Abstract
The endophytic microbiota can establish mutualistic or commensalistic interactions within the host plant tissues. We investigated the bacterial endophytic microbiota in three species of Mediterranean orchids (Neottia ovata, Serapias vomeracea, and Spiranthes spiralis) by metabarcoding of the 16S rRNA gene. We examined whether the different orchid species and organs, both underground and aboveground, influenced the endophytic bacterial communities. A total of 1,930 operational taxonomic units (OTUs) were obtained, mainly Proteobacteria and Actinobacteria, whose distribution model indicated that the plant organ was the main determinant of the bacterial community structure. The co-occurrence network was not modular, suggesting a relative homogeneity of the microbiota between both plant species and organs. Moreover, the decrease in species richness and diversity in the aerial vegetative organs may indicate a filtering effect by the host plant. We identified four hub OTUs, three of them already reported as plant-associated taxa (Pseudoxanthomonas, Rhizobium, and Mitsuaria), whereas Thermus was an unusual member of the plant microbiota. Core microbiota analysis revealed a selective and systemic ascent of bacterial communities from the vegetative to the reproductive organs. The core microbiota was also maintained in the S. spiralis seeds, suggesting a potential vertical transfer of the microbiota. Surprisingly, some S. spiralis seed samples displayed a very rich endophytic microbiota, with a large number of OTUs shared with the roots, a situation that may lead to a putative restoring process of the root-associated microbiota in the progeny. Our results indicate that the bacterial community has adapted to colonize the orchid organs selectively and systemically, suggesting an active involvement in the orchid holobiont.
Keywords: bacterial endophytes, orchids (Orchidaceae), microbiota, metabarcoding analysis, seeds
Introduction
Plants in natural and agricultural ecosystems interact with a multitude of microorganisms that colonize both the internal tissues and the outer surfaces of both underground and aboveground plant organs (Compant et al., 2019; Escudero-Martinez and Bulgarelli, 2019). These plant-associated microbial communities are collectively known as the plant microbiota (Marchesi and Ravel, 2015) and can theoretically comprise mutualistic, commensal and latent pathogenic strains in apparently healthy plants (Brader et al., 2017). Bacteria and fungi, the dominant components of the plant microbiota, can deeply influence plant growth and responses to stresses. For these reasons, several studies in the last decade focused on the structure and functions of the plant microbiota, with the aim to link them to plant fitness and crop productivity (Bulgarelli et al., 2013; Compant et al., 2019; Tian et al., 2020).
Endophytes can be defined as components of the plant microbiota that colonize internal healthy plant tissues and establish non-harmful relationships with their host (Hardoim et al., 2015). This definition includes mutualistic endophytes that form recognizable structures during plant colonization - such as mycorrhizal fungi in the roots of land plants and rhizobia in legume root nodules - whose interactions with the host plant have been extensively investigated because of their beneficial influence on plant growth, productivity and health (Das et al., 2017; Masson-Boivin and Sachs, 2018; Genre et al., 2020). However, there is ample evidence that many apparently commensalistic endophytes can also promote plant growth and defense (Santoyo et al., 2016; Hossain et al., 2017), but the ecology and functions of these beneficial endophytes is less understood. In particular, plant growth-promoting (PGP) endophytic bacteria can promote plant growth by mechanisms that include the release of phytohormones (Patten and Glick, 2002; Glick, 2012, 1995; Rashid et al., 2012; Duca et al., 2014), nitrogen fixation (Govindarajan et al., 2008; Urquiaga et al., 2012), improved acquisition of mineral nutrients (Malinowski et al., 2000; Rahman et al., 2005), production of growth-promoting compounds (Glick et al., 1998; Taghavi et al., 2009; Kumar et al., 2019) and increased stress tolerance (Kohler et al., 2008). Furthermore, part of the bacterial endophytic community showed tolerance toward heavy metals and pollutants, thus finding an important application for bioremediation (Guo et al., 2010; Chen et al., 2012; Stępniewska and Kuźniar, 2013). Bacterial endophytes can also protect the host plant against pathogen invasion and disease through direct biocontrol activities or by inducing plant systemic resistance (Berg and Koskella, 2018; de Vrieze et al., 2018).
The fact that only a minority of the environmental bacteria can be cultivated in the laboratory has been a limiting factor for environmental microbiologists, restricting knowledge about the interaction mechanisms between the microbiota and the plant. Next generation sequencing (NGS) technologies have allowed large-scale studies independent from the cultivation approach, providing details on taxonomic structure, abundance and potential physiological features of the microbial endophytes associated with plants (Knief, 2014). By using these molecular tools, it is possible to estimate the evolution and adaptation of the microbes associated with the host and to understand how they are related to each other (Khare et al., 2018; Lucaciu et al., 2019).
Orchidaceae is one of the largest plant families on Earth (Christenhusz and Byng, 2016). Orchids grow in a wide range of habitats and include epiphytes as well as terrestrial species. Most orchids are photosynthetic at adulthood, but about 200 species are fully achlorophyllous and rely on their mycorrhizal fungal partners for the provision of carbon (Hynson et al., 2013). Several forest orchid species develop green leaves but remain at least partly dependent on mycorrhizal fungi for carbon supply (Selosse and Roy, 2009). Irrespective of their trophic strategies, orchids are highly dependent on the relationship with mycorrhizal fungi, and their distribution is often correlated with the distribution of their mycorrhizal fungal partners (McCormick et al., 2018). Because of their importance in the survival of these endangered plants, most of the microbiological investigations on orchids have focused on their mycorrhizal associations, leaving aside the potential role of other microbial partners. Bacterial endophytes in orchids have been mainly investigated in commercially valuable species by isolation after surface sterilization of roots and/or leaves, and many isolates have been found to both increase seed germination and promote plant growth (Tsavkelova et al., 2007; Faria et al., 2013; da Silva et al., 2015; Gontijo et al., 2018; Herrera et al., 2020). By contrast, culture-independent approaches have been rarely applied to investigate the endophytic bacteria of orchids, and data are only available for the epiphytic orchids Dendrobium officinale and Dendrobium catenatum (Yu et al., 2013; Li et al., 2017). In these orchid species, endophytic bacteria belonging mainly to Proteobacteria were identified by means of PCR-DGGE and pyrosequencing, showing diversity in the roots and stems. In addition, meta-nifH sequencing suggested the presence of common diazotrophs in several D. catenatum tissues, which could play an important role in nitrogen supply (Li et al., 2017). Here, we have explored the endophytic bacterial communities of three terrestrial orchid species, namely Neottia ovata (L.) Bluff & Fingerh, Serapias vomeracea (Burm. F.) Briq. and Spiranthes spiralis (L.) Chevall. We investigated different plant organs in order to characterize the taxonomic diversity and the structure of the associated microbiota, and to identify a core microbiota. The same three orchid species were already found to host endophytes with a notable plant growth promoting potential, as assessed by cultivation and characterization of bacterial isolates (Alibrandi et al., 2020). Based on results on other plant species (Bulgarelli et al., 2013; Haruna et al., 2018), our aim was to verify the following hypotheses in orchids: (i) endophytic bacterial communities create distinctive phylogenetic structures in the various plant organs, and (ii) the reproductive organs (seeds) represent a reservoir of endophytes that can be vertically transmitted. To test these hypotheses, two comparisons were made: first, we compared the bacterial microbiota associated with different plant organs (roots, leaves, stems and capsules) of the three orchid species; second, we included the seed microbiota of S. spiralis (the only species for which seeds were available at the time of sampling) to analyze the distribution of unique, shared and core OTUs within this species.
This study provides new insights into the distribution of bacteria across different organs of terrestrial orchid species, and sheds light on the fraction of the bacterial community potentially transferred to the next plant generation by vertical transmission, pointing out a possible role of bacterial endophytes in orchid conservation.
Materials and Methods
Orchid Sampling and Surface-Sterilization
The plant species investigated in this work were Neottia ovata (L.) Bluff & Fingerh., Serapias vomeracea (Burm. f.) Briq. and Spiranthes spiralis (L.) Chevall., three Mediterranean orchid species that grow naturally in Italy. Samples of roots, stems, leaves and capsules of the three different orchids were harvested during flowering in the Liguria region (Italy), stored on ice in sterile containers, transported to the laboratory and kept at 4°C before being processed (within 2 days). Furthermore, seeds from surface sterilized capsules (see below for sterilization) of S. spiralis were also analyzed.
Plant samples from aboveground organs (stems, leaves and capsules) were surface–sterilized by stepwise immersion in 70% ethanol for 1 min, then in 2.5% sodium hypochlorite for 2 min and finally in 70% ethanol for 1 min, followed by five rinses in sterile distilled water (Alibrandi et al., 2018). Root samples were thoroughly rinsed with sterile water, sonicated and surface-sterilized with 95% ethanol for 20 s followed by immersion in 5% sodium hypochlorite for 3 min and washed seven times with sterile distilled water. Furthermore, we obtained the seeds of S. spiralis from surface-sterilized capsules. The surface-sterilized capsules were placed into a sterile falcon tube, sealed and seeds were recovered from the tube bottom once the capsules hatched.
Five biological replicates, each collected from different plant individuals, were used for each orchid organ, for a total of 65 samples. Samples were named according to the orchid species (NO for N. ovata, SV for S. vomeracea and SS for S. spiralis), and organs (R for root, L for leaf, St for stem, C for capsule, S for seed), the numbers indicating the biological replicate (Table 1).
TABLE 1.
Samples of the orchid species/organs investigated in this study.
| Sample name | Orchid species | Organ |
| (1–5)NO.pna.R | Neottia ovata | Root |
| (1–5)NO.pna.L | Neottia ovata | Leaf |
| (1–5)NO.pna.St | Neottia ovata | Stem |
| (1–5)NO.pna.C | Neottia ovata | Capsule |
| (1–5)SV.pna.R | Serapias vomeracea | Root |
| (1–5)SV.pna.L | Serapias vomeracea | Leaf |
| (1–5)SV.pna.St | Serapias vomeracea | Stem |
| (1–5)SV.pna.C | Serapias vomeracea | Capsule |
| (1–5)SS.pna.R | Spiranthes spiralis | Root |
| (1–5)SS.pna.L | Spiranthes spiralis | Leaf |
| (1–5)SS.pna.St | Spiranthes spiralis | Stem |
| (1–5)SS.pna.C | Spiranthes spiralis | Capsule |
| (1–5)SS.pna.S | Spiranthes spiralis | Seed |
For each sample, five biological replicates (collected from different plant individuals) were analyzed.
DNA Extraction
Total DNA was extracted separately from each organ sample (roots, leaves, stems, capsules and seeds) of each orchid species using the Qiagen DNeasy kit (QIAGEN®,Hilden, Germany) according to the manufacturer’s instructions. The resulting 65 DNA samples were checked for integrity by electrophoresis in a 1% agarose gel and stored at −20°C.
PNA Clamps
The endophytic bacterial microbiota was investigated by 16S rRNA gene metabarcoding (V3 – V4 regions). However, the molecular characterization of the microbiota inside host plant tissues was conditioned by the co-amplification of mitochondrial and plastid rRNA genes extracted from the eukaryotic plant cells. To reduce this issue, a suitable approach is the use of sequence-specific peptide nucleic acid (PNA) clamps, which bind to, and block, the amplification of host-derived DNA (Lundberg et al., 2013). Although there are universal PNAs available to block plant mitochondrial (mPNA) and plastid (pPNA) sequences, unfortunately they are not effective on a large number of plant species. In order to select the PNAs that could bind to the greatest number of contaminating sequences, an in silico analysis of mitochondrial and plastid sequences obtained from a preliminary sequencing was carried out. Several PNA clamps already tested on some plant species have been evaluated, and the best result in silico was obtained using the universal probe pPNA (5′-GGCTCAACCCTGGACAG-3′) according to Lundberg et al. (2013), and a newly designed mPNA probe (5′-CTACCGACGCTGGGG-3′), respectively, for plastid and mitochondrial sequences. To verify the effective reduction of host-derived sequences, four samples from root, stem, leaf and capsules of S. spiralis were amplified with or without the PNA clamps: the effect of PNA clamps on the relative abundance of bacterial, plastid and mitochondrial sequence reads was assessed by Student’s t-test using SPSS 20 (IBM Corporation, Armonk, NY, United Sates).
