Abstract
In March 2020, the SARS-CoV-2 virus outbreak was declared as a world pandemic by the World Health Organization (WHO). The only measures for controlling the outbreak are testing and isolation of infected cases. Molecular real-time polymerase chain reaction (PCR) assays are very sensitive but require highly equipped laboratories and well-trained personnel. In this study, a rapid point-of-need detection method was developed to detect the RNA-dependent RNA polymerase (RdRP), envelope protein (E), and nucleocapsid protein (N) genes of SARS-CoV-2 based on the reverse transcription recombinase polymerase amplification (RT-RPA) assay. RdRP, E, and N RT-RPA assays required approximately 15 min to amplify 2, 15, and 15 RNA molecules of molecular standard/reaction, respectively. RdRP and E RT-RPA assays detected SARS-CoV-1 and 2 genomic RNA, whereas the N RT-RPA assay identified only SARS-CoV-2 RNA. All established assays did not cross-react with nucleic acids of other respiratory pathogens. The RT-RPA assay’s clinical sensitivity and specificity in comparison to real-time RT-PCR (n = 36) were 94 and 100% for RdRP; 65 and 77% for E; and 83 and 94% for the N RT-RPA assay. The assays were deployed to the field, where the RdRP RT-RPA assays confirmed to produce the most accurate results in three different laboratories in Africa (n = 89). The RPA assays were run in a mobile suitcase laboratory to facilitate the deployment at point of need. The assays can contribute to speed up the control measures as well as assist in the detection of COVID-19 cases in low-resource settings.
In December 2019, a virus causing severe acute respiratory syndrome (SARS-CoV-2) was first identified in diseased patients in China. Genome sequencing and phylogenic analysis identified the virus as a close relative of SARS-CoV in the genus Betacornavirus of subfamily Coronavirinae within the family Coronaviridae.(1) The respiratory disease soon after was designated COVID-19 and rapidly spread first inside and then outside of China. The World Health Organization (WHO) announced COVID-19 as a global pandemic in March 2020.2 Currently, SARS-CoV-2 has infected more than 50 million individuals worldwide due to a high transmissibility rate and a low infectious dose.3 The only effective control measures are early identification and isolation of infected cases, contact tracing, as well as social distancing and compulsory use of facial masks in closed rooms.4
Real-time reverse transcription polymerase chain reaction (real-time RT-PCR) assays were quickly developed for the detection of SARS-CoV-2.5 Real-time RT-PCR is highly sensitive and specific. In general, the results can be produced in less than 3 h under optimal conditions. If local diagnostic capacity is not available, samples collected from suspected cases have to be transported to diagnostic laboratories providing suitable test capacities. In most of the cases, the test results can be provided within a timeframe ranging from 24 h up to 3 days. Noteworthy, in the early phase of the epidemic, diagnostic laboratories, e.g., in Germany, were overwhelmed with the number of incoming samples.6
RPA is conducted at a single constant temperature (∼42 °C) and results are produced in 15 min or less. The speed of the RPA is achieved via a more-than-exponential amplification based on a mixture of enzymes and proteins (recombinase, single-stranded DNA-binding protein, and strand-displacing polymerase).7 Polyethylenglycol (Carbowax20M) is used as a crowding agent in the reaction and contributes significantly to the amplification process by concentrating the proteins into smaller reaction volumes. Real-time data acquisition is possible using a fluorescent exo-probe.8 Recently, mathematical analysis methods have been developed to allow quantification.9,10
RPA assays have been successfully evaluated for detection of several emerging and neglected pathogens.11−19 It has been successfully implemented into a solar-powered mobile suitcase laboratory (Figure S1) and used as a point-of-need molecular diagnostic tool during the Ebola virus outbreak in West Africa.12
In this study, we developed and evaluated three real-time reverse transcription-RPA (RT-RPA) assays targeting the RNA-dependent RNA polymerase (RdRP), envelope protein (E), and nucleocapsid protein (N) genes of SARS-CoV-2. The limit of detection, cross-reactivity, and clinical performance were determined and compared with real-time RT-PCR.
Experimental Methods
Molecular RNA Standard and RPA Oligonucleotide
For assay validation, molecular RNA standards based on the RdRP, E, and N genes (accession number: NC_045512, nucleotides: 14977–15975, 26112–26479, and 28280–29536, respectively) were synthesized by GenExpress (Berlin, Germany). The E gene RNA was also used as a positive control for the RT-PCR. Oligonucleotides for the RdRP and E genes were updated from our previous design for SARS-CoV and MERS CoV (unpublished data), whereas the N gene amplicon was modified from a previously published article20 (Table 1). All oligonucleotides were synthesized by TIB MOLBIOL GmbH (Berlin, Germany).
Table 1. RT-RPA Assay Oligonucleotidesa.
| gene | oligonucleotide | sequence 5′-3′ |
|---|---|---|
| RdRP | forward | TATGCCATTAGTGCAAAGAATAGAGCTCGCAC |
| reverse | CAACCACCATAGAATTTGCTTGTTCCAATTAC | |
| exo-probe | TCCTCTAGTGGCGGCTATTGATTTCAATAAbTXfTTTGATGAAACTGTCTATTG-PH | |
| E | forward | GAAGAGACAGGTACGTTAATAGTTAATAGCGTA |
| reverse | AAAAAGAAGGTTTTACAAGACTCACGTTAACsA | |
| exo-probe | ATCGAAGCGCAGTAAGGATGGCTAGbTXfTAACTAGCAAGAATAC-PH | |
| N | forward | CCTCTTCTCGTTCCTCATCACGTAGTCGCAAC |
| reverse | AGTGACAGTTTGGCCTTGTTGTTGTTGGCCTT | |
| exo-probe | TAGAATGGCTGGCAATGGCGGTGATGCTGCbTXfTTGCTTTGCTGCTGCTT-PH |
BHQ1-dt (bT), tetrahydrofuran (X), Fam-dT (fT), phosphothioate backbone (s), and PH: 3′ phosphate to block elongation.
