Abstract
The role of physicians has always been to synthesize the data available to them to identify diagnostic patterns that guide treatment and follow response. Today, increasingly sophisticated machine learning algorithms may grow to support clinical experts in some of these tasks. Machine learning has the potential to benefit patients and cardiologists, but only if clinicians take an active role in bringing these new algorithms into practice. The aim of this review is to introduce clinicians who are not data-science experts to key concepts in machine learning that will allow them to better understand the field and evaluate new literature and developments. We then summarize the current literature in machine learning for cardiovascular disease, using both a bibliometric survey, with code publicly available to enable similar analysis for any research topic of interest, and select case studies. Finally, we list several ways clinicians can and must be involved in this emerging field.
Keywords: cardiology, machine learning, deep learning, artificial intelligence, literature search, bibliometric analysis
Condensed Abstract
The aim of this review is to introduce clinicians who are not data-science experts to key concepts in machine learning that will allow them to better understand the field and evaluate new literature and developments. We then summarize the current literature in machine learning for cardiovascular disease, using both a bibliometric survey, with code publicly available to enable similar analysis for any research topic of interest, and select case studies. Finally, we list several ways clinicians can and must be involved in this emerging field.
Introduction
Machine learning (ML)—the use of computer algorithms that can learn complex patterns from data—has significant potential to impact cardiology due to the number of diagnostic and management decisions that rely on digitized, patient-specific information such as ECGs, echocardiograms and more (1); and due to the growing amount and complexity of medical knowledge. The staggering volume of health care data—clinical notes, wearable and sensor data, medication lists, imaging, and much more—continues to increase astronomically, with two zettabytes (one zettabyte = one trillion gigabytes) estimated to be produced in 2020 (2). Simply put, “the complexity of medicine now exceeds the capacity of the human mind.” (3) As a result, our knowledge and interpretation of available data, our skills, and our practice may vary from clinician to clinician, sometimes failing to leverage all of medical knowledge. Designed, validated, and implemented appropriately, machine learning algorithms will help in acquiring, interpreting, and synthesizing healthcare data from disparate sources and putting it at our fingertips – as if we had an expert sub-specialist to call upon for every patient and every clinical situation.
As machine learning algorithms permeate into clinical cardiology, they may be deployed in multiple areas. Algorithms may help the front office schedule different patients with the appropriate amount of time based on their EHR data, or help primary care physicians better guide referrals to the cardiologist’s office. Machine learning will help in remote testing and monitoring of patients, guiding the data acquisition that will be sent to a clinic or hospital. Perhaps the computer-generated ECG reports will become more reliable, because machine learning algorithms are analyzing the signal. Ultrasound, nuclear, CT and MRI images will be better-protocolled and have less image noise or fewer artifacts. Automated cardiac measurements from echocardiograms, CT scans, and MRIs may become more accurate and reliable (4). Perhaps cardiac risk scores won’t be calculated from just a handful of variables, but from every piece of data in the patient’s chart. Perhaps ML algorithms will help us discover new and subtle patterns in data that may change how we care for our patients. Perhaps clinical guidelines may recommend ML-enabled testing for patient diagnosis and management. Taken together, these and other ML-based improvements may make for a revolution in cardiology. Despite this potential, and a growing body of literature (detailed below), machine learning’s real-world impact on clinical cardiologists and their patients has been quite limited to date(1,5).
To borrow from American poet and musician Gil Scott-Heron, this revolution will not be televised. By the time algorithms like those described above make it into the clinic, their presence will be largely invisible to the practitioner. Cardiologists’ active involvement in machine learning before it reaches the clinic is critical in order to help it reach its full potential for patient care. Cardiologists will be the ones to follow new developments in medical machine learning, serve as peer reviewers, and evaluate whether new work is impactful or incremental to patients and providers. Cardiologists will lead clinical trials evaluating efficacy and safety of machine learning algorithms, just as they do for drugs or medical devices. In the clinic, cardiologists will help decide, for example, whether to buy new ML-powered software to improve clinical operations or diagnosis. When a machine learning algorithm suggests an unusual result, cardiologists will have to decide whether that result is spurious or a truly novel finding that a physician would not see on his or her own. And they will have to explain their reliance on or rejection of ML-based test results to their patients. For all of these reasons, even cardiologists who are not machine learning experts can benefit from some basic tools and concepts with which to understand, evaluate, and, when appropriate, champion the incorporation of evidence-based machine learning research into their practice.
The goal of this review is to present these concepts for machine learning, to provide a way to survey the ever-growing body of literature in machine learning for cardiology, and to provide suggestions for how cardiologists who are not data science experts can participate in the machine learning revolution.
Machine learning: key concepts for busy clinicians
In this section we will provide a brief overview of six machine learning concepts that clinicians need to be familiar with to read clinical papers using machine learning methods, and eventually to facilitate the use of novel machine learning algorithms inside the clinic. Our goal is not to provide a comprehensive technical primer on machine learning algorithms, already well covered in several excellent resources (4–11).
Artificial intelligence (AI), simply defined as a computer system that is able to perform tasks that normally require human intelligence, is something cardiologists have been taking advantage of for decades virtually every day as hundreds of millions of ECGs are interpreted by computers worldwide every year (12,13). Here, we will use ECG analysis as a canonical example to illustrate several concepts in machine learning, because it is clinically familiar and has been well-studied by humans, by non-ML computer algorithms, and by ML algorithms.
Concept 1. Traditional rules-based algorithms apply rules to data, while machine learning algorithms learn patterns from data.
Computers can mimic human intelligence through rules-based algorithms, used in traditional computerized ECG interpretation (12), or, through machine learning (ML) algorithms. Rules-based algorithms make use of a set of rules explicitly programmed by humans to mimic the knowledge a cardiologist might use in reading the ECG, for example: determining the rate, identifying P-waves before every QRS, recognizing pathognomonic waveform changes. In this case, the rules used that lead to a computer’s ECG diagnosis are well understood.
In contrast to rules-based algorithms, machine learning algorithms that learn rules and patterns from the data fed to them, rather than having those rules explicitly programmed. This fundamental difference is responsible for both the excitement about machine learning algorithms’ potential to solve problems in cardiology beyond human capability—for example, detecting patients with ejection fraction ≤35% or detecting patients with hypoglycemia based on an ECG alone (14,15)—but it is also cause for the caution clinicians must have in evaluating and eventually implementing ML solutions.
There are several types of machine learning algorithms, from decision trees and support vector machines (SVMs), to highly complex, data-hungry algorithms called neural networks. Neural networks are used in deep machine learning (deep learning, or DL), and their ability to analyze large amounts of highly complex data—electronic health record (EHR) data, for example, or the collection of pixels that make up medical images—are especially exciting for cardiology applications. An ML algorithm is trained on data, yielding a trained ML model that can then be evaluated on never-before-seen test data. Several excellent reviews discuss different types of ML algorithms, including neural networks, in more detail (10,16–18).
Concept 2. ML algorithms can learn patterns from labeled examples: supervised learning
Machine learning algorithms can learn patterns from data in two main ways. The first approach is to provide data (e.g., a set of ECGs from patients visiting a clinic) along with a corresponding label for what the algorithm is meant to learn (in this example, the label would be the diagnosis for each single ECG). This approach is called supervised learning (14,19–21). Based on the labeled examples alone, the algorithm learns for itself the most important features of the ECG that drive its decision, and devises rules to exploit those features for diagnosis of new ECGs never seen before.
Supervised learning algorithms have the advantage of having a clear goal: predicting the label of interest. But the disadvantage of supervised ML algorithms is that their ability to find interesting patterns in the data is also constrained by those labels. As any clinician who has written question-and-answers for board prep or put together a self-assessment program (SAP) knows, choosing the right data for training and deciding on the correct answer or label is critically important to training and takes a lot of work. Similarly, a major challenge in supervised machine learning is the availability of data sets of adequate size that have correctly annotated labels of interest. This is not always straightforward. For example, data scientists may trust that when a patient has a specific diagnostic code in their EHR, such as acute venous thromboembolism (VTE), that the diagnostic code can serve as an accurate clinical label for VTE. However, studies have founded that a VTE diagnosis in the EHR has a positive predictive value as low a 31% for an outpatient diagnosis and 65% for an inpatient diagnosis (22). Therefore, correct labelling of datasets requires active curation by physicians, and will often require consensus from more than one physician.
