Abstract
Nudix hydrolases attract considerable attention for their wide range of specialized activities in all domains of life. One particular group of Nudix phosphohydrolases (DIPPs), through their metabolism of diphosphoinositol polyphosphates (PP-InsPs), regulates the actions of these polyphosphates upon bioenergetic homeostasis. In the current study we describe, at an atomic level, hitherto unknown properties of human DIPP1.We provide X-ray analysis of the catalytic core of DIPP1 in crystals complexed with either natural PP-InsPs, alternative PP-InsP stereoisomers, or non-hydrolysable methylene bisphosphonate analogues (“PCP-InsPs”). The conclusions that we draw from these data are interrogated by studying the impact upon catalytic activity upon mutagenesis of certain key residues. We present a picture of a V-shaped catalytic furrow with overhanging ridges constructed from flexible positively-charged side chains; within this cavity, the labile phosphoanhydride bond is appropriately positioned at the catalytic site by an extensive series of interlocking polar contacts which we analogize as ‘suspension cables’. We demonstrate functionality for a triglycine peptide within a β-strand which represents a non-canonical addition to the standard Nudix catalytic core structure. We describe pre-reaction enzyme/substrate states which we posit to reflect a role for electrostatic steering in substrate capture. Finally, through time-resolved analysis we uncover a chronological sequence of DIPP1/product post-reaction states, one of which may rationalize a role for InsP6 as an inhibitor of catalytic activity.
Keywords: cell-signaling, Nudix, phosphatase, reaction mechanism
Introduction
Nudix hydrolases are found in all classes of organisms (1-3). They were originally catalogued by virtue of their common feature of hydrolyzing a nucleoside diphosphate linked to some other moiety, X (3), but the range of their catalytic activities has since expanded. Representatives of this family are now known that each hydrolyze a focused subset from a wide range of organic pyrophosphates, such as the 7-methylguanosine caps at the 5′-end of mRNAs, nucleotide sugars, nucleoside di- and triphosphates, dinucleoside polyphosphates, and diphosphoinositol polyphosphates (PP-InsPs; see the list of abbreviations and (Figure 1). Most of the enzymes in this family possess a consensus catalytic sequence (or ‘Nudix box’) that is generally described as GX5EX5UXREX2EEXGU, where ‘U’ designates an aliphatic, hydrophobic residue. The latter consensus sequence (sometimes with minor modifications) creates a scaffold for metal binding and basic hydrolase chemistry (4), and it is generally considered that it is left to residues from outside of the Nudix box to define the substrate specificities of each of the individual enzymes (5).
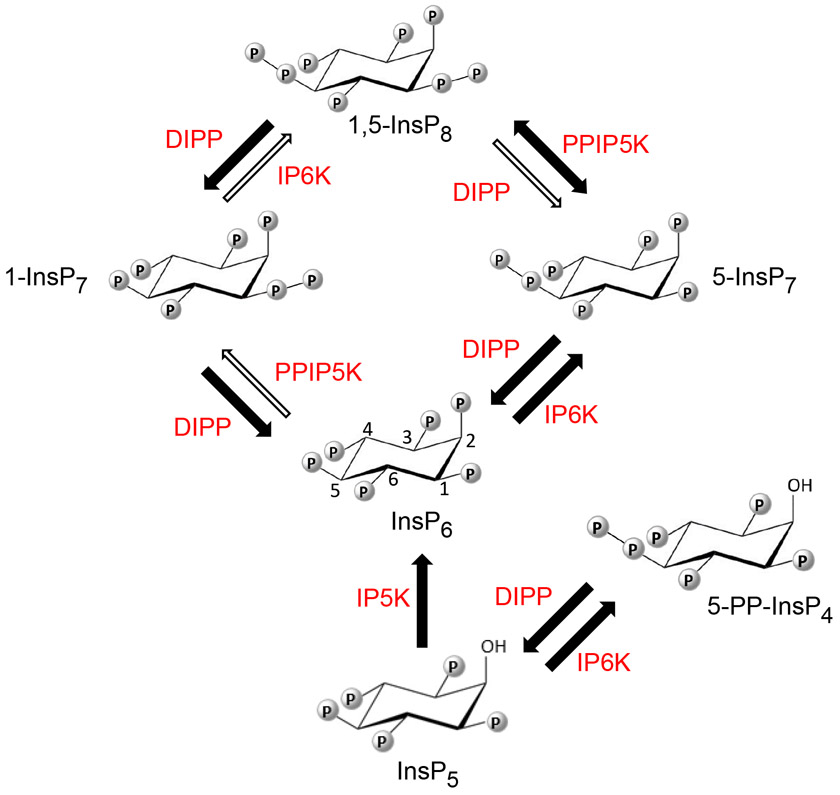
Figure 1.
The primary pathways of mammalian PP-InsP synthesis and metabolism. Thick black arrows and thin white arrows depict major and minor metabolic fluxes, respectively, so as to incorporate the concept (7, 17) that the pathway for 1,5-InsP8 turnover comprises a predominantly cyclical interconversion of InsP6 to 5-InsP7 to 1,5-InsP8 to 1-InsP7 to InsP6. Each arrow is paired with an abbreviation for the responsible enzyme: IP5K (inositol pentakisphosphate 2-kinase, E.C. 2.7.1.155); IP6K (inositol hexakisphosphate 5-kinase, E.C. 2.7.4.21); PPIP5K (diphosphoinositol pentakisphosphate 1-kinase, E.C. 2.7.1.155; this enzyme also possesses an active 1,5-InsP8 1-phosphatase activity, as illustrated by the ‘double-headed arrow’). DIPP = diphosphoinositol polyphosphate diphosphatase, E.C. 3.6.1.52). Locants are provided for the inositol ring of InsP6.
The Diphosphoinositol Polyphosphate Phosphohydrolases (DIPPs) represent a subgroup of the Nudix family that regulate the levels and hence the cell-signaling functions of the PP-InsPs (6, 7). Several DIPPs are expressed by the human genome: DIPP1/NUDT3 (6), DIPP2/NUDT4 (8), DIPP3α/NUDT10 and DIPP3β/NUDT11 (9, 10). DIPP orthologs are widely-distributed within the animal and fungal kingdoms (11). Human DIPP1 has been the most well-studied of these phosphatases. Several genome-wide association studies have linked obesity to changes in levels of DIPP1 expression (12). The expression level of the Drosophila ortholog of DIPP1 may play important roles during development (13); whole-animal knockouts impact fly insulin signaling and feeding behavior (12). It has also been suggested that elevated DIPP1 expression is a candidate for promoting a sarcopenia phenotype (14).
