Abstract
The human solute carrier 22A (SLC22A) family consists of 23 members, representing one of the largest families in the human SLC superfamily. Despite their pharmacological and physiological importance in the absorption and disposition of a range of solutes, eight SLC22A family members remain classified as orphans. In this study, we used a multifaceted approach to identify ligands of orphan SLC22A15. Ligands of SLC22A15 were proposed based on phylogenetic analysis and comparative modelling. The putative ligands were then confirmed by metabolomic screening and uptake assays in SLC22A15 transfected HEK293 cells. Metabolomic studies and transporter assays revealed that SLC22A15 prefers zwitterionic compounds over cations and anions. We identified eight zwitterions, including ergothioneine, carnitine, carnosine, gabapentin, as well as four cations, including MPP+, thiamine and cimetidine, as substrates of SLC22A15. Carnosine was a specific substrate of SLC22A15 among the transporters in SLC22A family. SLC22A15 transport of several substrates was sodium dependent and exhibited a higher Km for ergothioneine, carnitine and carnosine compared to previously identified transporters for these ligands. This is the first study to characterize the function of SLC22A15. Our studies demonstrate that SLC22A15 may play an important role in determining the systemic and tissue levels of ergothioneine, carnosine and other zwitterions.
Keywords: metabolomic, carnitine, ergothioneine, carnosine, transporter
1. INTRODUCTION
The human solute carrier 22A (SLC22A) family, which consists of 23 members, includes organic cation (OCT), organic anion (OAT) and zwitterion (OCTN) transporters. SLC22A family members mediate the transmembrane flux of numerous endogenous molecules and xenobiotics across the plasma membrane. Despite the large number of studies that have demonstrated the importance of members of the SLC22A family in drug disposition and response and as mediators of drug-drug interactions1–3, eight of the 23 members and their species orthologs do not have experimentally validated substrates (Fig. 1A) and remain classified as orphans. In this study, we used multiple methods ranging from mining genomewide association studies (GWAS) of serum metabolites to comparative structure modelling and isotopic uptake studies in transporter transfected cell lines to characterize SLC22A15.
Fig. 1.

Phylogeny tree analysis, structural analysis and plasma expression of SLC22A15. (A) Circular tree of the human SLC22A and SLC22B transporter family using 28 protein sequences. Six new members of the SLC22 family are included here. Blue font represents cation transporters. Red font represents anion transporters, and green font represents zwitterion transporters. Pink font represents sugar transporters. SLC22 family members with unknown substrates are in black font, including SLC22A15, which is the focus of this study. †Six new members in the SLC22 family. (B). Surface representations of the calculated electrostatic potential (−5 to 5 kT e-1) of the human SLC22A1, SLC22A6 and SLC22A4, which are characterized cation, anion and zwitterion transporters, respectively, and the human SLC22A15. The cross-section allows for visual inspection of the predicted binding pockets with blue color estimated as having positive charges and red being estimated as having negative charges. (C). Western blot of cellular components expressing SLC22A15 with C-terminal Myc-DDK Tag. SLC22A15 is expressed on the plasma membrane and the size is slightly above 52 kDa. When the plasma membrane fraction is deglycosylated (degly), the size is reduced to ~52 kDa.
There are increasing efforts to uncover the biological roles of SLC transporters using a multitude of new methodologies, including various in vitro model systems, transgenic animals, human genetic studies, and combinations of experimental and computational approaches4. For example, in recent studies, in vitro metabolomic screens have been used to identify the endogenous substrates of SLC22A1 (OCT1), which was previously thought to function primarily as a xenobiotic transporter5. Metabolomic GWAS were used to reveal important endogenous substrates of SLCO1B1 (OATP1B1), an important transporter in hepatic drug disposition6. Loss of function human genetic mutations as well as gene deletions in mice have revealed the biological roles of several transporters, including SLC10A7 in glycosaminoglycan synthesis and specifically in skeletal development7 and SLC22A14 in sperm motility and male fertility in mice8. GWAS of human disease or specific solutes have revealed the biological roles of transporters in uric acid disposition (e.g., SLC2A9)9 and in diabetes (e.g., SLC16A11)10. Despite these successes, it remains an enormous challenge to identify substrates of orphan transporters, even when they can be recombinantly expressed on the plasma membranes11, 12.
SLC22A15 is one of the orphan transporters in the SLC22A family without known substrates or inhibitors. Based on phylogenetic analyses, the transporter has been designated as a carnitine transporter13; however, no study has endeavored to experimentally identify its actual substrates or its transport mechanism. Interestingly, genetic polymorphisms in this transporter have been associated with a response to an anti-asthmatic drug, albuterol,14 and with tumor growth15, 16; yet, without knowledge of its substrates or function, the mechanisms for these associations remain undefined. In this study, we conducted a range of experiments, which provide important information on the substrates and inhibitors of SLC22A15 as well as on the transporter mechanism. With the substrates in mind, we speculate on the mechanisms which underlie associations between genetic polymorphisms in the transporter or change in its expression levels, and the clinical phenotypes. Further studies are needed to fully understand the role of the transporter in human health, disease and the inter-individual variation in drug response.
2. MATERIALS AND METHODS
Clustering of human SLC22A15 and other orthologs in the SLC22 family
The full-length reference sequences for all proteins of members in the SLC22 family were obtained from UniProt. A multiple sequence alignment was created using Clustal Omega, and on-line tool, https://www.ebi.ac.uk/Tools/msa/clustalo/. The resulting dendrogram was created using online tool, iTOL (https://itol.embl.de/).
Comparative structure modelling
Previously, our group has constructed various comparative structure models of members in the human SLC superfamily using known structures of homologous proteins as templates17–19. In this study, we created a model to calculate the electrostatic potential within the predicted SLC22A15 substrate binding pocket. We used the 2.6 Å crystal structure of the maltose-bound human GLUT3 (SLC2A3) in the outward-open conformation as the template (PDB ID 4ZWC)20. The maltose-bound human GLUT3 (SLC2A3) shares the major facilitator superfamily fold assignment21 and has a 18% sequence identity to SLC22A15. The sequence alignment between the template and SLC22A15 was obtained by a manual refinement of gaps in the output from the PROMALS3D server22. The maltose molecule from the crystal structure of human GLUT3 was copied from the template structure into a model as a rigid body. Using the “automodel” class of MODELLER 9.16, 100 models were generated for SLC22A1523. The models had acceptable normalized discrete optimized protein energy scores (zDOPE)24, 25. The top scoring models were selected for electrostatic potential analysis with the APBS electrostatics plugin26 within PyMOL 2.0.3 using the AMBER force field with default settings. Using the methods above, we also generated comparative models of SLC22A1, SLC22A6 and SLC22A4 targets, which have known preferred ion specificities.
Creation of transient and stable cell lines
Transient or stable cell lines recombinantly transfected with the cDNA of SLC22A15 or the empty vector (EV) cDNA were created in human embryonic kidney cells (HEK293) with the Flp-In integration system (Life Technologies Corporation, Carlsbad, CA, USA). The vector pCMV6-Entry containing the cDNA of SLC22A15 (NM_018420) was purchased from OriGene (OriGene Technologies Inc., Rockville, MD, USA). Lipofectamine LTX (Life Technologies Corporation, Carlsbad, CA, USA) was used as the transfection reagent for transiently or stably transfected cells. Appropriate DNA to LTX ratios were followed. Using a 48-well plate (seeding density 1.8×105 cells/well, next day transfection), we used 200ng of DNA and 0.4 μL of Lipofectamine LTX in Opti-MEM medium (Life Technologies Corporation) to transfect each well. Cells transfected transiently were ready to use for transport studies 36–48 hours after transfection. To achieve stable cell lines, HEK293 Flp-In cells were transfected with pCMV6-Entry containing the SLC22A15 cDNA with a C-terminal GFP tag. After 48 hours, positive clones were selected using fresh DMEM media with 500 ug/mL of Geneticin (Life Technologies Corporation), changed every two days for one week. HEK293 Flp-In stably expressing SLC22A15-GFP tagged cells were then sorted using the FACS Aria II operated by the University of California San Francisco (UCSF) Flow Cytometry and Cell Sorting core facilities. This process allowed selection of HEK293 Flp-In cells expressing SLC22A15-GFP positive cells.
Western blotting analysis of SLC22A15
Different expression vectors containing vector only (pCMV6-Entry) and SLC22A15 (containing reference sequence NM_018420.3) were transiently transfected in HEK293 Flp-In cells cultured in a 100 mm tissue culture plate. The expressed proteins contained a C-terminal Myc-DDK tag (Origene Technologies Inc., Rockville, MD). After 48 hours post-transfection, SLC22A15 protein levels were evaluated. The Plasma Membrane Protein Extraction Kit (Abcam plc., Cambridge, UK, catalog number: ab65400) was used to separate plasma membranes from intracellular membranes10. Transfected cells in a 2 mL homogenization buffer were homogenized using Dounce homogenizer method, in which a round glass pestle was manually driven into a glass tube. The plasma membrane fraction was resuspended in 50 μL buffer provided in the extraction kit. The cell samples were treated with the deglycosylating enzyme, PNGaseF (New England BioLabs, Ipswich, MA, USA, catalog number: NEB P0708L) per the manufacturer’s protocol. The samples were then ready for determination of protein content. The whole protein extracted from the above was separated on 10% SDS-PAGE gels (Life Technologies) and transferred to Polyvinylidene fluoride (PVDF) membrane (MilliporeSigma, Burlington, MA, USA). The membrane was blocked for 1 hour with Tris-buffered saline containing 0.05% Tween 20 and 5% nonfat milk (Bio-Rad Laboratories, Hercules, CA, USA). Primary antibody (Anti-DDK (FLAG) Monoclonal antibody, HRP) (OriGene Technologies, Inc.), directed against DDK (FLAG tag), was incubated in the 5% nonfat milk at 1:2000, overnight at 4 deg. Following antibody incubation, immunoblotting was performed using standard procedures, and the signals were detected by chemiluminescence reagents (GE Healthcare, Chicago, IL, USA).
Cloning of SLC22A15
To confirm that the whole transcript of SLC22A15 correctly matched to the reference sequence, we cloned the gene from total RNA of human brain and human kidney (Clontech, Mountain View, CA). SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to transcribe total RNA into cDNA. Primers that were used in the PCR to clone the SLC22A15 transcripts, NM_018420, are shown in Supplemental Table 1. The KOD Xtreme Hot Start DNA polymerase (MilliporeSigma, Burlington, MA, USA, catalog number: 71975–3) was used for PCR. The PCR cycling conditions used were: (i) initial activation at 94°C for 2 min, 35 cycles at (ii) denature at 98°C for 10 sec, (iii) annealing and extension at 68°C for 2 min. pcDNA5/FRT vector (Life Technologies Corporation, catalog number: V601020) was digested by BamHI and XhoI (NEB) and clean-up by QIAquick gel extraction kit (Qiagen, Hilden, Germany). Digested pcDNA5/FRT vector and SLC22A15 PCR product were mixed with In-Fusion HD Enzyme Premix, available from In-Fusion HD Cloning kit (Takara Bio USA, Inc., Mountain View, CA, USA, catalog number: 639642) and then incubated at 50°C for 15 min, followed by transformation of Stellar competent cells. The PCR product cloned into pcDNA5/FRT was sequenced by MCLAB (South San Francisco, CA, USA), to confirm the transcript, NM_018420, which contains 1644 bp.