RT-RPA Analytical Sensitivity and Specificity
Serial dilutions of the molecular standards (106–100 RNA molecules/per reaction) were screened in the respective RT-RPA assays. To determine the minimal number of RNA molecules per reaction detected in 95% of cases, a probit regression analysis of five datasets from the replicate RPA reactions of the complete standard dilution range was performed using STATISTICA software (StatSoft, Hamburg, Germany), and the graph was created by GraphPad Prism version 6.07 software (GraphPad Software, Inc., San Diego, California). The analytical specificity of the RT-RPA assays was tested with genomes of the viruses listed in Table 2. The nucleic acid extracts were provided by Charité Medical University (Berlin, Germany), Robert Koch Institute (Berlin, Germany), Landesgesundheitsamt Niedersachsen (Hannover, Germany), Quality Control for Molecular Diagnostics (Glasgow, Scotland), and the Friedrich-Loeffler-Institute (Greifswald-Insel Riems, Germany).
Table 2. Viral Genomes Analyzed for the Cross-Reactivity by the Three SARS-CoV-2 RT-RPA Assays.
| viral nucleic acid | RdRP | E | N |
|---|---|---|---|
| SARS-CoV-2 | + | + | + |
| SARS-CoV-1 | + | + | – |
| coronavirus 229E | – | – | – |
| coronavirus NL63 | – | – | – |
| coronavirus OC43 | – | – | – |
| MERS-Coronavirus | – | – | – |
| influenza A (H1N1 pdm09) | – | – | – |
| influenza A (H3N2) | – | – | – |
| influenza A (H5N1) | – | – | – |
| influenza A (H1N1 H275Y) | – | – | – |
| influenza B (Victoria) | – | – | – |
| influenza B (Yamagata) | – | – | – |
| parainfluenza virus 1 (patient isolate) | – | – | – |
| parainfluenza virus 2 (patient isolate) | – | – | – |
| parainfluenza virus 3 (patient isolate) | – | – | – |
| parainfluenza virus 4 (patient isolate) | – | – | – |
| respiratory syncytial virus A and B | – | – | – |
| human rhinovirus A 16 | – | – | – |
| human rhinovirus B 5 | – | – | – |
| human metapneumovirus A1 | – | – | – |
| human metapneumovirus B2 | – | – | – |
| adenovirus type 1 | – | – | – |
| adenovirus type 4 | – | – | – |
| adenovirus type 34 | – | – | – |
| A/Anhui/1/13 (H7N9) | – | – | – |
| A/ Chicken/Germany/79 “Taucha“ (H7N7) | – | – | – |
| A/Chicken/Brescia/19/02 (H7N7) | – | – | – |
| A/Cygnusolor/Germany/R1377/07 (H5N1) | – | – | – |
| newcastle disease virus clone 30 | – | – | – |
| infectious laryngotracheitis virus U76 | – | – | – |
| infectious bronchitis M41 | – | – | – |
RT-RPA Reaction Conditions
For RT-RPA, the TwistAmp exo kit (TwistDx, Cambridge, UK) was used in combination with lyophilized RevertAid reverse transcriptase (Life Technologies, Darmstadt, Germany). Per reaction, 29.5 μL of rehydration buffer, 2.5 μL of RevertAid reverse transcriptase (200 U/μL), 9.7 μL of H2O, 2.1 μL of forward primer (10 pmol/μL), 2.1 μL of reverse primer (20 pmol/μL), 0.6 μL of exo-probe (10 pmol/μL), 2.5 μL of 280 mM magnesium acetate, and 1 μL of the template were added into the lid of the reaction tube containing the freeze-dried pellet. The tube was closed, centrifuged, mixed, centrifuged, and placed immediately into the T8 (Axxin, Fairfield, Australia) isothermal fluorescence reader. The reaction was incubated at 42 °C for 15 min. A mixing step was conducted after 230 s for the RdRP and E RT-RPA assays and after 320 s for the N RT-RPA assay. The threshold time (TT) was calculated as the starting point of the amplification curve above the threshold of the negative control (water as template) in the first derivative analysis in the T8 Desktop software (Axxin, Fairfield, Australia).
Clinical Samples
The three RT-RPA assays were validated with leftover RNA extracts from suspected COVID-19 cases diagnosed at the Leipzig University Hospital by real-time RT-PCR (E based assay as screening test and RdRP as confirmatory test). A total of 18 positive and 18 negative samples (blinded) were tested. Diagnostic sensitivity and specificity, positive predictive value (PPV), and negative predictive value (NPV) for the RT-RPA assays were calculated using standard formulas against the real-time RT-PCR as reference test (Table 3).21 Preliminary testing of clinical samples was performed at the Institute Pasteur Dakar in Dakar (Senegal), laboratory at the Kumasi Centre for Collaborative Research in Tropical Medicine (Ghana), and Ain Shams and Cairo Universities (Egypt), in comparison to real-time RT-PCR assays used in these laboratories.