Four different PP-InsP substrates of DIPPs are produced by animal and yeast cells: 5-PP-InsP4, which is synthesized from Ins(1,3,4,5,6)P5, in addition to “1-InsP7” (i.e., 1-PP-InsP5), “5-InsP7” (5-PP-InsP5) and “1,5-InsP8” (1,5-[PP]2-InsP4) which are all synthesized from InsP6 (Figure 1). In mammalian cells, these PP-InsPs supervise fundamental aspects of metabolic and bioenergetic homeostasis (7), while 5-PP-InsP4 has a separate role in telomere maintenance (15, 16). Originally DIPPs were considered to switch-off PP-InsP signaling (6), but in the context of a PP-InsP metabolic cycle (Figure 1; (17)), DIPPs (rather than the PPIP5Ks) also assume primary responsibility for the synthesis and metabolism of 1-InsP7.
The PP-InsPs represent the most dense three-dimensional phosphate arrays found in nature (7), so consequently the DIPP active site is obliged to accommodate considerable steric bulk and more intense negative charge than any other Nudix hydrolase. Thus, it can be anticipated that unique adaptations by the DIPP family have evolved; indeed, the DIPP family have been assigned to an exclusive monophyletic clade (5). To understand the success of this evolutionary path at an atomic level, the tools of structural biology are very insightful. Such information also illuminates catalytic mechanisms, substrate recognition and reaction specificity, regulatory processes, while also offering templates for the rational design of selective inhibitors as research tools or for therapeutic applications.
In 2009, Thorsell et al (18) solved the crystal structure of human DIPP1 in complex with Mg2+ (an essential cofactor (6)) and the InsP6 reaction product; the analysis of the electron density of the InsP6 revealed a mixture of two ligand conformations. One of these two orientations of InsP6 was used as a template to model 3,5-InsP8 into the active site, from which a catalytic mechanism for PP-InsP hydrolysis was proposed (18). However, Thomas and Potter (19) have argued that the mechanistic proposals made by Thorsell et al (18) should be treated cautiously. For example, Thomas and Potter (19) noted there was no justification as to which of the two DIPP1-bound InsP6 conformations is the more catalytically relevant; second, the biologically-relevant structure of InsP8 was subsequently shown to be the 1,5-isomer (20, 21), not the 3,5-enantiomer assumed by Thorsell et al. Furthermore, it is necessary to identify catalytic mechanisms that can account for DIPP1-mediated dephosphorylation of all of its PP-InsP substrates (not only 1,5-InsP8 but also 5-PP-InsP4, 1-InsP7 and 5-InsP7 (Figure 1).
Subsequent to publication of the study by Thorsell et al (18), there have been two Protein Data Base entries (www.rcsb.org) that describe the core structures of a DIPP2 (5LTU) and a DIPP3 (3MCF), but neither crystal structure has bound PP-InsPs. Two further entries, 6PCK and 6PCL, describe DIPP1 crystals in complex with 1-InsP7 and 5-InsP7, respectively, but the position of the nucleophilic water molecule was not identified, and the accompanying publication (20) did not focus upon reaction mechanisms or substrate recognition.
We have now added significantly to our molecular insight into these and several other important aspects of DIPP activity, through comprehensive biophysical and structural analyses of the interactions of DIPP1 with a comprehensive variety of PP-InsPs, and by catalytic assays of several DIPP1 mutants. In addition, we have used metabolically-stable methylene bisphosphonate analogues, i.e., 5-PCP-InsP7 (i.e., 5-PCP-InsP5) and 1,5-PCP-InsP8 (1,5-[PCP]2-InsP4) (22) to obtain new information on DIPP1 pre-reactions states, and we have performed structural time course experiments to identify new post-reaction features that suggest possible functional significance of product-bound enzyme.
Materials and Methods
Protein Expression and Purification
The cDNA encoding residues 1-148 of human DIPP1 was purchased from Addgene (catalog # 42348). The recombinant plasmid was transformed into competent E. coli BL21(DE3). An overnight culture of the transformed E. coli cells was inoculated into nutrient-rich 2xYT medium at pH 7.5 which was cultured at 37 °C to an optical density of 0.7 at 595 nm. Isopropyl β-d-thiogalactopyranoside (0.1 mM) was then added and cultures were continued at 15 °C for 22 hours. The cells were harvested by centrifugation at 5000 x g for 10 min and disrupted using a Constant Cell Disruption System (Constant System Ltd) at 20 KPsi. Recombinant protein was then purified at 4 °C as follows: cell supernatant was first mixed with Ni-NTA agarose (Qiagen), then washed with buffer containing 300 mM NaCl, 20 mM Tris-HCl, pH 7.5, 20 mM imidazole, and the target protein was eluted with imidazole concentration of 250 mM. The eluate was loaded to HiTrap™ Heparin HP column (GE Healthcare) and eluted with 10 column volumes of a 50-2000 mM NaCl gradient in 20 mM Tris-HCl, pH 7.5. Finally, protein was applied to a Superdex™ 200 size exclusion column (GE Healthcare) and eluted with 150 mM NaCl, 20 mM Tris-HCl, pH 7.5. The final preparations were concentrated to 32.6 mg/ml and stored at −80 °C; purity was validated by 4-20% SDS-PAGE.
Enzyme assays
Phosphatase activity against 10 μM of each PP-InsP substrate was measured after 30 min assays in 100 μl reaction mixtures containing 70-1500 ng wild-type DIPP1 (or the indicated mutant), 20 mM HEPES, pH 7.2, 100 mM KCl, 0.8 mM MgCl2 and 20 μM Na2EDTA. Inorganic phosphate release was determined with a colorimetric assay as previously reported (23). The individual PP-InsPs were all chemically synthesized as described previously (24-28).
Crystallization
We first incubated 11.2 mg/ml DIPP1 protein plus 2 mM ligand on ice for 30 min. Then, unless otherwise stated below, crystal complexes were prepared by a two-step process at 20 °C. For step one, crystals were formed over a 2 day period by the hanging drop vapor diffusion method; hanging drops comprised 1.5 μl of protein/ligand solution plus 1.5 μl of the “co-crystallization medium” that was present in the reservoir, which comprised 10% (w/v) PEG 8000, 10% (v/v) isopropanol, 200 mM Li2SO4, 75 mM Na acetate and 25 mM HEPES, final pH 5.9, plus 2 mM ligand. For step two, crystal complexes were placed into a “soaking medium” (compositions are described below) for various additional times.