Concept 3. ML algorithms can learn patterns without labeled examples: unsupervised and reinforcement learning
The second approach that can be used is unsupervised learning, designed to discover the hidden patterns by analyzing data without a label. An unsupervised learning approach is similar to a medical student that is given a huge number of ECGs without any diagnosis. The student may not have been told what atrial fibrillation is or what a left bundle branch block looks like, or that pericarditis can cause PR segment depression, but he can still classify the examples in similar groups, selecting the most important ECG characteristics that he thinks differentiate one example from another. He may learn on his own that the bumps and squiggles in an ECG seem to follow a certain sequence and that they have different durations and morphologies, or he may notice an entirely novel characteristic not typically taught in the textbooks. In the same way, an algorithm can cluster available data in several groups, and can learn the data features that are most relevant in differentiating the examples.
Unsupervised learning holds several advantages in machine learning: it allows the algorithm to develop an understanding of the data that is unconstrained by labor-intensive and often variable human labels, and it allows the algorithm to come up with novel groupings and clusters of the data that a human being may not be aware of. These learned features as groupings can also be exploited by a subsequent supervised learning step (23), just as the medical student, having pre-digested the ECGs on his own, then has an easier time learning that certain patterns correspond to sinus arrhythmia or hyperkalemia.
Unsupervised learning approaches are common in the analysis of EHR or genetic data to automatically extract the most useful information (24) to identify distinct disease subgroups—of type 2 diabetes, for example (25)—or to find a new set of biomarkers that may be better predictors than standard biomarkers in defining distinct subsets of individuals with similar health status (26). Clustering—dividing any type of data in groups of similar data points based on the data’s characteristics—is indeed a successful application of unsupervised learning to clinical data (27,28). As above, the clusters generated may lead to novel groups of data points that may illuminate novel subgroups of disease, new biomarkers, or new predictors of a clinical outcome of interest. However, clustering often represents only the first step to group the data before a more insightful analysis. Importantly, clusters that may suggest new groupings of disease or other novel insights must be validated clinically, as it can be easy to set clustering parameters to create subgroups, or lump together groups that should be separate.
In addition to supervised and unsupervised learning, a third category of learning methods, reinforcement learning, is used when the ML algorithm is given iterative information about the outcome of its predictions in the environment as feedback that helps guide its future predictions. Imagine training an ML algorithm to dose intravenous heparin, for example. For this task, neither a supervised approach, such as labeling different doses of heparin as ‘good’ or ‘bad,’ nor an unsupervised approach (no label at all) would work well. Instead, in a reinforcement learning approach, the algorithm would have a goal—optimize partial thromboplastin time (PTT)—and would predict different heparin doses, receiving a positive or negative reward depending on how close to the goal PTT it got. That reward would then inform the algorithm’s next guess, and so forth. Reinforcement learning has been used with success to learn complex strategy games like Go (23), where the goals and the effect of the algorithm’s decision on the environment is clear, and where the algorithm can play Go millions of times, often losing the game until it learns. It has also been studied for some clinical tasks, such as guiding dofetilide dosing or ventilator settings (29,30). However, reinforcement learning may be of limited use in clinical tasks where the goal and the environment’s possible response are much more complex, and where ‘losing the game’ in a clinical setting during the learning phase of the algorithm is not ethically acceptable.
Concept 4. When ML algorithms learn rules that perform well on training data but fail on test data, they have failed to learn rules that are generalizable
This problem, called overfitting, happens when there is mismatch between the complexity of the ML algorithm and the size of the training dataset provided to it. Take for example a very complex algorithm trained on a small dataset: the algorithm learns rules so tailored to the specific training examples at hand that it has in effect “memorized” them instead of learning the general rules behind them. This means the model performance will appear to be very good, but then fail when deployed on larger datasets. The problem is even more severe in the presence of class imbalance, i.e., when one subgroup of the training data has only very few samples.
Overfitting is one of the most common problems encountered when using supervised machine learning algorithms, and the main limitation to their application to real-life clinical situations (31). Overfitting can happen to human learners as well, for example, the fellow who has studied 10 ECGs very closely but then fails the board exam. To date, however, a human learner is typically much better at generalizing knowledge from a small dataset than an ML algorithm.
It is common to estimate the complexity of an ML model based on the number of parameters in the trained model, which can be in the order of hundreds of millions for modern DL architectures, even when trained on just a few thousand training examples (32). Methods to mitigate overfitting include 1) reducing the model complexity, 2) increasing the number of examples in the training set (although this is often not possible) 3) limiting the number of iterations of the algorithm (i.e., training cycles) on the training data, 4) balancing the parameters learned in the model (regularization) to obtain a simpler model that underfits on the training data but generalizes better, and 5) using not one model but an ensemble of separate models to come to the desired prediction so that various overfitting effects of each singular model balance out (33).
Given the common and dangerous issue of overfitting, the performance of the trained model must be tested on cases that are completely independent from the training examples, and ideally, from test data from multiple external medical centers. In order to evaluate performance of any ML algorithm on a clinical task, full information on the data and methods used in training and testing must be reported (11).
Concept 5. Accuracy, interpretability and explainability of a machine learning algorithm
Simply put, diagnostic accuracy is defined by the proportion of correct predictions (true positives and true negatives) in a given test dataset. Supervised machine learning models can be more accurate on a given diagnostic task than the average clinician, as shown by deep learning algorithms matching the diagnostic accuracy of a team of 21 board-certified practicing cardiologists in the classification of 12 heart rhythm types from single-lead ECGs (19).
However accurate they may be, typically, the rules by which ML models have achieved their performance are not clear – this is especially true for highly complex neural networks. This lack of easy interpretability of the neural networks’ decision-making means that it can be difficult to verify the learned rules have truly generalized to real-life clinical situations (34).
It can also be difficult to learn from novel features or patterns the ML algorithms may have detected. Lack of interpretability raises some important questions for the clinician.
First, does it matter that we don’t know how a tool works, as long as we have validated that it works very well? Second, what is the level of testing an ML must go through to be considered safe for clinical situations, especially when we don’t know how it works?
While one would like an ML model to maximize both accuracy and interpretability, to date there is a tradeoff between the two, especially when using neural networks (deep learning), whose operations are often too complicated to analyze and interpret. As clinicians, we may need to choose whether to take advantage of a “black-box” algorithm with proven 99% accuracy, or an interpretable algorithm with recognizable features that lead to the model decision, but has only 80% accuracy (35). The debate on this topic is still open and it is one of the reasons for the slow adoption of current machine learning techniques in medicine. Possible solutions to this dilemma are the use of interpretable models substituting the black box algorithms (34), or the use of model-agnostic methods to explain the decision of the black box with local (case specific) and global (model specific) explanations (36). Clinicians will need to evaluate algorithms in clinical trials and incorporate their recommendations in guidelines documents (Table 3). A more futuristic approach involves the redesign of AI models towards a knowledge-driven reasoning-based approach, which may be extremely valuable in medicine, but these methods are currently under investigation in the computer science world, while their applicability in medicine is likely at least a decade away (37).
Table 3.
Roles for the non-ML expert in machine learning for cardiology
| Being an informed consumer of the literature and of cardiac ML research project |
| Collaborating with machine learning and data science experts to innovate clinical practice |
| Beta-testing AI-enabled products in the clinical setting |
| Advising one’s medical center, clinic, or institution on investments in data science and ML tools |
| Advocating for responsible data sharing at one’s institution and via professional societies |
| Advocating for harmonization and standardization of data formats and data processing across institutions |
| Ensuring that algorithms have been trained from data that adequately represents patients, acquisition methods and equipment, and other factors. |
| Participating in annotation of datasets as the clinical expert |
| Participating in validating model predictions as the clinical expert |
| Testing AI-enabled clinical products and/or novel insights in randomized controlled trials |
| Evaluating and incorporating AI-based decision making in updated clinical guidelines documents |
| Creating and promoting training opportunities (courses, fellowships) in data science for cardiology trainees and in clinical science for data scientists |
| Evaluating and updating cardiology training — what core competencies will remain a requirement, and what will be replaced by ML? |
Concept 6. Machine learning algorithms can be re-trained to include more data or different data types
Like a medical student can learn to interpret many different types of data—ECGs, chest X-rays, lab values—with proper training, and can improve the more data they see, machine learning algorithms are dynamic and can be re-trained with additional and/or different types of labeled data with minimal or no human supervision. For example, the same deep learning algorithms developed for classification of non-clinical images (dogs, cats, trees) have been adapted and re-used to detect congenital heart disease, diabetic retinopathy, cardiac views, skin lesions, and a host of other clinical tasks (20,38–40). This is quite important, as a machine learning algorithm does not need to be fundamentally redesigned when facing a new problem and dataset: the algorithms used to implement a classification task are fundamentally similar whether the data is ECG, ultrasound, or nuclear imaging, the algorithms used to implement a segmentation task are similar whether it is on CT or MRI data, and so forth.
