Abstract
BACKGROUND
Patients with PTEN hamartoma tumor syndrome (PHTS) have germline mutations in the tumor-suppressor gene encoding phosphatase and tensin homologue (PTEN). Such mutations have been associated with a hereditary predisposition to multiple types of cancer, including the Cowden syndrome. However, a majority of patients who have PHTS-related phenotypes have tested negative for PTEN mutations. In a previous study, we found that the E3 ubiquitin ligase WWP1 negatively regulates the function of PTEN.
METHODS
In a prospective cohort study conducted from 2005 through 2015, we enrolled 431 patients with wild-type PTEN who met at least the relaxed diagnostic criteria of the International Cowden Consortium. Patients were scanned for WWP1 germline variants. We used the Cancer Genome Atlas (TCGA) data set as representative of apparently sporadic cancers and the Exome Aggregation Consortium data set excluding TCGA (non-TCGA ExAC) and the noncancer Genome Aggregation Database (gnomAD) as representative of population controls without a reported cancer diagnosis. We established both in vitro and murine in vivo models to functionally characterize representative WWP1 variants.
RESULTS
The existence of germline WWP1 variants was first established in a family with wild-type PTEN who had oligopolyposis and early-onset colon cancers. A validation series indicated that WWP1 germline variants occurred in 5 of 126 unrelated patients (4%) with oligopolyposis as a predominant phenotype. Germline WWP1 variants, particularly the WWP1 K740N and N745S alleles, were enriched in patients who did not have PHTS but had prevalent sporadic cancers, including PTEN-related cancer types in TCGA (odds ratio, 1.5; 95% confidence interval, 1.1 to 2.1; P = 0.01). The prioritized WWP1 variants resulted in gain-of-function effects, which led to aberrant enzymatic activation with consequent PTEN inactivation, thereby triggering hyperactive growth-promoting PI3K signaling in cellular and murine models.
CONCLUSIONS
In this study involving patients with disorders resulting in a predisposition to the development of multiple malignant neoplasms without PTEN germline mutations, we confirmed the function of WWP1 as a cancer-susceptibility gene through direct aberrant regulation of the PTEN–PI3K signaling axis. (Funded by the National Institutes of Health and others.)
HEREDITARY CANCER SYNDROMES, which account for approximately 10% of all cancers, serve as powerful models for the practice of precision medicine.1 The identification of genes that are associated with seemingly rare cancer-predisposition syndromes often provides insights regarding aspects of normal development and pathways that can be targeted for the treatment of the more common sporadic forms of human cancers. One of these genes is PTEN (Online Mendelian Inheritance in Man database number, 601728), which encodes a dual-specificity lipid and protein phosphatase. This form of phosphatase has a classic tumor-suppressive function that is attributed largely to its ability to dampen the growth-promoting signaling cascade consisting of phosphatidylinositol 3-kinase (PI3K), AKT (also called protein kinase B), and mechanistic (previously called mammalian) target of rapamycin (mTOR).2,3
Regardless of the clinical diagnosis, patients who have germline PTEN mutations are described as having the PTEN hamartoma tumor syndrome (PHTS), which includes entities such as the Cowden syndrome, the Bannayan–Riley–Ruvalcaba syndrome (BRRS), the Proteus syndrome, and Proteus-like syndromes.4 Clinically, the identification of a germline PTEN mutation not only establishes a PHTS molecular diagnosis but also informs cancer risk assessment and PTEN-specific medical management of mutation-positive probands and family members.5,6 However, the fact that PTEN is mutated in only a subgroup of patients (approximately 25%)7 poses a clinical challenge and yet offers a working hypothesis that wild-type PTEN-related overgrowth and neoplasia syndromes have other causes.
WWP1 is an E3 ubiquitin ligase enzyme that has been shown to be overexpressed or amplified in multiple types of cancer.8–13 We recently found that WWP1 activation in animal and in vitro models inhibited PTEN function, which led to protumorigenic phenotypes.14 In the study described here, we sought to determine and characterize whether human WWP1 could be a broad cancer predisposition gene in neoplasia syndromes without PTEN mutations.
Methods
STUDY PATIENTS AND OVERSIGHT
From 2005 through 2015, we prospectively enrolled patients with PHTS-related neoplasias from 66 sites, including academic medical centers, cancer centers, and community clinics associated with the International Cowden Consortium worldwide.5,6 The study, which was sponsored by the National Institutes of Health and other granting agencies (including foundations), was approved by the institutional review board at the Cleveland Clinic. All the patients provided written informed consent.
Eligible patients met at least the relaxed operational diagnostic criteria of the International Cowden Consortium.15 Relaxed criteria are defined as full criteria minus one factor, with such patients being described as having a Cowden-like syndrome. (Details regarding these criteria are provided in the Methods section in Supplementary Appendix 1, available with the full text of this article at NEJM.org.) The Cowden syndrome is characterized by an elevated lifetime risk of a specific set of cancers, which are collectively called Cowden component cancers.5 All the patients underwent analysis for the presence of mutations and deletions in PTEN and SDHx (including SDHB, SDHC, and SDHD, which encode succinate dehydrogenase).6 Only patients without pathogenic germline mutations in PTEN or SDHx were included in the study (Table S1 in Supplementary Appendix 1).
GENETIC STUDIES
For the discovery series of patients, we performed whole-exome sequencing of peripheral-blood DNA obtained from 83 probands, as described in detail in Supplementary Appendix 1. We performed Sanger sequencing using region-specific mutational analysis based on polymerase-chain-reaction assays to validate variants that had been prioritized through whole-exome sequencing. For the extended validation series of patients, we performed high-resolution melting analysis to scan for WWP1 variants in an additional 348 patients. Suspected variants were validated with the use of Sanger sequencing. To compare frequencies among samples obtained from patients with various disorders and apparently healthy populations, we used the Cancer Genome Atlas (TCGA) data set as representative of apparently sporadic cancers and the Exome Aggregation Consortium data set excluding TCGA (non-TCGA ExAC) and the noncancer Genome Aggregation Database (gnomAD) as representative population controls without a reported cancer diagnosis.
IN VITRO FUNCTIONAL STUDIES
We performed in vitro assays in human-derived, commercially available cell lines, including 293T (embryonic kidney) cells, HCT116 (colorectal carcinoma) cells, and DLD-1 (colorectal adenocarcinoma) cells. We also used CRISPR (clustered regularly interspaced short palindromic repeats) techniques to generate knock-in WWP1 wild-type and mutant mice and derived mouse embryonic fibroblast cells. Stable cell lines were generated either through lentiviral transduction or through CRISPR technology. We performed Western blot analysis (denaturing and native), immunoprecipitation, in situ ubiquitination assays, and soft agar colony-formation assays on wild-type and mutant cells.14
IN VIVO XENOGRAFT MURINE MODELING
For assaying tumor growth in the xenograft model, 7-week-old male FOXN nude mice housed in specific pathogen-free environments were injected subcutaneously with stable HCT116 or DLD-1 derivatives. The care and treatment of animals were approved by the animal research committee at Beth Israel Deaconess Medical Center. Detailed protocols regarding the in vitro and in vivo studies are provided in Supplementary Appendix 1.