Table 3. Retrospective Samples University of Leipziga.
| |
result
tables |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
RT-RPA |
real-time RT-PCR |
|||||||||
| reference test real-time RT-PCRCT | analyzed test RT-RPA | sensitivity | specificity | PPV | NPV | n | RT-RPA | pos | neg | |
| <35 | RdRP | estimate: | 0.93 [0.69; 0.99] | 1 [0.82; 1.0] | 1 [0.77; 1.0] | 0.95 [0.76; 0.99] | 32 | pos | 13 | 0 |
| neg | 1 | 18 | ||||||||
| >35 | 95% CI: | 1 [0.5; 1.0] | 1 [0.84; 1.0] | 1 [0.51; 1.0] | 1 [0.84; 1.0] | 22 | pos | 4 | 0 | |
| neg | 0 | 18 | ||||||||
| <35 | E | estimate: | 0.93 [0.69; 0.99] | 0.5 [0.29; 0.71] | 0.59 [0.39; 0.77] | 0.9 [0.60; 0.98] | 32 | pos | 13 | 9 |
| neg | 1 | 9 | ||||||||
| >35 | 95% CI: | 0.5 [0.15; 0.85] | 0.5 [0.29; 0.71] | 0.18 [0.05; 0.48] | 0.82 [0.52; 0.95] | 22 | pos | 2 | 9 | |
| neg | 2 | 9 | ||||||||
| <35 | N | estimate: | 0.93 [0.66; 0.99] | 0.94 [0.74; 0.99] | 0.93 [0.69; 0.99] | 0.94 [0.74; 0.99] | 32 | pos | 13 | 1 |
| neg | 1 | 17 | ||||||||
| >35 | 95% CI: | 0.5 [0.15; 0.85] | 0.5 [0.29; 0.71] | 0.18 [0.05; 0.48] | 0.81 [0.52; 0.95] | 22 | pos | 2 | 9 | |
| neg | 2 | 9 | ||||||||
Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of RdRP, E, and N RT-RPA assays in comparison to real-time RT-PCR results (n = 36).
Real-Time RT-PCR
In Germany, the RNA extracts from patient materials were tested with a real-time PCR assay combining commercial SARS-CoV-2-E oligonucleotides (TIB MOLBIOL GmbH, Berlin, Germany) and Superscript III Platinum One-Step qRT-PCR kits (Life Technologies, Darmstadt, Germany) according to the manufacturer’s instructions using the following temperature profile: 50 °C for 15 min, 95 °C for 2 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s on an Agilent Technologies Stratagene Mx 3000p Real-Time PCR system. The same protocol was followed in Egypt but using the LightCycler Multiplex RNA Virus Master kit (Roche, Mannheim, Germany). At the laboratory in Dakar, RNA was extracted from 140 μL of swab VTM using the QIAamp viral RNA mini kit (Qiagen, Heiden, Germany). The samples were tested with the same SARS-CoV-2-E oligonucleotides as above using the Luna Universal One-Step RT-qPCR Kit (New England BioLabs, UK) according to the manufacturer’s instructions using the following temperature profile: 55 °C for 10 min, 95 °C for 1 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s on a CFX96 Real-Time PCR System (Bio-Rad, CA). At the Kumasi Centre for Collaborative Research in Ghana, RNA was extracted from swab VTM using the nucleic acid extraction and purification reagents series C—spin column method (Guangzhou LBP Medical Science & Technology Co. Ltd.). The RT-PCR used was the DA AN Gene PCR kit (Da an Gene Co. Ltd. Sun Yat-Sen University, Guangzhou, China) targeting the SARS-CoV 2 ORF1ab and N genes, according to the manufacturer’s instructions. Three microliters of the RNA extracts were used in the RT-PCR assays.
Results
Analytical Sensitivity and Specificity
To determine the analytical sensitivity of the RdRP, E, and N RT-RPA assays, each assay was tested with a serial dilution of 106–100 RNA molecules/reaction of the respective molecular RNA standard in five replicates. Molecular RNA standards with the concentration from 106 to 102 molecules/reaction were detected in all five RPA runs of the three RT-RPA-assays. The E and N RT-RPA identified 100 RNA molecules in 3/5 RT-RPA runs, while in RdRP RT-RPA, all five runs were positive. Only the RdRP RT-RPA amplified one RNA copy in 2/5 RT-RPA runs. With this dataset, probit regression analysis was performed and revealed a 95% detection limit of two RNA molecules for the RdRP RT-RPA assay, and 15 RNA molecules each for E and N RT-RPA assays (Figure 1).
Figure 1.
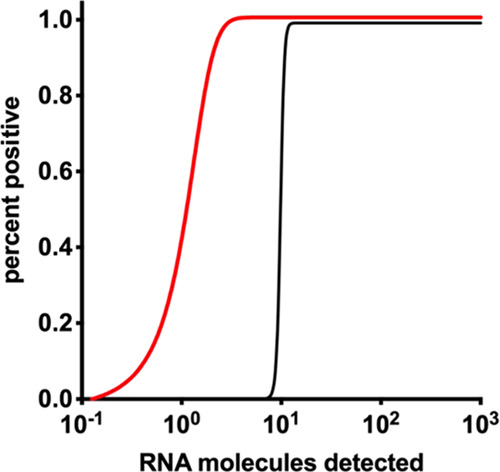
Probit regression analysis for RdRP, E, and N RT-RPA assays. The limit of detection in 95% of cases is two RNA molecules/reaction for the RdRP RT-RPA assay (red) and 15 RNA molecules per reaction each for E and N RT-RPA assays (black).
RdRP and E RT-RPA assays were able to amplify genomic RNA of both SARS-CoV-1 and -2, whereas the N RT-RPA assay recognizes SARS-CoV-2 RNA only. None of the assays shows cross-reactivity to nucleic acid extracts from other respiratory viruses listed in Table 2.
Clinical Samples
All extracted RNA samples provided by the University of Leipzig, which had been stored at −80 °C were first tested by the three RT-RPA assays and retested by real-time RT-PCR to confirm previous results. The RT-RPA assay targeting the RdRP gene produced the best clinical performance (Table 3). All RT-RPA assays detected samples with a CT > 30 at a maximum of 12 min (Figure 2).
Figure 2.