Library Preparation and Ion Torrent Sequencing of the Bacterial 16S rRNA Gene
The DNA obtained from all samples was used as template for the metabarcoding of the 16 rRNA gene by Ion Torrent sequencing. The hypervariable V4 and V5 regions of the 16S rRNA genes were amplified by PCR using the primers 520 F (5′-AYTGGGYDTAAAGNG-3′) and 907 R (5′-CCGTCAATTCMTTTRAGTTT-3′) (Claesson et al., 2009; Engelbrektson et al., 2010). Two PCR reactions were performed. In the first reaction, the DNA of all the samples was amplified including mPNA and pPNA clamps to reduce co-amplification of host plant DNA. PCR was carried out at a final volume of 15 μl including the following reagents: 10 ng of template DNA, 3 μl of 5x KAPAHiFi (KAPABiosystems, Woburn, MA, United States) buffer, KAPA dNTP mix 200 μM each, 5 pmol of each primer each and KAPAHiFi polymerase 0.3 units. The following PCR program was used: 3 min 95° C, followed by 35 cycles of (20 s 95°C – denaturation; 15 s 78°C – PNA annealing; 30 s 55°C – primer annealing; 30 s 72°C – elongation) and 5 min at 72°C. PCR products were visualized by agarose gel electrophoresis, and then used as template for the second PCR reaction. The second PCR was carried out with primer 520 F and 907 R comp, adapter (5′-CCATCTCATCCCT GCGTGTCTCCGACTCAG-3′) and barcodes, with KAPA2G Robust HotStart Ready-Mix (KAPA Biosystems) as described by Ambika Manirajan et al. (2016). Both PCRs were performed with a MyCycler TM (Bio-Rad). The PCR products were purified using QIAquick PCR purification kit (QIAGEN GmbH), and further purified with DNA purification beads NucleoMag NGS clean-up kit (MACHEREY-NAGEL GmbH & Co., KG) to remove contaminants (nucleotides, primers, adapters, enzymes, buffer additives, and salts). Amplicon concentration was estimated by measuring the fluorescence with a spectrophotometer. Ion Torrent sequencing was performed on pooled PCR products. Emulsion PCR (Ion PGM Hi-Q View OT2 kit, Life Technologies) was performed with Ion One Touch 2 (Life Technologies, Carlsbad, CA, United States). The quality of the final product was assessed with the Ion Sphere Quality Control Kit (Life Technologies, Carlsbad, CA, United States) and loaded on 318 chip for sequencing (Life Technologies), using an Ion PGM sequencer (Life Technologies, Carlsbad, CA, United States).
The sequences were submitted to the European Nucleotide Archive (ENA, www.ebi.ac.uk/ena) under the accession number PRJEB40289.
Bioinformatic Analysis of Metabarcoding Sequences
Raw sequence data were processed with QIIME, version 1.9 (Caporaso et al., 2010). Sequences were de-replicated and assigned to specific samples according to the corresponding barcodes, filtered by length and quality (length: 350–450 bp; quality threshold: 20) and grouped in operational taxonomic units (OTU) with uclust (Edgar, 2010) using a 97% identity threshold. Chimeric sequences were removed with Chimera Slayer (against the Silva base reference alignment). Reference sequences for each OTU were assigned to a taxonomic group by comparison with the Silva database (version 132). Singletons and contaminant sequences (mitochondrial and plastid) were eliminated.
To obtain a more accurate classification, phylogenetic trees were built by aligning the 16S rRNA gene sequences of relevant OTUs (such as the hub taxa) to the type strain sequences of the nearest genera, using the Muscle alignment implemented in the megaX software (Kumar et al., 2018). Phylogenetic trees were inferred with the maximum likelihood methods, using RaxML (maximum randomized axillary probability) as implemented in the open source CIPRES Science Gateway1. One-thousand bootstrap resamplings were generated, and values above 50% were shown at the branches. The trees were visualized and edited using the open source iTOL (Interactive tree of life2).
The composition of the microbiota was compared between the different orchid species and organs analyzed. Taxa relative abundances were calculated on the OTU table without any rarefaction; bar charts were plotted using the ggplott2 package in R software version 1.2.1335 (Gergs and Rothhaupt, 2015; Wickham, 2016).
The data sets were processed after different rarefactions: a single rarefaction (221 reads per sample), based on the sample with the least number of readings, was used for the comparison of the alpha diversity between the three orchid species, while three rarefactions were used for the comparison of organs within each species (2965, 1617, and 221 reads per sample for N. ovata, S. vomeracea and S. spiralis, respectively). Community richness (“observed OTUs” index), diversity (Shannon index) and phylogenetic diversity (PD index) were calculated using the QIIME script alpha_diversity.py; the R statistical software was used to perform ANOVA with Tukey HSD post hoc test (p < 0.05).
Differences in community structure between species and organs were determined by permutational multivariate ANOVA on Bray-Curtis similarity matrices after 9,999 permutations and plotting (Non-metric multidimensional scaling – NMDS – ordination). Analyses were performed using the Qiime script beta_diversity.py. NMDS and boxplots were calculated in R software version 1.2.1335 with the vegdist function in “vegan” and “ggplot2” packages, respectively. Differences at p < 0.05 were regarded as statistically significant.
Shared and Core OTUs
Recent studies (Bulgarelli et al., 2012; Lundberg et al., 2012) have defined a core range of species of the microbiota associated with plant species as taxonomic core microbiota. Bacterial OTUs shared between the different orchid species and between the different organs of each species were identified. In this work, the OTUs occurring in 100% of the species or organs were considered as belonging to the core microbiota, as calculated with the QIIME script compute_core_microbiome.py. This was considered as the “stable” fraction of the community suggesting possible key functional roles for the host. For visualization and interpretation of these data, Venn diagrams were generated using the free online tool of the Ghent University3.
Co-occurrence Network Analysis
To identify hub OTUs, a co-occurrence analysis was performed with the Co-occurrence Network Inference software (CoNet; Faust and Raes, 2016), using not-normalized data, as recommended, to reduce the compositional effect (Berry and Widder, 2014). To ensure statistical robustness, only OTUs with more than 50 reads and occurring in at least 20 samples were considered. Pairwise scores were calculated for four measures: Bray-Curtis and Kullback–Leibler similarities, and Pearson and Spearman correlations. One-hundred permutations (with row shuffling re-sampling and re-normalization for correlations), and 100 bootstrap scores, were generated. Unstable edges (outside the 2.5–97.5 percentiles of the bootstrap distribution) were eliminated. The individual p-values of the four measures were merged with the Brown’s method. Only edges with false discovery rate (FDR)-corrected p-values < 0.05 were retained, and only if they were supported by at least three of the four measures. The network layout was generated with the “edge-forced spring embedded” algorithm, which leads to unbiased networks showing interconnected nodes closer to each other and less-linked ones placed in the outside position. Network legends were created with the Cytoscape Add-on “Legend creator4”. Hub taxa were identified by scoring the degree (number of connections) against the betweenness centrality and the closeness centrality, according to Agler et al. (2016). The taxonomical identity of the hub OTUs was confirmed/improved by manual BLAST.
Results
Sequencing Output, OTU Clustering, and Effect of PNA Blockers
Ion Torrent sequencing generated 6,460,549 raw sequences. After length and quality filtering, 4,117,638 sequences remained, which were grouped into OTUs at 97% similarity level. The mitochondrial and plastid sequences and the singletons were removed to end up with 1,469,200 total sequences (Supplementary Table 1) grouped in 1,930 OTUs. Two samples (both corresponding to leaves of S. spiralis) were eliminated due to the low number of reads (Supplementary Table 1).
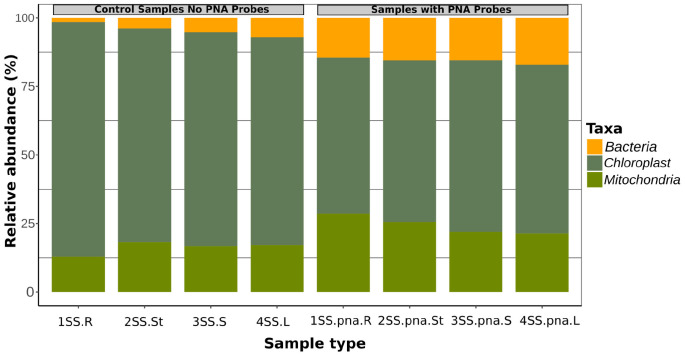
The addition of the PNA probes significantly reduced the relative abundance of plastid sequences with respect to the same samples without PNA probes (t-test, p = 0.0095), while mitochondrial sequences were slightly but not significantly higher (p = 0.052) (Figure 1). Relative abundance of bacterial sequences was significantly increased in the samples amplified with PNA probes (t-test, p = 0.00048; Figure 1).
FIGURE 1.
Relative fraction of sequences belonging to Bacteria, Plastids and Mitochondria in four orchid samples, as obtained by metabarcoding of 16S rRNA genes without PNA blockers, compared with the same samples with PNA.
Composition of the Bacterial Orchid Microbiota
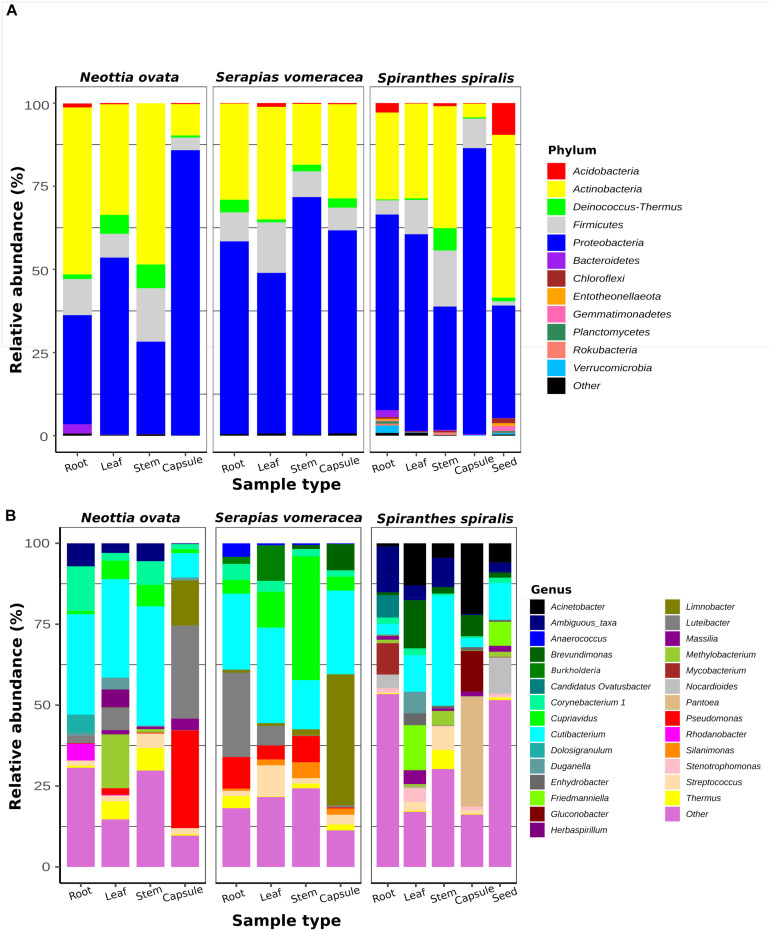
To identify the taxa associated with the orchid samples and to analyze their distribution in the different species and organs, two taxonomic levels (phylum and genus) were considered (Figure 2). The two most represented phyla were Proteobacteria and Actinobacteria, with Proteobacteria dominating (from 37 to 86%) in most orchid samples. Proteobacteria were particularly abundant in the S. vomeracea stems (71.40%) and in the capsules of N. ovata, S. vomeracea, and S. spiralis (85.72, 61.01, and 86.03%, respectively). Complementarily, Actinobacteria were dominant in N. ovata roots and stems (50.02 and 48.47%) and in S. spiralis seeds (48.95%). Firmicutes (from 1.26 to 16.83%) and Deinococcus-Thermus (from 0.29 to 7.14%) were less represented but distributed in all organs and species (Figure 2A). By contrast, Acidobacteria were rather infrequent, except in the S. spiralis seeds (9.59%) (Figure 2A).
FIGURE 2.
Bacterial taxa distribution in the orchid microbiota. Relative abundance of panel (A) predominant bacterial phyla (>1%) and (B) predominant genera (>0.5%), as obtained by 16S rRNA gene metabarcoding of orchid plants (n = 5).