STATISTICAL ANALYSIS
For the genetic analysis, we used OpenEpi software (Open Source Epidemiologic Statistics for Public Health) to calculate odds ratios for the presence of WWP1 variants (http://openepi.com/Menu/OE_Menu.htm). For analyses comparing various population groups, we used two-by-two tables to calculate odds ratios. We calculated 95% confidence intervals and corresponding P values using the mid-P exact test. An odds ratio of more than 1.0 implied that the incidence was greater in the population of interest than in the standard population. A P value of less than 0.05 was considered to indicate statistical significance.
RESULTS
IDENTIFICATION OF WWP1 AS A CANDIDATE GENE IN PTEN WILD-TYPE PROBANDS
The hypothesis that PTEN is mutated in only a subgroup of patients with PHTS-related neoplasias7 points toward other unidentified causes of cancer predisposition in patients with wild-type PTEN. To address this hypothesis, we evaluated WWP1, a HECT-type ubiquitin E3 ligase (one of three broad types of ubiquitin E3 ligases; HECT stands for homologous to the E6-AP carboxy terminus) that has recently been shown to reduce PTEN tumor-suppressive function through polyubiquitination-mediated inhibition of PTEN dimerization and membrane recruitment.14 If our hypothesis was correct, then the existence of germline WWP1 variants in patients with wild-type PTEN would imply impaired PTEN function, independent of PTEN mutational status.
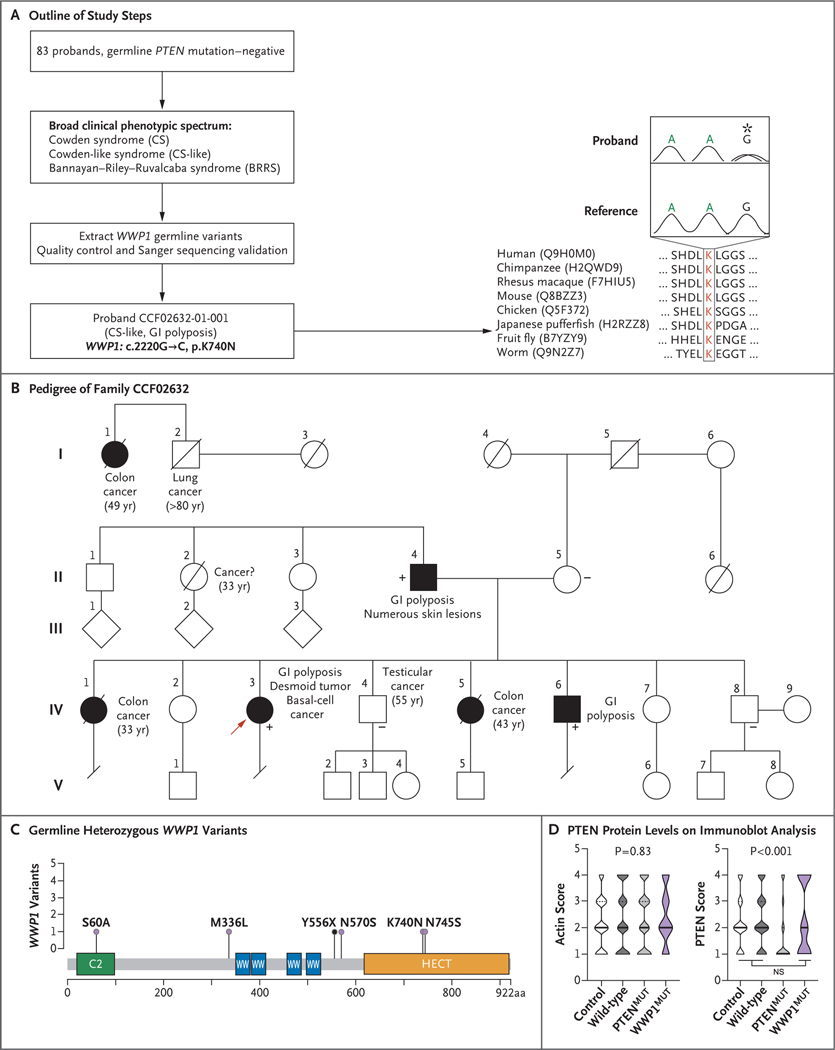
First, we investigated whole-exome sequencing data from 83 probands with wild-type PTEN who met at least the relaxed operational diagnostic criteria of the International Cowden Consortium (Table S1).15 After performing variant filtration, quality control, and Sanger sequencing validation, we initially identified one proband with a Cowden-like syndrome who had a germline heterozygous WWP1 c.2220G→C, p.K740N variant occurring at a highly evolutionarily conserved amino acid residue (Fig. 1A and Fig. S1). Patients with a Cowden-like syndrome do not meet the strict operational diagnostic criteria (i.e., minus one criterion); only 5% of these patients have germline PTEN mutations.16 The proband belonged to a family with a notable history of early-onset colon cancer and the occurrence of gastrointestinal oligopolyposis (Fig. 1B). The same germline WWP1 variant was found in two other affected family members (Family Members II-4 and IV-6) and was absent in three unaffected family members (Family Members II-5, IV-4, and IV-8), which indicated phenotype-dependent segregation (Figs. S2 and S3).
Figure 1. Identification of Germline Heterozygous WWP1 Variants in Cowden-like Syndrome.