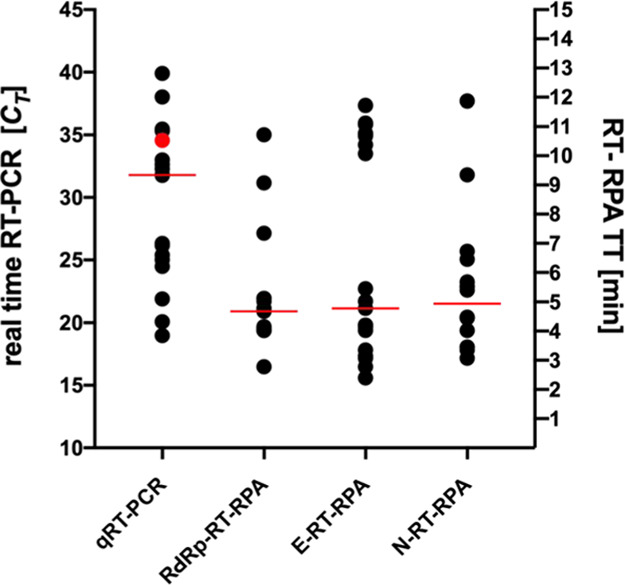
Results of 36 clinical samples analyzed with real-time RT-PCR for the E gene and RT-RPA assays for the RdRP, E, and N genes. CT is the cycle threshold, and TT is the threshold time. The red dot represents the real-time RT-PCR sample not detected by the RdRP RT-RPA, and the horizontal red lines indicate the median of TT or CT values.
In Dakar, Senegal, CT range 19.1–39.1 was scored positive by the RdRP RT-RPA assay, whereas the E and N assays showed poor performance (Table S1(i)). Initially, it was unclear why the RdRP RT-RPA assay performed poorly in the lab in Kumasi (Table S1(ii)), where only 7/11 scored positive (CT range 18.1–36.1) until it emerged that RNA extracts had been stored at −20 °C. In Egypt (Table S1(iii)), the RNA extracts were stored at −80 °C, but were subsequently transported to another laboratory, where they were refrozen (7/16 positive; CT range, 17.0–39.7). The RdRP RT-RPA results from Dakar were confirmed when testing was repeated in Egypt with extracts from fresh swabs (26/26). Similar results were achieved at the lab in Dakar where 16/16 samples albeit in a low CT range (19–39) scored positive by RdRP RT-RPA. We calculated the clinical sensitivity and specificity for all RdRP RT-RPA results from African labs, which resulted in similar values as those observed for the samples from Germany (Table 4); however, the RdRP RT-RPA assay showed 100% concordance with real-time RT-PCR results when the results from fresh extracts only (Table S1(i and iv)) were considered.
Table 4. Retrospective Samples of Four Laboratories in Africa (see Table S1)a.
| |
result tables |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
RT-RPA |
real-time RT-PCR |
|||||||||
| reference test real-time RT-PCRCT | analyzed test RT-RPA | sensitivity | specificity | PPV | NPV | n | RT-RPA | pos | neg | |
| <35 | RdRP | estimate: 95% CI: | 0.93 [0.87; 0.98] | 1.00 [0.84; 1] | 1.0 [0.93; 1] | 0.87 [0.768; 0.96] | 73 | pos | 50 | 0 |
| neg | 3 | 20 | ||||||||
| >35 | RdRP | estimate: 95% CI: | 0.31 [0.14; 0.55] | 1.0 [0.84;1] | 1.0 [0.57; 1] | 0.64 [0.47; 0.79] | 36 | pos | 5 | 0 |
| neg | 11 | 20 | ||||||||
All results were used (n = 89 samples) to calculate sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the RdRP RT-RPA assay in comparison to real-time RT-PCR assays.
Suitcase Lab
The assays were performed in the suitcase lab (Figure S1), which is a water- and dust-resistant case and fully equipped to conduct isothermal amplification assay at low-resource settings. The suitcase lab has a size of 62 + 49 + 30 cm. The power source is via a solar-powered battery (Goal Zero South Bluffdale, UT).19 To protect the lab technician while handling highly infectious samples, glovebox (Bodo Koennecke, Berlin, Germany) was used to achieve the first lysis step.16
Discussion
For molecular amplification targets, the CDC, Atlanta, recommends two regions in the N gene, while the WHO recommends two targets, e.g., the E gene followed by a SARS-CoV-2 confirmatory assay targeting RdRP.22
To identify a highly sensitive and specific SARS-CoV-2 RPA assay, three RT-RPA assays targeting the RdRP, E, and N genes of the SARS-CoV-2 genome were evaluated. The RdRP RT-RPA assay produced sensitivity and specificity similar to a real-time RT-PCR within less than 15 min. The speed and clinical accuracy of the RT-RPA assay make it an ideal candidate molecular assay at point of need.