Creation of pCMV6-Entry vector containing SLC22A15 cDNA with c-terminal GFP tagged.
In-Fusion assembly method was used to assemble the DNA of the GFP fragment downstream of the SLC22A15 cDNA. The KOD Xtreme Hot Start DNA polymerase (MilliporeSigma, Burlington, MA, USA, catalog number: 71975-3) and In-Fusion HD Cloning kit (Takara Bio USA, Inc., Mountain View, CA, USA, catalog number: 639642) were used for amplification and in-fusion assembly reaction, respectively. The pcDNA5/FRT vector containing the EGFP DNA (720 bp) (https://www.snapgene.com/resources/plasmid-files/?set=fluorescent_protein_genes_and_plasmids&plasmid=EGFP) and the pCMV6-Entry vector containing SLC22A15 were assembled to create the pCMV6-Entry expression vector containing SLC22A15 cDNA with a c-terminal GFP tagged. The primers that are used for this assembly are shown in Supplemental Table 1. The PCR conditions used for this assembly were: PCR and assembly procedures were identical with cloning SLC22A15 in previous section with only minor modifications. Anneal and extension length of pCMV6-Entry SLC22A15 and GFP were 7 min and 1 min, respectively.
Metabolomic study
We conducted a metabolomic profiling study to discover endogenous substrates of SLC22A15 by preparing samples of cell pellets and sending them to Metabolon, Inc. (Morrisville, NC, USA). HEK293 Flp-In cell pellets were prepared in five 100 mm culture dishes each of cells transiently expressing SLC22A15 cDNA (NM_018420) or vector only (pCMV6-Entry). Media was suctioned and replaced after 24 hours of transient transfection with DMEM-H21 phenol red free, media (Life Technologies, Corporation) containing 20% Fetal Bovine Serum, U.S.D.A. Approved Origin (Product Number 89510–186, Lot Number 356B16, VWR International, Radnor, PA, USA). After an additional 24 hours, all culture dishes were rinsed twice with cold PBS, and cells were removed using a scraping method to transfer to a 15-mL falcon tube. These tubes were then centrifuged (3000 rpm), supernatant discarded, and pellet stored in −80 °C prior to shipping to Metabolon Inc. Metabolites were determined using mass spectrometry (Metabolon DiscoveryHD™ metabolomics platform), as described in our previous publication17.
Metabolon returned the processed metabolomic data as described previously6, 17. Quantification and normalization were performed for each metabolite identified (~700) by Metabolon Inc. Using the normalized metabolites intensity value, we used Student’s t-tests to determine if there were significant differences in the levels of each metabolite between cells transiently expressing empty vector only (EV) and cells transiently expressing SLC22A15. This was followed by using a false discovery rate (FDR) to correct the p-values. Results of this metabolomic analysis can be viewed in Supplemental Table 3.
Transporter uptake studies
HEK293 Flp-In cells transiently expressing SLC22A15 or stably expressing SLC22A15-GFP tagged were seeded at a density of 100,000 cells/0.3 mL in poly-D-lysine 48-well plates for next day transient transfection or at a density of 200,000 cells/0.3 mL in poly-D-lysine 48-well plates for next day transporter uptake studies. On the day of the uptake study, cells were washed with Hanks’ Balanced Salt Solution (HBSS) and incubated for 10 minutes at 37°C. HBSS containing trace amounts of radioactive compound (Supplemental Table 2) (3H or 14C) with unlabeled compound (1 μM) or inhibitor were added to the cells for 15 min, unless described differently in the figure legend. After 15 min, cells were washed with ice cold HBSS twice and then dried before adding 0.75 mL lysis buffer (0.1N sodium hydroxide (NaOH) and 1% v/v SDS solution) to lyse the cells. After shaking the cells with lysis buffer for 60–90 min, 0.69 mL of cell lysate was transferred to scintillation fluid (EcoLite(+)™, MP Biomedicals, Santa Ana, CA, USA) for scintillation counting (Beckman Coulter LS6500). For the sodium dependence study, three different uptake buffers were used instead of the HBSS buffer: (i) sodium buffer (140 mM sodium chloride, 1.25 mM magnesium sulfate, 4.73 mM potassium chloride and 1.25 mM calcium chloride); (ii) lithium buffer (140 mM lithium chloride, 1.25 mM magnesium sulfate, 4.73 mM potassium chloride and 1.25 mM calcium chloride); or (iii) NMDG buffer (140 mM N-methyl-D-glucamine chloride, 1.25 mM magnesium sulfate, 4.73 mM potassium chloride and 1.25 mM calcium chloride). The pH of each buffer above was adjusted with NaOH to pH 7.4 for sodium buffer, or with KOH to pH 7.4 for lithium and NMDG buffers. For pH dependence experiments, HBSS buffer was adjusted to different pH’s (5.5, 7.4 and 8.5) by adding hydrochloric acid or NaOH. For each uptake study, the uptake values were corrected for protein concentration as determined with a BCA assay kit (Thermo Scientific, Waltham, MA, USA).
Kinetic studies of various SLC22A15 substrates
Kinetics studies of four substrates, ergothioneine, carnitine, carnosine and creatine, were performed in HEK293 Flp-In cells transiently transfected with SLC22A15. Experimental condition used in kinetic studies were similar to methods published previously by our group27, 28. We first assessed the time-dependent uptake of the substrates at 1 μM with trace amounts of radioactive compound. These kinetic studies were also performed on stable cell lines expressing GFP-tagged SLC22A15 to confirm that the results were similar to those in the transiently transfected cells. For each substrate, we varied the concentrations of the unlabeled compounds up to 3 mM. Five minutes (at 37°C) were selected for kinetic studies, as this fell within the linear range in the uptake versus time plot for each substrate. Each point represents the mean ± SD uptake in the transporter-transfected cells minus that in empty vector cells. The data were fit to a Michaelis–Menten equation and kinetic parameters were estimated. Plots were generated from a representative experiment of 3 to 4 independent studies.
Inhibition of SLC22A15-mediated ergothioneine uptake
HEK293 Flp-In cells stably transfected with GFP-tagged SLC22A15 were seeded at a density of 90,000 cells/0.1 mL in 96-well plates coated with poly-d-lysine (0.1 mg/mL), approximately 16 to 24 hours prior to the inhibition study. Prior to the inhibition studies, cells were washed with 2 × 0.1 mL of Hank’s buffered salt solution (HBSS) per well and then preincubated for 10 to 15 min in 0.1 mL of HBSS buffer (at 37°C). After preincubation, the HBSS was removed, and the cells were incubated with uptake buffer (1 μM unlabeled ergothioneine and trace amounts of [3H]-ergothioneine in HBSS) containing either 20 mM unlabeled ergothioneine as a positive control, 1% DMSO as negative control or 500 μM of the selected test compounds. For some test compounds, 100 μM was used in addition to 500 μM. Uptake of [3H]-ergothioneine was stopped after 15 min by washing twice with ice-cold HBSS (0.1 mL). MicroScint-20 (Perkin Elmer, Waltham, WA) (0.1 mL) was added to the 96-well plates and sealed with an adhesive plastic cover. The plates were placed on a shaker for 2 hours. The plates were read in a MicroBeta2 (Perkin Elmer). All values were determined in triplicate, and final values are expressed as % uptake relative to the negative control (1% dimethyl sulfoxide (DMSO)). Inhibition studies for each test compounds were evaluated at least one time and compounds were randomly selected to test two or three times to check for consistency. For inhibition of SLC22A15, SLC22A4 and SLC22A5 uptake by HEPES, we added the appropriate volume of a stock solution of HEPES 1 M (pH 7.4) (Teknova, California, USA) to HBSS to achieve various concentrations of HEPES (5 mM to 25 mM).
Transporter efflux study
Efflux of carnitine was performed using methods described in the literature29, 30. HEK293 Flp-In cells stably transfected with GFP-tagged SLC22A15 or EV were seeded at a density of 200,000 cells/0.3 mL in 48-well poly-d-lysine coated plates. Prior to the efflux study, cells were washed with 2 × 0.4 mL HBSS per well and then incubated in HBSS containing [14C]-carnitine with 20 μM unlabeled carnitine for 30 min to facilitate the intracellular accumulation of carnitine. Cells were washed once with cold HBSS (1 mL) and incubated in HBSS with and without 0.5 mM quinidine (for SLC22A15 stable cells). The efflux of [14C]-carnitine was determined by collecting the extracellular media and cells at indicated time points. The cells were lysed (in 0.75 mL of 0.1% SDS/0.1N NaOH) and radioactivity in the cell lysate and media was measured as described in a previous section (Transporter uptake studies). The radioactivity was normalized by the protein content of the cell lysate. Efflux studies for carnitine were evaluated twice and carnitine efflux at each time point in each experiment was determined in triplicate. The percent efflux of carnitine at indicated time point was calculated by the [14C] count in the media divide by the [14C] count in the media plus in the cell.
RNAseq study and data analysis
Total RNA from four samples of HEK293 Flp-In EV cells and four samples of HEK293 Flp-In SLC22A15-GFP tagged stably transfected cells were collected following the protocol by manufacturer (Qiagen RNeasy kit, Qiagen, Hilden, Germany). Isolated mRNA samples were frozen immediately and stored at −70 °C before shipping to the Novogene Co., Ltd. (https://en.novogene.com/, Sacramento, CA, USA) for mRNA sequencing, by the NovaSeq 6000 S4 (Illumina Inc., San Diego, USA). In brief, sequencing library construction included the following steps: RNA quality checking (Agilent Eukaryote Total RNA Nano Kit, Agilent Technologies, Santa Clara, USA), library construction (NEBNext Ultra II RNA Library Prep Kit for Illumina, New England Biolabs, Ipswich, USA), library purification (Beckman AMPure XP beads, Brea, USA), insert fragments test (Agilent High Sensitivity DNA Kit, Agilent Technologies), quantitative analysis of library (ABI 7500 real-time PCR instrument, Life Technologies, Carlsbad, USA; KAPA SYBR green fast universal 2× qPCR master mix, KAPA Biosystems, Boston, USA) and cBOT automatic cluster (TruSeq PE Cluster Kit v3-cBot-HS, San Diego, USA).