Panel A provides an outline of the steps that were performed in the WWP1 discovery study. The missense heterozygous WWP1 variant shows high evolutionary conservation of the affected amino acid residue across seven different species besides human. UniProt accession codes are presented in parenthesis for each species, and the affected amino acid is shown in red in the boxed area. Above the sequence alignments is the Sanger sequencing chromatogram from proband CCF02632–01-001; the asterisk indicates the location of the mutation. GI denotes gastrointestinal, and K lysine. Panel B shows the pedigree of the CCF02632 family with the presence of wild-type WWP1 (−) or the WWP1 K740N variant (+). Male family members are indicated by squares, female family members by circles, affected members by solid figures, unaffected members by open figures, and deceased members by a slash. The proband (IV-3) for whom whole-exome sequencing was performed is indicated by a red arrow. Roman numerals on the left represent the generation number, and Arabic numerals represent consecutively numbered family members within each generation. The age at the time of the cancer diagnosis, when known, is indicated in parentheses. Panel C shows germline heterozygous WWP1 variants that were identified in five unrelated probands with Cowden-like syndrome; K740N denotes the variant identified in the CCF02632 family. The illustration represents WWP1 protein functional domains. Arabic numerals correspond to amino acid numbers. Variants are depicted in the lollipop plot overlying the WWP1 protein structure. The black lollipop indicates a truncating variant. The frequency of each variant correlates with the height of the vertical line representing each lollipop. C2 denotes C2-domain, HECT homologous to the E6-AP carboxyl terminus domain, and WW WWP repeating motif. Panel D shows violin plots indicating PTEN protein levels on immunoblot analysis. Protein lysates were extracted from lymphoblastoid cell lines derived from 26 population controls, from patients with oligopolyposis (121 with wild-type PTEN and WWP1 and 5 patients with WWP1 mutations), and from 107 patients with PTEN mutations. Protein levels are divided across quartiles, with 1 indicating the lowest level and 4 indicating the highest. Solid black horizontal bars indicate median protein scores; dashed bars indicate quartiles. Actin was used as a loading control. P values are derived from Kruskal–Wallis one-way testing of analysis of variance comparing all four groups. For the PTEN score (plot at right), unpaired two-tailed t-tests indicate a significant difference only for mutant PTEN as compared with the other three groups. NS denotes not significant. In contrast to PTEN protein levels in cells from germline PTEN mutation carriers, these levels were similar among the patients with WWP1 variants, those with oligopolyposis who had wild-type PTEN or WWP1, and controls.
VALIDATION SERIES
In patients with PTEN mutations who have undergone colonoscopy, more than 90% have colorectal polyps that are typically found in a mix of histologic subtypes.17 Patients with PHTS with colorectal cancer tend to have either preexisting or coexisting colonic mixed polyposis. Gastrointestinal hamartomatous polyposis is also one of the classic manifestations of BRRS.18 We therefore expanded our analyses to scan for WWP1 germline variants in 126 patients with oligopolyposis and in 123 patients with BRRS who had wild-type PTEN. We identified 5 more unrelated patients with oligopolyposis and underlying WWP1 germline variants (Fig. 1C, Fig. S4, and Table S2). Lymphoblastoid cell lines derived from patients with WWP1 variants showed normal PTEN protein levels that were similar to levels in patients with wild-type PTEN (Fig. 1D). This finding was consistent with a mechanism of action for WWP1 that was independent of PTEN levels.14 We did not identify additional WWP1 variants in patients with BRRS or in a series of patients who did not meet the selection criteria of polyposis or BRRS but who had a high PHTS-related phenotypic burden. Overall, we identified germline heterozygous WWP1 variants in 5 of 126 patients (4%) with a Cowden-like syndrome with wild-type PTEN who had gastrointestinal oligopolyposis as a predominant phenotype. Details regarding these analyses are provided in Supplementary Appendix 1.
APPARENTLY SPORADIC CANCERS
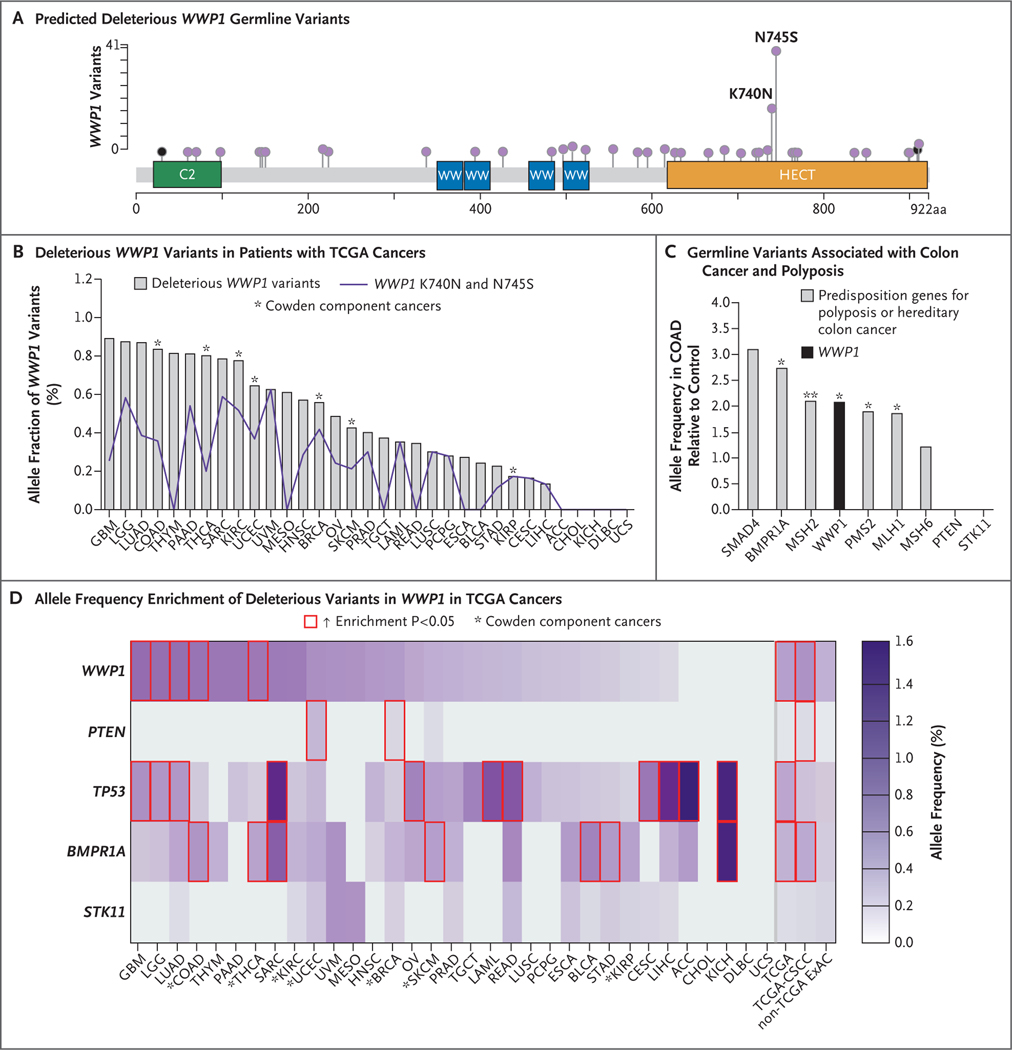
WWP1 is a proto-oncogene that has been shown to be somatically overexpressed or amplified in multiple cancer types.8–13 To determine whether WWP1 germline variants are overrepresented in the common apparently sporadic cancers, we analyzed the spectrum and frequencies of germline variants in 33 cancer types from TCGA, representing 10,389 cases (Table S3).19 We found predicted deleterious germline WWP1 variants in 28 of the 33 TCGA cancer types (Fig. 2A and 2B and Table S4 in Supplementary Appendix 2, which also includes Tables S5 through S9). The two most prevalent variants (c.2234A→G, p.N745S and c.2220G→C, p.K740N), which occurred in 22 of the 33 cancer types, accounted for 59 of 109 (54%) identified germline WWP1 variants (Fig. 2B and Table S4). These two variants occur in the catalytic C-terminal HECT domain of WWP1 and were also identified in two patients with Cowden-like syndrome who had oligopolyposis (Fig. 1C).