A given neural network can be trained ‘from scratch’ on different datasets for different tasks, or, it can be ‘pre-trained’ on a more general dataset and task to learn very basic data features before being ‘fine-tuned’ on e.g. a cardiology-specific dataset and task (38,40–45). This approach is called transfer learning, and it can be useful when the specialized medical data is rare. Similarly, a model that was trained on a certain cohort of medical data can also be further trained on additional medical data, for example from another hospital.
Finally, while ML algorithms can be used on different problems with very little tweaking needed, a model trained on a particular data type still has difficulty generalizing knowledge across different data types. For example, integrating the information about the left ventricle from multiple data types like an ECG, an echocardiogram, a nuclear study, and a cardiac catheterization is still a task that is much easier for a trained clinician than for a machine learning model. Such cross-modal synthesis of knowledge from different data types is a task where ML algorithms could have the potential to shine, and this is an active topic of research in computer science.
Machine learning in cardiology: a computational survey of the literature and case studies
With a conceptual framework for machine learning algorithms in place, we now present a survey of the literature on machine learning in cardiology, highlighting several applications. Because machine learning papers in cardiology are growing so quickly, we provide an open-source semi-automated method to survey all machine learning papers in cardiology that can be updated over time, as well as specific case studies to detail several use cases to date: denoising and image enhancement, feature extraction and representation, improving traditional algorithms with data repurposing, novel insights, and improving healthcare systems.
Research landscape
To provide an informal survey of the research landscape, we retrieved bibliometric information on all publications fitting search terms corresponding to machine learning and cardiology. These publications were annotated according to whether they were original research or reviews, editorials, or other commentary; as well as according to the machine learning problem(s) addressed, the machine learning method(s) used, the cardiology disease(s) studied, and the type(s) of medical data studied. These data were retrieved, analyzed, and visualized using the Python programming language and application programming interfaces (APIs) from NCBI PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) and preprint servers arXiv (https://arxiv.org/), bioRxiv (https://biorxiv.org), and medRxiv (https://medrxiv.org). This approach allowed a method for surveying the literature which can be easily updated and applied toward other search topics. Code is available at https://github.com/ArnaoutLabUCSF/cardioML/JACC_2020 for use and adaptation by the research community. (Figures in the published version of this manuscript use data through July 20, 2020.)
The past five years have seen over 3,000 papers on machine learning in cardiology published in PubMed (about 80% original research) or posted on the popular preprint servers arXiv and bioRxiv, where scientists are increasingly sharing their papers with each other and with the public before, during, and sometimes as an alternative to, the peer review process (46) (Figure 1). This represents tremendous growth, such that in 2020 nearly 1 in every 1,000 new papers in PubMed will be on artificial intelligence and/or machine learning in cardiology. Despite this uptick in publications, there are still several areas that are open for study. For peer-reviewed papers in PubMed, approximately two thirds involve atherosclerosis (including dyslipidemia and cerebrovascular disease), heart failure, or hypertension and other cardiac risk factors (Figure 2). Publications fairly evenly cover the major data types: electronic health record (EHR) data, electrocardiography (ECG), ultrasound (US), magnetic resonance imaging (MRI) and computed tomography (CT). Omics is also a popular subject. The two least expensive data types, X-rays and laboratory studies, have received less attention.
Figure 1. Marked growth in cardiac AI/ML studies.
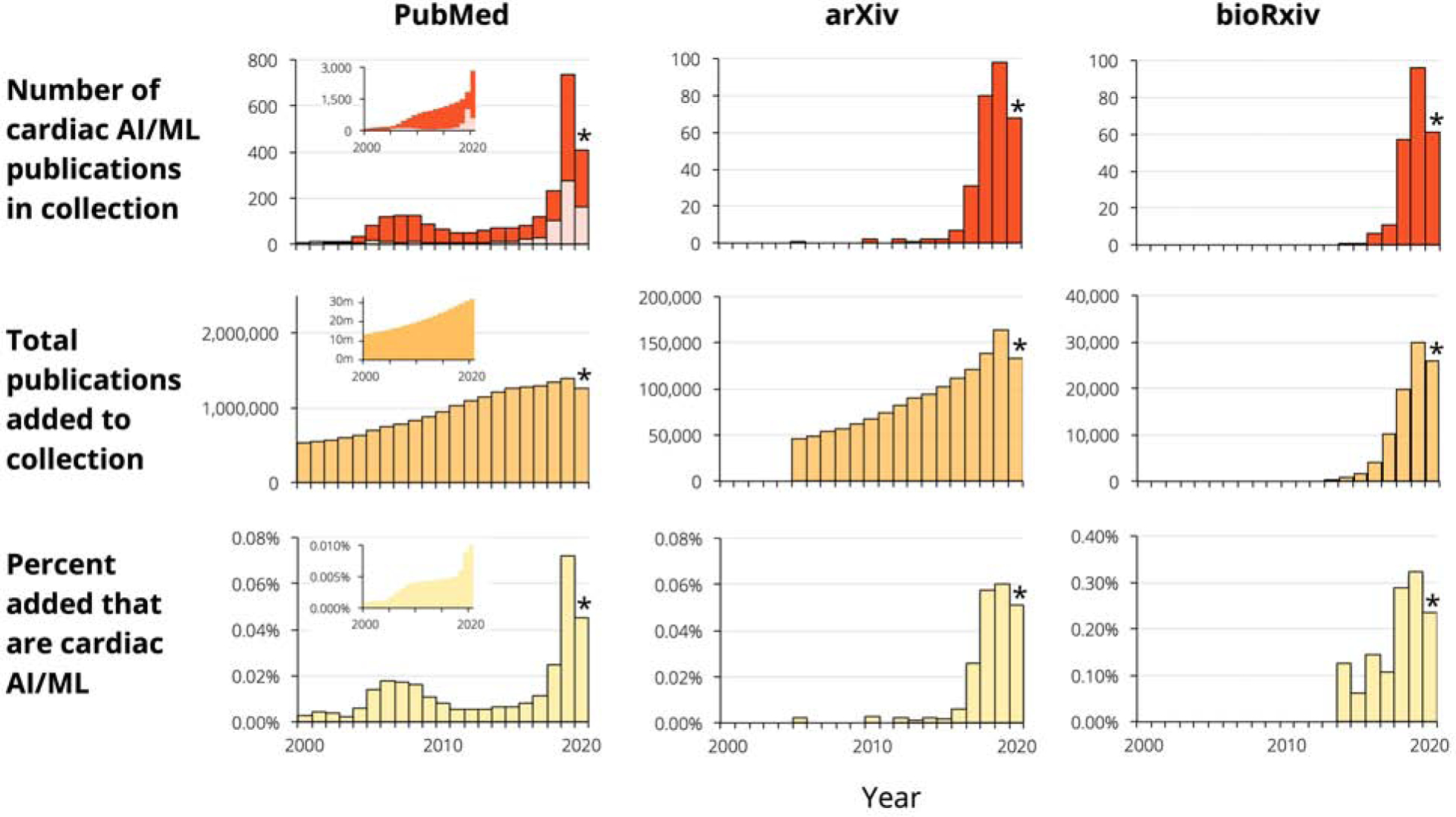
The plots in the first row show numbers of publications per year; red denotes original research, while pink denotes non-original articles such as reviews and commentary. The second row shows the total number of papers (any topic) added to each of the three collections, while the third row presents the percentage of papers on cardiac AI/ML. The insets for Pubmed in the first two rows show the cumulative number of papers across all previous years, while in the third row it shows the cumulative percentages considering all papers till that year. *2020 numbers are partial.
Figure 2. Content in cardiac AI/ML publications on PubMed.