Real-time RT-PCR is currently the method of choice for molecular laboratory screening of samples from suspected COVID-19 patients. Satisfactory results have been achieved from swab and saliva samples23 and also sputum, blood, urine, and stool have been successfully tested.24 The identification of asymptomatic carriers relies entirely on nasopharyngeal, oropharyngeal swab, or saliva testing.25
In general, state and commercial diagnostic laboratories in developed countries have all the infrastructure for real-time RT-PCR. A compilation of SARS-CoV-2 diagnostic kits hosted by Foundation for Innovative New Diagnostics (FIND) currently lists 108 commercial PCR kits and more than 200 in-house real-time RT-PCR kits.26 In emerging nations, usually only reference laboratories are adequately equipped to perform real-time RT-PCR16 and alternative testing formats are needed. For direct virus detection, the FIND webpage currently lists only 21 rapid antigen detection tests (RDT) with an IVD license.27 RDT are easy to use, very fast, and cheap to produce, but less sensitive and can produce a high false-negative rate.28 Since the pandemic spread to Africa and South America, there is an urgent need for molecular assays with performance and accuracy known from real-time RT-PCR and applicability in low-resource settings. Isothermal amplification assays have the potential to provide rapid direct virus detection in infrastructure limited settings.19
Although there has been a flurry of publications on LAMP assays, only two are commercially available with an IVD license.29 The pH drop occurring during the LAMP amplification reaction has been recently exploited for the detection of SARS-CoV-2 to allow simple phenol red- or cresol red-based colorimetric distinction between positives and negatives just by sight.30,31 In general, RT-LAMP assays have a run time of about 30 min and need devices to maintain an amplification temperature of 65 °C. The longer run time of this isothermal amplification, however, makes unspecific amplifications more likely and requires to define a cutoff for each LAMP assay.32 In contrast, RPA is very fast (3–15 min), requires a lower temperature, and has already shown to perform at high clinical sensitivity and specificity during the Ebola virus disease outbreak in Guinea.12
The RdRP RT-RPA assay described here shows great accuracy even with samples containing less than 10 target molecules corresponding to real-time RT-PCR CT values > 35 (Figure 2 and Table 3). In this respect, the performance of RdRP RT-RPA is comparable to several real-time RT-PCRs.33
The N gene is the main target of the CDC real-time RT-PCR22 and the reverse transcription recombinase-aided amplification (RT-RAA) assay,34 and both generated excellent clinical sensitivity and specificity. Both the N and E RT-RPA assays described here produced very good analytical sensitivity (15 copies per reaction); nevertheless, the clinical performances were poor, especially for samples with low viral loads. RPA oligonucleotide design has a great influence on the sensitivity of RPA assays, and RPA amplicons always need to be validated with patient material to come to a final assessment.
The E gene RT-RPA was disqualified because it produced too many false positives in a TT range of 10–12 min (Figure 2), i.e., at a low-target RNA load. Contamination as described for real-time RT-PCR developments was excluded since amplification was not observed in any of the negative controls.35,36 False-positive real-time RT-PCR results in clinical material with low viral loads have been observed for PCR assays.37 By comparing three RT-RPA assays and validating them with clinical samples, we were able to pick a highly accurate RPA assay, i.e., RdRP RT-RPA.
Our preliminary result indicates that the developed RdRP RT-RPA assay performs well across several laboratories if RNA is extracted from fresh swab samples (Table 3, S1(i and iv)) and can achieve concordance to real-time RT-PCR results. The observation on the quality of the RNA or sample confirms earlier observations made for the foot and mouth disease virus and for dengue virus RT-RPA assays.8,11 Therefore, to achieve the best results, RT-RPA should be used directly with RNA extracts and results obtained with stored samples may underestimate the performance of the assay. Moreover, the postextraction contamination with RNase must be avoided.
The limitation of the study is that several different real-time RT-PCR assays were used in the various laboratories; moreover, the sample size is small. Therefore, a larger study is needed to determine reliable clinical sensitivities and specificity in comparison to a real-time RT-PCR used across all participating labs. This has already been shown by a Chinese group, which tested 926 samples by RT-RAA, an RT-RPA equivalent using E.coli SSB instead of GP32 in the enzyme mixture.38 These results indicate that a good performance of the RPA assays with clinical samples can be expected. Inclusion of an internal positive control will increase the practicability of the developed RPA assay since the effect of inhibitors will be excluded in case of false-negative results. In addition, establishing a duplex RPA assay relaying on both RdRP and N genes will boost the assay clinical specificity.
The ID-Now system (Abbott) combines a simple extraction nonpurification protocol in combination with the isothermal nicking enzyme amplification reaction (NEAR). The total turnaround time is 17 min. The ID-Now showed great efficacy for Influenza A and B testing,39 but has a limited clinical sensitivity for SARS-CoV-2, especially for samples with CT value > 30,40 which drove the FDA to release an alert against using the ID-Now.41
We introduced few improvements during the development of the RPA assays for SARS-CoV-2. First, the best assay sensitivity was achieved using double concentration of the reverse primer. Coronaviruses have a positive-sense single-stranded RNA genome and more reverse primer produces more cDNA for the following amplification step.42,43 Second, during the oligonucleotide design, any sequences pairing to the human genome were strictly excluded to avoid problems as reported for some real-time RT-PCR assays.44 Third, to avoid primer dimer and nonspecific amplification, phosphothioate nucleotides were introduced in selected RPA primers.45
In addition to the amplification and detection, the virus inactivation and extraction are crucial steps in the diagnosis of SARS-CoV-2. To assure the safety of the healthcare worker or laboratory technician, sample inactivation is recommended in a BSL-2 cabinet and under field condition in a mobile glovebox.22 The German committee for biological reagents (ABAS) has, however, recently relaxed its regulations and a BSL-2 bench is now not required if swabs are immersed into an inactivating lysis buffer immediately after sampling while wearing PPE.46
For swabs collected from an Ebola-infected patient, we showed that the combination of detergent and heat in the SPEEDXTRACT protocol can be achieved in 10 min.12 Several rapid and simple extraction procedures that use a mix of detergents and heating have now been published for SARS-CoV-2 samples.47−49 Most of these methods generate crude extracts, and RPA is very robust in crude samples and works in the presence of agents with an inhibitory effect on PCR including 15 ± 25% of milk, 50 g/L hemoglobin, 4% V/V ethanol, and 0.5 U of heparin.50 Virus RNA was detectable in preparations containing 1:10 dilutions of crude human serum, urine, and tick pool homogenate.50−52 We are now investigating such an approach for integration into the suitcase laboratory.