Figure 2. Enrichment of Germline WWP1 Variants in 33 Cancer Types from the Cancer Genome Atlas (TCGA).
Panel A shows the predicted deleterious germline WWP1 variants that have been identified in patients with 33 TCGA cancer types. Variants are depicted in the lollipop plot overlying the WWP1 protein structure. Black lollipops indicate truncating variants. The frequency of the variant correlates with the height of the vertical line representing each lollipop. Panel B shows the percentage of deleterious WWP1 variants identified in patients with TCGA cancers. The purple line depicts the frequency of WWP1 K740N and N745S variants. Asterisks indicate Cowden syndrome component cancers represented in TCGA. Panel C shows germline variants in known predisposition genes for colon cancer and polyposis identified in the TCGA colorectal adenocarcinoma (COAD) data set. Allele frequencies are normalized to background allele frequencies in the Exome Aggregation Consortium (ExAC) data set excluding TCGA cancers. One asterisk indicates P<0.05 and two asterisks P<0.01 for the comparison between the two databases, as calculated by one-tailed mid-P exact testing. Panel D shows a heat map of allele frequency enrichment of deleterious variants in WWP1 across all 33 TCGA cancer types, as compared with known cancer predisposition genes. The red outlines indicate that the cancer type has significant enrichment (P<0.05 by one-tailed mid-P exact testing) for patients with deleterious germline variants of a particular gene, as compared with non-TCGA ExAC population controls without cancer. The last three columns represent collective summaries for each gene. CSCC denotes Cowden syndrome component cancers.
Our findings indicate that rare germline WWP1 variants (minor allele frequency [MAF], ≤0.1%) exist in patients with a Cowden-like syndrome who have oligopolyposis and in those with common sporadic cancers. This finding prompted us to investigate the frequency of WWP1 variants in patients without a reported cancer diagnosis. To this end, we first analyzed the non-TCGA ExAC data set20 representing 53,105 population controls without prevalent cancer. Implementing filtration criteria that were identical to those used in the analysis of TCGA patients, we identified predicted deleterious WWP1 variants (MAF, ≤0.1%) in 428 of 106,210 alleles (Table S5 in Supplementary Appendix 2). In comparison, TCGA patients with apparently sporadic cancers showed deleterious WWP1 variants in 109 of 20,778 alleles (Table S4). These data indicate a significantly higher frequency of deleterious WWP1 variants in a cancer-affected population (TCGA) than in population controls without cancer (non-TCGA ExAC) (odds ratio, 1.3; 95% confidence interval [CI], 1.1 to 1.6; P = 0.02).
Although we observed the WWP1 variants K740N and N745S in non-TCGA ExAC, these variants remained overrepresented in TCGA patients with cancer (odds ratio, 1.6; 95% CI, 1.2 to 2.1; P = 0.003). We further substantiated these observations in the subgroup of patients with apparently sporadic Cowden syndrome component cancers, in whom we noted a significantly higher frequency of WWP1 variants than that in the ExAC population controls (odds ratio, 1.5; 95% CI, 1.1 to 2.1; P = 0.01), particularly WWP1 K740N and N745S (odds ratio for the presence of either allele, 1.9; 95% CI, 1.2 to 2.8; P = 0.006). Similarly, an analysis of the noncancer subgroup (233,473 alleles) in the expanded gnomAD database revealed deleterious WWP1 variants (MAF, ≤0.1%) in 637 alleles (Table S6 in Supplementary Appendix 2). Thus, similar to the patients in the ExAC non-TCGA listings, patients with cancer in the TCGA database had significantly higher frequencies of WWP1 variants than those without reported cancer in the gnomAD database (odds ratio, 1.9; 95% CI, 1.6 to 2.4; P<1×10−7). This observation remained consistent for patients in TCGA with Cowden component cancers (odds ratio, 2.3; 95% CI, 1.7 to 3.0; P = 3×10−6).
COMPARISON WITH CLASSIC CANCER-PREDISPOSITION GENES
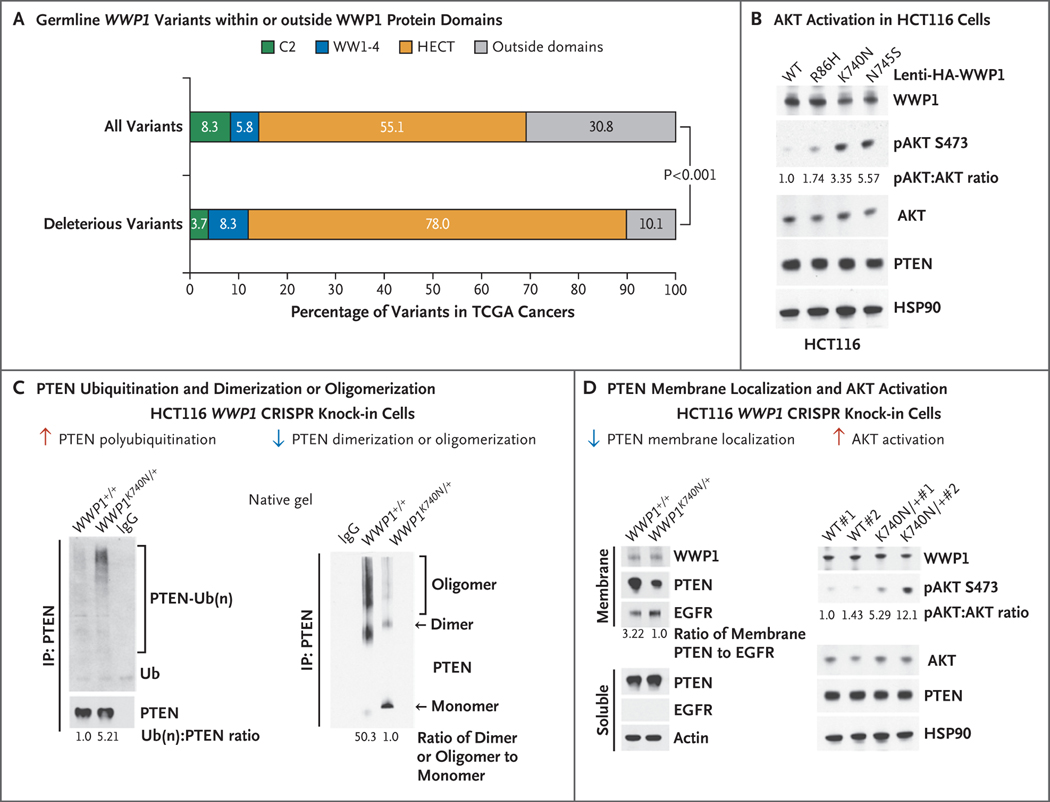
Next, we compared germline allele frequencies of WWP1 with those of genes that have been classically known to be associated with polyposis or a heredity predisposition to colon cancer, including SMAD4, BMPR1A, MSH2, PMS2, and others (Fig. 2C and Table S7 in Supplementary Appendix 2). An analysis of the colon adenocarcinoma subgroup in the TCGA database on the basis of its relevance to the predominant oligopolyposis phenotype revealed a similar and significant frequency of germline variants among WWP1 and the representative genes for polyposis and a genetic predisposition for colon cancer. Notably, even though PTEN is known to be associated with colon-cancer susceptibility, we did not identify germline PTEN mutations in this database. We then compared germline allele frequencies of WWP1 with those of classic cancer-susceptibility genes, including PTEN, TP53, BMPR1A, and STK11, across all 33 TCGA cancer types. We identified significantly higher frequencies of WWP1 variants in multiple cancer types, including two Cowden component cancers (colon adenocarcinoma and thyroid cancer), than in non-TCGA ExAC population controls5 (Fig. 2D and Tables S8 and S9 in Supplementary Appendix 2). As illustrative positive controls, we similarly identified higher frequencies of variants in other cancer-predisposition genes, including germline mutations in PTEN that have been associated with breast cancer and uterine cancer and in TP53 mutations that have been associated with adrenocortical carcinoma and sarcoma (Fig. 2D). Overall, these observations support a nonrandom enrichment of WWP1 germline variants among patients who have oligopolyposis, a hereditary predisposition for cancer, or particular malignant neoplasias. An analysis of all types of cancer in the study patients revealed that the majority of predicted deleterious germline WWP1 variants occurred within the HECT catalytic domain (odds ratio, 2.9; 95% CI, 1.7 to 5.1; P<0.001) (Fig. 3A), which led to our hypothesis that mutation-mediated catalytic hyperactivation of WWP1 could be the underlying mechanism for tumorigenesis.