Among genera, Cutibacterium was distributed in all samples (Figure 2B). This genus was abundant in the roots (31.02%), stems (36.83%) and leaves (30.40%) of N. ovata, and less represented in the capsules, in which Pseudomonas (30.18%) and Luteibacter (28.77%) were the dominant genera.
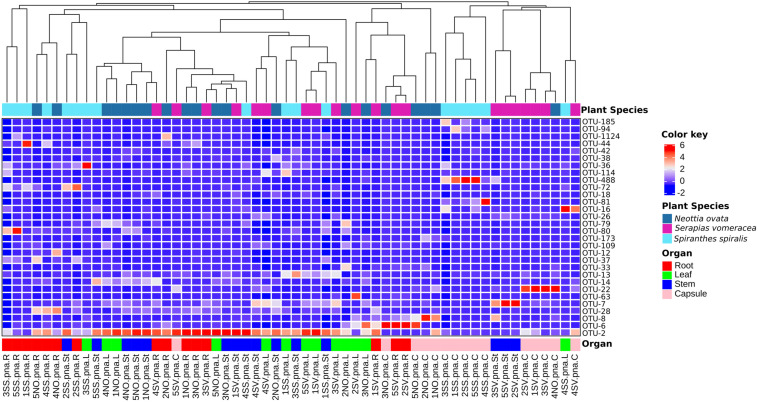
In S. vomeracea, Luteibacter (26.08% in the root), Cupriavidus (38.20% in the stem), Cutibacterium (29.50% in the leaf) and Limnobacter (40.53% in the capsule) were the most abundant genera. In S. spiralis, the distribution model mainly concerned Pantoea and Acinetobacter (33.83 and 21.79% in the capsule) and Cutibacterium (34.00% in the stem), while in leaves there was no clear distribution of dominant genera (Figure 2B). All minor genera (<0.5%) were mostly distributed in both roots and seed of S. spiralis (Figure 2B, indicated as “other”). The distribution and abundance of major OTUs (>0.5% of the total orchid microbiota) were used to create a heat map. The dendrogram resulting from single hierarchical clustering showed only a partial tendency of the samples to group by organ and/or species (Figure 3).
FIGURE 3.
Heatmap of major bacterial OTUs (>0.5%) in the orchid microbiota. Samples were clustered by orchid species (top row) or organs (bottom row). Legends to the right of the figures indicate color scheme for log transformed OTU abundance, species and organ samples.
Alpha- and Beta-Diversity
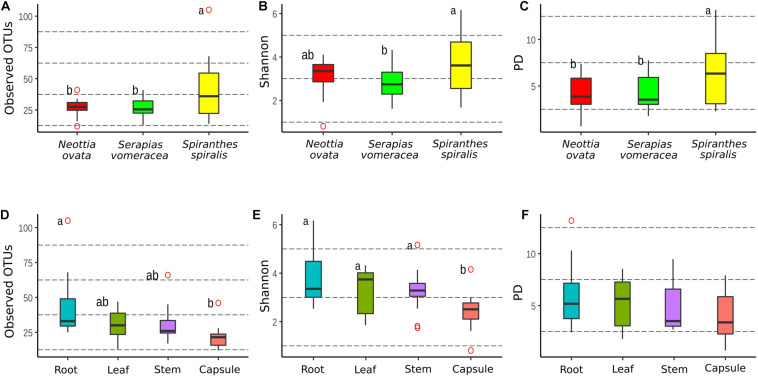
The dataset was processed to obtain three rarefied sub-datasets. A first sub-dataset was rarefied to 221 reads and used to compare the alpha diversity values of the three orchid species and the organs of S. spiralis, whereas two more sub-dataset were rarefied to 2,965 and 1,617 reads to compare the organs of N. ovata and S. vomeracea, respectively. Good’s coverage was >99% for all N. ovata samples (Supplementary Table 2), >96% for all S. vomeracea samples (Supplementary Table 3), and >85% for all S. spiralis samples except two ones with 76 and 82% (Supplementary Table 4). The alpha diversity of the microbiota in each sample was analyzed by considering the Observed OTUs (richness), Shannon (diversity) and Phylogenetic Diversity indices (Supplementary Tables 2–4). The S. spiralis microbiota showed the highest values in all indices, when compared with the other two orchid species (Figures 4A–C). When the different organs of the three species were considered, there was a trend of decreasing values in richness, diversity and phylogenetic diversity indices along a vertical progression from roots to capsules, with roots and capsules being significantly different for richness and capsule being significantly different from all other samples for diversity (Figures 4D–E).
FIGURE 4.
Comparison of alpha-diversity metrics between the microbiota of three Mediterranean orchid species. Richness (observed OTUs), diversity (Shannon) and phylogenetic diversity (PD) for different orchid species (A–C) and organs (D–F). ° indicates outliers. Different letters above the bars indicate significantly different means (Tukey test, p < 0.05); no letters (panel F) indicate no statistically significant differences.
Comparison of the same indices in the different organs within each individual orchid species showed some significant differences in N. ovata (Supplementary Figures 1A–C) and S. spiralis (Supplementary Figures 1G–I), but not in S. vomeracea (Supplementary Figures 1D–F). In particular, OTU richness was significantly higher in the root samples of N. ovata, as compared to stem and capsule, whereas root and leaf samples were different from capsule for the Shannon index. Finally, root and stem samples of N. ovata were significantly different in the phylogenetic diversity index. Root and capsule samples of S. spiralis were significantly different in both richness and diversity indices, but not for phylogenetic diversity (Supplementary Figures 1G–I). In particular, both OTU richness and diversity (Shannon index) were significantly higher in the root samples of S. spiralis, as compared to the capsules.
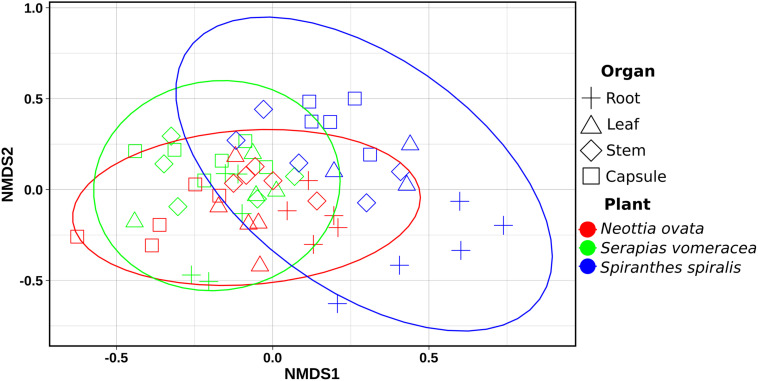
Non-metric multidimensional scaling (NMDS) ordination analysis showed a large overlap in the microbiota of S. vomeracea and N. ovata, whereas the S. spiralis samples, in particular the root samples, clustered separately (Figure 5). These results were confirmed by ADONIS pairwise comparisons of microbiota structure between orchid species and organs (Table 2). Although the homogeneity of the sample dispersion was reduced in the species groups, S. spiralis was significantly different from N. ovata and S. vomeracea. The sample dispersion was heterogeneous in the organ groups, which showed significant differences except for the stems-leaves comparison (Table 2). Analysis of Alpha- and beta-diversity was also performed without considering the different number of sequences per sample (non-rarefied dataset). The respective results showed no substantial differences with those obtained with the rarefied datasets (compare Supplementary Figure 2 with Figure 4, Supplementary Figure 3 with Supplementary Figure 1, Supplementary Figure 4 with Figure 5, and Supplementary Table 5 with Table 2).
FIGURE 5.
Non-metric Multidimensional Scaling (NMDS) Plot visualization of beta diversity, using the Bray–Curtis distances, separating samples by orchid species/organs. Plot ellipses represent the 95% confidence regions for species clusters (stress = 0.24).
TABLE 2.
ADONIS pairwise comparisons of microbiota structure between orchid species and organs, calculated on Bray–Curtis dissimilarity measures.
| Species comparison | ADONIS Test |
|
| R2 | p-value | |
| Serapias vomeracea vs. Neottia ovata | 0.0458 | 0.0608 |
| Serapias vomeracea vs. Spiranthes spiralis | 0.1023 | 0.0002 |
| Neottia ovata vs. Spiranthes spiralis | 0.0945 | 0.0002 |
| Organ comparison | ||
| Roots vs. Stems | 0.0831 | 0.0049 |
| Roots vs. Leaves | 0.0649 | 0.0436 |
| Roots vs. Capsules | 0.0897 | 0.0010 |
| Stems vs. Leaves | 0.0528 | 0.1304 |
| Stems vs. Capsules | 0.1140 | 0.0006 |
| Leaves vs. Capsules | 0.0897 | 0.0064 |
Significant differences are highlighted in bold.
Core Microbiota in the Three Orchid Species
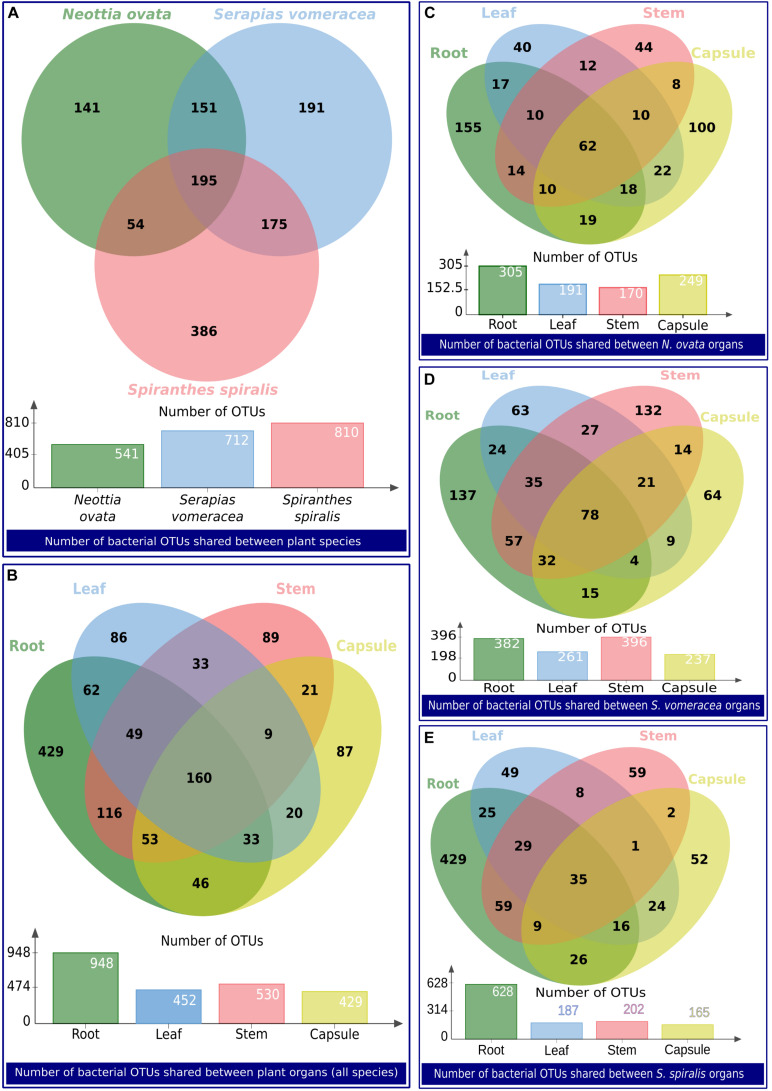
The numbers of unique and shared OTUs identified in the orchid species and/or organs are indicated in the Venn diagrams in Figure 6. Overall, 718 OTUs were unique to N. ovata, S. vomeracea and S. spiralis (141, 191, and 386, respectively), whereas 195 OTUs were shared between all three species (Figure 6A); the latter represent the “species core microbiota.” When the organs of all species were considered, 691 OTUs were found to be unique to roots, stems, leaves and capsules (429, 89, 86, and 87 OTUs, respectively), 298 and 144 were shared between two and three organs, respectively, and 160 were shared by all orchid organs (Figure 6B); the latter represent the “organ core microbiota.” Considering the microbiota of the different organs in each orchid species, the numbers of unique and shared OTUs are indicated for N. ovata (Figure 6C), S. vomeracea (Figure 6D) and S. spiralis (Figure 6E). These shared OTUs corresponded to a similar percentage of the total identified OTUs for N. ovata (11.46%) and S. vomeracea (10.95%), whereas a lower percentage (4.25%) was found in S. spiralis.
FIGURE 6.
Venn diagrams showing the number of bacterial OTUs shared between groups of samples. The groups of samples are indicated as a label below each diagram.