For crystal complexes with either 5-InsP7, 5-PP-InsP4 or 4-InsP7, the soaking medium for the second step comprised 200 mM LiCl, 20% (w/v) PEG 8000, 20% (v/v) isopropanol, 20 mM MgCl2, 80 mM NaF, and 100 mM HEPES, pH 7.0 and 2 mM ligand; the soaking time was 2 days. Additionally, for in crystallo analysis of the reaction, preformed DIPP/5-InsP7 crystal complexes were transferred into ligand-free soaking buffer that contained 200 mM LiCl, 20% (w/v) PEG 8000, 20% (v/v) isopropanol, 20 mM MgCl2, 100 mM HEPES, pH 7.0; aliquots were removed and cryo-trapped at various times in liquid nitrogen.
For 1,5-PCP-InsP8 (synthesized as previously described; (22)) the soaking buffer was 150 mM Li2SO4, 20% (w/v) PEG 8000, 10% (v/v) isopropanol, 25 mM HEPES, 20 mM MgCl2 and 75mM Na acetate, pH 5.9; the soaking time was 1 day. For 1-InsP7 and 2-InsP7, their concentrations were increased to 4 mM, and these were added to preformed crystal complexes contained InsP6, in soaking buffer that contained 200 mM Li2SO4, 25% (w/v) PEG 8000, 10% (v/v) isopropanol, 20 mM MgCl2 and 80 mM NaF, 25 mM HEPES, 75 mM Na acetate, pH 5.9.
Separately, preformed crystals that we attempted to co-crystallize with 1,5-InsP8 were frozen within 30 sec transferred from soaking buffer that contained 400 mM LiCl, 30% (w/v) PEG 8000, 20% (v/v) isopropanol, 20 mM MgCl2, 4 mM 1,5-InsP8, and 100mM HEPES pH 7.0 and 80 mM NaF, whereupon the reaction product, 1-InsP7, was captured in the crystal complex.
The crystals in complex with 5-PCP-IP7 were obtained solely by co-crystallization in buffer containing 200 mM Li2SO4, 18% (w/v) PEG 8000, 5% (v/v) isopropanol, 100mM Na acetate pH 4.5. No subsequent soaking was performed.
After crystal soaking, the isopropanol concentration was adjusted to 20%, if appropriate, and then samples were snap-frozen in liquid nitrogen. We note here that, using the procedures described above, we were unable to obtain DIPP crystals in complex with either 3-InsP7 or 6-InsP7.
Data collection, structure determination, and refinement.
Diffraction data were collected using APS beam lines 22-ID and 22-BM. All data were processed with the program HKL2000 (29). The initial complex structures of DIPP1 were determined by direct Fourier synthesis using phase calculated from the reported DIPP1 structure (PDB: 2FVV), and refined with the equivalent test sets. All other isomorphous crystals structures were determined with the same approach. The structure was manually rebuilt with COOT (30), and refined with REFMAC from the CCP4 package (30). The molecular graphics representations were prepared with the program PyMol (Schrödinger, LLC) (23). Atomic coordinates and structure factors have been deposited with the Protein Data Bank with accession codes 6WO7, 6WO8,6WO9, 6WOA, 6WOB, 6WOC, 6WOD, 6WOE, 6WOF, 6WOG, 6WOH, and 6WOI (and see Supplementary Table S1).
Isothermal Titration Calorimetry.
Calorimetry experiments were performed using a Microcal PEAQ-ITC (Malvern Panalytical) with 10 to 100 μM recombinant wild-type DIPP1 (or the indicated mutant) in the sample cell and 100 to 1600 μM of ligand in the syringe, each of which were maintained at 25 °C in buffer containing 20 mM Tris-HCl, pH 7.2, 150 mM KCl, 0.05 mM EDTA, with or without 0.8 mM MgCl2 as indicated. The 204 μL sample cell and the syringe were cleaned before each run. Thermograms were constructed from 19 injections, each of which involved 2 μL ligand delivered for 4 s, with an equilibration time of 150-300 s between each injection. The stirring speed was set to 750 rpm. More details are provided in the supplementary figures and legends. Data were fitted to a single-binding site model using the analysis software provided by the manufacturer. At least three runs were performed for each condition.
Results and Discussion
Extending the range of PP-InsP substrates for DIPP1.
Previous studies have compared human DIPP1 activities towards 1-InsP7, 5-InsP7 and 1,5-InsP8, and one of the consistent conclusions has been that the enzyme exhibits a preference for 1-InsP7 over 5-InsP7 (22, 31). That observation is in agreement with our current data: 1-InsP7 exhibits a 2-fold higher Vmax value than 5-InsP7 (p < 0.001; Figure 2). In addition, we also describe significant DIPP1 activities against all six possible InsP7 isomers; the enzyme can even accommodate the axial 2-diphosphate group (Figure 2). Clearly, the placement of the diphosphate group around the inositol ring has only moderate impact on the maximal rates of PP-InsP hydrolysis. Thus, non-canonical PP-InsP isomers (e.g., such as those in Dictyostelids with diphosphates at positions 4 and/or 6 (32)), can be expected to be readily hydrolyzed by corresponding DIPP orthologs. A further observation is that 1,5-InsP8 and 5-PP-InsP4 join 1-InsP7 in the category of preferred substrates, even though 5-PP-InsP4 is distinct in not having a 1-diphosphate.
Figure 2.
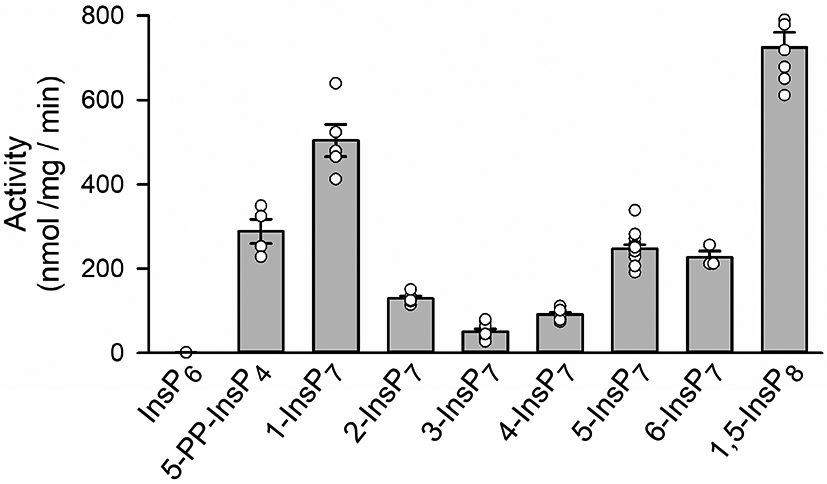
DIPP1 phosphatase activities towards various PP-InsPs. Phosphatase assays were performed as described in the Methods Section. Individual data points are shown, as well as mean values (the vertical bars) and standard errors. The 2-fold difference in rates of hydrolysis of 1-InsP7 compared to and 5-InsP7 is statistically significant (p < 0.001; determined by one-way ANOVA).