Figure 3. Increase in PTEN Polyubiquitination, Membrane Dissociation, and Subsequent AKT Activation with WWP1 Gain-of-Function Mutations.
Panel A shows the distribution of germline WWP1 variants within or outside WWP1 protein domains. Amino acid boundaries of each structural domain were extracted from UniProt (WWP1_HUMAN, Q9H0M0) as follows: C2 (5–98), WW1 (349–382), WW2 (381–414), WW3 (456–489), WW4 (496–529), and HECT (588–922). Panel B shows the activation of AKT (also called protein kinase B) in colorectal carcinoma cells (HCT116), with stable expression of the indicated constructs. Total lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then probed with the indicated antibodies. Lenti-HA denotes lentiviral HA-tagged plasmids. Panel C shows an analysis of HCT116 cells with WWP1+/+ or CRISPR knock-in WWP1K740N/+ with respect to endogenous PTEN ubiquitination (blot at left) and dimerization and oligomerization (blot at right) by immunoprecipitation (IP) of PTEN, followed by Western blotting with the use of the indicated antibodies. Ub(n) denotes the number of ubiquitin chains. Panel D shows Western blot analysis of membrane and soluble fractions isolated from HCT116 cells with WWP1+/+ and WWP1K740N/+ with respect to PTEN membrane localization (blot at left) and AKT activation (blot at right). In the blot at left, epidermal growth factor receptor (EGFR) served as a membrane marker and actin as the internal control. In the blot at right, total lysates were resolved by SDS-PAGE and then probed with indicated antibodies.
FUNCTIONAL CHARACTERIZATION OF WWP1 VARIANTS
To address our hypothesis, we first analyzed both colon and kidney cell lines as relevant to the observed human phenotypes. We first assessed whether representative germline variants affect WWP1 catalytic activity toward PTEN polyubiquitination, based on the identification of the HECT-type E3 ubiquitin ligase WWP1 as a physical interactor that functionally triggers atypical nondegradative K27-linked polyubiquitination of PTEN to suppress its dimerization, membrane recruitment, and tumor suppressive function.14 We prioritized the three most frequently mutated WWP1 variants: R86H (C2 domain) along with K740N and N745S (both in the HECT domain) (Table S4). As proof of principle, we prioritized WWP1 R86H on the basis of frequency alone, even though the variant did not pass computational filters for deleteriousness. (Gain-of-function mutations are difficult to predict with computational filters, since functional effects can be understood only in the context of the particular biologic features of the gene.21) Overexpression of wild-type WWP1 in human embryonic kidney 293T (HEK293T) cells increased PTEN polyubiquitination, whereas the expression of individual WWP1 mutants (R86H, K740N, and N745S) further exacerbated this effect without affecting PTEN protein levels; these findings indicate that these mutants play a gain-of-function role in triggering PTEN polyubiquitination (Fig. S5A). The expression of WWP1 mutants suppressed PTEN dimerization or oligomerization in a process that was similar to wild-type WWP1 overexpression or amplification,14 as evaluated in native gel analyses (Fig. S5B). This expression in turn triggered downstream AKT activation in both HCT116 and DLD-1 colon cancer cells with endogenous wild-type PTEN (Fig. 3B and Fig. S5C).
Next, to study the aberrant PTEN regulation by WWP1 mutants endogenously, we used CRISPR technology to generate endogenous knock-in WWP1K740N/+ HCT116 cells, which reproduced the mutant heterozygous genotype observed in human patients (Fig. S6). In vivo ubiquitination assays revealed that endogenous K740N knock-in WWP1 mutant protein robustly elevated PTEN polyubiquitination, which in turn inhibited PTEN dimerization or oligomerization (Fig. 3C). In support of our hypothesis, WWP1 K740N knock-in suppressed PTEN membrane localization and in turn promoted AKT activation, as evaluated by cell fractionation and Western blotting analyses (Fig. 3D). To further mimic physiologic conditions (i.e., the constitutional heterozygous mutant state), we also generated CRISPR knock-in K736N mutant mice (corresponding to the K740N variant in humans) and further isolated primary mouse embryonic fibroblasts. We specifically prioritized the WWP1 K740N variant to generate the knock-in mice since it represents the variant that is segregated in our family with Cowden-like syndrome (Fig. 1B) and is one of the most frequently mutated WWP1 variants in apparently sporadic cancers (Table S4). Western blot analyses of protein lysates derived from mouse embryonic fibroblasts with the Wwp1K736N/+ genotype showed stronger activation of the PI3K–AKT–mTOR pathway than mouse embryonic fibroblasts with the wild-type Wwp1+/+ genotype, as revealed by higher levels of phosphorylation of AKT and ribosomal protein S6 (Fig. S7).