Conclusions
With the current clinical performance of the developed RdRP RT-RPA assay, the test provides a promising approach for the use at point of need. It could be deployed at local hospitals, healthcare centers, and walk-through test centers. The robust suitcase laboratory concept would work in infrastructure limited settings. Currently, the limited commercial availability of RT-RPA reagents is an obstacle for wide implementation of RT-RPA for in vitro diagnostic use. License-free RT-RPA reagents would be a boost for diagnostics in infrastructure-poor settings.
Acknowledgments
The clinical evaluation in Ghana and Senegal was supported by EDCTP grant RIA2020EF-2937. The reference viral RNA extracts from cell culture used for the cross-detection studies were part of the Bill & Melinda Gates Foundation (grant ID INV-005971) to Charite—Universitätsmedizin Berlin. The findings and conclusions contained within are those of the authors and do not necessarily reflect positions or policies of the Bill & Melinda Gates Foundation.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c04779.
Mobile suitcase lab (Figure S1) and raw data of screening clinical samples in African settings (Table S1) (PDF)
Author Contributions
A.A.E.W. and P.P. contributed equally to this work. Conceptualization was performed by A.A.E.W., P.P., F.T.H., and M.W. A.A.E.W., P.P., M.M., C.P., D.R., S.B.-F., J.K., O.B., M.F., M.M.D., M.G., N.D., M.K., M.N., O.L., A.A.S., M.A.S., O.F., U.T., U.G.L., and M.W. contributed to data curation. Formal analysis was conducted by A.A.E.W., P.P., M.M., C.P., D.R., J.K., O.B., M.F., M.M.D., M.G., M.K., H.A., M.E., A.Z., G.I., and M.W. A.A.E.W., P.P., M.M., C.P., D.R., S.B.-F., and M.W. performed the investigation. Methodology was designed by A.A.E.W., P.P., V.M.C., M.F, F.T.H., U.T., U.G.L., and M.W. Resources were obtained by N.D., O.F., A.A.S., M.A.S., V.M.C., O.L., F.T.H., U.T., and U.G.L. A.A.E.W., P.P., D.R., and M.W. contributed to software. A.A.E.W., N.D., O.F., A.A.S., F.T.H., U.T., U.G.L., and M.W. supervised the study. A.A.E.W., P.P., and M.W. contributed to validation, visualization, and writing of the original draft. Review and editing was done by all authors.
The authors declare the following competing financial interest(s): All authors except Marco Kaiser and Olfert Landt are in the public research sector. Mentioned authors are employed by GenExpress and/or Tib MolBiol, manufacturer of molecular standard or oligonucleotides. This does not alter the authors adherence to all the scientific policies on sharing data and materials.
Supplementary Material
References
- The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. WHO announces COVID-19 outbreak apandemic. https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic (accessed June 25, 2020).
- Li R.; Pei S.; Chen B.; Song Y.; Zhang T.; Yang W.; Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C. C.; Cheng K. K.; Lam T. H.; Migliori G. B. Mask wearing to complement social distancing and save lives during COVID-19. Int. J. Tuberc. Lung Dis. 2020, 24, 556–558. 10.5588/ijtld.20.0244. [DOI] [PubMed] [Google Scholar]
- Corman V. M.; Landt O.; Kaiser M.; Molenkamp R.; Meijer A.; Chu D. K.; Bleicker T.; Brunink S.; Schneider J.; Schmidt M. L.; Mulders D. G.; Haagmans B. L.; van der Veer B.; van den Brink S.; Wijsman L.; Goderski G.; Romette J. L.; Ellis J.; Zambon M.; Peiris M.; Goossens H.; Reusken C.; Koopmans M. P.; Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. Coronavirus and the race to distribute reliable diagnostics. Nat. Biotechnol. 2020, 38, 382–384. 10.1038/d41587-020-00002-2. [DOI] [PubMed] [Google Scholar]
- Piepenburg O.; Williams C. H.; Stemple D. L.; Armes N. A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd El Wahed A.; El-Deeb A.; El-Tholoth M.; Abd El Kader H.; Ahmed A.; Hassan S.; Hoffmann B.; Haas B.; Shalaby M. A.; Hufert F. T.; Weidmann M. A portable reverse transcription recombinase polymerase amplification assay for rapid detection of foot-and-mouth disease virus. PLoS One 2013, 8, e71642 10.1371/journal.pone.0071642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M. I.; Klemer D. P.; Fuller S. L.; Chang J. H.; Klemer D. R.; Putnam M. L. Mathematical modeling of a real-time isothermal amplification assay for Erwinia amylovora. Eng. Rep. 2019, 1, e12047 10.1002/eng2.12047. [DOI] [Google Scholar]
- Moody C.; Newell H.; Viljoen H. A mathematical model of recombinase polymerase amplification under continuously stirred conditions. Biochem. Eng. J. 2016, 112, 193–201. 10.1016/j.bej.2016.04.017. [DOI] [Google Scholar]