Overall topology of the DIPP1/PP-InsP crystal complex.
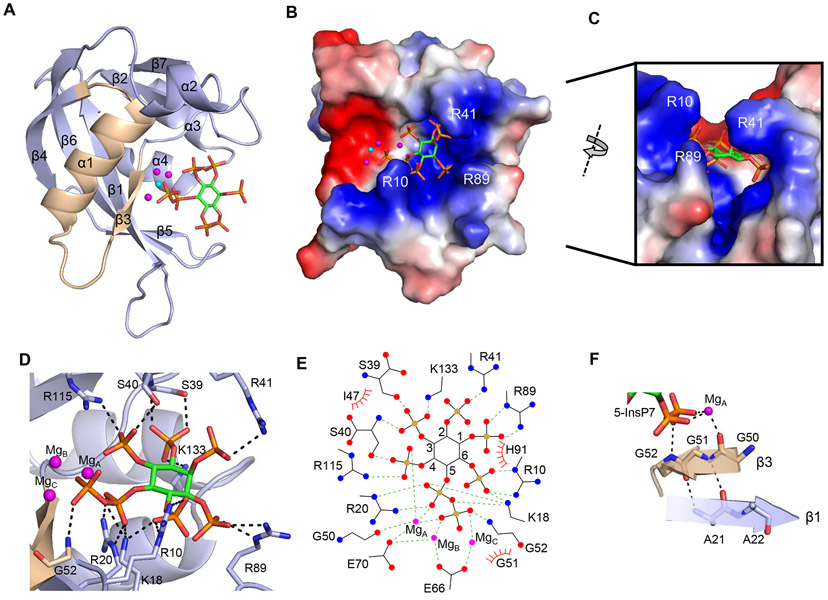
In view of the high catalytic activity of DIPP1 (Figure 2) we generated crystals in the presence of F− to inhibit phosphatase activity and preserve substrates in the complexes. Among the PP-InsPs in mammalian cells (Figure 1), 5-InsP7 is the most abundant (7); here, we show the human DIPP1 structure in complex with 5-InsP7 at 1.4 Å resolution (Figure 3A-F ; Supplementary Table S1). We have confirmed an earlier finding by Thorsell et al (18) who identified three catalytically-relevant Mg2+: MgA, MgB and MgC (Figure 3A, D, E; Figure 4A, B). In agreement with previous studies (18, 20), DIPP1 is shown to exhibit the canonical α-β-α Nudix fold, within which we picture the eponymous catalytic motif largely as a loop-helix-loop, but with contributions from flanking β−3 and β−4 strands (Figure 3A; Supplementary Figure S1). Previous studies also include some residues from the N-terminal β-strand in their structural overview of the Nudix consensus catalytic sequence (4, 5).
Figure 3.
Crystal structure of DIPP1. A, Ribbon plot of DIPP11-148 in complex with 5-InsP7. The protein’s Nudix motif is wheat-colored for emphasis. The fluoride ion is show as a cyan sphere. The three magnesium ions are shown as magenta spheres. 5-InsP7 is depicted as a stick model, with carbon colored light green, phosphorous is orange, and oxygen is red. B, Electrostatic surface plot with blue and red coloration to denote positive and negative electrostatic potentials, respectively, at physiological pH; also shown is the 5-InsP7 ligand, magnesium ions and fluoride. C, A portion of the structure in panel B is rotated, to provide a transverse view of the substrate biding furrow, topped with overhanging ridges from the sidechains of Arg10, Arg41 and Arg89. D, A description of residues (stick format) that have polar interactions with 5-InsP7 (< 3.2 Å; broken black lines) when it is positioned within the active site. E, ligplot describing all interactions between DIPP1, Mg, and 5-InsP7. F, Polar contact networks of the β3 strand that begins the Nudix motif DIPP1.
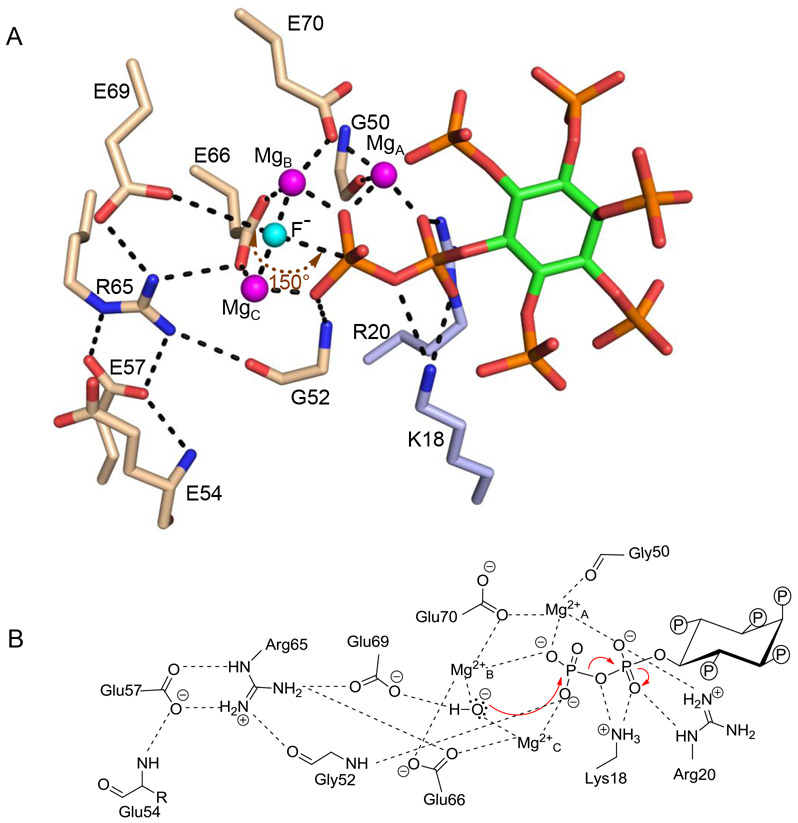
Figure 4.