RNA-Seq data processing includes the following steps: (i) checking data quality and removing excess adaptors; (ii) mapping the high-quality short sequence reads to the human (hg19) reference genome; (iii) transcript assembling using the TopHat software (version 2.0.9); and (iv) gene expression quantification (in fragments per kilobase transcript per million mapped reads or FPKM) using Cufflinks.
Gene expression profiling by RNA-Seq is a powerful approach to identifying the genes that are regulated by the SLC22A15 transfected gene. Identification of differentially expressed genes (DEGs) was performed in this study by NovoGene Co., Ltd. Differential expression analysis between two groups (two biological replicates per condition) was performed using the DESeq2 R package (1.14.1). DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting p-values were adjusted using the Benjamini and Hochberg’s approach for controlling the False Discovery Rate (FDR). Genes with an adjusted P-value <0.05 found by DESeq2 were assigned as differentially expressed. The fold change (FC) of each gene was the log transformation (base 2) of the specific value (SLC22A15/EV) of FPKM. If FC > 0, the gene is upregulated and if FC < 0, the gene is downregulated.
3. RESULTS
3.1. Multiple sequence alignment and comparative structure modeling suggest that SLC22A15 is a zwitterion transporter
There are six new members in the SLC22 family, including one in SLC22A family and five in the atypical SLC22B subfamily (Synaptic Vesicle glycoprotein, SV)21, 31 (http://slc.bioparadigms.org/, https://opendata.cemm.at/gsflab/slcontology/) (Fig. 1A). For the purposes of the phylogenetic tree generation, we included all 23 members of the SLC22A family and the five new members of the SLC22B subfamily. The multiple sequence analysis indicated that human SLC22A15 is distinct from known organic cation, anion and zwitterion transporters in SLC22 family, but closest to the zwitterion transporters, SLC22A4, SLC22A5 and SLC22A16 (Fig. 1A). The sequence analysis expands on results from the Nigam laboratory13, 32, 33, which also showed that SLC22A15 was nearest to SLC22A4, SLC22A5 and SLC22A16.
Orthogonally, due to the growing list of publicly available atomic-resolution structures of human and non-human SLC transporters, it is now possible to model the structure of a number of human SLC transporters. In line with our previous work, we created a comparative structure model of SLC22A15 to visualize the electrostatic potential within the predicted substrate binding pocket17, we observed a mixture of positive and negative charges within the binding site, resembling that of SLC22A4, suggesting an affinity for zwitterionic compounds (Fig. 1B).
3.2. SLC22A15 is expressed on the plasma membrane
According to UniProt, SLC22A15 protein contains 547 amino acids (https://www.uniprot.org/uniprot/Q8IZD6). We confirmed that the transcript encoding the SLC22A15 protein is present using PCR in the total brain cDNA and that the nucleotide sequences matched to the reference sequence (RefSeq, NM_018420.3). Western blotting of the plasma membrane protein fraction isolated from HEK293 Flp-In cells transiently transfected with SLC22A15 showed that the transporter has a plasma membrane protein expression and the expected molecular size of about 60 kDa. The apparent molecular size of the protein is reduced after treatment with deglycosylating enzymes. As expected, the total cell lysate and cytoplasma fraction also expressed the SLC22A15 Myc-DDK tag protein, but at much lower levels (Fig. 1C). The expression of SLC22A15 on the plasma membrane is consistent with known SLC22A family members1, 34.
3.3. Metabolomic studies reveal zwitterions as substrates of SLC22A15
We used metabolomic methods to identify metabolites that are significantly different between cells transiently expressing SLC22A15 compared to empty vector (EV) transfected cells. This approach has been successful in identifying potential new substrates of membrane transporters5, 17, 35. After exposing the cells to cell culture media plus fetal bovine serum, 717 metabolites were detectable using the Metabolon Inc. LC/MS-MS platforms (Supplemental Table 3). Among the top 10 metabolites that were most significantly different between SLC22A15 expressing cells and EV transfected cells, six were zwitterions (3-hydroxybutyrylcarnitine, creatine, creatine phosphate, propionylcarnitine, carnosine and (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (HEPES)), three were lipids and one was a nucleotide. Among the significant metabolites (cut-off adjusted p-value < 0.01), five zwitterions are known substrates of members in the SLC22 family (ergothioneine, tryptophan betaine, and carnitine) or other SLC families (creatine and carnosine) (Fig. 2A). Ten acylcarnitine metabolites were significantly different between SLC22A15 and EV transfected cells with adjusted p-value < 0.01 (Fig. 2B), consistent with multiple sequence alignments that showed that SLC22A15 is closest to SLC22A4, SLC22A5 and SLC22A16, which are known to transport zwitterions including ergothioneine, carnitine and acetylcarnitine. Interestingly, creatine and carnosine, though zwitterions, are not known to be substrates of any members of the SLC22 family. However, creatine is a substrate of SLC6A8 and carnosine is a substrate of SLC15A236, 37. Many lipophilic long chain molecules, such as monoacylglycerol and long chain fatty acids are among the significant metabolites with adjusted p-value < 0.01 (Fig. 2C, D).
Fig. 2.

Metabolomic study showing the three major groups of metabolites that are significantly different between SLC22A15 and EV cells. (A) zwitterions, (B) acylcarnitines, (C) monoacylglycerols. These metabolites reached the significant threshold of p-adjusted value < 0.01 (Supplemental Table 1). (D) Chemical structures of the zwitterions, four acylcarnitine and four monoacylglycerols.
3.4. Transporter assays support the functional role of SLC22A15 as a transporter of organic zwitterions and cations
3.4.1. Substrates of SLC22A15
A diverse set of 23 cationic, anionic and zwitterionic compounds that are known substrates of SLC22A family members were tested in HEK293 Flp-In cells transfected with SLC22A15 and EV (Supplemental Table 2). Consistent with the phylogenetic and metabolomic analyses, the experiments indicated that SLC22A15 prefers zwitterions (Fig. 3A) including ergothioneine, carnosine, carnitine and creatine. From our diverse set of screens, we also identified several organic cations (1-methyl-4-phenylpyridinium (MPP+) and tetraethylammonium (TEA)) that were substrates of SLC22A15 (Fig. 3A). The top three metabolites that had the highest fold-uptake over empty vector were the zwitterions: ergothioneine, carnosine and betaine.
Fig. 3.

Uptake of zwitterions and cations in HEK293 Flp-In cells expressing SLC22A15 or other zwitterion transporters. (A) Uptake of six zwitterions and two cations that are canonical substrates of SLC22 family members or significant in the in vitro metabolomic study (see Fig. 2). Uptake of these compounds was significantly different between EV and SLC22A15 transiently transfected cells. †Compounds that were found to be significantly different between EV and SLC22A15 transfected cells in the metabolomic study (Fig. 2). The bars represent the mean uptake values from one experiment (± S.D. fold over EV) from three replicate wells. (B) Uptake of zwitterions or cations in cells expressing EV (white bars), SLC22A4 (grey bars), SLC22A5 (dark grey bars) and SLC22A15 (black bars). Uptake, expressed as fold uptake over uptake in EV, was performed in transiently transfected cells (HEK293 Flp-In). The bars represent the mean uptake values from one experiment (± S.D. fold over EV) from three to four replicate wells. All uptake values are significantly higher than EV cells, except for those label n.s. (not significant). Significance were determined by one-way analysis of variance followed by Dunnett’s multiple comparisons test (by comparisons to EV as control). (C) Kinetics of uptake of four zwitterions for SLC22A15. A representative curve for each substrate is shown in this figure. Kinetic parameters of uptake of ergothioneine, carnosine, carnitine and creatine for SLC22A15 are listed in Supplemental Table 4. The mean and S.D. of the kinetic parameters from two to three experiments are shown in Supplemental Table 4. (D) Sodium dependence studies of the uptake of four zwitterions in SLC22A15 overexpressing cells. Cells were pre-incubated with sodium buffer, lithium buffer or NMDG-chloride buffer and uptake was performed using the respective buffer. The figure shows a representative plot from one experiment (mean ± S.D. fold over EV) in triplicate wells. (E) Uptake of zwitterions and cations in HEK293 Flp-In cells stably expressing SLC22A15-GFP in the presence (grey bars) and absence of quinidine (black bars represent uptake in SLC22A15 expressing cells and white bars represent uptake in EV cells). Phenylalanine (1 mM) was used to inhibit endogenous amino acid transporter, SLC7A5 (LAT1). The data represent the uptake of metabolites or prescription drugs that are zwitterions or cations in SLC22A15 transfected cells compared to EV (mean ± SD). (F) [14C]-Carnitine efflux from SLC22A15 stably expressing cells and empty vector (EV) cells. SLC22A15 cells and EV cells were preloaded with trace amount of [14C]-carnitine and 20 μM carnitine for 30 min. Subsequently, cells were washed twice with cold HBSS. The efflux of [14C]-carnitine was measured following addition of HBSS buffer to the SLC22A15 and EV cells. Quinidine (0.5 mM) was included in the HBSS buffer (black squares) to inhibit SLC22A15.
3.4.2. Substrate selectivity of SLC22A15 compared to SLC22A4 and SLC22A5
With the exception of carnosine and creatine, the other zwitterions and cations shown in Fig. 3A, are substrates for organic cation transporters (SLC22A1, SLC22A2 and SLC22A3) and novel organic cation transporters (SLC22A4 and SLC22A5). We determined the selectivity of these metabolites for SLC22A15 by comparing their uptakes in cells expressing SLC22A15, SLC22A4 or SLC22A5 (Fig. 3B). Among these eight metabolites, some were substrates for all three transporters (betaine, ergothioneine and TEA), some for SLC22A15 and one other transporter (dimethylglycine and carnitine) and some were specific for SLC22A15 (carnosine, creatine, MPP+). Since the discovery of SLC22A4 as an ergothioneine transporter in 200538, no other ergothioneine transporter has been reported. SLC22A5 showed a small but significant uptake of [3H]-ergothioneine (2.5-fold increase compared to EV cells, p< 0.05 using a Student’s t-test between EV and SLC22A5), however, a much greater fold uptake of ergothioneine was observed with the SLC22A15-expressing cells. Using one-way ANOVA analysis and multiple comparison, only SLC22A4 and SLC22A15 showed significant uptake of ergothioneine over EV cells (Fig. 3B). Since the identification of SLC22A5 as a carnitine transporter39, three other carnitine transporters in human SLC22 family have been reported: SLC22A1640, 41, SLC22A442 and SLC22A129. Our data (Fig. 3B) and data from others43, 44 suggest that SLC22A4 does not transport carnitine. In our studies, SLC22A15 transports ergothioneine and carnitine, although ergothioneine seems to be preferred over carnitine (Fig. 3B). Betaine, which is a known substrate of SLC22A445 and SLC22A546, was also a substrate of SLC22A15. Dimethylglycine, known to interact with SLC22A547, showed significant uptake in SLC22A15 transiently transfected cells (Fig. 3B), consistent with its significant accumulation in our metabolomic study (Supplemental Table 3, adjusted p-value = 0.02). This is the first study to show that a member of the SLC22 family transports carnosine and creatine. Both exhibited significant accumulation in our transporter assay as well as in our metabolomic screen in SLC22A15 transfected cells (Fig. 3A, B, Supplemental Table 3). MPP+ and TEA, known substrates of organic cation transporters in the SLC22 family exhibited weak uptake by SLC22A15 (1.5–2 fold over empty vector) (Fig. 3B). In addition, our results showed that SLC22A16 transports the zwitterions, dimethylglycine and betaine (Supplemental Fig. 1A) and creatine (Supplemental Fig. 1B), but not carnosine and MPP+ (Supplemental Fig. 1B).