Shared OTUs were taxonomically diversified (Supplementary Table 6), and no dominant phyla could be identified: relative frequencies reflected those of the total community (Figure 2). Mainly plant-associated genera formed the core microbiota of the three orchid species, also including typically beneficial taxa, such as Agromyces, Bacillus, Bradyrhizobium, Burkholderia, Comamonas, Luteibacter, Methylobacterium, Pseudomonas, Rhizobium, Sphingomonas, Stenotrophomonas, and Variovorax, among others (Supplementary Table 6). Further taxa were members of Enterobacteriaceae as well as other typically human-associated taxa (Cutibacterium, Escherichia-Shigella, Neisseria, and Staphylococcus). Typical phytopathogens were absent.
Co-occurrence Network Analysis
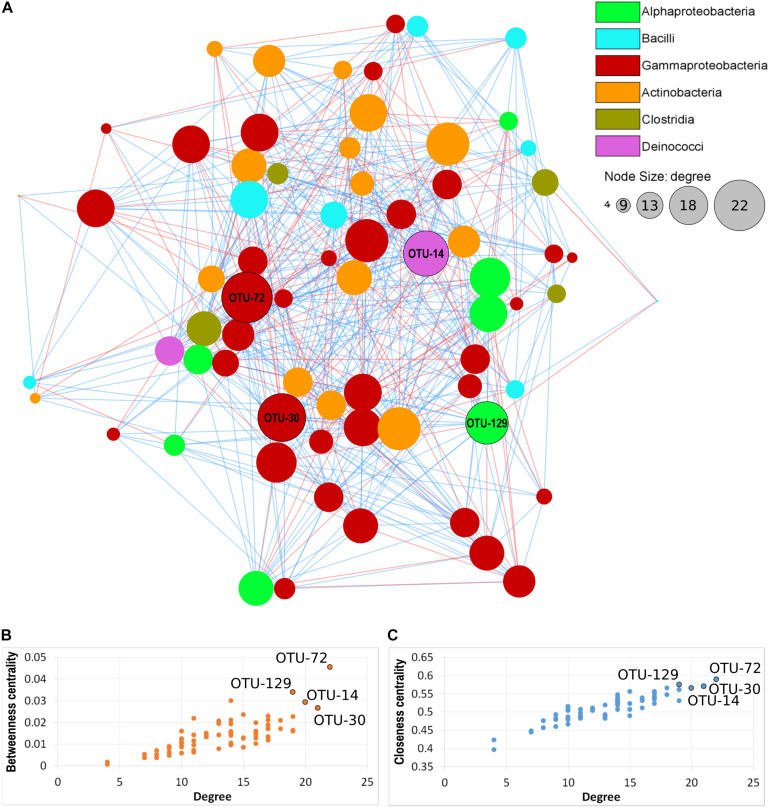
Seventy nodes (corresponding to OTUs) composed the bacterial interaction network; each node showed 4–22 edges (significant correlations). In total, there were 465 correlations (345 positives and 120 negative). Seventeen correlations were significant for three of the four distance/correlation methods calculated, while 448 were significant for all four methods. The network was not modular (Figure 7A), thus indicating a relative homogeneity of the microbiota across orchid species and organs. Based on connectivity scores (Figures 7B,C), four OTUs were identified as “hub taxa” (Figure 7, labeled nodes). After manual BLAST, they were identified at the genus level as Mitsuaria, Pseudoxanthomonas, Thermus, and Rhizobium. Three species have been described in the genus Mitsuaria, and the phylogenetic analysis of OTU72 showed that it was closer to M. noduli and M. chitinosanitabida than to the recently described M. chitinovorans (Sisinthy and Gundlapally, 2020; Supplementary Figure 5).
FIGURE 7.
Microbial co-occurrence network of the orchid microbiota. (A) Nodes represent OTUs, edges represent positive (blue lines) or negative (red lines) correlations. Node size indicates the degree (number of connections) and node color indicate the taxonomical class, according to the legend. Labeled nodes represent the hub OTUs. (B,C) Connectivity scores of the nodes, based on degree, betweenness centrality and closeness centrality, used to identify the hub OTUs (labeled nodes).
Three of the hub OTUs had more positive than negative correlations, while OTU-72 (Mitsuaria) had the same number (11) of positive and negative correlations. These hub OTUs represented 0.04, 0.37, 0.54, and 2.62 (Rhizobium, Pseudoxanthomonas, Mitsuaria, and Thermus, respectively) of the total microbiota. Mitsuaria and Thermus were included in the core microbiota of all species, whereas Pseudoxanthomonas and Rhizobium were included in the core of S. vomeracea and S. spiralis, respectively.
Occurrence of Endophytic Bacteria in Seeds of Spiranthes spiralis
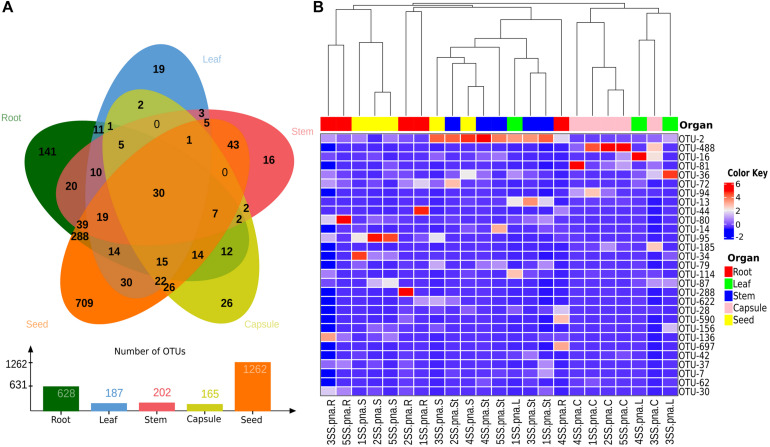
To investigate the possibility of a vertical transmission of bacterial components of the orchid microbiota, seeds were obtained from surface sterilized S. spiralis capsules, hatched inside sterile falcon tubes. Quite surprisingly, metabarcoding of the seed microbiota yielded the highest number of OTUs (1,262), when compared with the other organs of S. spiralis (Figure 8). Overall, 911 OTUs were unique to the roots, stems, leaves, capsules, and seeds (141, 16, 19, 26, and 709, respectively), whereas 30 OTUs represented the core microbiota of S. spiralis. Interestingly, the seeds shared a notably higher number of OTUs with the roots (288) than with other organs (43 with stems, 30 with leaves and 26 with capsules).
FIGURE 8.
Analyses of shared and major bacterial OTUs in the Spiranthes spiralis microbiota. (A) Venn diagram showing unique and shared OTUs between vegetative and reproductive organs (root, leaf, stem, capsule, seed). (B) Heatmap showing bacterial OTUs > 0.5% of the total sequences; samples were clustered by orchid organs (top row). Color Key from blue to red indicates increasing abundance in natural logarithm of counts, with OTUs along the Y-axis and samples along X-axis.
Discussion
Because of their dependence on specific biotic interactions, aboveground with pollinators and belowground with mycorrhizal fungi, orchids may be predisposed to become endangered, especially in a scenario of global climate change (Dearnaley et al., 2012). In addition to these well-studied interactions, orchids also establish multiple interactions with a pervasive plant microbiota that colonizes the interior of both below- and aboveground tissues. Endophytic bacteria likely play important functions for their host, and many bacterial endophytes isolated from orchid tissues showed plant growth promoting potential. For example, endophytic bacteria isolated from the tropical orchid Dendrobium nobile could promote orchid seed germination (Tsavkelova et al., 2016), and bacterial strains isolated from Cymbidium sp. could promote orchid acclimatization and/or growth (Faria et al., 2013; Gontijo et al., 2018). Although these observations suggest important roles of the bacterial orchid microbiota, very little is known about orchid-associated microbial communities, the level of specificity of their interactions with the host, their distribution in the plant and their modes of transmission across individuals and generations.
Here, we have explored the association of three terrestrial orchid species with their endophytic microbiota. Our analysis targeted the different taxonomic groups of bacteria that populated both vegetative and reproductive orchid organs, which, to the best of our knowledge, have not yet been studied in depth. By using an NGS-based metabarcoding approach, we have revealed different taxonomic groups in the microbiota of N. ovata, S. vomeracea, and S. spiralis, which could potentially play key roles by interacting with the host. The results confirmed our first hypothesis that the microbiota is organized in distinctive phylogenetic structures in various plant species and organs. Analysis of alpha and beta-diversity suggested that organs were the main determinant of the bacterial composition of the microbiota in orchids. There was in fact no evidence of clustering of bacterial communities according to plant species, and N. ovata and S. vomeracea were highly similar despite the very different habitat they colonize. N. ovata was collected in woods, whereas S. vomeracea is a meadow orchid species that was collected on poor grassland, a habitat more similar to the one supporting S. spiralis growth. This is in contrast to studies performed on different Agave species (Coleman-Derr et al., 2016) but is coherent with a recent study on the diversity patterns of endophytic bacterial communities of three rainforest plant species, where an extensive overlap in bacterial communities was observed between different plant organs and plant species (Haruna et al., 2018). Significant differences between plant organs were actually detected in our orchids, with a clear trend of decreasing diversity and richness along the vertical axis, that became significant when the two extremes (capsules and roots) were compared. This situation suggests an active role of the orchid plants in selecting specific bacterial taxa, more and more specialized while moving from the below-ground to the aerial parts. Bacterial root endophytes likely derive from the soil and rhizosphere microbiota (Compant et al., 2010). Once they enter the root tissues, bacterial endophytes can spread and colonize aerial plant parts, although endophytic bacteria can also enter the leaves through the stomata (Senthilkumar et al., 2011).
Endophytic bacteria in plant organs are not randomly distributed, and some dominant phyla appear to be tissue- or organ-specific. Numerous studies, conducted with different plant organs, revealed significant differences in the composition of the plant endophytic microbiota (Bodenhausen et al., 2013; Romero et al., 2014; Akinsanya et al., 2015; Maropola et al., 2015; Sarria-Guzmán et al., 2016; Li et al., 2017; Tian and Zhang, 2017; Yang et al., 2017). In the three orchid species, Proteobacteria and Actinobacteria were the dominant groups, irrespective of the plant species, but with differences in the distribution in the plant organs. In addition, the distribution of the bacterial genera did not determine a clear clustering by species/organ. This suggests a good overlap of the key members of the community in the different orchid species, some of which could be involved in essential processes, such as nitrogen fixation (Sun et al., 2004; Moyes et al., 2016; Puri et al., 2016).
Among the most representative genera, Cutibacterium has been identified, which is widely known as a commensal and is part of the human skin microbiota. However, a recent study, in support of our data, demonstrated the presence of Cutibacterium as an endophyte of roots and leaves in the wheat cultivars Triticum spelta cv Rokosz and Triticum aestivum cv Hondia (Kuźniar et al., 2020). It is also very interesting, as reported by Shaffer et al. (2018), that this genus has been classified as a hyphal endosymbiont of a leaf fungal endophyte (Fusarium concolor) from Hybanthus prunifolius. This aspect deserves to be deepened by further studies to understand the functional roles that could possibly mediate tripartite interactions with orchid mycorrhizal fungi.
The other most representative genera, distributed differently in the three orchid species, were Pseudomonas, Luteibacter, Cupriavidus, Limnobacter, Pantoea, and Acinetobacter which are included among the genera most commonly found as bacterial endophytes (Marquez-Santacruz et al., 2010; Romero et al., 2014; Shi et al., 2014). Pseudomonas species, thanks to their metabolic versatility and ubiquity, colonize several natural habitats by adopting a variety of lifestyles. This genus has been identified as an endosymbiont group in Australian terrestrial orchids (Wilkinson et al., 1989, 1994), and in the roots of epiphytic orchids (Tsavkelova et al., 2004).
The genus Pantoea is a diverse group of bacteria commonly found in association with plants. Pantoea endophytes were found mainly in plants such as vine, pea, rice and barley (Bell et al., 1995; Elvira-Recuenco and van Vuurde, 2000; Mano et al., 2007; Rahman et al., 2018).