Reaction mechanism for 5-InsP7 hydrolysis by DIPP1. Catalytically-relevant, polar interactions (< 3.2 Å; broken lines) between fluoride (cyan sphere), magnesium ions (magenta spheres), 5-InsP7 (stick model, carbon is light green, phosphate is orange, and oxygen is red) and amino-acid residues (nitrogen is blue, oxygen is red, and carbons are color coded to indicate Nudix-motif (wheat) and non-Nudix (blue) origins). By assuming F− occupies the precise position of the catalytic water, a 150° angle is estimated for the in-line nucleophilic attack. B, A proposed reaction mechanism for 5-InsP7 hydrolysis, including water as the nucleophile. Also depicted are long- and short-range polar contacts that we analogize as suspension cables to position substrate in the catalytic pocket.
Our results draw attention to several structural features that have escaped the attention of previous X-ray crystallographic analyses of DIPP1. For example, we argue that the entire β−3 strand, beginning with Gly50, can be considered a fundamental element of the phosphatase chemistry of the DIPP1 Nudix motif, through its coordination of a catalytically-critical Mg2+ (Figure 3A, F; Figure 4A, B). The adjacent Gly51 helps maintain the protein fold by clamping the β−3 sheet to the antiparallel β−1 sheet through backbone-to-backbone hydrogen bonds with Ala21 (Figure 3F). The absence of a side chain at Gly51 is another structural adaptation that permits the close approach of the substrate’s 5-β-phosphate and its coordination with MgC and Gly52 (Figure 3F; Figure 4A, B). These proposed structural and catalytic roles of the Gly tripeptide are validated by previous mutagenic studies (33).
Furthermore, our surface plots (Figure 3B) describe a strongly electropolarized, 17 Å-deep furrow that traverses one face of the protein. The negatively charged surface of this groove hosts Nudix motif catalytic residues - Gly50, Gly52, Glu66, and Glu70. The contiguous positively charged region fashions a PP-InsP binding pocket that, in cross-section, can be viewed as being V-shaped and topped with overhanging ridges, all largely comprised of residues with long and flexible side-chains: Arg10, Lys18, Arg20, Arg41, Arg89, His91, Arg115, and Lys133 (Figure 3B,C). Interestingly, all but Lys18 and Arg20 are scattered through various loop structures: one at the N-terminus, and those linking β2-β3, β5-β6, α2-β7 and α3-α4 (Supplementary Figure S1). Thus, substrate selectivity is enforced through a significant number of positively charged residues that contact various points around the phosphate-congested inositol ring. As such, our data provide an important exception to the proposed general rule that substrate specificity of individual Nudix enzymes is predominantly enforced by a single ‘X-loop’ (5), that corresponds to the β5-β6 loop of DIPP1 (Supplementary Figure S1).
The DIPP1 reaction mechanism.
In 2009, Thorsell et al (18) solved the structure of a DIPP1/ Mg2+/InsP6 crystal complex; in modeling studies they replaced this InsP6 with the non-physiological 3,5-InsP8 in order to propose a reaction mechanism for DIPP1-catalyzed PP-InsP hydrolysis. We set out to clarify and extend these proposals, by analyzing DIPP1 crystals in complex with biologically relevant PP-InsPs. Through a structural analysis of a DIPP1/ Mg2+/F−/5-InsP7 crystal complex (Figure 4A, B; Supplementary Figure S2), we describe a network of catalytically-relevant polar contacts that is more intense than that previously formulated (18): for example, we uncover the catalytic significance of Gly52, and additional polar contacts involving MgA and MgB. Armed with this and other new information, we put forward the view that the diphosphate of 5-InsP7 is appropriately positioned at the catalytic site by extensive series of interlocking polar contacts that we analogize as ‘suspension cables’. All are ultimately anchored by Glu54, which links to Glu57, which in turn has two contacts with Arg65: one through the latter’s Nε and another through one Nη, then Gly52, and finally the β-phosphate. The second Nη from Arg65 contacts Glu66, from which emerge two ‘cables’ of contacts, one through MgC which interacts with the β-phosphate, while the other connects Glu66 to MgB; the latter not only contacts the β-phosphate, but also links through Glu70 and MgA to coordinate with both α- and β-phosphates (Figure 4A, B). Within this series of polar interactions we position a nucleophilic water, based on its proposed usurpation by F− (a DIPP1 inhibitor (6)) during crystallization (Figure 4A, B). Our data are also consistent with this water being activated by a binuclear complex of MgB and MgC (Figure 4A, B), as initially proposed by Thorsell et al (18). Since our crystal complexes contain the natural 5-InsP7 substrate, we can also describe that the angle of attack of the putative nucleophilic water (150°; Figure 4A) is acceptably close to the optimum value of 180° for an in-line mechanism (2), and its 2.45 Å separation from the β-phosphate is within hydrogen-bonding range. Our data also support one other proposal by Thorsell et al: coordinated by Glu70 and the backbone carboxylate of Gly50, MgA promotes the scissile bond breakage (Figure 4A, B). The proposed importance to catalysis of Arg65 and Glu69 was confirmed by separately mutating each residue to Ala, which significantly decreased the specific activities towards 1-InsP7, 5-InsP7 and 1,5-InsP8 (Table 1, 2).
Table 1.