3.4.3. Kinetics of uptake of substrates of SLC22A15
We selected ergothioneine, carnitine, carnosine and creatine for kinetic studies. The uptakes of these substrates were dependent on time, (Supplemental Fig. 1C) and we selected the five minute time mark at which to evaluate kinetic properties of each of the substrates. The uptake kinetics of [3H]-ergothioneine, [14C]-carnitine, [3H]-carnosine and [14C]-creatine exhibited saturable characteristics at high concentrations with Km values (mean ± SD) of 354 ± 95 μM, 99 ± 94 μM, 510 ± 175 μM, 291 ± 124 μM, respectively (Fig. 3C, Supplemental Table 4). Overall, the Km of ergothioneine for SLC22A15 was higher compared to the Km of ergothioneine for SLC22A4 (21 μM)38. Similarly, carnitine had a higher Km for SLC22A15 than it did for SLC22A5 (5 μM)39. In addition, the Km of carnosine and creatine for SLC22A15 were also higher compared to Km of carnosine for SLC15A2 (43 μM)48 and Km of creatine for SLC6A8 (46 μM)49. Collectively, these data suggest that SLC22A15 serves as a low-affinity transporter for its substrates.
3.4.4. Dependence of SLC22A15-mediated transport on sodium and pH
The SLC22A15 mediated uptake of ergothioneine, carnosine, carnitine and creatine were dependent on sodium, though lithium was also able to drive the uptake of these compounds (Fig. 3D). When replacing sodium chloride (140 mM) with lithium chloride (140 mM), the SLC22A15-mediated uptake of ergothioneine, carnitine and carnosine did not change, unlike SLC22A4-mediated ergothioneine and SLC22A5-mediated carnitine uptake (Supplemental Fig. 1D). Replacing sodium with NMDG (140 mM) resulted in substantial reduction of uptake of ergothioneine, carnitine, and carnosine. Interestingly, when sodium chloride was replaced with lithium chloride or NMDG-chloride, the uptake of creatine was significantly reduced in the EV cells (Fig. 3D). The lower uptake of creatine in EV cells in the absence of sodium was likely due to loss of function of an endogenously expressed sodium-dependent creatine transporter, for example, SLC6A8, which is expressed in abundance in HEK293 cells (https://www.proteinatlas.org/ENSG00000130821-SLC6A8/cell). There was a small effect (<1.5-fold difference from pH 7.4) of varying pH (pH 5.5, pH 7.4 and pH 8.5) on the SLC22A15-mediated uptake of either carnosine or ergothioneine (Supplemental Fig. 1E).
3.4.5. Murine Slc22a15
Next, we determined whether the murine Slc22a15 transported ergothioneine, carnitine, carnosine and creatine. Among the four metabolites tested, only ergothioneine showed significant uptake in Slc22a15 transiently expressing HEK293 Flp-In cells compared to EV (Supplemental Fig. 1F). However, the uptake of creatine by Slc22a15 was significantly higher in Slc22a15 transfected cells compared to EV, when lithium replaced sodium in the buffer (Supplemental Fig. 1F).
3.4.6. Validation of substrates in stable cell lines expressing SLC22A15
We validated the previously identified substrates in stable cell lines expressing empty vector and SLC22A15-GFP tagged (Fig. 3E). Overall, the trend for uptake of various substrates was similar between transiently transfected cells and stable cell lines, and the uptake was inhibited by quinine in both. The other zwitterions and cations that exhibited significantly increased uptake in SLC22A15-GFP cells compared to EV were ergothioneine, carnosine, betaine, glycylsarcosine (a zwitterionic dipeptide), acetylcarnitine (zwitterion), thiamine (cation), gabapentin (zwitterionic drug) and cimetidine (cationic drug) (Fig. 3E). Among these compounds, glycylsarcosine was not previously identified as a substrate of the SLC22 family members.
3.4.7. Efflux of carnitine
In the metabolomic screen, several metabolites (e.g., carnitine and acylcarnitines) had lower accumulation in the SLC22A15 transfected cells compared with the EV cells (Fig. 2), consistent with the transporter mediating their efflux. Indeed, our experiments indicated that SLC22A15 was able to efflux carnitine and this process was inhibited by quinidine (Fig. 3F). Members of the SLC22A family are facilitative transporters that can translocate substrates across biological membranes in accordance with the electrochemical gradient. For example, SLC22A1 (OCT1) effluxes carnitine and acylcarnitine29 and SLC22A5 (OCTN2) mediates the influx and efflux of zwitterionic metabolites30.
3.5. Further screening reveals additional zwitterion and cation compounds as substrates or inhibitors of SLC22A15
3.5.1. Inhibition studies
We selected 84 compounds to test as inhibitors of SLC22A15-mediated [3H]-ergothioneine uptake including additional metabolites involved in the metabolism of ergothioneine, carnitine and niacin; various substrates and inhibitors of other SLC22 family members; and various drugs and synthetic molecules (Fig. 4, Supplemental Table 5). In general we observed that at high concentrations (500 μM) (i) metabolites that have similar backbone structure to ergothioneine (e.g. hypaphorine, hercynine) and carnitine (acetylcarnitine, propionylcarnitine) inhibited >50% of SLC22A15-mediated ergothioneine uptake (Fig. 4A); (ii) carnosine, and creatine, which are substrates of SLC22A15, reduced SLC22A15-mediated ergothioneine uptake (Fig. 4A); (iii) pyrilamine, cimetidine, gabapentin, tryptophan and ondansetron, as well as fluorescence substrates of OCTs, ASP+ (4-Di-1-ASP (4-(4-(Dimethylamino)styryl) -N-Methylpyridinium Iodide)) and ASP2+, inhibited SLC22A15 by ≥ 50% (Fig. 4B); (iv) 9 known inhibitors of OCTs and OCTNs inhibited SLC22A15-mediated ergothioneine uptake whereas inhibitors of OATs were poor inhibitors of SLC22A15 (Fig. 4B); and (v) a single fluoroquinolone antibiotic, levofloxacin, inhibited SLC22A15-mediated uptake (Fig. 4C);. Levofloxacin exists as a zwitterionic species at pH 7.4 and has previously been observed to be an inhibitor of OCT1 (SLC22A1)50. Various drugs and metabolites including albuterol, ketamine and scopolamine and various zwitterionic metabolites (e.g. enalaprilat, ramiprilat) did not inhibit SLC22A15-mediated uptake Fig. 4C, Supplemental Table 5). In general, the essential amino acids did not inhibit SLC22A15 even at high concentrations (500 μM), however gamma-aminobutyric acid, GABA, inhibited SLC22A15 by >50% at 500 μM (Fig. 4C, Supplemental Table 5). Since Gly-Sar is a substrate of SLC22A15, we selected a few di- and tri-peptides that are known substrates of SLC15 family members51. At 500 μM, Gly-Sar inhibited SLC22A15 by 20%. Similar inhibition was observed with Gly-Gly (500 μM) and slightly more inhibition was observed with Gly-Gly-His (500 μM). Gly-Gly-Gly, however, exhibited no inhibitory effects at 500 μM (Fig. 4C). Overall, the limited inhibition screen of endogenous compounds and drugs showed that compounds with the following chemical features exhibit strong inhibitory effects on SLC22A15 mediated uptake: (i) quaternary amine adjacent to a carboxylic acid (e.g. ergothioneine, betaine, hypaphorine, carnitine, trigonelline); and (ii) lipophilic compounds, which are also inhibitors of OCTs and OCTNs (e.g. quinidine, verapamil, chloroquine, amitriptyline). Future work evaluating a larger number of compounds as potential inhibitors of SLC22A15-mediated ergothioneine uptake is needed to understand the chemical features of SLC22A15 inhibitors.
Fig. 4.

Inhibition of SLC22A15-mediated uptake of [3H]-ergothioneine by different classes of compounds in HEK293 Flp-In cells stably expressing SLC22A15. Compounds used in these inhibition studies include (A) metabolites identified in the metabolomic study (Fig. 2) and other compounds in their metabolic pathways; (B) substrates and/or inhibitors of OCTs, OCTNs and OATs; (C) other zwitterion and cation drugs. Figure shows a representative plot from one experiment (mean ± S.D. from three replicate wells). All compounds were screened at 500 μM (black bar), except a few compounds at 100 μM (blue bar); (D) SLC22A15-mediated uptake of substrates in the presence of varying concentrations of HEPES, a commonly used buffering agent in uptake media. Uptake in control (EV) cells is also included in the graph; (i) SLC22A15-mediated uptake of four substrates in the presence and absence of HEPES. Two experiments were performed in transiently transfected cells. Data shown are representative of one experiment from triplicate wells (mean ± SD); (E) Percent uptake of [3H]-ergothioneine at various concentration of HEPES. IC50 of HEPES is 4.8 mM. †Compounds that were found to be significantly different between EV and SLC22A15 transfected cells in the metabolomic study.
3.5.2. The effect of HEPES, a commonly used buffering agent, on SLC22A15-mediated transport
HEPES, a commonly used buffering agent and a zwitterion, was one of the top compounds identified in metabolomic study (Fig. 2, Supplemental Table 3). We investigated the effect of HEPES, at 25 mM, the maximum concentration used in transporter assays. Typically, HEPES is used at 5 mM, 10 mM or 25 mM. At 25 mM of HEPES, the SLC22A15-mediated uptake of carnosine, betaine and MPP+ was abolished (Fig. 4D). The IC50 of HEPES for inhibition of SLC22A15-mediated ergothioneine was 5.8 mM ± 1.4 (mean ± SD) (Fig. 4E). We did not observe significant inhibition of SLC22A4 and SLC22A5 at the same concentrations of HEPES (5 mM to 25 mM) (Supplemental Fig. 1G).
3.6. Overexpression of SLC22A15 in HEK293 cells resulted in differentially expressed genes involved in cell cycle regulation, cellular senescence and cancer.