Their potential role as PGP for host plants is well documented, such as ACC-deaminase activity (Zhang et al., 2011), hormone production (Tsavkelova et al., 2007; Chalupowicz et al., 2009), and siderophores (Loaces et al., 2011). As demonstrated by our recent study (Alibrandi et al., 2020), endophytes isolated from the same orchid species investigated in the current work, belonging to Pseudomonas and Pantoea genera, can use different mechanisms to promote plant growth. Strains classified in the genus Luteibacter were found in close association with plants (Johansen et al., 2005; Aamot et al., 2017) and lichens (Cardinale et al., 2006). In Luteibacter genus, some strains were also characterized as endohyphal bacteria within a range of ascomycete fungi (Arendt et al., 2016; Araldi-Brondolo et al., 2017). These bacterial / fungal complexes modulate several fungal phenotypes, including the possibility that some fungi act as plant growth promoters (Arendt et al., 2016; Araldi-Brondolo et al., 2017). This genus was mainly distributed in the two orchid species (N. ovata and S. vomeracea) that showed no significant differences in microbiota structure, suggesting a species-dependent colonization for this genus.
Members of the Limnobacter genus have often been detected in various environments, such as surface seawater, deep ocean, human intestine and volcanic deposits (Lu et al., 2008; Eloe et al., 2010; Rigsbee et al., 2011; Vedler et al., 2013). However, Limnobacter was found in maize seeds (cultivar Ngonda 108) at different stages of growth (Liu et al., 2013). We have identified Limnobacter as the dominant genus in N. ovata capsules, indicating that these bacteria tend to colonize preferentially the aerial parts of the orchids.
Cupriavidus species have been commonly isolated in different habitats from soil, water, plant nodules and human medical specimens (Coenye et al., 1999, 2003; Chen et al., 2001; Goris et al., 2001; Sato et al., 2006). It is known that some strains isolated from different mimosa species growing in distinct regions of China, Costa Rica, Taiwan, and Papua New Guinea (Chen et al., 2003; Barrett and Parker, 2006; Elliott et al., 2009; Liu et al., 2011) induce the formation of root nodules in legume plants. The Cupriavidus genus was found mainly in N. ovata and S. vomeracea (more abundant in the stems) and was distributed in both above- and below-ground organs. This result suggests that members of this genus are able to colonize the whole plant regardless of the organ.
Acinetobacter includes heterogeneous bacteria, typically free-living saprophytes and ubiquitous, associated with various habitats, e.g., soil, water, wastewater, humans, used kitchen sponges, food and animals (Visca et al., 2011; Doughari et al., 2011; Jung and Park, 2015; Cardinale et al., 2017). Acinetobacter is also known as an endophytic bacterium and plant growth promoter. A. calcoaceticus strains isolated from coffee plant tissues were capable of producing phosphatases and siderophores (Silva et al., 2012). Similarly, A. johnsonii isolated from surface-sterilized roots of Beta vulgaris showed the ability to increase the absorption of N, P, K, and Mg and produced the phytohormone auxin. Through these traits, this PGP strain was able to promote the growth of sugar beet by increasing the yield of sucrose and fructose (Shi et al., 2011). We found the Acinetobacter genus distributed in all organs of S. spiralis, which indicates a species preference for colonization independently of the plant organ.
Co-occurrence analysis identified four hub OTUs, two of them being typical plant-associated taxa, namely Pseudoxanthomonas and Rhizobium. Rhizobium typically forms nitrogen fixing symbioses with legume roots but is also able to promote plant growth in non-legume plants (García-Fraile et al., 2012). Mitsuaria is a genus within the Betaproteobacteriales that includes Gram-negative, obligate aerobic, oxidase, and catalase positive bacteria (Amakata et al., 2005). To date, only three species have been identified: M. chitosanitabida, isolated from soil (Amakata et al., 2005), M. noduli, isolated from root nodules of plants growing in soils contaminated with heavy metals (Fan et al., 2018), and very recently M. chitinivorans isolated from tube well water (Sisinthy and Gundlapally, 2020). Mitsuaria isolates have been also identified as plant-associated microbes both in the rhizosphere and as plant endophytes of both monocotyledons and dicotyledons (Nascimento et al., 2015; Huang et al., 2017; Fan et al., 2018). Mitsuaria has been reported to have biocontrol effects against phytopathogens, such as Rhizoctonia solani and Pythium aphanidermatum in tomato and soybean (Benítez et al., 2009). This ability may relate to the unique chitinase and chitosanase that are characteristics of Mitsuaria (Sisinthy and Gundlapally, 2020). A suppressive effect of Mitsuaria species was also demonstrated against bacterial diseases caused by Ralstonia pseudosolanacearum (Marian et al., 2018) and drought stress in Arabidopsis thaliana (Huang et al., 2017). It is worth noting that Mitsuaria is part of the core microbiota of all three orchid species tested (Supplementary Table 6); the multiple relationships of the genus Mitsuaria with the other members of the orchid microbiota could generate additional synergistic effects for the benefit of the host, as recently suggested in a study on tomato plants (Marian et al., 2019).
Thermus is a genus of Gram-negative thermophilic bacteria belonging to the phylum Deinococcus-Thermus. As the name suggests, one of its peculiar characteristics is the ability to live in environments with extreme temperatures. Thermus-like bacteria have been found in both shallow and deep-sea marine hydrothermal systems, as well as in low saline sulfate sources (Huber and Stetter, 2004). It was therefore surprising to find it as an orchid endophyte, and more as a hub taxon. To date, there are no studies of Thermus bacteria-plant interaction, and therefore this aspect deserves attention for future studies.
On the one hand, the core microbiota included the most abundant OTUs, most of which were related to plant-beneficial taxa. However, it was interesting that the OTUs shared between roots and other organs concerned only less abundant OTUs, suggesting an important role also for the less abundant bacteria. Some microbial taxa that occur in low abundance are called “satellite taxa” (Hanski, 1982; Magurran and Henderson, 2003) and can be defined mainly based on their local abundance and specificity to the habitat (Jousset et al., 2017). It was recently suggested that taxa occurring in low abundance could manage to counteract unwanted microbial invasions in soil communities (Mallon et al., 2015) and largely contributed to the production of volatile antifungal compounds that finally protected plants from soil borne phytopathogens (Hol et al., 2015).
Seeds can host the microbiota potentially inheritable by the next plant generation, thus restoring the microbiota imprinting from the parental plants. The seed microbiota has attracted increasing interest in the last years. Seed-associated bacteria have been detected in seeds of various crop species, including rice (Bacilio-Jiménez et al., 2001; Cottyn et al., 2001; Hardoim et al., 2012), maize (Liu et al., 2013), tobacco (Mastretta et al., 2009), coffee (Vega et al., 2005), quinoa (Pitzschke, 2016), common bean (López-López et al., 2010), grapevine (Compant et al., 2011), barley (Rahman et al., 2018), pumpkin (Fürnkranz et al., 2012) and alfalfa (Charkowski et al., 2001), as well as in wild plants such as cardon cactus (Pachycereus pringlei) (Puente et al., 2009), Eucalyptus spp. (Ferreira et al., 2008), Norwegian spruce (Picea abies) (Cankar et al., 2005) and the South American tree Anadenanthera colubrina (Alibrandi et al., 2018). Surprisingly, the analysis of the S. spiralis seed microbiota showed that these reproductive organs were the niche of the highest number of OTUs, about twice that of the roots (organs usually associated with a greater microbial diversity). This finding contradicts several studies, which suggested that seed endophytic microbiota tends to be paucispecific, generally limited to few genera, mainly Bacillus, Pseudomonas, Paenibacillus, Micrococcus, Staphylococcus, Pantoea, and Acinetobacter (Truyens et al., 2015). Despite showing the highest microbiota diversity, the S. spiralis seeds further filtered the microbiota core (from 35 to 30 OTUs) and confirmed our second hypothesis that the seeds may represent a reservoir of endophytes that can be vertically transmitted. The most unexpected result was the large number of OTUs shared between the seeds and the roots (Figure 8), that was much higher than the fraction of the microbiota shared between the seeds and the capsules. It was curious to find that capsules and seeds (which are inside the capsules) only shared 26 OTUs, 10 times less than those shared between roots and seeds. As these shared OTUs were not identified in the other above-ground orchid tissues, the most likely explanation is that they may originate from outside the plants. A possible hypothesis on the origin of such abundant seed microbiota in S. spiralis is via the pollen. Plant pollens are a rich source of microbes (Ambika Manirajan et al., 2016, 2018), and bacteria experimentally introduced into flowers were retrieved in the seed microbiome (Mitter et al., 2017). Therefore, pollen-associated bacteria are considered to contribute to the assemblage of the seed microbiota (Rodríguez et al., 2018). Furthermore, pollination in orchid plants does not rely on individual pollen grains, but on the so-called “pollinia” (clusters of tightly aggregated pollen grains), which might increase the microbial load at each pollination event. Another possible origin is the aerosol generated by the rain when it hits the soil (Joung et al., 2017). In this case, the aerosol droplets carrying soil microbes could reach aerial parts, adhering to the sticky flower tissues and being then collected in the flower cavities more than on other plant surfaces, where rains can flush them away rapidly (Allard et al., 2020). This interesting finding should be further investigated in other sampling sites, years and on other orchid species, to generalize whether Orchidaceae can be considered as a plant family with an abundant and root-like seed microbiome. Further studies will be also required to verify the actual vertical transmission of seed-associated microbes in orchids, as well as their role in orchid growth and health.
Conclusion
In this study, we analyzed the total endophytic bacterial microbiota associated to three terrestrial orchid species. We found that:
-
-
Plant organs influenced the diversity and structure of the endophytic community more than plant species.
-
-
The systemic colonization of the orchid microbiota followed a common pattern, with a reduction of diversity from the roots to the capsules, which indicates an active role of the plant in the process of microbiome selection.
-
-
The conspicuous diversity of the seed microbiota of Spiranthes spiralis, which notably overlapped with that of the roots, suggests a role of orchid seeds as a vector of microbes to restore the rhizosphere microbiome. This aspect further supports the evidence of a beneficial role of the orchid microbiota for the orchid host.
This work contributes to increase our knowledge on the diversity of the bacterial microbiota of terrestrial orchids, opening new hitherto unexplored scenarios for orchid holobiont studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB40289.
Author Contributions
PA performed the experiments, analyzed the data, and wrote the manuscript. SS contributed to the experiments and data interpretation, and critically reviewed the manuscript. SP conceived the study, supervised the experiments, validated the data, and wrote and critically reviewed the manuscript. MC conceived the study, supervised the experiments and the data analysis, and wrote and critically reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jacopo Calevo and Miriam Bazzicalupo (Turin, Italy) for their precious help with sampling and Bellinda Schneider (Giessen) for the support with IonTorrent sequencing. We thank Matteo Chialva, Francesco Venice, and Alessandro Silvestri (Turin, Italy) for their important contribution on the use of R software and the preparation for archiving of high-throughput sequencing data.