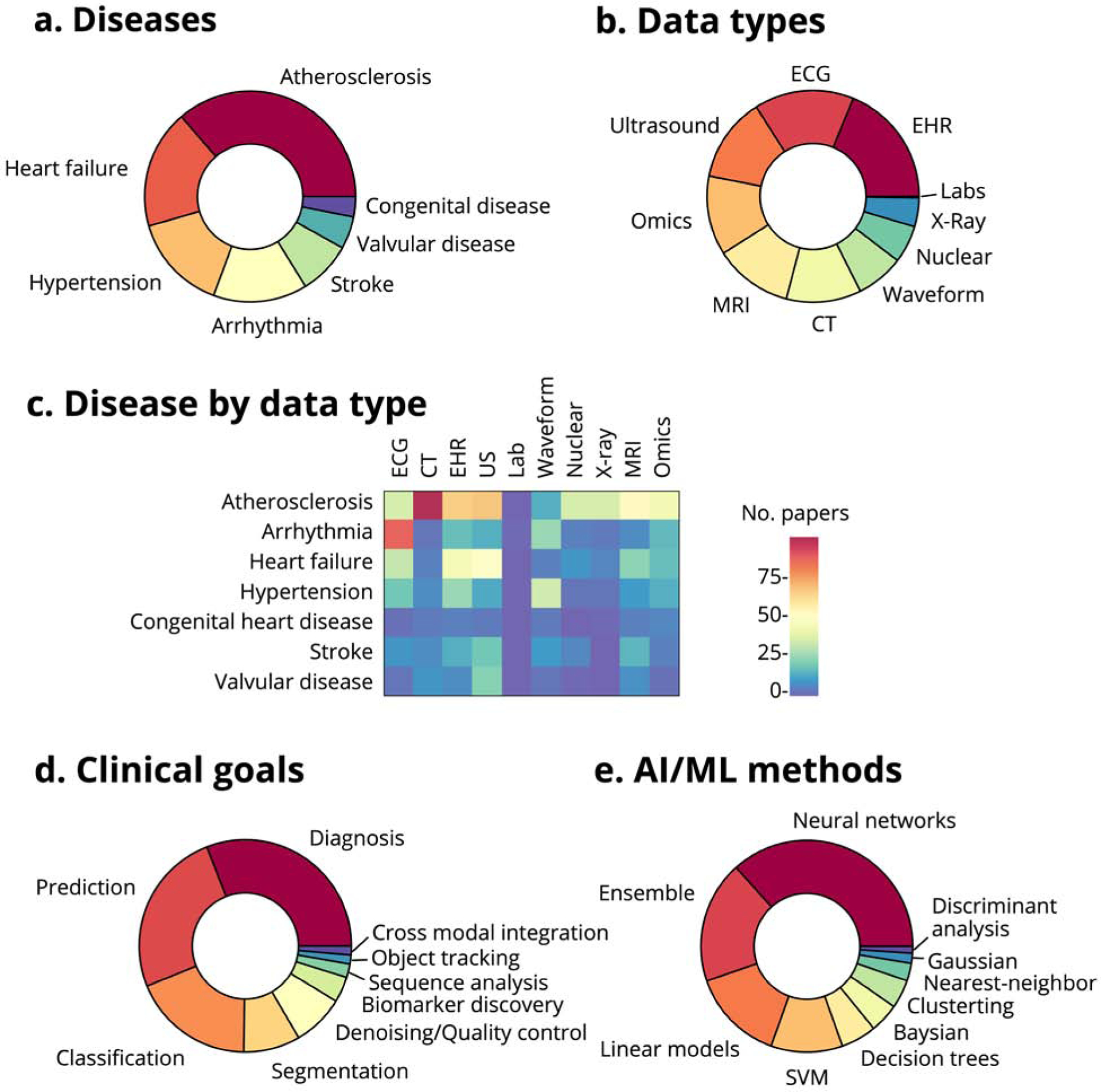
(a) Distribution of publications by disease category. (b) Distribution of publications by data modality. (c) Number of papers studying various cardiac disease categories by data type (waveform data includes catheterization, arterial pulse waveforms, plethysmography, and other waveform data but does not include ECG; atherosclerosis includes dyslipidemia, peripheral vascular disease and cerebrovascular disease). (d) Distribution of publications by goal. (e) Distribution of publications by machine learning method.
The vast majority of publications involving CT looked at atherosclerosis; most heart failure studies used the EHR and US; and, predictably, most studies of arrhythmia used ECG data (Figure 2). Three-quarters of these papers are about diagnosis, prediction, or classification, with relatively less attention to object tracking and novel biomarker discovery. Consistent with trends across artificial intelligence and machine learning, most cardiac AI/ML studies use neural networks (or an ensemble of methods that usually include neural networks), with a minority exclusively using more traditional techniques like linear models, SVMs, and decision trees.
For papers published in PubMed, the leaders in research output in cardiac AI/ML are the United States (especially California, Massachusetts, and New York) and the United Kingdom, followed by China, Germany, and the Netherlands, with other developed countries contributing materially as well (Figure 3). This work is very broadly distributed, with high per-capita contributions from all the developed countries but also work across much of the developing world. Collaborations in cardiac AI/ML span the globe and are similarly multi-institutional within the United States.
Figure 3. An international and collaborative effort.
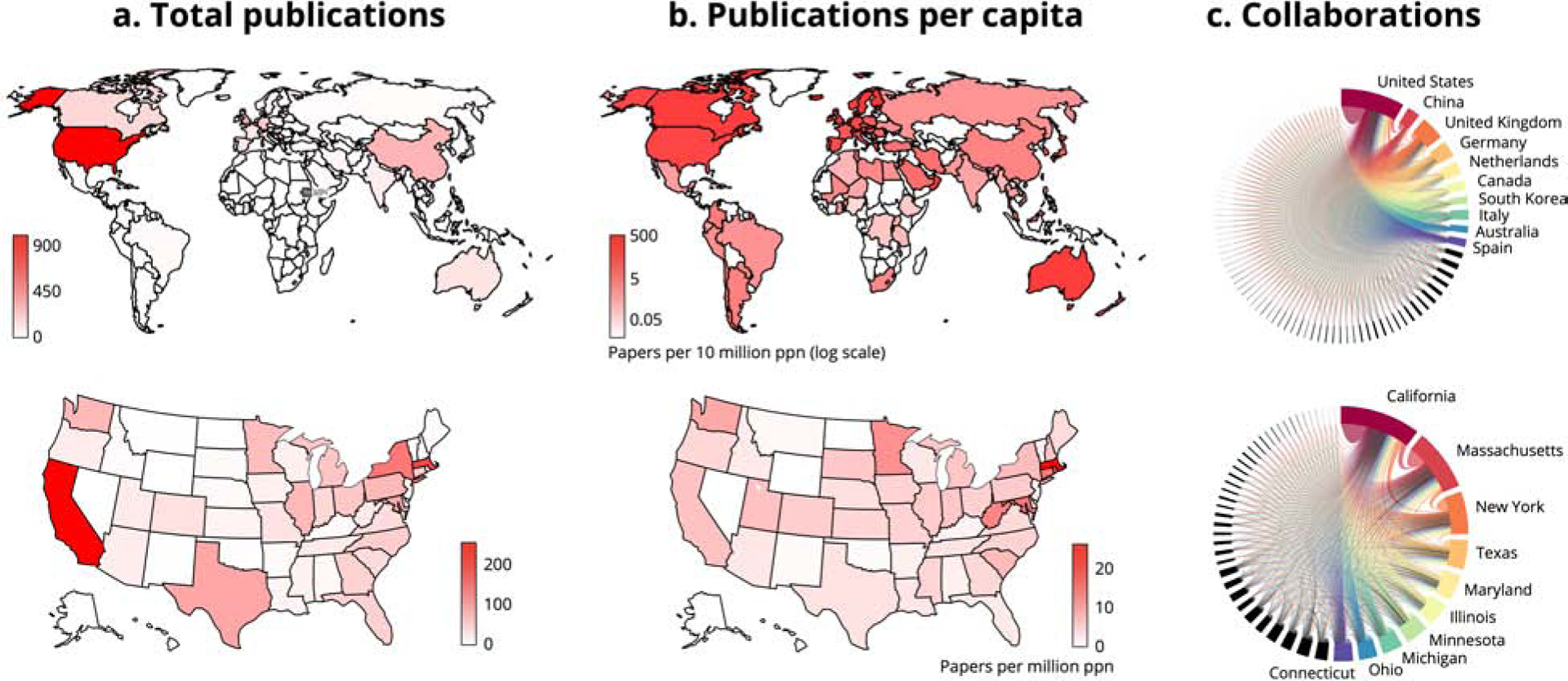
(a) Total publications on cardiac AI/ML in PubMed, worldwide and in the United States (note log scale). (b) Publications per capita worldwide and in the United States (note log scale). (c) Collaborations measured by author locations on each publication, presented worldwide and statewide (for clarity, only top ten countries and/or states are labeled).
Case studies of machine learning in action
In terms of disease topics and data modalities studied, the survey shows which have been more and less studied. Building from the machine learning Concepts referenced above, we developed a toolbox of questions and considerations when evaluating the impact and rigor of machine learning studies (Table 1). With this in mind, we have noted several interesting applications of machine learning to cardiology. Because of the largely “data-agnostic” nature of machine learning algorithms (Concept 6), we organize these case studies by machine learning use case, rather than by clinical data type or disease.
Table 1.