We performed transcriptomic analysis to identify differentially expressed genes between SLC22A15 overexpressing cells and EV cells. In total, there were 7,696 genes that were differentially expressed (p-adjusted < 0.05), of which 6,533 genes had a FPKM > 1 in the cells. This data set is available at https://doi.org/10.6084/m9.figsh are.12478865. A volcano plot allowed us to identify the most biologically significant genes (Fig. 5). As expected, the SLC22A15 transcript exhibited the largest difference between the overexpressing and EV cells (−log10P-value = 8.7, p<10−300, https://doi.org/10.6084/m9.figsh are.12478865). We next performed a functional classification and pathway analysis of 7,696 differentially expressed genes. This dataset is available at https://doi:10.6084/m9.figshare.12478910. Based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, the top three most enriched pathways were cell cycle (p-adjusted = 0.0014), microRNAs in cancer (p-adjusted = 0.008) and cellular senescence (p-adjusted = 0.02) (https://doi.org/10.6084/m9.figsh are.12478865). Furthermore, the differentially expressed genes were significantly enriched (p-value adjusted 0.0001) for biological process terms related to cellular process, cellular component biogenesis and metabolomic process: ribonucleoprotein complex biogenesis, G1/S transition of mitotic cell cycle, ribosome biogenesis, ncRNA processing, cell cycle G1/S phase transition and autophagy (https://doi:10.6084/m9.figshare.12478910). Recent studies suggest that SLC22A15 plays a role in tumor growth15, 16, consistent with our data indicating that the top differentially expressed genes are involved in cancer pathways. Interestingly, one of the top differentially expressed genes, GSTP1 (−log10P-value = 2.5, p<10−300), encodes a typical antioxidant enzyme to neutralize endogenous reactive oxygen species. The expression levels of a second top gene, HSPA1A (−log10P-value = 2.0, p<10−300), can be reduced by carnosine52 and increased by ergothioneine and carnitine53.
Fig. 5.
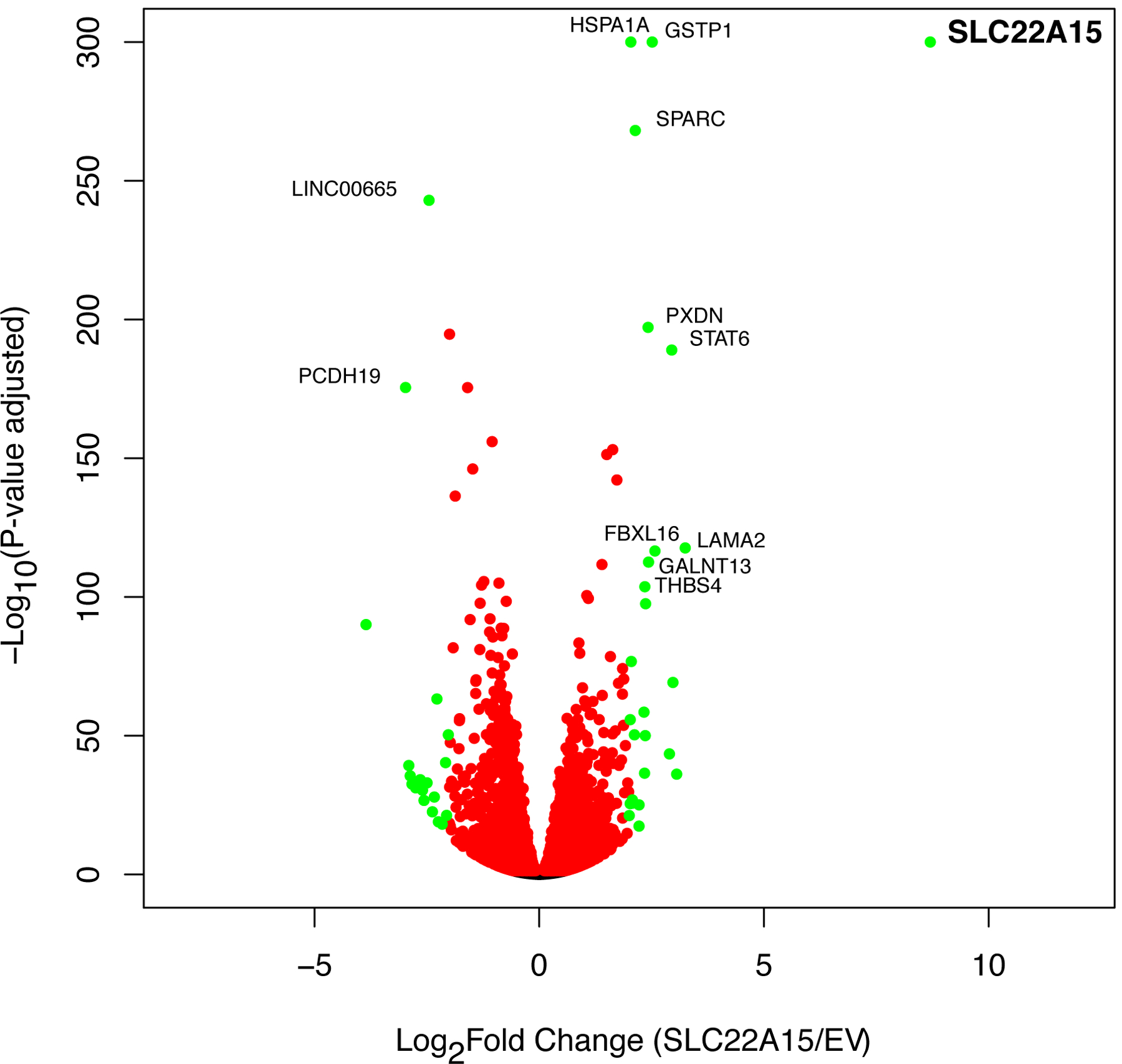
A volcano plot showing the 7,696 genes which are differentially expressed (measured with RNAseq) between EV cells and SLC22A15 cells at p-value adjusted < 0.05. The fold-changes (log2) of transcript expression were calculated between HEK293 Flp-In EV/SLC22A15-GFP cells, and the –log10(p-value adjusted) were plotted against the log2(fold change) for the 7,696 transcripts. Each dot represents mean values (n = 4). The red dots represent p-adjusted (FDR) < 0.05, while the green dots represent p-adjusted (FDR) < 0.05 and log2fold > 2- or < −2-fold changes in expression. Twelve genes that are labeled have p-adjusted < 1×10−100 and log2fold > 2.0- or < −2.0-fold changes in expression. The top-most significant genes, SLC22A15, GSTP1 and HSPA1A have p-value adjusted < 1×10−300. These three genes are plotted as p=1×10−300 in this figure.
4. DISCUSSION
Members of the SLC22A family, the organic ion family, include the human organic cation (OCT), organic anion (OAT) and zwitterion (OCTN) transporters. Because many transporters in this family are highly expressed in the intestine, liver or kidney, they play significant roles in the absorption, distribution and elimination of various endogenous molecules and xenobiotics including prescription drugs (e.g. SLC22A1 to SLC22A8). As a result, organic cation (SLC22A1 and SLC22A2) and anion (SLC22A6 and SLC22A8) transporters are target sites for drug-drug interactions3, 54 and their interactions with candidate drugs are examined as part of the drug development process. Despite their importance, one-third of the proteins in the SLC22A family have no assigned substrates and are designated as orphan transporters. Interestingly, though several of the orphan transporters in the SLC22A family have been studied in knockout mouse models or in cells, their substrates have not been identified8, 11, 55. Our study focused on identifying the substrates of one of the orphan SLC22A family members, SLC22A15, which was termed FLIPT1 in earlier studies suggesting that the protein may play a role in carnitine flux13. However, no study has experimentally examined the ligand specificity of the transporter. The goal of our study was to use various computational and experimental methods, ranging from structural modelling to metabolomic and transporter uptake assays, to characterize the substrate specificity and transporter mechanism of SLC22A15. Our key findings are (a) SLC22A15 preferentially transports zwitterions, and notably ergothioneine, carnosine and carnitine, though it tolerates organic cations and (b) that the transporter is sodium dependent. Below we discuss each of these findings.
SLC22A15 preferentially transports zwitterions.
Though the transporter transports carnitine, as previously predicted13, it seems to preferentially transport other zwitterions, e.g. ergothioneine and carnosine, and weakly interact with organic cations, e.g. TEA, MPP+, cimetidine (Fig. 2, Fig. 3). Although most of its substrates are also substrates of other SLC22 family members including the major zwitterion transporters, SLC22A4 and SLC22A5, and the organic cation transporters, SLC22A1, SLC22A2 and SLC22A3, carnosine, glycylsarcosine and creatine were not previously reported as substrates of any member of the SLC22A family. These three zwitterionic compounds are well-known substrates of the SLC15A and SLC6A families36, 37, 51. Compared with the other zwitterion transporters in the SLC22A family, SLC22A15 exhibited a higher Km for its substrates, including ergothioneine, carnosine, creatine and carnitine (Fig. 3). These results suggest that the transporter may act to regulate the levels of its substrates at high concentrations, especially in tissues in which it is co-expressed with other higher-affinity SLC transporters for the same substrates. High Km (or so-called low affinity) transporters may serve in the influx or efflux of compounds in cells, taking a more important role at higher substrate concentrations when the high affinity transporters become saturated. Efflux roles may be particularly important for compounds that exhibit cellular toxicities at high levels, especially for those that are particularly hydrophilic. Low affinity transporters may also work with high affinity transporters for the same substrates in tissues in which the concentrations of the substrates vary. For example, the low affinity transporter, PEPT1 (SLC15A1) acts to salvage oligopeptides at high concentrations in the early proximal tubule56, whereas PEPT2 (SLC15A2), which is a higher affinity oligopeptide transporter, acts in the later proximal tubule to salvage its substrates at lower levels57. In tissues in which the complementary higher affinity transporter is absent, SLC22A15 may provide the sole means of entry or escape of its substrates. For example, OCT1 appears to act in the liver, a tissue with a low abundance of the high affinity carnitine transporter (OCTN2), as an efflux transporter for carnitine, which is synthesized in the liver. A survey of publicly available databases indicates that SLC22A15 is ubiquitously expressed throughout all tissues and tends to overlap in its expression pattern with SLC22A4, suggesting that it is playing a role in the regulation of ergothioneine levels. Whereas SLC22A4 plays a major role in regulating ergothioneine levels in red blood cells, SLC22A15 is found at significantly higher levels in all regions of the brain compared to SLC22A4, suggesting that it may play a role in the brain (Supplemental Fig. 2B, C). SLC22A15 also has the highest expression overall in the bone marrow (Supplemental Fig. 2A). In contrast, SLC22A4 has the highest expression in whole blood compared to all other zwitterion transporters, with SLC22A15 ranking second highest (GTEx reports 26.17 and 6.8 TPM, respectively).