Funding. PA was supported by a Ph.D. fellowship by the Italian Ministry of Education, University and Research (MIUR), and by a mobility grant by the German Exchange Service (DAAD), grant number 57381332.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.604964/full#supplementary-material
References
- Aamot H. U., Hofgaard I. S., Lysøe E. (2017). Complete genome sequence of Luteibacter rhizovicinus strain LJ96T, isolated from the rhizosphere of barley (Hordeum vulgare L.) in Denmark. Genom. Data 11 104–105. 10.1016/j.gdata.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agler M. T., Ruhe J., Kroll S., Morhenn C., Kim S.-T., Weigel D., et al. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14:e1002352. 10.1371/journal.pbio.1002352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinsanya M. A., Goh J. K., Lim S. P., Ting A. S. (2015). Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. Genom. Data 6 159–163. 10.1016/j.gdata.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibrandi P., Cardinale M., Rahman M. D. M., Strati F., Ciná P., de Viana M. L., et al. (2018). The seed endosphere of Anadenanthera colubrina is inhabited by a complex microbiota, including Methylobacterium spp. and Staphylococcus spp. with potential plant-growth promoting activities. Plant Soil 422 81–99. 10.1007/s11104-017-3182-4 [DOI] [Google Scholar]
- Alibrandi P., Lo Monaco N., Calevo J., Voyron S., Puglia A. M., Cardinale M., et al. (2020). Plant growth promoting potential of bacterial endophytes from three terrestrial mediterranean orchid species. Plant Biosyst. 10.1080/11263504.2020.1829731 [DOI] [Google Scholar]
- Allard S. M., Ottesen A. R., Micallef S. A. (2020). Rain induces temporary shifts in epiphytic bacterial communities of cucumber and tomato fruit. Sci. Rep. 10:1765 10.1038/s41598-020-58671-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amakata D., Matsuo Y., Shimono K., Park J. K., Yun C. S., Matsuda H., et al. (2005). Mitsuaria chitosanitabida gen. nov., sp. nov., an aerobic, chitosanase-producing member of the ‘Betaproteobacteria.’. Int. J. Syst. Evol. Microbiol. 55(Pt 5), 1927–1932. 10.1099/ijs.0.63629-0 [DOI] [PubMed] [Google Scholar]
- Ambika Manirajan B., Maisinger C., Ratering S., Rusch V., Schwiertz A., Cardinale M., et al. (2018). Diversity, specificity, co-occurrence and hub taxa of the bacterial–fungal pollen microbiome. FEMS Microbiol. Ecol. 94:fiy112. 10.1093/femsec/fiy112 [DOI] [PubMed] [Google Scholar]
- Ambika Manirajan B., Ratering S., Rusch V., Schwiertz A., Geissler-Plaum R., Cardinale M., et al. (2016). Bacterial microbiota associated with flower pollen is influenced by pollination type, and shows a high degree of diversity and species-specificity. Environ. Microbiol. 18 5161–5174. 10.1111/1462-2920.13524 [DOI] [PubMed] [Google Scholar]
- Araldi-Brondolo S. J., Spraker J., Shaffer J. P., Woytenko E. H., Baltrus D. A., Gallery R. E., et al. (2017). Bacterial endosymbionts: master modulators of fungal phenotypes. in The Fungal Kingdom, eds J. Heitman, B. Howlett, P. Crous, E. Stukenbrock, T. James, N. Gow (Washington, DC:ASM Press), 981–1004. 10.1128/microbiolspec.FUNK-0056-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt K. R., Hockett K. L., Araldi-Brondolo S. J., Baltrus D. A., Arnold A. E. (2016). Isolation of endohyphal bacteria from foliar ascomycota and in vitro establishment of their symbiotic associations. Appl. Env. Microbiol. 82 2943–2949. 10.1128/AEM.00452-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacilio-Jiménez M., Aguilar-Flores S., del Valle M. V., Pérez A., Zepeda A., Zenteno E. (2001). Endophytic bacteria in rice seeds inhibit early colonization of roots by Azospirillum brasilense. Soil Biol. Biochem. 33 167–172. 10.1016/S0038-0717(00)00126-7 [DOI] [Google Scholar]
- Barrett C. F., Parker M. A. (2006). Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. Nodule Bacteria on two Mimosa spp. in Costa Rica. Appl. Environ. Microbiol. 72 1198–1206. 10.1128/AEM.72.2.1198-1206.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C. R., Dickie G. A., Harvey W. L. G., Chan J. W. Y. F. (1995). Endophytic bacteria in grapevine. Can. J. Microbiol. 41 46–53. 10.1139/m95-006 [DOI] [Google Scholar]
- Benítez M.-S., McSpadden Gardener B. B. (2009). Linking sequence to function in soil bacteria: sequence-directed isolation of novel bacteria contributing to soilborne plant disease suppression. Appl. Env. Microbiol. 75 915–924. 10.1128/AEM.01296-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M., Koskella B. (2018). Nutrient- and dose-dependent microbiome-mediated protection against a plant pathogen. Curr. Biol. 28 2487.e3–2492.e3. 10.1016/j.cub.2018.05.085 [DOI] [PubMed] [Google Scholar]
- Berry D., Widder S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5:219. 10.3389/fmicb.2014.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen N., Horton M. W., Bergelson J. (2013). Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8:e56329. 10.1371/journal.pone.0056329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G., Compant S., Vescio K., Mitter B., Trognitz F., Ma L.-J., et al. (2017). Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 55 61–83. 10.1146/annurev-phyto-080516-035641 [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Rott M., Schlaeppi K., van Themaat E. V. L., Ahmadinejad N., Assenza F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488 91–95. 10.1038/nature11336 [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., van Themaat E. V. L., Schulze-Lefert P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64 807–838. 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- Cankar K., Kraigher H., Ravnikar M., Rupnik M. (2005). Bacterial endophytes from seeds of Norway spruce (Picea abies L. Karst). FEMS Microbiol. Lett. 244 341–345. 10.1016/j.femsle.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale M., Puglia A. M., Grube M. (2006). Molecular analysis of lichen-associated bacterial communities. FEMS Microbiol. Ecol. 57 484–495. 10.1111/j.1574-6941.2006.00133.x [DOI] [PubMed] [Google Scholar]
- Cardinale M., Kaiser D., Lueders T., Schnell S., Egert M. (2017). Microbiome analysis and confocal microscopy of used kitchen sponges reveal massive colonization by Acinetobacter, Moraxella and Chryseobacterium species. Sci. Rep. 7:5791. 10.1038/s41598-017-06055-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupowicz L., Barash I., Panijel M., Sessa G., Manulis-Sasson S. (2009). Regulatory interactions between quorum-sensing, auxin, cytokinin, and the Hrp regulon in relation to gall formation and epiphytic fitness of pantoea agglomerans pv. gypsophilae. Mol. Plant Microbe Int. 22 849–856. 10.1094/MPMI-22-7-0849 [DOI] [PubMed] [Google Scholar]
- Charkowski A. O., Sarreal C. Z., Mandrell R. E. (2001). Wrinkled alfalfa seeds harbour more aerobic bacteria and are more difficult to sanitize than smooth seeds. J. Food Prot. 64 1292–1298. 10.4315/0362-028X-64.9.1292 [DOI] [PubMed] [Google Scholar]
- Chen W.-M., Laevens S., Lee T. M., Coenye T., De Vos P., Mergeay M., et al. (2001). Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. Evol. Microbiol. 51 1729–1735. 10.1099/00207713-51-5-1729 [DOI] [PubMed] [Google Scholar]
- Chen W.-M., Moulin L., Bontemps C., Vandamme P., Béna G., Boivin-Masson C. (2003). Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J. Bacteriol. 185 7266–7272. 10.1128/jb.185.24.7266-7272.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-M., Tang Y. Q., Mori K., Wu X. L. (2012). Distribution of culturable endophytic bacteria in aquatic plants and their potential for bioremediation in polluted waters. Aquat. Biol. 15 99–110. 10.3354/ab00422 [DOI] [Google Scholar]
- Christenhusz M. J. M., Byng J. W. (2016). The number of known plants species in the world and its annual increase. Phytotaxa 261 201–217. 10.11646/phytotaxa.261.3.1 24943631 [DOI] [Google Scholar]
- Claesson M. J., O’Sullivan O., Wang Q., Nikkilä J., Marchesi J. R., Smidt H., et al. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. 10.1371/journal.pone.0006669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T., Falsen E., Vancanneyt M., Hoste B., Govan J. R. W., Kersters K., et al. (1999). Classification of Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. Int. J. Syst. Evol. Microbiol. 49 405–413. 10.1099/00207713-49-2-405 [DOI] [PubMed] [Google Scholar]
- Coenye T., Vandamme P., LiPuma J. J. (2003). Ralstonia respiraculi sp. nov., isolated from the respiratory tract of cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 53 1339–1342. 10.1099/ijs.0.02440-0 [DOI] [PubMed] [Google Scholar]
- Coleman-Derr D., Desgarennes D., Fonseca-Garcia C., Gross S., Clingenpeel S., Woyke T., et al. (2016). Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 209 798–811. 10.1111/nph.13697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S., Clément C., Sessitsch A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42 669–678. 10.1016/j.soilbio.2009.11.024 [DOI] [Google Scholar]
- Compant S., Mitter B., Colli-Mull J. G., Gangl H., Sessitsch A. (2011). Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb. Ecol. 62 188–197. 10.1007/s00248-011-9883-y [DOI] [PubMed] [Google Scholar]
- Compant S., Samad A., Faist H., Sessitsch A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19 29–37. 10.1016/j.jare.2019.03.004.C [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottyn B., Regalado E., Lanoot B., De Cleene M., Mew T. W., Swings J. (2001). Bacterial populations associated with rice seed in the tropical environment. Phytopathology 91 282–292. 10.1094/phyto.2001.91.3.282 [DOI] [PubMed] [Google Scholar]
- da Silva J. A. T., Tsavkelova E. A., Zeng S., Ng T. B., Parthibhan S., Dobránszki J., et al. (2015). Symbiotic in vitro seed propagation of Dendrobium: fungal and bacterial partners and their influence on plant growth and development. Planta 242 1–22. 10.1007/s00425-015-2301-9 [DOI] [PubMed] [Google Scholar]
- Das K., Prasanna R., Saxena A. K. (2017). Rhizobia: a potential biocontrol agent for soilborne fungal pathogens. Folia Microbiol. 62 425–435. 10.1007/s12223-017-0513-z [DOI] [PubMed] [Google Scholar]
- de Vrieze M., Germanier F., Vuille N., Weisskopf L. (2018). Combining different potato-associated Pseudomonas strains for improved biocontrol of Phytophthora infestans. Front. Microbiol. 9:2573. 10.3389/fmicb.2018.02573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearnaley J. D. W., Martos F., Selosse M. A. (2012). “Orchid mycorrhizas: molecular ecology, physiology, evolution and conservation aspects,” in Fungal Associations. The Mycota. A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research, Vol. 9 ed. Hock B. (Heidelberg: Springer; ), 207–230. 10.1007/978-3-642-30826-0_12 [DOI] [Google Scholar]
- Doughari H. J., Ndakidemi P. A., Human I. S., Benade S. (2011). The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes Environ. 26 101–112. 10.1264/jsme2.ME10179 [DOI] [PubMed] [Google Scholar]
- Duca D., Lorv J., Patten C. L., Rose D., Glick B. R. (2014). Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek 106 85–125. 10.1007/s10482-013-0095-y [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Elliott G. N., Chou J.-H., Chen W.-M., Bloemberg G. V., Bontemps C., Martínez-Romero E., et al. (2009). Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ. Microbiol. 11 762–778. 10.1111/j.1462-2920.2008.01799.x [DOI] [PubMed] [Google Scholar]
- Eloe E. A., Shulse C. N., Fadrosh D. W., Williamson S. J., Allen E. E., Bartlett D. H. (2010). Compositional differences in particle-associated and free-living microbial assemblages from an extreme deep-ocean environment. Environ. Microbiol. Rep. 3 449–458. 10.1111/j.1758-2229.2010.00223.x [DOI] [PubMed] [Google Scholar]
- Elvira-Recuenco M., van Vuurde J. W. L. (2000). Natural incidence of endophytic bacteria in pea cultivars under field conditions. Can. J. Microbiol. 46 1036–1041. 10.1139/w00-098 [DOI] [PubMed] [Google Scholar]