- Abd El Wahed A.; Patel P.; Faye O.; Thaloengsok S.; Heidenreich D.; Matangkasombut P.; Manopwisedjaroen K.; Sakuntabhai A.; Sall A. A.; Hufert F. T.; Weidmann M. Recombinase Polymerase Amplification Assay for Rapid Diagnostics of Dengue Infection. PLoS One 2015, 10, e0129682 10.1371/journal.pone.0129682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye O.; Faye O.; Soropogui B.; Patel P.; El Wahed A. A.; Loucoubar C.; Fall G.; Kiory D.; Magassouba N.; Keita S.; Konde M. K.; Diallo A. A.; Koivogui L.; Karlberg H.; Mirazimi A.; Nentwich O.; Piepenburg O.; Niedrig M.; Weidmann M.; Sall A. A. Development and deployment of a rapid recombinase polymerase amplification Ebola virus detection assay in Guinea in 2015. Eurosurveillance 2015, 20, 30053 10.2807/1560-7917.ES.2015.20.44.30053.. [DOI] [PubMed] [Google Scholar]
- Frimpong M.; Ahor H. S.; Wahed A. A. E.; Agbavor B.; Sarpong F. N.; Laing K.; Wansbrough-Jones M.; Phillips R. O. Rapid detection of Mycobacterium ulcerans with isothermal recombinase polymerase amplification assay. PLoS Neglected Trop. Dis. 2019, 13, e0007155 10.1371/journal.pntd.0007155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissenkötter J.; Hansen S.; Bohlken-Fascher S.; Ademowo O. G.; Oyinloye O. E.; Bakarey A. S.; Dobler G.; Tappe D.; Patel P.; Czerny C. P.; Abd El Wahed A. Development of a pan-rickettsial molecular diagnostic test based on recombinase polymerase amplification assay. Anal. Biochem. 2018, 544, 29–33. 10.1016/j.ab.2017.12.018. [DOI] [PubMed] [Google Scholar]
- Patel P.; Abd El Wahed A.; Faye O.; Pruger P.; Kaiser M.; Thaloengsok S.; Ubol S.; Sakuntabhai A.; Leparc-Goffart I.; Hufert F. T.; Sall A. A.; Weidmann M.; Niedrig M. Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid Detection of the Chikungunya Virus. PLoS Neglected Trop. Dis. 2016, 10, e0004953 10.1371/journal.pntd.0004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann M.; Faye O.; Faye O.; Abd El Wahed A.; Patel P.; Batejat C.; Manugerra J. C.; Adjami A.; Niedrig M.; Hufert F. T.; Sall A. A. Development of Mobile Laboratory for Viral Hemorrhagic Fever Detection in Africa. J. Infect. Dis. 2018, 218, 1622–1630. 10.1093/infdis/jiy362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd El Wahed A.; Patel P.; Heidenreich D.; Hufert F. T.; Weidmann M. Reverse transcription recombinase polymerase amplification assay for the detection of middle East respiratory syndrome coronavirus. PLoS Curr. 2013, 5 10.1371/currents.outbreaks.62df1c7c75ffc96cd59034531e2e8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal D.; Ghosh P.; Khan M. A.; Hossain F.; Bohlken-Fascher S.; Matlashewski G.; Kroeger A.; Olliaro P.; Abd El Wahed A. Mobile suitcase laboratory for rapid detection of Leishmania donovani using recombinase polymerase amplification assay. Parasites Vectors 2016, 9, 281 10.1186/s13071-016-1572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd El Wahed A.; Weidmann M.; Hufert F. T. Diagnostics-in-a-Suitcase: Development of a portable and rapid assay for the detection of the emerging avian influenza A (H7N9) virus. J. Clin. Virol. 2015, 69, 16–21. 10.1016/j.jcv.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrmann O.; Bachmann I.; Spiegel M.; Schramm M.; El Wahed A. A.; Dobler G.; Dame G.; Hufert F. T. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (exo-IQ). Clin Chem. 2020, hvaa116 10.1093/clinchem/hvaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman D. G.; Bland J. M. Diagnostic tests 2: Predictive values. BMJ 1994, 309, 102 10.1136/bmj.309.6947.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. W.; Schmitz J. E.; Persing D. H.; Stratton C. W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512-20 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.; Cui B.; Duan X.; Zhang P.; Zhou X.; Yuan Q. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int. J. Oral Sci. 2020, 12, 11 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Xu Y.; Gao R.; Lu R.; Han K.; Wu G.; Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M.; Yokoe D. S.; Havlir D. V. Asymptomatic Transmission, the Achilles’ Heel of Current Strategies to Control Covid-19. N. Engl. J. Med. 2020, 382, 2158–2160. 10.1056/NEJMe2009758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIND WHO Find Evaluation Update: SARSCOV-2 Molecular Diagnostics. https://www.finddx.org/covid-19/sarscov2-eval-molecular/ (accessed June 25, 2020).
- FIND-WHO FIND Evaluation Update: SARSCOV-2 Immunoassays. https://www.finddx.org/covid-19/sarscov2-eval-immuno/ (accessed June 25, 2020).
- FDA Coronavirus (COVID-19) Update: FDA AuthorizesFirst Antigen Test to Help in the Rapid Detection of the Virus thatCauses COVID-19 in Patients. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-antigen-test-help-rapid-detection-virus-causes (accessed June 25, 2020).
- FIND-WHO SARS-COV-2 Diagnostic Pipeline. https://www.finddx.org/covid-19/pipeline/?avance=all&type=Manual+NAT&test_target=RNA&status=all§ion=molecular-assays&action=default#diag_tab (accessed June 25, 2020).