Considerations When Evaluating Machine Learning Manuscripts or Projects.
| Questions to ask | Examples |
|---|---|
| What is the problem being addressed? Is solving the problem impactful for medicine? | High value: An unrecognized or unsolved problem; a problem where clinical practice has been shown to fall short. Intermediate value: A solution exists, but the new solution provides much better accuracy, reproducibility or time-efficiency, or can work in a different environment or patient group from the current standard. Low value: A solution for something that is not a significant clinical problem, or, a robust, well-benchmarked solution already exists. |
| What is the current state of the art and how does it perform? | Does a highly accurate, scalable, and efficient solution already exist? Is there no good current solution in clinical practice? |
| Does the problem need a machine learning solution? | Is it a problem of complex pattern finding in complex/nonlinear data? Will the use of machine learning significantly improve the performance with respect to a standard rule-based algorithm? |
| What benchmark is the machine learning solution trying to beat? | High clinical impact: beating current standard of care (human expertise, prevailing risk prediction model), or, no benchmark may exist for an entirely novel model. Intermediate impact: a benchmark established by a prior ML model. Low impact: benchmark exists but is not referenced. |
| If supervised learning is used, what is the ground truth, or gold-standard, label? | Strong ground-truth label: a gold-standard diagnosis, such as pathologic diagnosis, as the basis for a disease label. Intermediate label: Blinded and/or independent votes from expert clinicians; electronic health record data (depends on the quality of the EHR data). Weak label: ICD codes alone, or other surrogates that are known to have poor sensitivity and specificity for the condition under study. |
| If unsupervised learning is used, what methods will be used to validate the patterns learned by the model? | A common example of unsupervised learning is in clustering data into subgroups without a priori knowledge of whether those subgroups are meaningful. In this case, clinically relevant methods of sampling and measuring differences among the learned subgroups are important. |
| Is the training dataset appropriate for the task at hand? | Classification problems typically need a large amount of training data to avoid overfitting; the only method to verify that the model does not overfit the training data is to verify that the testing set is completely independent than the training set, and that the model performs well in the testing set. |
| Are the validation and test datasets independent from the training dataset? | Examples of dependent features would include two QRS morphologies from two ECGs from the same patient, two image slices from the same CT scan, or two blood glucose measurements from the same patient. Putting one in the training dataset, and the other in the test set will make test set performance falsely high. Instead, training and test datasets should be split in a manner that retains sample independence. |
| Are all datapoints being processed or manipulated in the same manner? | Often, training data needs to be pre-processed in order to be ready for machine learning. However, it must all be processed in the same way, or else the model runs the risk of learning the manipulations made to one subgroup of data compared to another, rather than learning the meaningful patterns in the data. |
| How is the clinical use case being formulated? | Whether as a classification problem, a segmentation problem, a time series problem, or something else, the machine learning formulation of the problem should be relevant to the clinical task. |
| Are methods clear and reproducible? | All information on data preprocessing steps, machine learning algorithms used, and the parameters of those algorithms, should be presented. Code can be included, but it must then be tested to run as described. |
| Are results reported both for the algorithm output and for the clinical problem of interest? | Performance metrics from machine learning algorithms (precision, recall, Dice score, and others) are important. However, results should also be reported in terms clinically relevant to the task (e.g. sensitivity, specificity, number needed to treat, time to diagnosis, etc.). Often, the costs for a false positive or a false negative error are quite different, so it is important to choose a working point that keeps into account these different costs. Metrics should include confidence intervals and p values to demonstrate whether they are statistically significantly different from the chosen benchmarks. |
Denoising and image enhancement.
Across several modalities, machine learning has been applied to the clinical problem of image post-processing and de-noising. In ultrasound (47,48), CT (49,50), MRI (51,52), and nuclear imaging (53,54) the complex patterns of noise encountered in clinical imaging have lent themselves to neural network-based denoising. In terms of clinical utility, machine learning has the potential to decrease the time and labor involved in image post-processing, and to reduce inter-operator, inter-vendor, and inter-institution differences in data processing. In the case of X-ray and CT imaging, neural network-based image analysis may allow high-quality imaging with reduced radiation dose compared to the current standard. Currently, most machine learning approaches to denoising have taken a supervised learning approach, where models are being trained to approximate proprietary denoising software as the ground-truth label (48), or are trained by adding noise to input data. Several studies in the literature have been trained on phantoms and relatively small amounts of clinical data. This means that current neural network denoisers may still be learning relatively simple noise patterns rather than the true extent of variability in clinical imaging. The performance of denoising networks has been primarily measured in terms of comparing input images to their ground-truth labels, which is a necessary and important performance metric; however, measuring changes in diagnostic performance based on machine-learning-based image denoising is an important next step to validating their utility in clinical practice. Some clinical trials are underway to test image denoising on a larger scale (55,56)
Feature extraction and representation.
Supervised and unsupervised machine learning is also being used to represent important features of clinical data into simpler, more compact, more uniform formats: this process is called feature extraction. For example, free-text clinical notes can be analyzed and represented by the list of diseases and procedures mentioned; an ECG can be analyzed and represented by a small group of numbers that summarize the intervals, axis, and QRS morphology; an ultrasound image can be represented by the structures detected in that image. Machine learning algorithms can also represent data in ways that are not intuitive to a human, but that are nevertheless simpler and more useful to computers. The automatic extraction of these features via complex non-linear combination of the input data is indeed the key for the superior classification performance of modern deep learning algorithms (21,32). These extracted features can allow for comparing data across institutions (data harmonization), and combining features extracted from different data types can enable a multi-modal representation of a particular patient or disease. Therefore, models that effectively extract important clinical features from different data types in a scalable fashion are fundamentally important to powering larger and more complex machine learning studies in the near future.
To date, features learned from neural networks and other ML algorithms have been used in cardiology in myriad classification tasks, such as classifying cardiac views from different imaging modalities (20,57) or classifying the presence or absence of disease from image, ECG, text, ascultation, or lab data (58–61); segmentation of cardiac structures and/or abnormalities (41,62,63) (Table 2), and tissue characterization (64–66) of imaging, text, and ECG data. Several machine learning algorithms that have been cleared by the FDA, while proprietary in nature, are likely based on classification and/or segmentation algorithms (examples shown in Table 2). Training models to perform these foundational tasks has often required considerable data labeling; therefore, this work will continue to benefit from parallel research on leveraging small datasets (20,67,68) as well as synthesizing larger datasets from smaller ones (69,70).
Table 2.
Select FDA-cleared machine learning products for cardiology
| Company | Product | Indication |
|---|---|---|
| AliveCor | AliveCor Heart Monitor | atrial fibrillation detection |
| Apple | Apple Watch | atrial fibrillation detection |
| Arterys | CardioDL | cardiac MRI measurement |
| Caption Health | EchoMD AutoEF, Guidance | echocardiogram LVEF measurement, guidance |
| Canon | Advanced Intelligent Clear-IQ Engine (AiCE)* | general biomedical image denoising |
| Eko Devices | Eko Analysis Software | audiogram interpretation |
| FitBit | ECG App | atrial fibrillation detection |
| PhysIQ | Heart Rhythm and Respiration Module (HRRM) | ECG, vital signs, cardiac function |
| Qompium | FibriCheck | atrial fibrillation detection |
| Shenzhen Carewell Electronics | AI-ECG Platform & Tracker | ECG interpretation |
| Subtle Medical | SubtlePET*, SubtleMR* | general biomedical image denoising |
| Ultromics | EchoGo Core | echocardiogram measurements |
| Zebra Medical Vision | HealthCCS | coronary calcium score |
these products are not specifically cardiovascular, but provide general tools for image denoising
Improving traditional algorithms and data repurposing for advanced insights.
Machine learning is being used to improve upon traditional risk prediction algorithms using available registry data (71–74). Machine learning can enable development of biomarkers that otherwise would be highly labor-intensive to calculate or were not consistently scores on retrospective imaging, such as the quantitation of epicardial adipose tissue on imaging studies to improve risk prediction for myocardial infarction (75); this work is now being studied in a clinical trial (76). Researchers are also using machine learning to test whether advanced insights are possible from data types not typically used for those insights: examples include using clinical data to predict genetic variants, predicting pulmonary-to-systolic blood flow ratios from chest X-rays, fall risk prediction from wearable home sensors, and calculating a calcium score from chest CTs or from perfusion imaging (77–81). These studies benefit when a robust ground-truth label and clinical performance benchmark are present from traditional sources (e.g. genetic sequencing, right heart catheterization, clinical falls data, and standard calcium scores for the examples mentioned here).
Novel insights.