- Engelbrektson A., Kunin V., Wrighton K. C., Zvenigorodsky N., Chen F., Ochman H., et al. (2010). Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 4 642–647. 10.1038/ismej.2009.153 [DOI] [PubMed] [Google Scholar]
- Escudero-Martinez C., Bulgarelli D. (2019). Tracing the evolutionary routes of plant–microbiota interactions. Curr. Opin. Microbiol. 49 34–40. 10.1016/j.mib.2019.09.013 [DOI] [PubMed] [Google Scholar]
- Fan M.-C., Nan L.-J., Zhu Y.-M., Chen W.-M., Wei G.-H., Lin Y.-B. (2018). Mitsuaria noduli sp. nov., isolated from the root nodules of Robinia pseudoacacia in a lead–zinc mine. Int. J. Syst. Evol. Microbiol. 68:87. 10.1099/ijsem.0.002459 [DOI] [PubMed] [Google Scholar]
- Faria D. C., Dias A. C. F., Melo I. S., de Carvalho Costa F. E. (2013). Endophytic bacteria isolated from orchids and their potential to promote plant growth. World J. Microbiol. Biotechnol. 29 217–221. 10.1007/s11274-012-1173-4 [DOI] [PubMed] [Google Scholar]
- Faust K., Raes J. (2016). CoNet app: inference of biological association networks using Cytoscape. F1000Res 5:1519. 10.12688/f1000research.9050.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A., Quecine M. C., Lacava P. T., Oda S., Azevedo J. L., Araújo W. L. (2008). Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 287 8–14. 10.1111/j.1574-6968.2008.01258.x [DOI] [PubMed] [Google Scholar]
- Fürnkranz M., Lukesch B., Müller H., Huss H., Grube M., Berg G. (2012). Microbial diversity inside pumpkins: microhabitat-specific communities display a high antagonistic potential against phytopathogens. Microb. Ecol. 63 418–428. 10.1007/s00248-011-9942-4 [DOI] [PubMed] [Google Scholar]
- García-Fraile P., Carro L., Robledo M., Ramírez-Bahena M.-H., Flores-Félix J.-D., Fernández M. T., et al. (2012). Rhizobium promotes non-legumes growth and quality in several production steps: towards a biofertilization of edible raw vegetables healthy for humans. PLoS One 7:e38122. 10.1371/journal.pone.0038122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A., Lanfranco L., Perotto S., Bonfante P. (2020). Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 18 649–660. 10.1038/s41579-020-0402-3 [DOI] [PubMed] [Google Scholar]
- Gergs R., Rothhaupt K.-O. (2015). Invasive species as driving factors for the structure of benthic communities in Lake Constance, Germany. Hydrobiologia 746 245–254. 10.1007/s10750-014-1931-4 [DOI] [Google Scholar]
- Glick B. R. (1995). The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41 109–117. 10.1139/m95-015 [DOI] [Google Scholar]
- Glick B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Sci. Cairo 2012:963401. 10.6064/2012/963401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R., Penrose D. M., Li J. (1998). A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J. Theor. Biol. 190 63–68. 10.1006/jtbi.1997.0532 [DOI] [PubMed] [Google Scholar]
- Gontijo J. B., Andrade G. V. S., Baldotto M. A., Baldotto L. E. B. (2018). Bioprospecting and selection of growth-promoting bacteria for Cymbidium sp. orchids. Sci. Agric. 75 368–374. 10.1590/1678-992x-2017-0117 [DOI] [Google Scholar]
- Goris J., De Vos P., Coenye T., Hoste B., Janssens D., Brim H., et al. (2001). Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. 1998 emend. Int. J. Syst. Evol. Microbiol. 51 1773–1782. 10.1099/00207713-51-5-1773 [DOI] [PubMed] [Google Scholar]
- Govindarajan M., Balandreau J., Kwon S. W., Weon H. Y., Lakshminarasimhan C. (2008). Effects of the inoculation of Burkholderia vietnamensis and related endophytic diazotrophic bacteria on grain yield of rice. Microb. Ecol. 55 21–37. 10.1007/s00248-007-9247-9 [DOI] [PubMed] [Google Scholar]
- Guo H., Luo S., Chen L., Xiao X., Xi Q., Wei W., et al. (2010). Bioremediation of heavy metals by growing hyperaccumulator endophytic bacterium Bacillus sp. L14. Bioresour. Technol. 101 8599–8605. 10.1016/j.biortech.2010.06.085 [DOI] [PubMed] [Google Scholar]
- Hanski I. (1982). Dynamics of regional distribution: the core and satellite species hypothesis. Oikos 38 210–221. 10.2307/3544021 [DOI] [Google Scholar]
- Hardoim P. R., Hardoim C. C. P., van Overbeek L. S., van Elsas J. D. (2012). Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One 7:e30438. 10.1371/journal.pone.0030438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim P. R., van Overbeek L. S., Berg G., Pirttilä A. M., Compant S., Campisano A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79 293–320. 10.1128/mmbr.00050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruna E., Zin N. M., Kerfahi D., Adams J. M. (2018). Extensive overlap of tropical rainforest bacterial endophytes between soil, plant parts, and plant species. Microb. Ecol. 75 88–103. 10.1007/s00248-017-1002-2 [DOI] [PubMed] [Google Scholar]
- Herrera H., Sanhueza T., Novotná A., Charles T. C., Arriagada C. (2020). Isolation and identification of endophytic bacteria from mycorrhizal tissues of terrestrial orchids from Southern Chile. Diversity 12:55 10.3390/d12020055 [DOI] [Google Scholar]
- Hol W. H. G., Garbeva P., Hordijk C., Hundscheid M. P. J., Gunnewiek P. J. A. K., van Agtmaal M., et al. (2015). Non-random species loss in bacterial communities reduces antifungal volatile production. Ecology 96 2042–2048. 10.1890/14-2359.1 [DOI] [PubMed] [Google Scholar]
- Hossain M., Sultana F., Islam S. (2017). “Plant growth-promoting fungi (PGPF): phytostimulation and induced systemic resistance,” in Plant-Microbe Interactions in Agro-Ecological Perspectives: Microbial Interactions and Agro-Ecological Impacts, Vol. 2 eds Singh D. P., Singh H. B., Prabha R. (Singapore: Springer; ), 135–191. 10.1007/978-981-10-6593-4_6 [DOI] [Google Scholar]
- Huang X.-F., Zhou D., Lapsansky E. R., Reardon K. F., Guo J., Andales M. J., et al. (2017). Mitsuaria sp. and Burkholderia sp. from Arabidopsis rhizosphere enhance drought tolerance in Arabidopsis thaliana and maize (Zea mays L.). Plant Soil 419 523–539. 10.1007/s11104-017-3360-4 [DOI] [Google Scholar]
- Huber R., Stetter K. O. (2004). The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. New York, NY: Springer-Verlag. [Google Scholar]
- Hynson N. A., Madsen T. P., Selosse M. A., Adam I. K. U., Ogura-Tsujita Y., Roy M., et al. (2013). “The physiological ecology of mycoheterotrophy,” in Mycoheterotrophy, ed. Merckx V. (New York, NY: Springer; ). [Google Scholar]
- Johansen J. E., Binnerup S. J., Kroer N., Mølbak L. (2005). Luteibacter rhizovicinus gen. nov., sp. nov., a yellow-pigmented gammaproteobacterium isolated from the rhizosphere of barley (Hordeum vulgare L.). Int. J. Syst. Evol. 55 2285–2291. 10.1099/ijs.0.63497-0 [DOI] [PubMed] [Google Scholar]
- Joung Y. S., Ge Z., Buie C. R. (2017). Bioaerosol generation by raindrops on soil. Nat. Commun. 8:14668. 10.1038/ncomms14668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset A., Bienhold C., Chatzinotas A., Gallien L., Gobet A., Kurm V., et al. (2017). Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 11 853–862. 10.1038/ismej.2016.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J., Park W. (2015). Acinetobacter species as model microorganisms in environmental microbiology: current state and perspectives. Appl. Microbiol. Biotechnol. 99 2533–2548. 10.1007/s00253-015-6439-y [DOI] [PubMed] [Google Scholar]
- Khare E., Mishra J., Arora N. K. (2018). Multifaceted interactions between endophytes and plant: developments and prospects. Front. Microbiol. 9:2732. 10.3389/fmicb.2018.02732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief C. (2014). Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 5:216. 10.3389/fpls.2014.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler J., Hernández J. A., Caravaca F., Roldán A. (2008). Plant-growth-promoting rhizobacteria and arbuscular mycorrhizal fungi modify alleviation biochemical mechanisms in water-stressed plants. Funct. Plant Biol. 35 141–151. 10.1071/FP07218 [DOI] [PubMed] [Google Scholar]
- Kumar A., Patel J. S., Meena V. S., Ramteke P. W. (2019). Plant growth-promoting rhizobacteria: strategies to improve abiotic stresses under sustainable agriculture. J. Plant Nutr. 42 1402–1415. 10.1080/01904167.2019.1616757 [DOI] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuźniar A., Włodarczyk K., Grz̨dziel J., Woźniak M., Furtak K., Wolińska A., et al. (2020). New insight into the composition of wheat seed microbiota. Int. J. Mol. Sci. 21:4634. 10.3390/ijms21134634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li O., Xiao R., Sun L., Guan C., Kong D., Hu X. (2017). Bacterial and diazotrophic diversities of endophytes in Dendrobium catenatum determined through barcoded pyrosequencing. PLoS One 12:e0184717. 10.1371/journal.pone.0184717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Y., Wu W., Wang E. T., Zhang B., Macdermott J., Chen W. X. (2011). Phylogenetic relationships and diversity of β-rhizobia associated with Mimosa species grown in Sishuangbanna, China. Int. J. Syst. Evol. Microbiol. 61 334–342. 10.1099/ijs.0.020560-0 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zuo S., Zou Y., Wang J., Song W. (2013). Investigation on diversity and population succession dynamics of endophytic bacteria from seeds of maize (Zea mays L., Nongda108) at different growth stages. Ann. Microbiol. 63 71–79. 10.1007/s13213-012-0446-3 [DOI] [Google Scholar]
- Loaces I., Ferrando L., Fernández Scavino A. (2011). Dynamics, diversity and function of endophytic siderophore-producing bacteria in rice. Microb. Ecol. 61 606–618. 10.1007/s00248-010-9780-9 [DOI] [PubMed] [Google Scholar]
- López-López A., Rogel M. A., Ormeño-Orrillo E., Martínez-Romero J., Martínez-Romero E. (2010). Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst. Appl. Microbiol. 33 322–327. 10.1016/j.syapm.2010.07.005 [DOI] [PubMed] [Google Scholar]
- Lu H., Fujimura R., Sato Y., Nanba K., Kamijo T., Ohta H. (2008). Characterization of Herbaspirillum- and Limnobacter-related strains isolated from young volcanic deposits in Miyake-Jima Island, Japan. Microb. Environ. 23 66–72. 10.1264/jsme2.23.66 [DOI] [PubMed] [Google Scholar]
- Lucaciu R., Pelikan C., Gerner S. M., Zioutis C., Köstlbacher S., Marx H., et al. (2019). A bioinformatics guide to plant microbiome analysis. Front. Plant Sci. 10:1313. 10.3389/fpls.2019.01313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg D., Lebeis S., Paredes S. H., Yourstone S., Gehring J., Malfatti S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488 86–90. 10.1038/nature11237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg D. S., Yourstone S., Mieczkowski P., Jones C. D., Dangl J. L. (2013). Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10 999–1002. 10.1038/nmeth.2634 [DOI] [PubMed] [Google Scholar]
- Magurran A. E., Henderson P. A. (2003). Explaining the excess of rare species in natural species abundance distributions. Nature 422 714–716. 10.1038/nature01547 [DOI] [PubMed] [Google Scholar]
- Malinowski D. P., Alloush G. A., Belesky D. P. (2000). Leaf endophyte Neotyphodium coenophialum modifies mineral uptake in tall fescue. Plant Soil 227 115–126. [Google Scholar]
- Mallon C. A., Poly F., Le Roux X., Marring I., van Elsas J. D., Salles J. F. (2015). Resource pulses can alleviate the biodiversity–invasion relationship in soil microbial communities. Ecology 96 915–926. 10.1890/14-1001.1 [DOI] [PubMed] [Google Scholar]
- Mano H., Tanaka F., Nakamura C., Kaga H., Morisaki H. (2007). Culturable endophytic bacterial flora of the maturing leaves and roots of rice plants (Oryza sativa) cultivated in a paddy field. Microb. Environ. 22 175–185. 10.1264/jsme2.22.175 26758818 [DOI] [Google Scholar]
- Marchesi J. R., Ravel J. (2015). The vocabulary of microbiome research: a proposal. Microbiome 3:31. 10.1186/s40168-015-0094-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marian M., Morita A., Koyama H., Suga H., Shimizu M. (2019). Enhanced biocontrol of tomato bacterial wilt using the combined application of Mitsuaria sp. TWR114 and nonpathogenic Ralstonia sp. TCR112. J. Gen. Plant Pathol. 85 142–154. 10.1007/s10327-018-00834-6 [DOI] [Google Scholar]