- Huang W. E.; Lim B.; Hsu C. C.; Xiong D.; Wu W.; Yu Y.; Jia H.; Wang Y.; Zeng Y.; Ji M.; Chang H.; Zhang X.; Wang H.; Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961. 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R.; Wu X.; Wan Z.; Li Y.; Jin X.; Zhang C. A Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 2826 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hin S.; Lopez-Jimena B.; Bakheit M.; Klein V.; Stack S.; Fall C.; Sall A. A.; Enan K.; Mustafa M.; Gillies L.; Rusu V.; Goethel S.; Paust N.; Zengerle R.; Frischmann S.; Weidmann M.. Fully automated point-of-care differential diagnosis of acute febrile illness. Submitted 2020. [DOI] [PMC free article] [PubMed]
- van Kasteren P. B.; van der Veer B.; van den Brink S.; Wijsman L.; de Jonge J.; van den Brandt A.; Molenkamp R.; Reusken C.; Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.; Ge Y.; Zhao K.; Zhu X.; Chen Y.; Wu B.; Zhu F.; Zhu B.; Cui L. A reverse-transcription recombinase-aided amplification assay for the rapid detection of N gene of severe acute respiratory syndrome coronavirus 2(SARS-CoV-2). Virology 2020, 549, 1–4. 10.1016/j.virol.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A. N.; Kessel B. False positives in reverse transcription PCR testing for SARS-CoV-2. medRxiv 2020, 20080911 10.1101/2020.04.26.20080911. [DOI] [Google Scholar]
- Bustin S. A.; Nolan T. RT-qPCR Testing of SARS-CoV-2: A Primer. Int. J. Mol. Sci. 2020, 21, 3004 10.3390/ijms21083004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falasca F.; Sciandra I.; Di Carlo D.; Gentile M.; Deales A.; Antonelli G.; Turriziani O. Detection of SARS-COV N2 Gene: Very low amounts of viral RNA or false positive?. J. Clin. Virol. 2020, 133, 104660 10.1016/j.jcv.2020.104660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Cai K.; He X.; Shen X.; Wang J.; Liu J.; Xu J.; Qiu F.; Lei W.; Cui L.; Ge Y.; Wu T.; Zhang Y.; Yan H.; Chen Y.; Yu J.; Ma X.; Shi H.; Zhang R.; Li X.; Gao Y.; Niu P.; Tan W.; Wu G.; Jiang Y.; Xu W.; Ma X. Multiple-centre clinical evaluation of an ultrafast single-tube assay for SARS-CoV-2 RNA. Clin. Microbiol. Infect. 2020, 26, 1076–1081. 10.1016/j.cmi.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar N.; Michael J.; Doran K.; Montgomery E.; Selvarangan R. Comparison of the ID Now Influenza A & B 2, Cobas Influenza A/B, and Xpert Xpress Flu Point-of-Care Nucleic Acid Amplification Tests for Influenza A/B Virus Detection in Children. J. Clin. Microbiol. 2020, 58, e01611-19 10.1128/JCM.01611-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W.; Smith E.; Manji R.; Schron D.; Berry G. J. Clinical Evaluation of Three Sample-To-Answer Platforms for the Detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58, 1–7. 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA Coronavirus (COVID-19) Update: FDA Informs Public About Possible Accuracy Concerns with Abbott ID NOW Point-of-Care Test. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-informs-public-about-possible-accuracy-concerns-abbott-id-now-point (accessed June 25, 2020).
- Barratt K.; Mackay J. F. Improving real-time PCR genotyping assays by asymmetric amplification. J. Clin. Microbiol. 2002, 40, 1571–1572. 10.1128/JCM.40.4.1571-1572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I. M.; Jacob K. C.; Woolhouse D.; Waller K.; Syrmis M. W.; Whiley D. M.; Siebert D. J.; Nissen M.; Sloots T. P. Molecular assays for detection of human metapneumovirus. J. Clin. Microbiol. 2003, 41, 100–105. 10.1128/JCM.41.1.100-105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillonel T.; Scherz V.; Jaton K.; Greub G.; Bertelli C. Letter to the editor: SARS-CoV-2 detection by real-time RT-PCR. Eurosurveillance 2020, 25, 2000880 10.2807/1560-7917.ES.2020.25.21.2000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerra A. Phosphorothioate primers improve the amplification of DNA sequences by DNA polymerases with proofreading activity. Nucleic Acids Res. 1992, 20, 3551–3554. 10.1093/nar/20.14.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (BAuA), B. f. A. u. A. Empfehlungen zu Arbeitsschutzmaßnahmen bei der Point-Of-Care-SARS-CoV-2 Diagnostik. (accessed Nov 8, 2020).
- Fomsgaard A. S.; Rosenstierne M. W. An alternative workflow for molecular detection of SARS-CoV-2 - escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Eurosurveillance 2020, 25, 2000398 10.2807/1560-7917.es.2020.25.14.2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lista mJ.; Page R.; Sertkaya H.; Matos P.; Ortiz-Zapater E.; Maguire T. J. A.; Poulton K.; O’Byrne A.; Bouton C.; Dickenson R. E.; Ficarelli M.; Howard M.; Betancor G.; Galao R. P.; Pickering S.; Signell A. W.; Wilson H.; Cliff P.; Ik M. T. K.; Patel A.; MacMahon E.; Cunningham E.; Doores K.; Agromayor M.; Martin-Serrano J. M.; Perucha E.; Mischo H. E.; Shankar-Hari M.; Batra R.; Edgeworth J.; Malim M. H.; Neil S.; Martinez-Nunez R. T.. Resilient SARS-CoV-2 diagnostics workflows including viral heat inactivation medRxiv 2020, 10.1101/2020.04.22.20074351. [DOI] [PMC free article] [PubMed]
- Kuiper J. W. P.; Baade T.; Kremer M.; Kranaster R.; Irmisch L.; Schuchmann M.; Zander J.; Marx A.; Hauck C. R. Detection of SARS-CoV-2 from raw patient samples by coupled high temperature reverse transcription and amplification. PLoS One 2020, 15, e0241740 10.1371/journal.pone.0241740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonney L. C.; Watson R. J.; Afrough B.; Mullojonova M.; Dzhuraeva V.; Tishkova F.; Hewson R. A recombinase polymerase amplification assay for rapid detection of Crimean-Congo Haemorrhagic fever Virus infection. PLoS Neglected Trop. Dis. 2017, 11, e0006013 10.1371/journal.pntd.0006013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher R. K.; Stewart G.; Boissinot M.; Bergeron M. G. Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 2016, 62, 947–58. 10.1373/clinchem.2015.245829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher R. K.; Stewart G.; Boissinot M.; Bergeron M. G. Isothermal recombinase polymerase amplification assay applied to the detection of group B streptococci in vaginal/anal samples. Clin. Chem. 2014, 60, 660–666. 10.1373/clinchem.2013.213504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