Apparent ligand binding affinities and estimated catalytic efficiencies for DIPP1
| Kd (μM) No Mg2+ |
Kd (μM) +0.8 mM Mg2+ |
Kcat sec−1 |
Kcat/KM 106 mol−1 sec−1 |
|
|---|---|---|---|---|
| 5-PP-InsP4 | 7.61 ± 1.62 | 0.251 ± 0.031 | 0.095±0.009 | 0.38 |
| 1-InsP7 | 21.97 ± 4.83 | 0.092 ± 0.007 | 0.165±0.013 | 1.79 |
| 2-InsP7 | 0.58 ± 0.07 | 0.090 ± 0.017 | 0.042±0.002 | 0.46 |
| 4-InsP7 | 4.03 ± 1.16 | 0.140 ± 0.030 | 0.029±0.002 | 0.21 |
| 5-InsP7 | 2.29 ± 0.45 | 0.124 ± 0.025 | 0.081±0.003 | 0.65 |
| 1,5-InsP8 | 4.54 ±0 .44 | 0.158 ± 0.009 | 0.237±0.011 | 1.50 |
| InsP6 | 11.16 ± 1.90 | 0.096 ± 0.006 | N/A | N/A |
| 1-InsP7+ F− | not tested | 0.065±0.006 | N/A | N/A |
| 5-InsP7+ F− | not tested | 0.087±0.020 | N/A | N/A |
Apparent binding affinities were calculated by isothermal titration calorimetry (see Methods Section and Supplemental Figure S3). These data, and the reaction rates described by Fig. 2, were used to estimate catalytic efficiencies, assuming apparent Kd = Km. N/A = not applicable, as in each case there is negligible catalytic activity (also see Fig. 2). Note that inhibition of catalysis by the addition of fluoride has little (30%) impact upon the apparent Kd determinations for two rapidly-metabolize substrates, 1-InsP7 and 5-InsP7; thus we have assumed that ligand-binding determinations are not significantly affected by the heat of the reactions.
Table 2.
Catalytic activities for Arg65Ala and Glu69Ala DIPP1 mutants
| 1-InsP7 | 5-InsP7 | 1,5-InsP8 | ||||
|---|---|---|---|---|---|---|
| nmol·mg−1·min−1 | Mutant/WT | nmol·mg−1·min−1 | Mutant/WT | nmol·mg−1·min−1 | Mutant/WT | |
| R65A | 34.9±3.1 | 0.07 | 6.8±0.6 | 0.03 | 96.5±6.1 | 0.13 |
| E69A | 19.8±1.6 | 0.04 | 2.9±0.4 | 0.01 | 53.3±2.4 | 0.07 |
Enzyme activities were measured at substrate concentrations of 10 μM in the presence of 0.8 mM Mg2+.
It has previously been argued that the Nudix motif hosts all of the catalytically-relevant DIPP1 amino-acid residues (18). In contrast, our new data indicate Lys18 stabilizes the α-phosphate, while Arg20 has catalytically-relevant bidental interactions with the α-phosphate of every substrates’ diphosphate (Figure 3, 5). It is also possible that either of these two residues could also function as a proton donor to stabilize the leaving oxygen atom (Figure 4A, B). In support of these ideas, we found that Lys18Ala and Arg20Ala mutants of DIPP1 both showed impaired catalytic efficiency (Table 1,3; Supplementary Figure S3, 4).
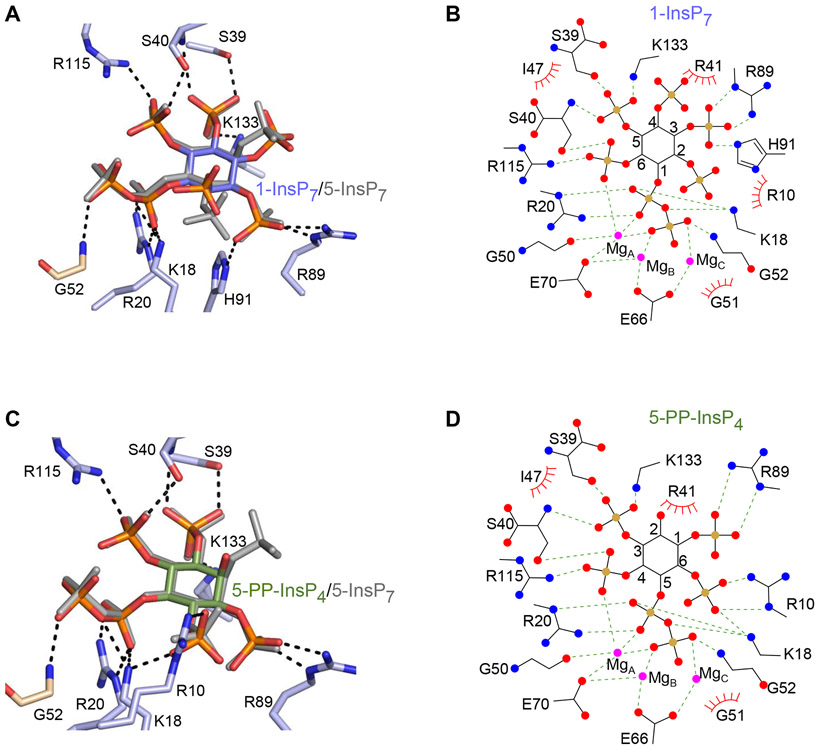
Figure 5.
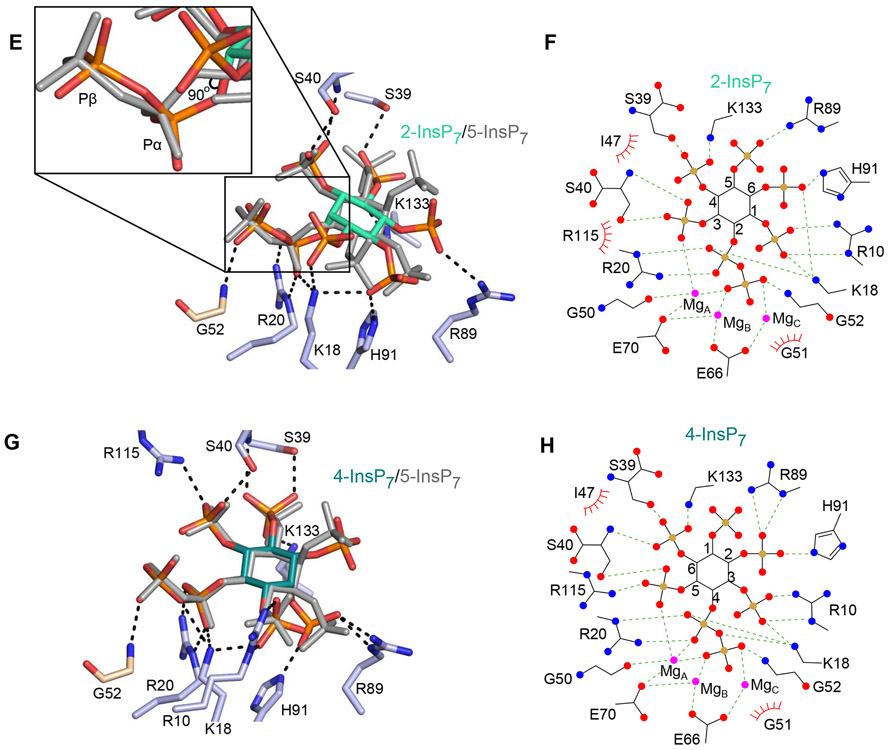
DIPP1 binding of multiple PP-InsP ligands. Descriptions of amino acid residues (stick format) that have polar interactions with the indicated PP-InsP (< 3.2 Å; broken lines). All data were obtained by either co-crystallization of protein and ligand, or by ligand soaking into preformed protein crystals. Residues belonging to the Nudix motif are wheat-colored. Nitrogen is colored blue, oxygen is red and phosphorous is orange. Inositol ring carbons are color-coded as described below; the positions of each PP-InsP ligand are shown relative to 5-InsP7, which is depicted in light gray. Corresponding color-colored ligplots describe interactions between DIPP1, Mg (colored magenta), and the indicated PP-InsP; inositol carbons are numbered, polar contacts are denoted as broken lines, and Van der Waals contacts are shown as “eyelash” graphics. A, 1-InsP7 (slate blue carbons) and B, corresponding ligplot. C, 5-PP-InsP4 (avocado green carbons) and D, corresponding ligplot. E, 2-InsP7 (turquoise carbons), with magnified inset to 90° displacement of the C-O bond relative to that of 5-InsP7 and F, corresponding ligplot. G, 4-InsP7 (teal carbons) and H, corresponding ligplot. See Supplementary Figure 2A,B for simulated annealing omit difference maps.