Ergothioneine is a dietary-derived amino acid synthesized by fungi and a few bacterial species. It has been described as a potent and unique antioxidant due to its impressive stability at physiological pH and the fact that it does not readily auto-oxidize as other antioxidants do (Grigat et al., 2007). Studies have established ergothioneine’s ability to react with reactive oxygen species, including singlet oxygen, hydroxyl radicals, hypochlorous acid and peroxynitrite58–60. Ergothioneine accumulates at high concentrations in areas subject to oxidative stress, specifically the brain, bone marrow, erythrocytes and cornea of the eye61. While ergothioneine deficiencies have not been reported, some studies show a decline in ergothioneine plasma levels in patients with neurodegenerative diseases such as Parkinson’s disease and cognitive deficits, suggesting that ergothioneine might play a role in protecting the brain from neuronal injury62, 63. Prior to this work, SLC22A4 previously was the only known ergothioneine transporter, and appears to play a major role in ergothioneine distribution in the liver and kidney and in red blood cells38. However, compared to SLC22A15, SLC22A4 is expressed at substantially lower levels in the brain (Supplemental Fig. 2). Further, studies in Slc22a4 knockout mice demonstrate that the ergothioneine levels in the brain remain unchanged in wildtype and Slc22a4−/− after IV administration of ergothioneine64, suggesting that another transporter is facilitating ergothioneine distribution in the brain. SLC22A15 may be playing a major role in the disposition of ergothioneine in the brain, though further studies are clearly needed. Supporting its role in ergothioneine disposition, several genetic variants in SLC22A15 have been associated with ergothioneine serum levels65. This dataset is available at https://doi:10.6084/m9.figshare.12478916). Both SLC22A4 and SLC22A15 are highly expressed in whole blood (Supplemental Fig. 2), though their relative expression in erythrocytes needs further study.
Like ergothioneine, carnosine, a dipeptide, has antioxidant effects, however, it is also an anti-glycating and metal ion sequestering agent66, 67. Carnosine is largely derived from the diet (e.g. red and white meat) though it may be synthesized in various tissues and broken down in the blood by carnosinase (CNDP1, CNDP2). Carnosine levels are high in the brain66, 67, consistent with expression patterns of SLC22A15. According to single RNAseq data, SLC22A15 is highly expressed in oligodendrocytes compared with neurons, astrocytes, microglia and endothelial cells68 (see http://www.brainrnaseq.org/, https://cells.ucsc.edu/?ds=autism&gene=SLC22A15, https://cells.ucsc.edu/?ds=allen-celltypes%2Fhuman-cortex&gene=SLC22A15 ), whereas SLC15A2, another carnosine transporter, is specifically expressed in astrocytes and microglia68. Beneficial effects of carnosine in neurological diseases and autism have been noted69, and interestingly, genetic polymorphisms in SLC22A15 locus are associated with autism (see https://doi:10.6084/m9.figshare.12478916). In addition to carnosine’s effects in the brain, the dipeptide also has effects in cancer reducing tumor growth16 and cell proliferation70–74. These data are consistent with our gene expression studies indicating that SLC22A15 may be involved in cell cycle, autophagy, cellular senescence and cancer (https://doi.org/10.6084/m9.figsh are.12478910).
Carnitine and creatine, are also substrates of SLC22A15 and other transporters, but with higher affinities (lower Km’s) for other transporters. For carnitine, SLC22A5 represents the corresponding lower Km transporter, and for creatine, SLC6A8 is a highly specific low Km creatine transporter75. Unlike SLC22A15 which has a low expression levels across many tissues, both SLC22A5 and SLC6A8 are highly expressed in many human tissues (Supplemental Fig. 2C). In the metabolomic study, we observed that levels of monoacylglycerols and fatty acids were significantly higher in SLC22A15 transfected cells (Supplemental Table 3), possibly reflecting the fact that these molecules are intermediate and end products of carnitine mediated fatty acid oxidation76, 77. In several GWAS, polymorphisms in or near SLC22A15 have been associated with monoacylglycerol levels (e.g. 1-palmitoleoylglycerol) and triacylglycerol levels (e.g. TAG 52:1, TAG 48:2)78, 79 as well as with fat-related traits (e.g. body mass index, trunk fat mass, whole body fat mass)80, consistent with a role of SLC22A15 in the flux of carnitine or carnitine derivatives. In our study, at concentrations of 100 μM monoacylglycerols were not able to inhibit SLC22A15-mediated ergothioneine uptake (Supplemental Fig. 1H, Fig. 2C, D). SLC22A15 may work together with SLC22A5 in tissues where both are expressed, playing roles in intracellular carnitine disposition, and may be particularly important in Carnitine Transporter Deficiency (CTD), an autosomal recessive disease, characterized by missense mutations in SLC22A542. Patients with CTD are treated with high doses of carnitine, which must enter tissues to mediate fatty acid oxidation81.
SLC22A15 is sodium dependent.
We found that SLC22A15 mediated transport of ergothioneine, carnosine, carnitine and creatine was sodium dependent, similar to SLC22A4 and SLC22A5, which are also sodium dependent transporters38, 39. These data support the idea that SLC22A15 is a sodium-driven plasma membrane transporter. Sodium dependent transporters can concentrate their substrates intracellularly, which is consistent with the high intracellular levels of ergothioneine compared to plasma levels. For example, ergothioneine levels in plasma are 0.5–3 μM and are much higher in erythrocytes (5.6–30 μM)82, 83, consistent with a sodium dependent mechanism for both SLC22A4 and SLC22A15. In the brain of rats fed ergothioneine, levels are about 7 μg/g compared with 1.3 μg/mL in plasma84. Carnosine, carnitine and creatine are also among the metabolites that are enriched in erythrocytes compared to plasma85. There are several examples of high and low Km transporters, which may both be sodium dependent. For example the sodium glucose transporter, SLC5A1 (SGLT1) is a high affinity, low capacity transporter for glucose, whereas its paralog, SLC5A2 (SGLT2), is a low affinity, high capacity Na+-glucose transporter86. These transporters work together in various tissues to acquire and salvage glucose.
In summary, we de-orphaned SLC22A15 and showed several zwitterions, including potent antioxidants are substrates of the transporter. With the identification of its substrates, kinetic properties and transport mechanism, our results will hopefully motivate future studies to understand the mechanisms for its biological roles in health and disease, including its roles in cancer, neurological disease and fatty acid metabolism.
Supplementary Material
Supplemental Table 5. Percent uptake of [3H]-ergothioneine mediated by stably expressing SLC22A15-GFP tagged in the presence of various compounds.
Supplemental Table 3. Results from metabolomic study using Metabolon Inc. platform. Metabolites were determined using mass spectrometry (Metabolon DiscoveryHD™ metabolomics platform).
Supplemental Table 4. Kinetic parameters of four zwitterions uptake by SLC22A15.
Supplemental Table 2. List of 23 radiolabeled compounds used for initial screening to validate metabolomic results and also to test other substrates of SLC22A family for uptake by SLC22A15 cells.
Supplemental Table 1. List of primers used for cloning SLC22A15, for creation of SLC22A15-GFP tagged and for sequencing.
ACKNOWLEDGEMENT
This study was supported in part by grants from the National Institutes of Health (R01 DK108722 and R01GM117163). The authors would like to acknowledge the following contract research companies for their excellent services: (i) Metabolon Inc. (Durham, North Carolina, USA) for performing non-targeted liquid chromatography-tandem mass spectrometry (LC-MS/MS) and gas chromatography/mass spectrometry platform; (ii) Novogene Co., Ltd (Sacramento, California, USA) for the RNAseq sequencing and data analysis services; (iii) University of California, San Francisco Laboratory for Cell Analysis for providing cell sorting services, this unit is supported by the National Cancer Institute Cancer Center Support Grant (P30CA082103). Results from the RNAseq data analyses are available and deposited in the figshare (DOI: 10.6084/m9.figshare.12478865 for transcriptomic levels and DOI:10.6084/m9.figshare.12478910 for gene and pathway enrichment analyses). Data compiled from the publicly available datasets to support the genetic associations of SLC22A15 with human disease, traits, metabolites levels and eQTLs are available and deposited in the figshare (DOI: 10.6084/m9.figshare.12478916).