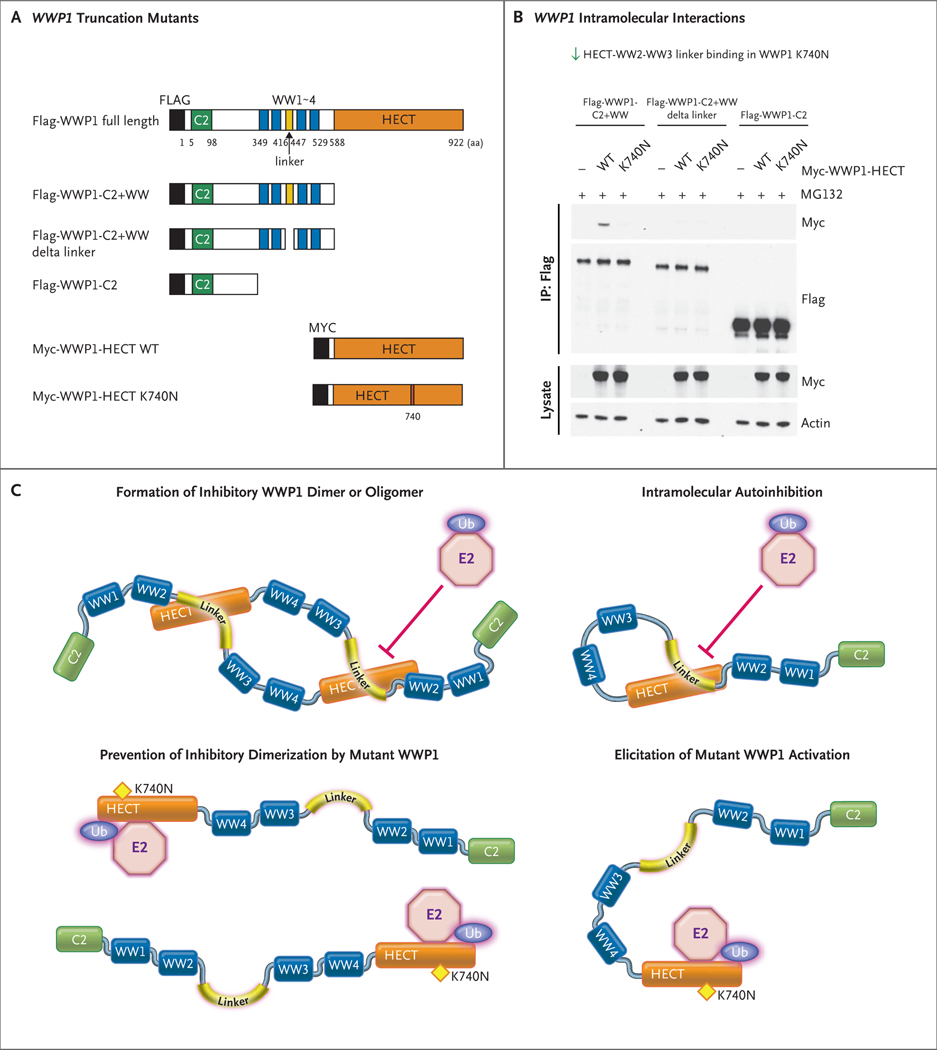
WWP1, which belongs to the NEDD4 family of ubiquitin E3 ligases, contains a C2 domain and four WW domains, along with the catalytic HECT domain. Previous studies have shown that members of the NEDD4 family of E3 ubiquitin ligases are characterized by an autoinhibitory regulatory mechanism that is mediated by interactions between its different domains,22 such as the interaction between the HECT domain and the WW2–WW3 linker region in WWP2 (another E3 ubiquitin ligase closely related to WWP1)23 and interaction between the C2 domain and the HECT domain in SMURF2 or SMURF1.24–26 Chen et al. found that deletion of the WW2–WW3 linker activated full-length WWP1.23 Therefore, one possibility is that the K740N mutation within the HECT domain disrupts the autoinhibitory interaction of this domain with the WW2–WW3 linker, which results in uncontrolled activation and gain-of-function effects. We found that the wild-type WWP1 HECT domain interacts with the N-terminal region of WWP1 (including the C2–WW1–4 domains), whereas the K740N mutation within the HECT domain decreased its binding with the N-terminal domain (Fig. 4A and 4B). Moreover, the deletion of the WW2–WW3 linker or inclusion of the C2 domain alone robustly decreased the respective interactions with the HECT domain, which corroborates the essential role of the WW2–WW3 linker and the dispensable role of the C2 domain in mediating these interactions (Fig. 4A and 4B). Functionally, as compared with wild-type WWP1, deletion of the WW2–WW3 linker triggered WWP1 catalytic activity, as revealed by its autoubiquitination and AKT activation (Fig. S8A and S8B). Native gel analyses revealed that the K740N mutation of WWP1 suppressed its ability to dimerize and oligomerize (Fig. S8C). Hence, several lines of evidence suggest that the WWP1 K740N mutation may promote a gain of catalytic activity by relieving the autoinhibitory physical interaction between its HECT domain and the WW2–WW3 linker region (Fig. 4C).
Figure 4. Promotion of WWP1 Catalytic Activity by WWP1 K740N through Disruption of Interaction between HECT and WW2–WW3 Linker Domains.
Panel A shows the truncation mutants of WWP1 with or without the K740N mutation within the HECT domain. Both Flag and Myc are short peptides used to tag proteins. Panel B shows an immunoprecipitation analysis of the interaction between multiple Flag-WWP1 truncation mutants and individual Myc-tagged HECT domains with or without the K740N mutation. Immunoprecipitation (IP) of Flag-WWP1 truncation mutants was performed with Flag antibody, followed by probing with Myc-tag antibody to detect the differential interaction between the HECT and WW2–WW3/C2 domains with or without the K740N mutation. Panel C shows a model of how the WWP1 E3 ligase activity is activated by the K740N variant by the disruption of WWP1 autoinhibition mediated by either homodimer or intramolecular processes. Ubiquitin conjugation enzyme, also known as E2, performs the second step in the ubiquitination process to coordinate with ubiquitin ligase for the transfer of ubiquitin moieties to the substrates. In turn, the inhibition of WWP1 activity is mediated by the interaction of its HECT domain with the WW2–WW3 linker domain.