- Marian M., Nishioka T., Koyama H., Suga H., Shimizu M. (2018). Biocontrol potential of Ralstonia sp. TCR112 and Mitsuaria sp. TWR114 against tomato bacterial wilt. Appl. Soil Ecol. 128 71–80. 10.1016/j.apsoil.2018.04.005 [DOI] [Google Scholar]
- Maropola M. K. A., Ramond J.-B., Trindade M. (2015). Impact of metagenomic DNA extraction procedures on the identifiable endophytic bacterial diversity in Sorghum bicolor (L. Moench). J. Microbiol. Methods 112 104–117. 10.1016/j.mimet.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Marquez-Santacruz H. A., Hernandez-Leon R., Orozco-Mosqueda M. C., Velazquez-Sepulveda I., Santoyo G. (2010). Diversity of bacterial endophytes in roots of Mexican husk tomato plants (Physalis ixocarpa) and their detection in the rhizosphere. Genet. Mol. Res. 9 2372–2380. 10.4238/vol9-4gmr921 [DOI] [PubMed] [Google Scholar]
- Masson-Boivin C., Sachs J. L. (2018). Symbiotic nitrogen fixation by rhizobia: the roots of a success story. Curr. Opin. Plant Biol. 44 7–15. 10.1016/j.pbi.2017.12.001 [DOI] [PubMed] [Google Scholar]
- Mastretta C., Taghavi S., van der Lelie D., Mengoni A., Galardi F., Gonnelli C., et al. (2009). Endophytic bacteria from seeds of Nicotiana tabacum can reduce cadmium phytotoxicity. Int. J. Phytoremed. 11 251–267. 10.1080/15226510802432678 [DOI] [Google Scholar]
- McCormick M. K., Whigham D. F., Canchani-Viruet A. (2018). Mycorrhizal fungi affect orchid distribution and population dynamics. New Phytol. 219 1207–1215. 10.1111/nph.15223 [DOI] [PubMed] [Google Scholar]
- Mitter B., Pfaffenbichler N., Flavell R., Compant S., Antonielli L., Petric A., et al. (2017). A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 8:11. 10.3389/fmicb.2017.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes A. B., Kueppers L. M., Pett-Ridge J., Carper D. L., Vandehey N., O’Neil J., et al. (2016). Evidence for foliar endophytic nitrogen fixation in a widely distributed subalpine conifer. New Phytol. 210 657–668. 10.1111/nph.13850 [DOI] [PubMed] [Google Scholar]
- Nascimento S. B., Lima A. M., Borges B. N., de Souza C. R. (2015). Endophytic bacteria from Piper tuberculatum Jacq.: isolation, molecular characterization, and in vitro screening for the control of Fusarium solani f. sp piperis, the causal agent of root rot disease in black pepper (Piper nigrum L.). Genet. Mol. Res. 14 7567–7577. 10.4238/2015.July.3.32 [DOI] [PubMed] [Google Scholar]
- Patten C. L., Glick B. R. (2002). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Env. Microbiol. 68 3795–3801. 10.1128/aem.68.8.3795-3801.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A. (2016). Developmental peculiarities and seed-borne endophytes in quinoa: omnipresent, robust bacilli contribute to plant fitness. Front. Microbiol. 7:2. 10.3389/fmicb.2016.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente M. E., Li C. Y., Bashan Y. (2009). Endophytic bacteria in cacti seeds can improve the development of cactus seedlings. Environ. Exp. Bot. 66 402–408. 10.1016/j.envexpbot.2009.04.007 [DOI] [Google Scholar]
- Puri A., Padda K. P., Chanway C. P. (2016). Evidence of nitrogen fixation and growth promotion in canola (Brassica napus L.) by an endophytic diazotroph Paenibacillus polymyxa P2b-2R. Biol. Fertil. Soils 52 119–125. 10.1007/s00374-015-1051-y [DOI] [Google Scholar]
- Rahman H., Sabreen S., Alam S., Kawai S. (2005). Effects of nickel on growth and composition of metal micronutrients in barley plants grown in nutrient solution. J. Plant Nutr. 28 393–404. 10.1081/PLN-200049149 [DOI] [Google Scholar]
- Rahman M. D. M., Flory E., Koyro H.-W., Abideen Z., Schikora A., Suarez C., et al. (2018). Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.). Syst. Appl. Microbiol. 41 386–398. 10.1016/j.syapm.2018.02.003 [DOI] [PubMed] [Google Scholar]
- Rashid S., Charles T. C., Glick B. R. (2012). Isolation and characterization of new plant growth-promoting bacterial endophytes. Appl. Soil Ecol. 61 217–224. 10.1016/j.apsoil.2011.09.011 [DOI] [Google Scholar]
- Rigsbee L., Agans R., Foy B. D., Paliy O. (2011). Optimizing the analysis of human intestinal microbiota with phylogenetic microarray. FEMS Microbiol. Ecol. 75 332–342. 10.1111/j.1574-6941.2010.01009.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez C. E., Mitter B., Barret M., Sessitsch A., Compant S. (2018). Commentary: seed bacterial inhabitants and their routes of colonization. Plant Soil 422 129–134. 10.1007/s11104-017-3368-9 [DOI] [Google Scholar]
- Romero F. M., Marina M., Pieckenstain F. L. (2014). The communities of tomato (Solanum lycopersicum L.) leaf endophytic bacteria, analyzed by 16S-ribosomal RNA gene pyrosequencing. FEMS Microbiol. Lett. 351 187–194. 10.1111/1574-6968.12377 [DOI] [PubMed] [Google Scholar]
- Santoyo G., Moreno-Hagelsieb G., del Carmen, Orozco-Mosqueda M., Glick B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183 92–99. 10.1016/j.micres.2015.11.008 [DOI] [PubMed] [Google Scholar]
- Sarria-Guzmán Y., Chávez-Romero Y., Gómez-Acata S., Montes-Molina J. A., Morales-Salazar E., Dendooven L., et al. (2016). Bacterial communities associated with different Anthurium andraeanum L. plant tissues. Microbes. Environ. 31 321–328. 10.1264/jsme2.ME16099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Nishihara H., Yoshida M., Watanabe M., Rondal J. D., Concepcion R. N., et al. (2006). Cupriavidus pinatubonensis sp. nov. and Cupriavidus laharis sp. nov., novel hydrogen-oxidizing, facultatively chemolithotrophic bacteria isolated from volcanic mudflow deposits from Mt. Pinatubo in the Philippines. Int. J. Syst. Evol. 56 973–978. 10.1099/ijs.0.63922-0 [DOI] [PubMed] [Google Scholar]
- Selosse M. A., Roy M. (2009). Green plants that feed on fungi: facts and questions about mixotrophy. Trends Plant Sci. 14 64–70. 10.1016/j.tplants.2008.11.004 [DOI] [PubMed] [Google Scholar]
- Senthilkumar M., Anandham R., Madhaiyan M., Venkateswaran V., Sa T. (2011). “Endophytic bacteria: perspectives and applications in agricultural crop production,” in Bacteria in Agrobiology: Crop Ecosystems, ed. Maheshwari D. (Berlin: Springer; ). [Google Scholar]
- Shaffer J. P., Zalamea P.-C., Sarmiento C., Gallery R. E., Dalling J. W., Davis A. S., et al. (2018). Context-dependent and variable effects of endohyphal bacteria on interactions between fungi and seeds. Fungal Ecol. 36 117–127. 10.1016/j.funeco.2018.08.008 [DOI] [Google Scholar]
- Shi Y., Lou K., Li C. (2011). Growth promotion effects of the endophyte Acinetobacter johnsonii strain 3-1 on sugar beet. Symbiosis 54 159–166. 10.1007/s13199-011-0139-x [DOI] [Google Scholar]
- Shi Y., Yang H., Zhang T., Sun J., Lou K. (2014). Illumina-based analysis of endophytic bacterial diversity and space-time dynamics in sugar beet on the north slope of Tianshan mountain. Appl. Microbiol. Biotechnol. 98 6375–6385. 10.1007/s00253-014-5720-9 [DOI] [PubMed] [Google Scholar]
- Silva H. S. A., Tozzi J. P. L., Terrasan C. R. F., Bettiol W. (2012). Endophytic microorganisms from coffee tissues as plant growth promoters and biocontrol agents of coffee leaf rust. Biol. Control 63 62–67. 10.1016/j.biocontrol.2012.06.005 [DOI] [Google Scholar]
- Sisinthy S., Gundlapally S. R. (2020). Mitsuaria chitinivorans sp. nov. a potential candidate for bioremediation: emended description of the genera Mitsuaria, Roseateles and Pelomonas. Arch. Microbiol. 202 1839–1848. 10.1007/s00203-020-01905-z [DOI] [PubMed] [Google Scholar]
- Stępniewska Z., Kuźniar A. (2013). Endophytic microorganisms—promising applications in bioremediation of greenhouse gases. Appl. Microbiol. Biotechnol. 97 9589–9596. 10.1007/s00253-013-5235-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., An S., Yang L., Wang Z. (2004). Chemical properties of the upper tailings beneath biotic crusts. Ecol. Eng. 23 47–53. 10.1016/j.ecoleng.2004.07.001 [DOI] [Google Scholar]
- Taghavi S., Garafola C., Monchy S., Newman L., Hoffman A., Weyens N., et al. (2009). Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl. Environ. Microbiol. 75 748–757. 10.1128/AEM.02239-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Lin X., Tian J., Ji L., Chen Y., Tran L.-S. P., et al. (2020). Research advances of beneficial microbiota associated with crop plants. Int. J. Mol. Sci. 21:1792. 10.3390/ijms21051792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X.-Y., Zhang C.-S. (2017). Illumina-based analysis of endophytic and rhizosphere bacterial diversity of the coastal halophyte Messerschmidia sibirica. Front. Microbiol. 8:2288. 10.3389/fmicb.2017.02288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truyens S., Weyens N., Cuypers A., Vangronsveld J. (2015). Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 7 40–50. 10.1111/1758-2229.12181 [DOI] [Google Scholar]
- Tsavkelova E. A., Cherdyntseva T. A., Botina S. G., Netrusov A. I. (2007). Bacteria associated with orchid roots and microbial production of auxin. Microbiol. Res. 162 69–76. 10.1016/j.micres.2006.07.014 [DOI] [PubMed] [Google Scholar]
- Tsavkelova E. A., Cherdyntseva T. A., Netrusov A. I. (2004). Bacteria associated with the roots of epiphytic orchids. Microbiology 73 710–715. 10.1007/s11021-005-0013-z [DOI] [PubMed] [Google Scholar]
- Tsavkelova E. A., Egorova M. A., Leontieva M. R., Malakho S. G., Kolomeitseva G. L., Netrusov A. I. (2016). Dendrobium nobile Lindl. seed germination in co-cultures with diverse associated bacteria. Plant Growth Regul. 80 79–91. 10.1007/s10725-016-0155-1 [DOI] [Google Scholar]
- Urquiaga S., Xavier R. P., de Morais R. F., Batista R. B., Schultz N., Leite J. M., et al. (2012). Evidence from field nitrogen balance and 15N natural abundance data for the contribution of biological N2 fixation to Brazilian sugarcane varieties. Plant Soil 356 5–21. 10.1007/s11104-011-1016-3 [DOI] [Google Scholar]
- Vedler E., Heinaru E., Jutkina J., Viggor S., Koressaar T., Remm M., et al. (2013). Limnobacter spp. as newly detected phenol-degraders among Baltic Sea surface water bacteria characterised by comparative analysis of catabolic genes. Syst. Appl. Microbiol. 36 525–532. 10.1016/j.syapm.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Vega F. E., Pava-Ripoll M., Posada F., Buyer J. S. (2005). Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 45 371–380. 10.1002/jobm.200410551 [DOI] [PubMed] [Google Scholar]
- Visca P., Seifert H., Towner K. J. (2011). Acinetobacter infection – an emerging threat to human health. IUBMB Life 63 1048—1054. 10.1002/iub.534 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2016). Getting Started with ggplot2. In: ggplot2. Use R!. Cham: Springer. [Google Scholar]
- Wilkinson K. G., Dixon K. W., Sivasithamparam K. (1989). Interaction of soil bacteria, mycorrhizal fungi and orchid seed in relation to germination of Australian orchids. New Phytol. 112 429–435. 10.1111/j.1469-8137.1989.tb00334.x [DOI] [Google Scholar]
- Wilkinson K. G., Dixon K. W., Sivasithamparam K., Ghisalberti E. L. (1994). Effect of IAA on symbiotic germination of an Australian orchid and its production by orchid-associated bacteria. Plant Soil 159 291–295. 10.1007/BF00009292 [DOI] [Google Scholar]
- Yang R., Liu, P., Ye W. (2017). Illumina-based analysis of endophytic bacterial diversity of tree peony (Paeonia Sect. Moutan) roots and leaves. Braz. J. Microbiol. 48 695–705. 10.1016/j.bjm.2017.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhou X.-F., Yang S.-J., Liu W.-H., Hu X.-F. (2013). Design and application of specific 16S rDNA-targeted primers for assessing endophytic diversity in Dendrobium officinale using nested PCR-DGGE. Appl. Microbiol. Biotechnol. 97 9825–9836. 10.1007/s00253-013-5294-y [DOI] [PubMed] [Google Scholar]
- Zhang Y., He L., Chen Z., Wang Q., Qian M., Sheng X. (2011). Characterization of ACC deaminase-producing endophytic bacteria isolated from copper-tolerant plants and their potential in promoting the growth and copper accumulation of Brassica napus. Chemosphere 83 57–62. 10.1016/j.chemosphere.2011.01.041 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB40289.