ABBREVIATIONS:
- ASP+
4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide
- EV
empty vector
- FLIPT1
Fly-like putative transporter 1
- GWAS
genome-wide association studies
- HBSS
Hank’s buffered salt solution
- HEK293
human embryonic kidney cells
- HEPES
(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid)
- IC50
concentration of a compound to inhibit half of the activity
- Km
the concentration of substrates to reach half of the maximum uptake rate (Vmax)
- MPP+
1-methyl-4-phenylpyridinium
- OAT
organic anion transporter
- OCT
organic cation transporter
- OCTN
organic zwitterions/cation transporter
- PEPT
peptide transporter
- SLC
solute carrier
- SLCO
solute carrier family of organic anion transporting polypeptides
- SNP
single nucleotide polymorphism
REFERENCES
- 1.Koepsell H Organic Cation Transporters in Health and Disease. Pharmacol Rev. January 2020;72(1):253–319. doi: 10.1124/pr.118.015578 [DOI] [PubMed] [Google Scholar]
- 2.Wright SH. Molecular and cellular physiology of organic cation transporter 2. Am J Physiol Renal Physiol. December 1 2019;317(6):F1669–F1679. doi: 10.1152/ajprenal.00422.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Transporter Consortium, Giacomini KM, Huang SM, et al. Membrane transporters in drug development. Nat Rev Drug Discov. March 2010;9(3):215–36. doi: 10.1038/nrd3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Superti-Furga G, Lackner D, Wiedmer T, et al. The RESOLUTE consortium: unlocking SLC transporters for drug discovery. Nat Rev Drug Discov. April 7 2020;doi: 10.1038/d41573-020-00056-6 [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Shu Y, Liang X, et al. OCT1 is a high-capacity thiamine transporter that regulates hepatic steatosis and is a target of metformin. Proc Natl Acad Sci U S A. July 8 2014;111(27):9983–8. doi: 10.1073/pnas.1314939111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goswami S, Yee SW, Xu F, et al. A Longitudinal HbA1c Model Elucidates Genes Linked to Disease Progression on Metformin. Clin Pharmacol Ther. November 2016;100(5):537–547. doi: 10.1002/cpt.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubail J, Huber C, Chantepie S, et al. SLC10A7 mutations cause a skeletal dysplasia with amelogenesis imperfecta mediated by GAG biosynthesis defects. Nat Commun. August 6 2018;9(1):3087. doi: 10.1038/s41467-018-05191-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maruyama SY, Ito M, Ikami Y, et al. A critical role of solute carrier 22a14 in sperm motility and male fertility in mice. Sci Rep. November 4 2016;6:36468. doi: 10.1038/srep36468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vitart V, Rudan I, Hayward C, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. April 2008;40(4):437–42. doi: 10.1038/ng.106 [DOI] [PubMed] [Google Scholar]
- 10.Rusu V, Hoch E, Mercader JM, et al. Type 2 Diabetes Variants Disrupt Function of SLC16A11 through Two Distinct Mechanisms. Cell. June 29 2017;170(1):199–212 e20. doi: 10.1016/j.cell.2017.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett KM, Liu J, Hoelting C, Stoll J. Expression and analysis of two novel rat organic cation transporter homologs, SLC22A17 and SLC22A23. Mol Cell Biochem. June 2011;352(1–2):143–54. doi: 10.1007/s11010-011-0748-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes CF, Godoy JR, Doring B, et al. The novel putative bile acid transporter SLC10A5 is highly expressed in liver and kidney. Biochem Biophys Res Commun. September 14 2007;361(1):26–32. doi: 10.1016/j.bbrc.2007.06.160 [DOI] [PubMed] [Google Scholar]
- 13.Eraly SA, Nigam SK. Novel human cDNAs homologous to Drosophila Orct and mammalian carnitine transporters. Biochem Biophys Res Commun. October 11 2002;297(5):1159–66. doi: 10.1016/s0006-291x(02)02343-4 [DOI] [PubMed] [Google Scholar]
- 14.Drake KA, Torgerson DG, Gignoux CR, et al. A genome-wide association study of bronchodilator response in Latinos implicates rare variants. J Allergy Clin Immunol. February 2014;133(2):370–8. doi: 10.1016/j.jaci.2013.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okada R, Koshizuka K, Yamada Y, et al. Regulation of Oncogenic Targets by miR-99a-3p (Passenger Strand of miR-99a-Duplex) in Head and Neck Squamous Cell Carcinoma. Cells. November 28 2019;8(12)doi: 10.3390/cells8121535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu G, Qian M, Lu L, et al. O-GlcNAcylation of YY1 stimulates tumorigenesis in colorectal cancer cells by targeting SLC22A15 and AANAT. Carcinogenesis. January 30 2019;doi: 10.1093/carcin/bgz010 [DOI] [PubMed] [Google Scholar]
- 17.Yee SW, Stecula A, Chien HC, et al. Unraveling the functional role of the orphan solute carrier, SLC22A24 in the transport of steroid conjugates through metabolomic and genome-wide association studies. PLoS Genet. September 2019;15(9):e1008208. doi: 10.1371/journal.pgen.1008208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geier EG, Schlessinger A, Fan H, et al. Structure-based ligand discovery for the Large-neutral Amino Acid Transporter 1, LAT-1. Proc Natl Acad Sci U S A. April 2 2013;110(14):5480–5. doi: 10.1073/pnas.1218165110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlessinger A, Geier E, Fan H, et al. Structure-based discovery of prescription drugs that interact with the norepinephrine transporter, NET. Proc Natl Acad Sci U S A. September 20 2011;108(38):15810–5. doi: 10.1073/pnas.1106030108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng D, Sun P, Yan C, et al. Molecular basis of ligand recognition and transport by glucose transporters. Nature. October 15 2015;526(7573):391–6. doi: 10.1038/nature14655 [DOI] [PubMed] [Google Scholar]
- 21.Perland E, Fredriksson R. Classification Systems of Secondary Active Transporters. Trends Pharmacol Sci. March 2017;38(3):305–315. doi: 10.1016/j.tips.2016.11.008 [DOI] [PubMed] [Google Scholar]
- 22.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. April 2008;36(7):2295–300. doi: 10.1093/nar/gkn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. December 5 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 24.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. November 2006;15(11):2507–24. doi: 10.1110/ps.062416606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolinsky TJ, Czodrowski P, Li H, et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. July 2007;35(Web Server issue):W522–5. doi: 10.1093/nar/gkm276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jurrus E, Engel D, Star K, et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. January 2018;27(1):112–128. doi: 10.1002/pro.3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou L, Stecula A, Gupta A, et al. Molecular Mechanisms for Species Differences in Organic Anion Transporter 1, OAT1: Implications for Renal Drug Toxicity. Mol Pharmacol. July 2018;94(1):689–699. doi: 10.1124/mol.117.111153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang X, Chien HC, Yee SW, et al. Metformin Is a Substrate and Inhibitor of the Human Thiamine Transporter, THTR-2 (SLC19A3). Mol Pharm. December 7 2015;12(12):4301–10. doi: 10.1021/acs.molpharmaceut.5b00501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HI, Raffler J, Lu W, et al. Fine Mapping and Functional Analysis Reveal a Role of SLC22A1 in Acylcarnitine Transport. Am J Hum Genet. October 5 2017;101(4):489–502. doi: 10.1016/j.ajhg.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohashi R, Tamai I, Nezu Ji J, et al. Molecular and physiological evidence for multifunctionality of carnitine/organic cation transporter OCTN2. Mol Pharmacol. February 2001;59(2):358–66. doi: 10.1124/mol.59.2.358 [DOI] [PubMed] [Google Scholar]
- 31.Meixner E, Goldmann U, Sedlyarov V, et al. A substrate-based ontology for human solute carriers. Mol Syst Biol. July 2020;16(7):e9652. doi: 10.15252/msb.20209652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engelhart DC, Granados JC, Shi D, et al. Systems Biology Analysis Reveals Eight SLC22 Transporter Subgroups, Including OATs, OCTs, and OCTNs. Int J Mol Sci. March 5 2020;21(5)doi: 10.3390/ijms21051791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engelhart DC, Azad P, Ali S, Granados JC, Haddad GG, Nigam SK. Drosophila SLC22 Orthologs Related to OATs, OCTs, and OCTNs Regulate Development and Responsiveness to Oxidative Stress. Int J Mol Sci. March 15 2020;21(6)doi: 10.3390/ijms21062002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrissey KM, Stocker SL, Wittwer MB, Xu L, Giacomini KM. Renal transporters in drug development. Annu Rev Pharmacol Toxicol. 2013;53:503–29. doi: 10.1146/annurev-pharmtox-011112-140317 [DOI] [PubMed] [Google Scholar]
- 35.Masuo Y, Ohba Y, Yamada K, et al. Combination Metabolomics Approach for Identifying Endogenous Substrates of Carnitine/Organic Cation Transporter OCTN1. Pharm Res. October 2 2018;35(11):224. doi: 10.1007/s11095-018-2507-1 [DOI] [PubMed] [Google Scholar]
- 36.Kamal MA, Jiang H, Hu Y, Keep RF, Smith DE. Influence of genetic knockout of Pept2 on the in vivo disposition of endogenous and exogenous carnosine in wild-type and Pept2 null mice. Am J Physiol Regul Integr Comp Physiol. April 2009;296(4):R986–91. doi: 10.1152/ajpregu.90744.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uemura T, Ito S, Masuda T, et al. Cyclocreatine Transport by SLC6A8, the Creatine Transporter, in HEK293 Cells, a Human Blood-Brain Barrier Model Cell, and CCDSs Patient-Derived Fibroblasts. Pharm Res. March 2 2020;37(3):61. doi: 10.1007/s11095-020-2779-0 [DOI] [PubMed] [Google Scholar]
- 38.Grundemann D, Harlfinger S, Golz S, et al. Discovery of the ergothioneine transporter. Proc Natl Acad Sci U S A. April 5 2005;102(14):5256–61. doi: 10.1073/pnas.0408624102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamai I, Ohashi R, Nezu J, et al. Molecular and functional identification of sodium ion-dependent, high affinity human carnitine transporter OCTN2. J Biol Chem. August 7 1998;273(32):20378–82. doi: 10.1074/jbc.273.32.20378 [DOI] [PubMed] [Google Scholar]
- 40.Enomoto A, Wempe MF, Tsuchida H, et al. Molecular identification of a novel carnitine transporter specific to human testis. Insights into the mechanism of carnitine recognition. J Biol Chem. September 27 2002;277(39):36262–71. doi: 10.1074/jbc.M203883200 [DOI] [PubMed] [Google Scholar]
- 41.Okabe M, Unno M, Harigae H, et al. Characterization of the organic cation transporter SLC22A16: a doxorubicin importer. Biochem Biophys Res Commun. August 5 2005;333(3):754–62. doi: 10.1016/j.bbrc.2005.05.174 [DOI] [PubMed] [Google Scholar]
- 42.Nezu J, Tamai I, Oku A, et al. Primary systemic carnitine deficiency is caused by mutations in a gene encoding sodium ion-dependent carnitine transporter. Nat Genet. January 1999;21(1):91–4. doi: 10.1038/5030 [DOI] [PubMed] [Google Scholar]
- 43.Tschirka J, Kreisor M, Betz J, Grundemann D. Substrate Selectivity Check of the Ergothioneine Transporter. Drug Metab Dispos. June 2018;46(6):779–785. doi: 10.1124/dmd.118.080440 [DOI] [PubMed] [Google Scholar]
- 44.Grigat S, Harlfinger S, Pal S, et al. Probing the substrate specificity of the ergothioneine transporter with methimazole, hercynine, and organic cations. Biochem Pharmacol. July 15 2007;74(2):309–16. doi: 10.1016/j.bcp.2007.04.015 [DOI] [PubMed] [Google Scholar]