In the above examples, the insights detected can be reasonably expected to be found in the data type studied. For example, even though we use blood chemistry to predict hyperkalemia precisely, we know that signs of electrolyte imbalance can be evident on ECG. Researchers are using similar approaches to detect novel insight in unexpected data types, finding patterns in data that are not evident to the trained clinician. For example, studies have reported detecting adverse drug reactions from social media, detecting gender from a fundoscopic retinal image, or detecting coronary microvascular resistance from ocular vessel exam (82–84). Especially when initial studies are published from small datasets, there is a possibility that the models are overfitting (Concept 4) rather than finding a true insight. Such findings must be validated rigorously in multi-center datasets and clinical trials, and computational experiments such as saliency and attention mapping must be performed to help understand what data features the machine learning models are detecting.
Healthcare systems.
Finally, researchers are putting machine learning to work on systems-level problems in healthcare, including deployment of AI algorithms for cardiovascular disease screening, and remote monitoring of patient medication adherence and optimization (85–89). The former trial, still underway, studies how AI-derived ECG results to screen for low ejection fraction are delivered to primary care physicians (89). A combination of clinical and administrative data can be used to predict optimal patient scheduling and procedure protocoling, as well as hospital readmission rates, transfer into and out of the intensive care unit (ICU), and to reduce false alarms in the ICU (90–93).
The use cases above demonstrate how widely machine learning approaches may change the practice of clinical cardiology. Publications to date still feature largely small and single-center datasets and demonstrate often modest gains over standard-of-care clinical performance, however, the quality and impact of machine learning in cardiology continues to gain ground, and as these proofs-of-concept mature, we may find them applied to clinical cardiology in the near future.
Challenges and a call to action for clinicians
From the above, we see that there is an opportunity for advanced pattern-finding from machine learning algorithms to help clinicians, especially as medical data is increasing in amount and complexity. We see that in cardiology, machine learning research is already well underway, with several interesting proofs-of-concept from the research community and some proprietary solutions introduced by industry. We have also seen that several important concepts in ML research design—including thoughtful selection of use cases and performance metrics, meticulous annotation and curation of datasets, and broad testing and validation to ensure generalizability of results—must be considered and implemented carefully in order to solve real-world problems in clinical cardiology.
We believe that machine learning will become part of the clinician’s toolbox, at the point of care (in electronic health records and risk calculator algorithms), ‘under the hood’ in diagnostic and therapeutic devices (such as ECG, CT, pacemakers, insulin pumps), and in scheduling and protocolling medical tests and appointments. While some cardiologists will choose to specialize in machine learning and data science, cardiologists who are not necessarily ML experts also have several important roles to play in the responsible incorporation of machine learning into cardiology (Table 3) including participating in annotation and curation of data as a clinical expert, advocating for ML solutions that solve important clinical problems for our patients without racial, gender, or other forms of bias, and running rigorous clinical trials of machine learning algorithms on large patient cohorts. We hope that cardiologists as active participants and informed users will help ensure that machine learning will live up to its potential to improve the field.
Central Illustration. Marked growth in cardiac AI/ML studies span several disease focus areas and several data types.
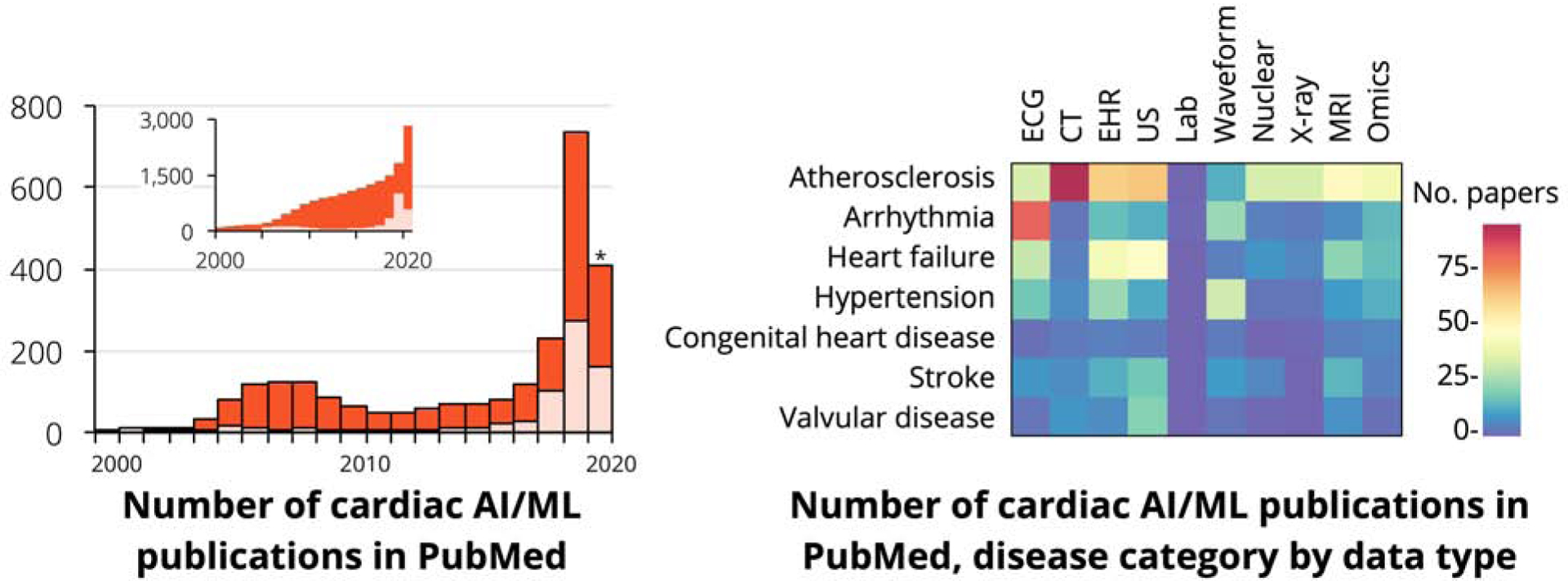
Publications on machine learning in cardiology continue to grow (inset shows cumulative number of cardiac AI/ML papers). Several disease categories and data modalities have been studied in these publications, but several areas remain open for further exploration. Asterisk indicates partial information for the year 2020.
Highlights.
Machine learning algorithms can find sophisticated patterns in medical data and have the potential to improve cardiovascular care.
Cardiologists must take an active role in shaping how machine learning is used in cardiovascular practice and research.
To empower cardiologists in this role, we provide a framework to help critically evaluate developments in machine learning.
We also provide an open-source bibliometric survey of the machine learning in cardiology.
Acknowledgments:
We thank Dr. S. Steinhubl for critical reading of the manuscript.
Funding Sources: R.A. and R.A. were supported by the National Institutes of Health (R01HL15039401), Department of Defense (W81XWH-19-1-0294) and the American Heart Association (17IGMV33870001). R.A. is additionally supported by the Chan Zuckerberg Biohub. Resources at Scripps Research (G.Q.) were provided by the National Institutes of Health (UL1TR002550 from the National Center for Advancing Translational Sciences, NCATS).
Abbreviations
- AI
artificial intelligence
- API
application programming interface
- CT
computed tomography
- ECG
electrocardiogram
- EHR
electronic health record
- ICU
intensive care unit
- ML
machine learning
- MRI
magnetic resonance imaging
- NCBI
National Center for Biotechnology Information
- PTT
partial thromboplastin time
- SAP
self-assessment program
- SVM
support vector machine
- US
ultrasound
- VTE
venous thromboembolism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have no relevant disclosures.
References
- 1.Shameer K, Johnson KW, Glicksberg BS, Dudley JT, Sengupta PP. Machine learning in cardiovascular medicine: are we there yet? Heart 2018;104:1156–1164. [DOI] [PubMed] [Google Scholar]
- 2.Harnessing the Power of Data in Health: Stanford Medicine 2017 Health Trends Report. June 2017. https://med.stanford.edu/content/dam/sm/sm-news/documents/StanfordMedicineHealthTrendsWhitePaper2017.pdf Accessed March 20 2020.