Table 3.
Activities and ligand binding affinities for DIPP1 mutants
| 1-InsP7 | 5-InsP7 | 1,5-InsP8 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| nmol· mg−1· min−1 |
Kd
(μM) |
Kcat/KM 106 mol−1 sec−1 |
nmol· mg−1· min−1 |
Kd
(μM) |
Kcat/KM 106 mol−1 sec−1 |
nmol· mg−1· min−1 |
Kd
(μM) |
Kcat/KM 106 mol−1 sec−1 |
|
| WT | 504±38 | 0.092±0.007 | 1.79 | 247±10 | 0.124±0.025 | 0.65 | 725±36 | 0.158±0.009 | 1.50 |
| K18A | 480±37 | 2.12±0.032 | 0.07 | 281±32 | 2.48±0.25 | 0.04 | 826±26 | 4.99±0.62 | 0.05 |
| R20A | 30.6±3.5 | Nd | Nd | 9.2±0.7 | Nd | Nd | 84.6±1.6 | Nd | Nd |
| R41A | 907±109 | 0.43±0.11 | 0.69 | 295±67 | 0.42±0.06 | 0.23 | 824±51 | 0.46±0.05 | 0.59 |
| H42A | 600±56 | 0.29±0.05 | 0.68 | 306±25 | 0.67±0.08 | 0.15 | 804±66 | 0.51±0.10 | 0.52 |
| R45A | 761±78 | 0.29±0.03 | 0.86 | 352±25 | 0.39±0.05 | 0.30 | 594±40 | 0.36±0.05 | 0.54 |
| H42A + R45A | 658±28 | 1.07±0.07 | 0.20 | 314±10 | 1.3±0.16 | 0.08 | 356±29 | 1.06±0.09 | 0.11 |
Enzyme activities were measured at substrate concentrations of 10 μM in the presence of 0.8 mM Mg2+. Binding affinities were calculated by isothermal titration calorimetry in the presence of 0.8 mM Mg2+. Nd=Not detected due to insufficient heat generation (Figure S4).
Alternate binding modes for various PP-InsP substrates.
We next sought to rationalize the high reactivity of DIPP1 towards multiple PP-InsPs (see Figure 2). To this end, we obtained DIPP1 crystals in complexes with two additional human PP-InsPs, 1-InsP7, and 5-PP-InsP4 (Figure 5 A-D), and two non-natural PP-InsPs, 2-InsP7 and 4-InsP7 (Figure 5 E-H). Each of these are novel structural complexes, except for DIPP1/1-InsP7 (see 6PCK and (20)). However, 6PCK does not identify either the nucleophilic water or its F− mimic, and the 2Fo-Fc electron density maps, contoured at 1.0 σ, were only sufficient to identify one of the three magnesium atoms that we clearly defined.
Despite our using F− to inhibit DIPP1, residual phosphatase activity was such that during our attempts to form DIPP1/1,5-InsP8 crystal complexes, the substrate was dephosphorylated to 1-InsP7 (Supplementary Figure S2A). Nevertheless, this orientation in the catalytic site of 1-InsP7 derived from 1,5-InsP8 can be observed from its omit map to be virtually identical to that for a DIPP1/1-InsP7 complex that was independently derived by directly soaking this substrate into the crystals only a minor difference in the orientation of the 4-phosphate was observed (Supplementary Figure S2A);. This variability is not catalytically relevant, as in neither orientation does the 4-phosphate form polar interactions with any amino-acid residues in DIPP1 (Figure 5A).
Using 5-InsP7 as a reference point, we overlaid the positions of the other PP-InsPs in their enzyme/crystal complexes. We observed that the pyrophosphate groups and each of the inositol rings are also very close to superimposable (Figure 5A-H ). This situation partly arises out of the mobility of amino acid side chains that participate in substrate recognition, but also relevant is a unique stereochemical property of the inositol ring: its 2-5 axis of symmetry. This is the basis for a long-standing hypothesis (34) that alternate inositol phosphates can be accepted into a particular protein’s binding pocket in alternate orientations (or ‘‘modes’’); one inositol phosphate mimics another simply through a presentation of its obverse orientation and/or a rotation of the inositol ring, so as to offer to the docking site some essentially similar recognition features, in a manner that maintains the ring’s overall staggered-chair conformation. Nevertheless, we were surprised that the axial 2-diphosphate (i.e., that in 2-InsP7) can be accommodated into the active site of DIPP1 (Figure 5E, F). A 180° vertical ring flip and a 22° rotation are sufficient to coincide the relative positions of the enzyme-bound diphosphate groups in 2- and 5-InsP7. The individual C-O bonds of the two diphosphate groups are 90° displaced, but in 2-InsP7 the β-phosphate adopts a syn conformation relative to the α-phosphate rather than the anti conformation observed for the other PP-InsPs, which enables the two β-phosphates to rotate back into virtually the same relative positions in the catalytic site (Figure 5E).