- 45.Urban TJ, Yang C, Lagpacan LL, et al. Functional effects of protein sequence polymorphisms in the organic cation/ergothioneine transporter OCTN1 (SLC22A4). Pharmacogenet Genomics. September 2007;17(9):773–82. doi: 10.1097/FPC.0b013e3281c6d08e. [DOI] [PubMed] [Google Scholar]
- 46.Wagner CA, Lukewille U, Kaltenbach S, et al. Functional and pharmacological characterization of human Na(+)-carnitine cotransporter hOCTN2. Am J Physiol Renal Physiol. September 2000;279(3):F584–91. doi: 10.1152/ajprenal.2000.279.3.F584 [DOI] [PubMed] [Google Scholar]
- 47.Todesco L, Bur D, Brooks H, et al. Pharmacological manipulation of L-carnitine transport into L6 cells with stable overexpression of human OCTN2. Cell Mol Life Sci. May 2008;65(10):1596–608. doi: 10.1007/s00018-008-8065-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiang J, Hu Y, Smith DE, Keep RF. PEPT2-mediated transport of 5-aminolevulinic acid and carnosine in astrocytes. Brain Res. November 29 2006;1122(1):18–23. doi: 10.1016/j.brainres.2006.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dodd JR, Birch NP, Waldvogel HJ, Christie DL. Functional and immunocytochemical characterization of the creatine transporter in rat hippocampal neurons. J Neurochem. November 2010;115(3):684–93. doi: 10.1111/j.1471-4159.2010.06957.x [DOI] [PubMed] [Google Scholar]
- 50.Parvez MM, Kaisar N, Shin HJ, Jung JA, Shin JG. Inhibitory Interaction Potential of 22 Antituberculosis Drugs on Organic Anion and Cation Transporters of the SLC22A Family. Antimicrob Agents Chemother. November 2016;60(11):6558–6567. doi: 10.1128/AAC.01151-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith DE, Clemencon B, Hediger MA. Proton-coupled oligopeptide transporter family SLC15: physiological, pharmacological and pathological implications. Mol Aspects Med. Apr-Jun 2013;34(2–3):323–36. doi: 10.1016/j.mam.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hipkiss AR, Cartwright SP, Bromley C, Gross SR, Bill RM. Carnosine: can understanding its actions on energy metabolism and protein homeostasis inform its therapeutic potential? Chem Cent J. February 25 2013;7(1):38. doi: 10.1186/1752-153X-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakrak O, Kerem M, Bedirli A, et al. Ergothioneine modulates proinflammatory cytokines and heat shock protein 70 in mesenteric ischemia and reperfusion injury. J Surg Res. January 2008;144(1):36–42. doi: 10.1016/j.jss.2007.04.020 [DOI] [PubMed] [Google Scholar]
- 54.Zamek-Gliszczynski MJ, Taub ME, Chothe PP, et al. Transporters in Drug Development: 2018 ITC Recommendations for Transporters of Emerging Clinical Importance. Clin Pharmacol Ther. November 2018;104(5):890–899. doi: 10.1002/cpt.1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito S, Honda G, Fujino Y, Ogata S, Hirayama-Kurogi M, Ohtsuki S. Knockdown of Orphan Transporter SLC22A18 Impairs Lipid Metabolism and Increases Invasiveness of HepG2 Cells. Pharm Res. January 11 2019;36(3):39. doi: 10.1007/s11095-018-2565-4 [DOI] [PubMed] [Google Scholar]
- 56.Shen H, Smith DE, Yang T, Huang YG, Schnermann JB, Brosius FC 3rd., Localization of PEPT1 and PEPT2 proton-coupled oligopeptide transporter mRNA and protein in rat kidney. Am J Physiol. May 1999;276(5):F658–65. doi: 10.1152/ajprenal.1999.276.5.F658 [DOI] [PubMed] [Google Scholar]
- 57.Liu W, Liang R, Ramamoorthy S, et al. Molecular cloning of PEPT 2, a new member of the H+/peptide cotransporter family, from human kidney. Biochim Biophys Acta. May 4 1995;1235(2):461–6. doi: 10.1016/0005-2736(95)80036-f [DOI] [PubMed] [Google Scholar]
- 58.Franzoni F, Colognato R, Galetta F, et al. An in vitro study on the free radical scavenging capacity of ergothioneine: comparison with reduced glutathione, uric acid and trolox. Biomed Pharmacother. September 2006;60(8):453–7. doi: 10.1016/j.biopha.2006.07.015 [DOI] [PubMed] [Google Scholar]
- 59.Rougee M, Bensasson RV, Land EJ, Pariente R. Deactivation of singlet molecular oxygen by thiols and related compounds, possible protectors against skin photosensitivity. Photochem Photobiol. April 1988;47(4):485–9. doi: 10.1111/j.1751-1097.1988.tb08835.x [DOI] [PubMed] [Google Scholar]
- 60.Motohashi N, Mori I. Thiol-induced hydroxyl radical formation and scavenger effect of thiocarbamides on hydroxyl radicals. J Inorg Biochem. March 1986;26(3):205–12. doi: 10.1016/0162-0134(86)80042-3 [DOI] [PubMed] [Google Scholar]
- 61.Cheah IK, Halliwell B. Ergothioneine; antioxidant potential, physiological function and role in disease. Biochim Biophys Acta. May 2012;1822(5):784–93. doi: 10.1016/j.bbadis.2011.09.017 [DOI] [PubMed] [Google Scholar]
- 62.Cheah IK, Feng L, Tang RMY, Lim KHC, Halliwell B. Ergothioneine levels in an elderly population decrease with age and incidence of cognitive decline; a risk factor for neurodegeneration? Biochem Biophys Res Commun. September 9 2016;478(1):162–167. doi: 10.1016/j.bbrc.2016.07.074 [DOI] [PubMed] [Google Scholar]
- 63.Hatano T, Saiki S, Okuzumi A, Mohney RP, Hattori N. Identification of novel biomarkers for Parkinson’s disease by metabolomic technologies. J Neurol Neurosurg Psychiatry. March 2016;87(3):295–301. doi: 10.1136/jnnp-2014-309676 [DOI] [PubMed] [Google Scholar]
- 64.Sugiura T, Kato S, Shimizu T, et al. Functional expression of carnitine/organic cation transporter OCTN1/SLC22A4 in mouse small intestine and liver. Drug Metab Dispos. October 2010;38(10):1665–72. doi: 10.1124/dmd.110.032763 [DOI] [PubMed] [Google Scholar]
- 65.Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. June 2014;46(6):543–550. doi: 10.1038/ng.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghodsi R, Kheirouri S. Carnosine and advanced glycation end products: a systematic review. Amino Acids. September 2018;50(9):1177–1186. doi: 10.1007/s00726-018-2592-9 [DOI] [PubMed] [Google Scholar]
- 67.Wu G Important roles of dietary taurine, creatine, carnosine, anserine and 4-hydroxyproline in human nutrition and health. Amino Acids. March 2020;52(3):329–360. doi: 10.1007/s00726-020-02823-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Sloan SA, Clarke LE, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. January 6 2016;89(1):37–53. doi: 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chmielewska K, Dzierzbicka K, Inkielewicz-Stepniak I, Przybylowska M. Therapeutic potential of carnosine and its derivatives in the treatment of human diseases. Chem Res Toxicol. March 23 2020;doi: 10.1021/acs.chemrestox.0c00010 [DOI] [PubMed] [Google Scholar]
- 70.Hwang B, Shin SS, Song JH, Choi YH, Kim WJ, Moon SK. Carnosine exerts antitumor activity against bladder cancers in vitro and in vivo via suppression of angiogenesis. J Nutr Biochem. December 2019;74:108230. doi: 10.1016/j.jnutbio.2019.108230 [DOI] [PubMed] [Google Scholar]
- 71.Menon K, Mousa A, de Courten B. Effects of supplementation with carnosine and other histidine-containing dipeptides on chronic disease risk factors and outcomes: protocol for a systematic review of randomised controlled trials. BMJ Open. March 22 2018;8(3):e020623. doi: 10.1136/bmjopen-2017-020623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mikula-Pietrasik J, Ksiazek K. L-Carnosine Prevents the Pro-cancerogenic Activity of Senescent Peritoneal Mesothelium Towards Ovarian Cancer Cells. Anticancer Res. February 2016;36(2):665–71. [PubMed] [Google Scholar]
- 73.Renner C, Zemitzsch N, Fuchs B, et al. Carnosine retards tumor growth in vivo in an NIH3T3-HER2/neu mouse model. Mol Cancer. January 6 2010;9:2. doi: 10.1186/1476-4598-9-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Z, Miao L, Wu X, et al. Carnosine Inhibits the Proliferation of Human Gastric Carcinoma Cells by Retarding Akt/mTOR/p70S6K Signaling. J Cancer. 2014;5(5):382–9. doi: 10.7150/jca.8024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Christie DL. Functional insights into the creatine transporter. Subcell Biochem. 2007;46:99–118. doi: 10.1007/978-1-4020-6486-9_6 [DOI] [PubMed] [Google Scholar]
- 76.Hoppel C The role of carnitine in normal and altered fatty acid metabolism. Am J Kidney Dis. April 2003;41(4 Suppl 4):S4–12. doi: 10.1016/s0272-6386(03)00112-4 [DOI] [PubMed] [Google Scholar]
- 77.Knottnerus SJG, Bleeker JC, Wust RCI, et al. Disorders of mitochondrial long-chain fatty acid oxidation and the carnitine shuttle. Rev Endocr Metab Disord. March 2018;19(1):93–106. doi: 10.1007/s11154-018-9448-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Long T, Hicks M, Yu HC, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. April 2017;49(4):568–578. doi: 10.1038/ng.3809 [DOI] [PubMed] [Google Scholar]
- 79.Rhee EP, Ho JE, Chen MH, et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. July 2 2013;18(1):130–43. doi: 10.1016/j.cmet.2013.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kichaev G, Bhatia G, Loh PR, et al. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am J Hum Genet. January 3 2019;104(1):65–75. doi: 10.1016/j.ajhg.2018.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waber LJ, Valle D, Neill C, DiMauro S, Shug A. Carnitine deficiency presenting as familial cardiomyopathy: a treatable defect in carnitine transport. J Pediatr. November 1982;101(5):700–5. doi: 10.1016/s0022-3476(82)80294-1 [DOI] [PubMed] [Google Scholar]
- 82.Mitsuyama H, May JM. Uptake and antioxidant effects of ergothioneine in human erythrocytes. Clin Sci (Lond). October 1999;97(4):407–11. [PubMed] [Google Scholar]
- 83.Wang LZ, Thuya WL, Toh DS, et al. Quantification of L-ergothioneine in human plasma and erythrocytes by liquid chromatography-tandem mass spectrometry. J Mass Spectrom. March 2013;48(3):406–12. doi: 10.1002/jms.3150 [DOI] [PubMed] [Google Scholar]
- 84.Briggs I Ergothioneine in the central nervous system. J Neurochem. January 1972;19(1):27–35. doi: 10.1111/j.1471-4159.1972.tb01250.x [DOI] [PubMed] [Google Scholar]
- 85.Chaleckis R, Murakami I, Takada J, Kondoh H, Yanagida M. Individual variability in human blood metabolites identifies age-related differences. Proc Natl Acad Sci U S A. April 19 2016;113(16):4252–9. doi: 10.1073/pnas.1603023113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wright EM, Loo DD, Hirayama BA. Biology of human sodium glucose transporters. Physiol Rev. April 2011;91(2):733–94. doi: 10.1152/physrev.00055.2009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 5. Percent uptake of [3H]-ergothioneine mediated by stably expressing SLC22A15-GFP tagged in the presence of various compounds.
Supplemental Table 3. Results from metabolomic study using Metabolon Inc. platform. Metabolites were determined using mass spectrometry (Metabolon DiscoveryHD™ metabolomics platform).
Supplemental Table 4. Kinetic parameters of four zwitterions uptake by SLC22A15.
Supplemental Table 2. List of 23 radiolabeled compounds used for initial screening to validate metabolomic results and also to test other substrates of SLC22A family for uptake by SLC22A15 cells.
Supplemental Table 1. List of primers used for cloning SLC22A15, for creation of SLC22A15-GFP tagged and for sequencing.