- 3.Obermeyer Z, Lee TH. Lost in Thought - The Limits of the Human Mind and the Future of Medicine. N Engl J Med 2017;377:1209–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Litjens G, Ciompi F, Wolterink JM et al. State-of-the-Art Deep Learning in Cardiovascular Image Analysis. JACC Cardiovasc Imaging 2019;12:1549–1565. [DOI] [PubMed] [Google Scholar]
- 5.Sardar P, Abbott JD, Kundu A, Aronow HD, Granada JF, Giri J. Impact of Artificial Intelligence on Interventional Cardiology: From Decision-Making Aid to Advanced Interventional Procedure Assistance. JACC Cardiovasc Interv 2019;12:1293–1303. [DOI] [PubMed] [Google Scholar]
- 6.Sengupta PP. Intelligent platforms for disease assessment: novel approaches in functional echocardiography. JACC Cardiovasc Imaging 2013;6:1206–11. [DOI] [PubMed] [Google Scholar]
- 7.Dey D, Slomka PJ, Leeson P et al. Artificial Intelligence in Cardiovascular Imaging: JACC State-of-the-Art Review. J Am Coll Cardiol 2019;73:1317–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med 2019;380:1347–1358. [DOI] [PubMed] [Google Scholar]
- 9.Nicol ED, Norgaard BL, Blanke P et al. The Future of Cardiovascular Computed Tomography: Advanced Analytics and Clinical Insights. JACC Cardiovasc Imaging 2019;12:1058–1072. [DOI] [PubMed] [Google Scholar]
- 10.Chang A Intelligence-Based Medicine: Artificial Intelligence and Human Cognition in Clinical Medicine and Healthcare. 1st Edition ed: Academic Press, 2020. [Google Scholar]
- 11.Norgeot B, Quer G, Beaulieu-Jones BK et al. Minimum information about clinical artificial intelligence modeling: the MI-CLAIM checklist. Nat Med 2020;26:1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kossmann CE. Electrocardiographic Analysis by Computer. JAMA 1965;191:922–4. [DOI] [PubMed] [Google Scholar]
- 13.Smulyan H The Computerized ECG: Friend and Foe. Am J Med 2019;132:153–160. [DOI] [PubMed] [Google Scholar]
- 14.Attia ZI, Kapa S, Lopez-Jimenez F et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat Med 2019;25:70–74. [DOI] [PubMed] [Google Scholar]
- 15.Porumb M, Stranges S, Pescapè A, Pecchia L. Precision Medicine and Artificial Intelligence: A Pilot Study on Deep Learning for Hypoglycemic Events Detection based on ECG. Sci Rep 2020;10:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henglin M, Stein G, Hushcha PV, Snoek J, Wiltschko AB, Cheng S. Machine Learning Approaches in Cardiovascular Imaging. Circ Cardiovasc Imaging 2017;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deo RC. Machine Learning in Medicine. Circulation 2015;132:1920–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sengupta PP, Shrestha S, Berthon B et al. Proposed Requirements for Cardiovascular Imaging-Related Machine Learning Evaluation (PRIME): A Checklist: Reviewed by the American College of Cardiology Healthcare Innovation Council. JACC Cardiovasc Imaging 2020;13:2017–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannun AY, Rajpurkar P, Haghpanahi M et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med 2019;25:65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madani A, Arnaout R, Mofrad M. Fast and accurate view classification of echocardiograms using deep learning. NPJ Digit Med 2018;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Attia ZI, Noseworthy PA, Lopez-Jimenez F et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 2019;394:861–867. [DOI] [PubMed] [Google Scholar]
- 22.Fang MC, Fan D, Sung SH et al. Validity of Using Inpatient and Outpatient Administrative Codes to Identify Acute Venous Thromboembolism: The CVRN VTE Study. Med Care 2017;55:e137–e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson KW, Torres Soto J, Glicksberg BS et al. Artificial Intelligence in Cardiology. J Am Coll Cardiol 2018;71:2668–2679. [DOI] [PubMed] [Google Scholar]
- 24.Miotto R, Li L, Kidd BA, Dudley JT. Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Sci Rep 2016;6:26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Cheng WY, Glicksberg BS et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med 2015;7:311ra174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shomorony I, Cirulli ET, Huang L et al. An unsupervised learning approach to identify novel signatures of health and disease from multimodal data. Genome Med 2020;12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Y, Ahmad FS, Shah SJ. Tensor Factorization for Precision Medicine in Heart Failure with Preserved Ejection Fraction. J Cardiovasc Transl Res 2017;10:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katz DH, Deo RC, Aguilar FG et al. Phenomapping for the Identification of Hypertensive Patients with the Myocardial Substrate for Heart Failure with Preserved Ejection Fraction. J Cardiovasc Transl Res 2017;10:275–284. [DOI] [PubMed] [Google Scholar]
- 29.Yu C, Liu J, Zhao H. Inverse reinforcement learning for intelligent mechanical ventilation and sedative dosing in intensive care units. BMC Med Inform Decis Mak 2019;19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy AE, Biswas M, Weber R et al. Applications of machine learning in decision analysis for dose management for dofetilide. PLoS One 2019;14:e0227324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med 2018;15:e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gadaleta M, Rossi M, Topol EJ, Steinhubl SR, Quer G. On the Effectiveness of Deep Representation Learning: the Atrial Fibrillation Case. Computer (Long Beach Calif) 2019;52:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overfitting in Machine Learning: What It Is and How to Prevent It. http://elitedatascience.com/overfitting-in-machine-learning Accessed March 30 2020.
- 34.Rudin C Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nature Machine Intelligence 2019;1:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topol EJ. Deep medicine : how artificial intelligence can make healthcare human again. First edition ed. New York: Basic Books, 2019. [Google Scholar]
- 36.Molnar C Interpretable machine learning. A Guide for Making Black Box Models Explainable. 2019. https://christophm.github.io/interpretable-ml-book/ Accessed March 29 2020 [Google Scholar]
- 37.Marcus G The Next Decade in AI: Four Steps Towards Robust Artificial Intelligence. arXiv e-prints, 2020:arXiv:2002.06177. [Google Scholar]
- 38.Esteva A, Kuprel B, Novoa RA et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gulshan V, Peng L, Coram M et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016;316:2402–2410. [DOI] [PubMed] [Google Scholar]
- 40.Arnaout R, Curran L, Zhao Y, Levine J, Chinn E, Moon-Grady A. Expert-level prenatal detection of complex congenital heart disease from screening ultrasound using deep learning. medRxiv 2020:2020.06.22.20137786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen HH, Liu CM, Chang SL et al. Automated extraction of left atrial volumes from two-dimensional computer tomography images using a deep learning technique. Int J Cardiol 2020;316:272–278. [DOI] [PubMed] [Google Scholar]
- 42.Gessert N, Lutz M, Heyder M et al. Automatic Plaque Detection in IVOCT Pullbacks Using Convolutional Neural Networks. IEEE Trans Med Imaging 2019;38:426–434. [DOI] [PubMed] [Google Scholar]
- 43.Matsumoto T, Kodera S, Shinohara H et al. Diagnosing Heart Failure from Chest X-Ray Images Using Deep Learning. Int Heart J 2020;61:781–786. [DOI] [PubMed] [Google Scholar]
- 44.Tadesse GA, Zhu T, Liu Y et al. Cardiovascular disease diagnosis using cross-domain transfer learning. Conf Proc IEEE Eng Med Biol Soc 2019;2019:4262–4265. [DOI] [PubMed] [Google Scholar]
- 45.Toba S, Mitani Y, Yodoya N et al. Prediction of Pulmonary to Systemic Flow Ratio in Patients With Congenital Heart Disease Using Deep Learning-Based Analysis of Chest Radiographs. JAMA Cardiol 2020;5:449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdill RJ, Blekhman R. Tracking the popularity and outcomes of all bioRxiv preprints. bioRxiv 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diller GP, Lammers AE, Babu-Narayan S et al. Denoising and artefact removal for transthoracic echocardiographic imaging in congenital heart disease: utility of diagnosis specific deep learning algorithms. Int J Cardiovasc Imaging 2019;35:2189–2196. [DOI] [PubMed] [Google Scholar]
- 48.Huang O, Long W, Bottenus N et al. MimickNet, Mimicking Clinical Image Post-Processing Under Black-Box Constraints. IEEE Trans Med Imaging 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blendowski M, Bouteldja N, Heinrich MP. Multimodal 3D medical image registration guided by shape encoder-decoder networks. Int J Comput Assist Radiol Surg 2020;15:269–276. [DOI] [PubMed] [Google Scholar]
- 50.Benz DC, Benetos G, Rampidis G et al. Validation of deep-learning image reconstruction for coronary computed tomography angiography: Impact on noise, image quality and diagnostic accuracy. J Cardiovasc Comput Tomogr 2020. [DOI] [PubMed] [Google Scholar]
- 51.Qin C, Schlemper J, Caballero J, Price AN, Hajnal JV, Rueckert D. Convolutional Recurrent Neural Networks for Dynamic MR Image Reconstruction. IEEE Trans Med Imaging 2019;38:280–290. [DOI] [PubMed] [Google Scholar]
- 52.Küstner T, Armanious K, Yang J, Yang B, Schick F, Gatidis S. Retrospective correction of motion-affected MR images using deep learning frameworks. Magn Reson Med 2019;82:1527–1540. [DOI] [PubMed] [Google Scholar]
- 53.Doris MK, Otaki Y, Krishnan SK et al. Optimization of reconstruction and quantification of motion-corrected coronary PET-CT. J Nucl Cardiol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lassen ML, Beyer T, Berger A et al. Data-driven, projection-based respiratory motion compensation of PET data for cardiac PET/CT and PET/MR imaging. J Nucl Cardiol 2019. [DOI] [PubMed] [Google Scholar]
- 55.Johns Hopkins U, Canon Medical S. Comparison of Magnetic Resonance Coronary Angiography (MRCA) With Coronary Computed Tomography Angiography (CTA). 2021.