XENOGRAFT MURINE MODELING
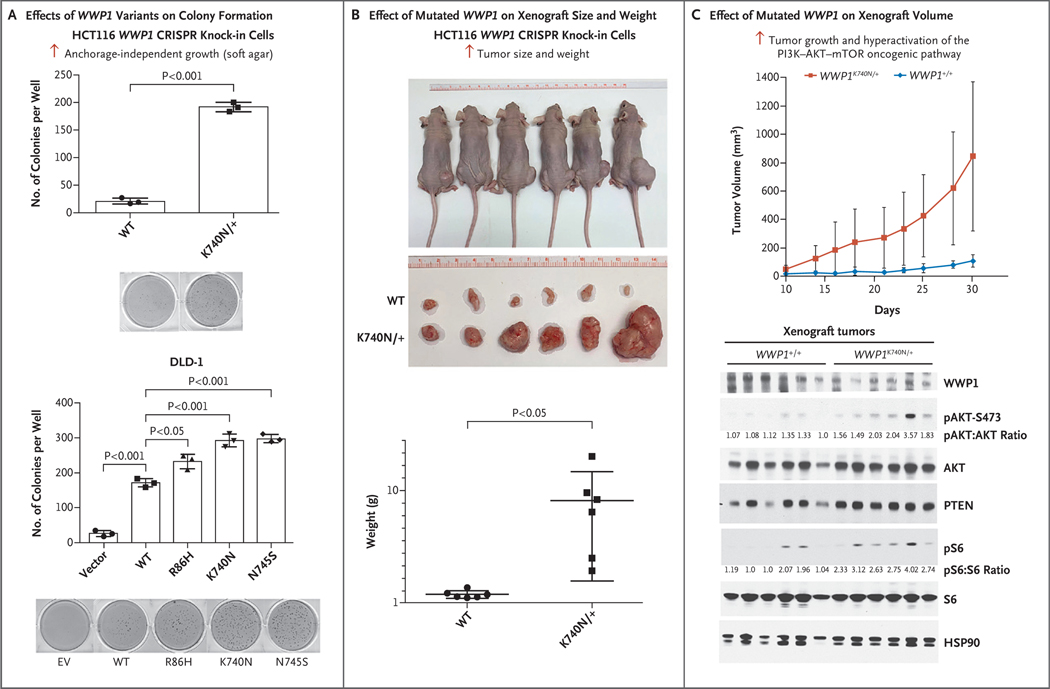
To determine the influence of WWP1 mutations on tumorigenesis and tumor growth in vivo, we next compared the oncogenic potential of wild-type WWP1 with endogenous CRISPR knock-in WWP1K740N/+ mutant cells. In support of our working model, the endogenous WWP1 K740N knock-in mutation, which strongly increased PTEN polyubiquitination (Fig. 3C), resulted in a higher level of anchorage-independent growth than wild-type WWP1 (Fig. 5A). Similarly, the overexpression of various germline WWP1 mutants, including R86H, K740N, and N745S, in DLD-1 colon cancer cells also markedly increased their tumorigenic potential in vitro and tumor growth in vivo in terms of both tumor weight and size, as evaluated in analyses involving soft agar and xenograft tumor methods, respectively (Fig. 5A and Fig. S9). In addition, tumors with the endogenous WWP1 K740N variant (WWP1K740N/+) grew much faster than those expressing wild-type WWP1 in a xenograft tumor model system, as revealed by analyses of tumor growth, weight, and size (Fig. 5B and 5C). Western blot analyses of tumors derived from cells of WWP1+/+ or WWP1K740N/+ genotypes showed the stronger oncogenic activity of the WWP1 K740N mutant in triggering hyperactivation of the PI3K–AKT–mTOR oncogenic pathway (Fig. 5C).
Figure 5. Characterization of Mutated WWP1 in Vitro and in Vivo.
Panel A shows the effects of WWP1 germline variants on colony-forming ability in soft agar in HCT116 cells (top graph) and in a colorectal adenocarcinoma cell line (DLD-1) (bottom graph). The numbers of colonies are quantified and presented as means of triplicate experiments. Panel B shows tumor xenograft assays performed by subcutaneously implanting HCT116 cells with either WWP1+/+ or WWP1K740N/+ genotypes in a group of six mice. Wild-type tumors were implanted on the left side of the mice, and tumors that stably expressed the K740N variant were implanted on the right side. The mean weight of the resulting tumors was much higher in mice implanted with the K740N genotype. Panel C shows mean tumor volumes as a function of the time in days in the same group of mice. After the mice were euthanized, the tumors were dissected and were analyzed by means of Western blotting with the use of the indicated antibodies. In all three panels, I bars indicate standard deviations.
DISCUSSION
Heritable mutations in multiple genes for both adenomatous and hamartomatous polyposis have been identified over the years. However, the genetic basis for predisposition to oligopolyposis is elusive. We found that germline WWP1 variants occur in patients with wild-type PTEN with characteristic PHTS-like phenotypes, particularly oligopolyposis. Patients with WWP1 germline mutations include those with Cowden-like syndrome (Table S1), with the shared clinical diagnosis of at least five gastrointestinal polyps, including at least one hamartomatous polyp or a polyp that is hyperplastic or serrated. In addition, all the patients were found to have wild-type genes that are known to be associated with a polyposis syndrome (PTEN, BMPR1A, SMAD4, ENG, APC, and STK11).17,27 Thus, mutations in WWP1 may represent a clear genetic cause of oligopolyposis, in which we found a mechanistic pathogenesis for intestinal neoplasia and cancer.
We further found that germline WWP1 variants were also markedly enriched in patients affected by prevalent, apparently sporadic human cancers. Notable enrichment of WWP1 variants was observed in PTEN-related Cowden syndrome component cancers,5 particularly colorectal adenocarcinoma and thyroid cancer. These data suggest that WWP1 may represent a broad cancer predisposition gene that warrants further investigation in PTEN-independent contexts. Although further data are required before recommending routine WWP1 genetic testing in the hereditary cancer clinic, the most prevalent and functionally characterized WWP1 K740N and N745S mutations may be considered in patients with wild-type PTEN who have a Cowden-like syndrome and an unexplained personal or family history of oligopolyposis or early-onset colorectal cancers.
