google Scholar]
- 10. Song B.; Zhan H.; Bian Q.; Li J. Knockdown of CUL4B inhibits proliferation and promotes apoptosis of colorectal cancer cells through suppressing the Wnt/β-catenin signaling pathway. Int. J. Clin. Exp. Pathol. 8:10394–10402; 2015. [PMC free article] [PubMed] [Google Scholar]
- 11. Yang Y.; Liu R.; Qiu R.; Zheng Y.; Huang W.; Hu H.; Ji Q.; He H.; Shang Y., Gong Y. CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene 34:104–118; 2015. [DOI] [PubMed] [Google Scholar]
- 12. Mok M. T.; Cheng A. S. CUL4B: A novel epigenetic driver in wnt/β-catenin-dependent hepatocarcinogenesis. J. Pathol. 236:1–4; 2015. [DOI] [PubMed] [Google Scholar]
- 13. Jiang T.; Tang H.-m.; Wu Z.-h.; Chen J.; Lu S.; Zhou C.-z.; Yan D.-w.; Peng Z.-h. Cullin 4b is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Med. Oncol. 30:1–8; 2013. [DOI] [PubMed] [Google Scholar]
- 14. Yuan J.; Han B.; Hu H.; Qian Y.; Liu Z.; Wei Z.; Liang X.; Jiang B.; Shao C.; Gong Y. CUL4B activates Wnt/β-catenin signalling in hepatocellular carcinoma by repressing Wnt antagonists. J. Pathol. 235:784–795; 2015. [DOI] [PubMed] [Google Scholar]
- 15. Dong J.; Wang X.; Yao J.; Li G.; Li X. Decreased CUL4B expression inhibits malignant proliferation of glioma in vitro and in vivo. Eur. Rev. Med. Pharm. Sci. 19:1013–1021; 2015. [PubMed] [Google Scholar]
- 16. Chen Z.; Shen B.-L.; Fu Q.-G.; Wang F.; Tang Y. X.; Hou C.-L.; Chen L. CUL4B promotes proliferation and inhibits apoptosis of human osteosarcoma cells. Oncol. Rep. 32:2047–2053; 2014. [DOI] [PubMed] [Google Scholar]
- 17. Yilmaz M.; Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metast. Rev. 28:15–33; 2009. [DOI] [PubMed] [Google Scholar]
- 18. Kang Y.; Massagué J. Epithelial-mesenchymal transitions: Twist in development and metastasis. Cell 118:277–279; 2004. [DOI] [PubMed] [Google Scholar]
- 19. Huber M. A.; Kraut N.; Beug H. Molecular requirements for epithelial–mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 17:548–558; 2005. [DOI] [PubMed] [Google Scholar]
- 20. Uematsu K.; He B.; You L.; Xu Z.; McCormick F.; Jablons D. M. Activation of the Wnt pathway in non small cell lung cancer: Evidence of dishevelled overexpression. Oncogene 22:7218–7221; 2003. [DOI] [PubMed] [Google Scholar]
- 21. Mazieres J.; He B.; You L.; Xu Z.; Jablons D. M. Wnt signaling in lung cancer. Cancer Lett. 222:1–10; 2005. [DOI] [PubMed] [Google Scholar]
- 22. Tennis M.; Van Scoyk M.; Winn R. A. Role of the Wnt signaling pathway and lung cancer. J. Thoracic Oncol. 2:889–892; 2007. [DOI] [PubMed] [Google Scholar]
- 23. Luu H. H.; Zhang R.; Haydon R. C.; Rayburn E.; Kang Q.; Si W.; Park J. K.; Wang H.; Peng Y.; Jiang W. Wnt/β-catenin signaling pathway as novel cancer drug targets. Curr. Cancer Drug Targets 4:653–671; 2004. [DOI] [PubMed] [Google Scholar]
- 24. Stewart D.J. Wnt signaling pathway in non–small cell lung cancer. J. Natl. Cancer Inst. 106(1):djt356; 2013. [DOI] [PubMed] [Google Scholar]
- 25. Miao Y.; Wang L.; Zhang X.; Xu X.; Jiang G.; Fan C.; Liu Y.; Lin X.; Yu J.; Zhang Y. Promoter methylationmediated silencing of b-catenin enhances invasiveness of non-small cell lung cancer and predicts adverse prognosis. PLoS One 9:e112258; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Retera J.; Leers M.; Sulzer M. A.; Theunissen P. The expression of beta-catenin in non-small-cell lung cancer: A clinicopathological study. J. Clin. Pathol. 51:891–894; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]