- 56.University of Z. Deep-Learning Image Reconstruction in CCTA. 2019.
- 57.Ostvik A, Smistad E, Aase SA, Haugen BO, Lovstakken L. Real-Time Standard View Classification in Transthoracic Echocardiography Using Convolutional Neural Networks. Ultrasound Med Biol 2019;45:374–384. [DOI] [PubMed] [Google Scholar]
- 58.Fries JA, Varma P, Chen VS et al. Weakly supervised classification of aortic valve malformations using unlabeled cardiac MRI sequences. Nat Commun 2019;10:3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papworth Hospital NHSFT. Cardiovascular Acoustics and an Intelligent Stethoscope. 2021.
- 60.Eko Devices I Heart Failure Monitoring With Eko Electronic Stethoscopes. 2021.
- 61.Akron Children’s H, Thomas C. Dispenza MD, Bockoven John R.. The Accuracy of an Artificially-intelligent Stethoscope.
- 62.Leclerc S, Smistad E, Pedrosa J et al. Deep Learning for Segmentation Using an Open Large-Scale Dataset in 2D Echocardiography. IEEE Trans Med Imaging 2019;38:2198–2210. [DOI] [PubMed] [Google Scholar]
- 63.Du X, Yin S, Tang R, Zhang Y, Li S. Cardiac-DeepIED: Automatic Pixel-Level Deep Segmentation for Cardiac Bi-Ventricle Using Improved End-to-End Encoder-Decoder Network. IEEE J Transl Eng Health Med 2019;7:1900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cilla M, Pérez-Rey I, Martínez MA, Peña E, Martínez J. On the use of machine learning techniques for the mechanical characterization of soft biological tissues. Int J Numer Method Biomed Eng 2018;34:e3121. [DOI] [PubMed] [Google Scholar]
- 65.Lee J, Prabhu D, Kolluru C et al. Fully automated plaque characterization in intravascular OCT images using hybrid convolutional and lumen morphology features. Sci Rep 2020;10:2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pazinato DV, Stein BV, de Almeida WR et al. Pixel-Level Tissue Classification for Ultrasound Images. IEEE J Biomed Health Inform 2016;20:256–67. [DOI] [PubMed] [Google Scholar]
- 67.Wong KCL, Syeda-Mahmood T, Moradi M. Building medical image classifiers with very limited data using segmentation networks. Med Image Anal 2018;49:105–116. [DOI] [PubMed] [Google Scholar]
- 68.Guo F, Ng M, Goubran M et al. Improving cardiac MRI convolutional neural network segmentation on small training datasets and dataset shift: A continuous kernel cut approach. Med Image Anal 2020;61:101636. [DOI] [PubMed] [Google Scholar]
- 69.Duchateau N, Sermesant M, Delingette H, Ayache N. Model-Based Generation of Large Databases of Cardiac Images: Synthesis of Pathological Cine MR Sequences From Real Healthy Cases. IEEE Trans Med Imaging 2018;37:755–766. [DOI] [PubMed] [Google Scholar]
- 70.Beaulieu-Jones BK, Wu ZS, Williams C et al. Privacy-Preserving Generative Deep Neural Networks Support Clinical Data Sharing. Circ Cardiovasc Qual Outcomes 2019;12:e005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kakadiaris IA, Vrigkas M, Yen AA, Kuznetsova T, Budoff M, Naghavi M. Machine Learning Outperforms ACC / AHA CVD Risk Calculator in MESA. J Am Heart Assoc 2018;7:e009476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Al’Aref SJ, Maliakal G, Singh G et al. Machine learning of clinical variables and coronary artery calcium scoring for the prediction of obstructive coronary artery disease on coronary computed tomography angiography: analysis from the CONFIRM registry. Eur Heart J 2020;41:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Myers PD, Huang W, Anderson F, Stultz CM. Choosing Clinical Variables for Risk Stratification Post-Acute Coronary Syndrome. Sci Rep 2019;9:14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tokodi M, Schwertner WR, Kovács A et al. Machine learning-based mortality prediction of patients undergoing cardiac resynchronization therapy: the SEMMELWEIS-CRT score. Eur Heart J 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eisenberg E, McElhinney PA, Commandeur F et al. Deep Learning-Based Quantification of Epicardial Adipose Tissue Volume and Attenuation Predicts Major Adverse Cardiovascular Events in Asymptomatic Subjects. Circ Cardiovasc Imaging 2020;13:e009829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Commandeur F, Slomka PJ, Goeller M et al. Machine learning to predict the long-term risk of myocardial infarction and cardiac death based on clinical risk, coronary calcium, and epicardial adipose tissue: a prospective study. Cardiovasc Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang Y, Hirdes JP, Dubin JA, Lee J. Fall Risk Classification in Community-Dwelling Older Adults Using a Smart Wrist-Worn Device and the Resident Assessment Instrument-Home Care: Prospective Observational Study. JMIR Aging 2019;2:e12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Toba S, Mitani Y, Yodoya N et al. Prediction of Pulmonary to Systemic Flow Ratio in Patients With Congenital Heart Disease Using Deep Learning-Based Analysis of Chest Radiographs. JAMA Cardiol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pina A, Helgadottir S, Mancina RM et al. Virtual genetic diagnosis for familial hypercholesterolemia powered by machine learning. Eur J Prev Cardiol 2020:2047487319898951. [DOI] [PubMed] [Google Scholar]
- 80.van Velzen SGM, Lessmann N, Velthuis BK et al. Deep Learning for Automatic Calcium Scoring in CT: Validation Using Multiple Cardiac CT and Chest CT Protocols. Radiology 2020;295:66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dekker M, Waissi F, Bank IEM et al. Automated calcium scores collected during myocardial perfusion imaging improve identification of obstructive coronary artery disease. Int J Cardiol Heart Vasc 2020;26:100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poplin R, Varadarajan AV, Blumer K et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2018;2:158–164. [DOI] [PubMed] [Google Scholar]
- 83.Rezaei Z, Ebrahimpour-Komleh H, Eslami B, Chavoshinejad R, Totonchi M. Adverse Drug Reaction Detection in Social Media by Deepm Learning Methods. Cell J 2020;22:319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hospices Civils de L Can we Predict COronary Resistance By EYE Examination ? (COREYE). 2020. [Google Scholar]
- 85.Optima Integrated H, University of California SF. Tailored Drug Titration Using Artificial Intelligence. 2021.
- 86.AiCure, Montefiore Medical C. Using Artificial Intelligence to Measure and Optimize Adherence in Patients on Anticoagulation Therapy. 2016.
- 87.University of M, Agency for Healthcare R, Quality. Improving Adherence and Outcomes by Artificial Intelligence-Adapted Text Messages. 2016.
- 88.Optima Integrated H, University of California SF. Piloting Healthcare Coordination in Hypertension. 2017.
- 89.Yao X, McCoy RG, Friedman PA et al. ECG AI-Guided Screening for Low Ejection Fraction (EAGLE): Rationale and design of a pragmatic cluster randomized trial. Am Heart J 2020;219:31–36. [DOI] [PubMed] [Google Scholar]
- 90.Carlin CS, Ho LV, Ledbetter DR, Aczon MD, Wetzel RC. Predicting individual physiologically acceptable states at discharge from a pediatric intensive care unit. J Am Med Inform Assoc 2018;25:1600–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chu J, Dong W, He K, Duan H, Huang Z. Using neural attention networks to detect adverse medical events from electronic health records. J Biomed Inform 2018;87:118–130. [DOI] [PubMed] [Google Scholar]
- 92.Hever G, Cohen L, O’Connor MF, Matot I, Lerner B, Bitan Y. Machine learning applied to multi-sensor information to reduce false alarm rate in the ICU. J Clin Monit Comput 2020;34:339–352. [DOI] [PubMed] [Google Scholar]
- 93.Au-Yeung WM, Sahani AK, Isselbacher EM, Armoundas AA. Reduction of false alarms in the intensive care unit using an optimized machine learning based approach. NPJ Digit Med 2019;2:86. [DOI] [PMC free article] [PubMed] [Google Scholar]