We found that representative WWP1 variants provide mechanistic gain of function by relieving the autoinhibitory interaction between their WW2–WW3 linker and HECT domains. This process results in aberrant enzymatic activation of the WWP1 E3 ligase, with consequent ubiquitination and inactivation of PTEN to trigger hyperactivation of PI3K signaling. Although we found both in vitro and in vivo evidence of the deleterious effects of the three most frequently mutated prioritized variants (R86H, K740N, and N745S), the functional significance and associated mechanisms of the remaining variants have yet to be determined. Thus, we identified WWP1 as a proto-oncogenic neoplasia-susceptibility gene through aberrant regulation of PTEN–PI3K signaling, independent of germline PTEN mutational status. These findings have important implications for cancer prevention and therapy, since WWP1 and the pathway it regulates are potential therapeutic targets,14 whereas targeting PTEN has not been feasible in clinical practice to date.28
Before mechanism resolution, WWP1 had been shown to be overexpressed or amplified in multiple tumor types, which suggested that it had a role as an oncogene.8–13 It was only recently that WWP1 was shown to be a switch that could be targeted to reactivate PTEN.14 WWP1 depletion resulted in reduced oncogenic PI3K–AKT signaling in mouse embryonic fibroblast cells with Pten mutations or heterozygous deletions; the presence of such mutations causes PHTS.4 Indole-3-carbinol, a compound found in cruciferous vegetables, was found to be a natural and potent WWP1 inhibitor.14 Thus, our findings suggest that patients with wild-type PTEN with germline WWP1 variants may also benefit from the modulation of the WWP1–PTEN axis as a preventative or therapeutic measure. Clinical trials are anticipated to determine the effective dose and efficacy in humans.
Supplementary Material
CLINICAL TRIAL REGISTRATION.
The Journal requires investigators to register their clinical trials in a public trials registry. The members of the International Committee of Medical Journal Editors (ICMJE) will consider most reports of clinical trials for publication only if the trials have been registered. Current information on requirements and appropriate registries is available at www.icmje.org/about-icmje/faqs/.
Acknowledgments
Supported by grants (R01 CA82328 and R35 CA197529, to Dr. Pandolfi) from the National Institutes of Health, by grants (P01 CA124570 and R01 CA118989, to Dr. Eng) from the National Cancer Institute, and by grants (all to Dr. Eng) from the American Cancer Society Clinical Research Professorship, Breast Cancer Research Foundation, and Doris Duke Distinguished Clinical Scientist Award. Dr. Lee was supported in part by the Postdoctoral Research Abroad Program Fellowship, the Taiwan National Science Council, and a postdoctoral training award (W81XWH-16-1-0249) from the Department of Defense Prostate Cancer Research Program; Dr. Yehia, in part by the Cancer Genomic Medicine Clinical Fellowship Training Program of the Ambrose Monell Foundation at the Cleveland Clinic Genomic Medicine Institute; and Dr. Kishikawa, by the Overseas Research Fellowships of the Japan Society for the Promotion of Science and a research fellowship of the Uehara Memorial Foundation.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank the patients who contributed to this study; the Genomic Medicine Biorepository of the Cleveland Clinic Genomic Medicine Institute; our database and clinical research teams; and Hannah Chen and Marilyn Seyfi for their technical and bioinformatics assistance, respectively.
REFERENCES
- 1.Rahman N. Realizing the promise of cancer predisposition genes. Nature 2014;505:302–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 1998;273:13375–8. [DOI] [PubMed] [Google Scholar]
- 3.Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 1998;95:29–39. [DOI] [PubMed] [Google Scholar]
- 4.Yehia L, Ngeow J, Eng C. PTEN-opathies: from biological insights to evidence-based precision medicine. J Clin Invest 2019;129:452–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan M-H, Mester JL, Ngeow J, Rybicki LA, Orloff MS, Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res 2012;18:400–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngeow J, Stanuch K, Mester JL, Barnholtz-Sloan JS, Eng C. Second malignant neoplasms in patients with Cowden syndrome with underlying germline PTEN mutations. J Clin Oncol 2014;32:1818–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yehia L, Eng C. 65 Years of the double helix: one gene, many endocrine and metabolic syndromes: PTEN-opathies and precision medicine. Endocr Relat Cancer 2018;25:T121–T140. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Sun X, Guo P, et al. Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene 2007;26:2386–94. [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Zhou Z, Ross JS, Zhou W, Dong J-T. The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer 2007;121:80–7. [DOI] [PubMed] [Google Scholar]
- 10.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 2008;14:10–21. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Q, Cao X, Yuan F, Li G, Tong T. Knockdown of WWP1 inhibits growth and induces apoptosis in hepatoma carcinoma cells through the activation of caspase3 and p53. Biochem Biophys Res Commun 2014;448:248–54. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Wu Z, Ma Z, Liu H, Wu Y, Zhang Q. WWP1 as a potential tumor oncogene regulates PTEN-Akt signaling pathway in human gastric carcinoma. Tumour Biol 2015;36:787–98. [DOI] [PubMed] [Google Scholar]
- 13.Sanarico AG, Ronchini C, Croce A, et al. The E3 ubiquitin ligase WWP1 sustains the growth of acute myeloid leukaemia. Leukemia 2018;32:911–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YR, Chen M, Lee JD, et al. Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science 2019;364(6441):eaau0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eng C. Will the real Cowden syndrome please stand up: revised diagnostic criteria. J Med Genet 2000;37:828–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh DJ, Coulon V, Lunetta KL, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet 1998;7:507–15. [DOI] [PubMed] [Google Scholar]
- 17.Heald B, Mester J, Rybicki L, Orloff MS, Burke CA, Eng C. Frequent gastrointestinal polyps and colorectal adenocarcinomas in a prospective series of PTEN mutation carriers. Gastroenterology 2010;139:1927–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorlin RJ, Cohen MM Jr, Condon LM, Burke BA. Bannayan-Riley-Ruvalcaba syndrome. Am J Med Genet 1992;44:307–14. [DOI] [PubMed] [Google Scholar]
- 19.Huang K-L, Mashl RJ, Wu Y, et al. Pathogenic germline variants in 10,389 adult cancers. Cell 2018; 173(2):355.e14–370.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balasubramanian S, Fu Y, Pawashe M, et al. Using ALoFT to determine the impact of putative loss-of-function variants in protein-coding genes. Nat Commun 2017;8:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 2009;10:398–409. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, Jiang H, Xu W, et al. A tunable brake for HECT ubiquitin ligases. Mol Cell 2017;66:345.e6–357.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogunjimi AA, Briant DJ, Pece-Barbara N, et al. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell 2005;19:297–308. [DOI] [PubMed] [Google Scholar]
- 25.Wiesner S, Ogunjimi AA, Wang H-R, et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 2007;130:651–62. [DOI] [PubMed] [Google Scholar]
- 26.Wan L, Zou W, Gao D, et al. Cdh1 regulates osteoblast function through an APC/C-independent modulation of Smurf1. Mol Cell 2011;44:721–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngeow J, Heald B, Rybicki LA, et al. Prevalence of germline PTEN, BMPR1A, SMAD4, STK11, and ENG mutations in patients with moderate-load colorectal polyps. Gastroenterology 2013;144:1402–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y-R, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol 2018;19:547–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.