One other point of note is that His91 participates in binding of 1-InsP7 through polar contacts with the 3-phosphate (Figure 5A). These interactions are much stronger than the Van der Waals interactions of His91 with 5-InsP7 (Figure 3E). This observation is consistent with a previous report that the His91Leu DIPP1 mutant exhibits substantially decreased activity towards 1,5-InsP8, yet 5-InsP7 hydrolysis was only mildly reduced (33). Other than that, the residues that participate in substrate recognition are largely the same for each ligand (Figure 5E, F). Moreover, for each of our DIPP1/PP-InsP complexes, there is conservation of positioning of each substrate, the metal cofactors, the nucleophilic water, and hence the bonding geometries with key catalytic residues (Figure 5E, F, and see PDB files listed in Supplementary Table S1). Consequently, similar reaction kinetics can be anticipated for every one of these PP-InsPs. Indeed, the specificity constants (Kcat/KM) for each of these PP-InsP substrates varied within only an 8.5-fold range (Table 1; Supplementary Figure S3). We hypothesize that the selection pressure has been for this family of enzymes to evolve with high activity, rather than substrate selectivity.
Pre-reaction states identified with PCP-InsPs
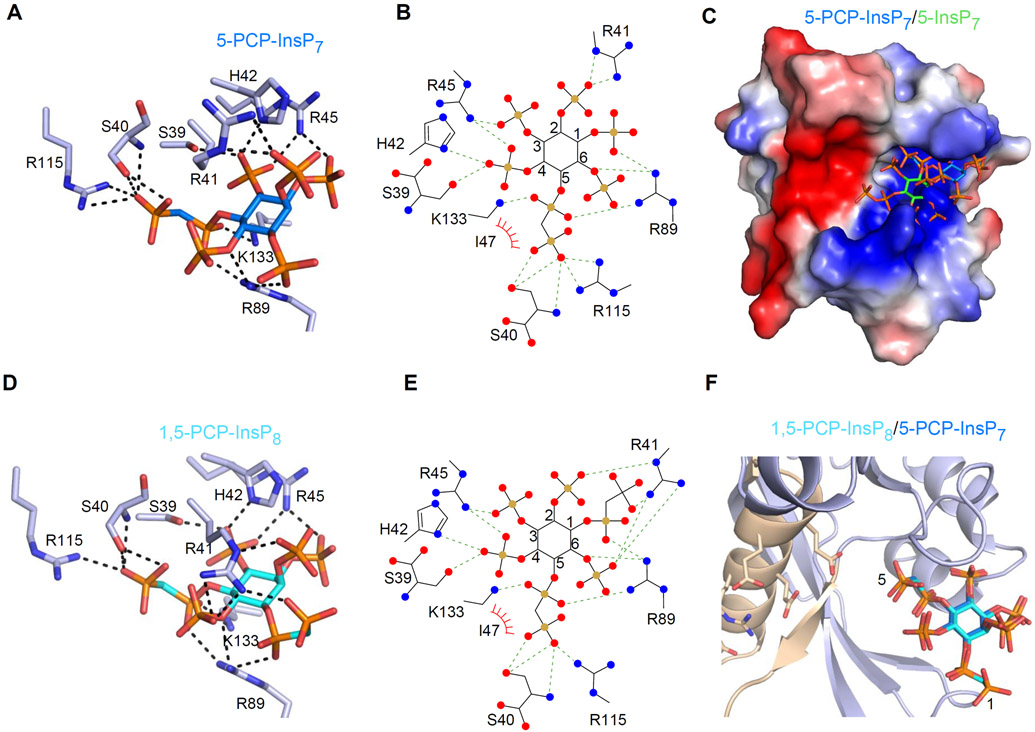
We have previously described structural analyses of the PPIP5K2 kinase domain (see Figure 1) in complex with metabolically-stable PP-InsP bioisosteres; this approach led us to identify pre-reaction enzyme states (27). We have now derived and analyzed the first DIPP1 crystal complexes that contain either of two methylene bisphosphonate analogues i.e., 5-PCP-InsP7 or 1,5-PCP-InsP8 (Figure 6A-F; Supplementary Figure S2B). These analogues are known not to be hydrolyzed by DIPP1 (22, 35). Interestingly, the positions of these two analogues within the crystals are virtually superimposable (Figure 6F); we calculated the RMSD for the two ligands to be only 0.5 Å. Their inositol rings are 5.3 Å more distal from the active site compared to 5-InsP7, and also rotated by 42° (Figure 6C). We hypothesize that these complexes correspond to pre-reaction states, perhaps reflecting substrate capture from the bulk phase by the process of electrotrostatic steering (36). These residues include His42 and Arg45 (Figure 6A, B, D, E), which hitherto were not known to play any role in substrate binding. A third residue, Arg41, also has polar interactions with the PCP-InsPs (Figure 6A, B, D, E), in addition to its ionic and Van der Waals interactions with natural substrates in the catalytic pocket (Figure 3E; 5B, D). It is of further note that the 5-PCP group of both ligands is oriented towards the active site by polar interactions with Ser40, Lys133 and Arg115. These three residues have an alternate function - binding to the 3- and 4-phosphates - once the 5-InsP7 substrate has fully entered the catalytic pocket.
Figure 6.
Interactions of DIPP1 with 5-PCP-InsP7 and 1,5-PCP-InsP8. A and B, DIPP1/5-PCP-InsP7 interactions in cartoon and ligplot formats, respectively. Residues are shown in as light blue sticks. Nitrogen is dark blue, oxygen is red, phosphorous is orange, and inositol ring carbons are azure in the cartoon, and black (with carbons numbered) in the ligplot. Polar contacts are denoted as broken lines; Van der Waals contacts are shown as “eyelash” graphics. C, The positions of 5-PCP-InsP7 and 5-InsP7 (green inositol ring) are superimposed upon an electrostatic surface plot, with blue and red coloration to denote positive and negative electrostatic potentials, respectively, at physiological pH. D and E, DIPP1/1,5-PCP-InsP8 interactions in cartoon and ligplot formats, respectively, color coded as in panel A, except the inositol ring in the cartoon is colored cyan. F, superimposition of 5-PCP-InsP7 and 1,5-PCP-InsP8. See Supplementary Figure 2A for simulated annealing omit difference maps.
It is intriguing that the crystal complexes with the PP-InsP analogues, 5-PCP-InsP7 or 1,5-PCP-InsP8, do not contain Mg2+ ions (Figure 6A-F), even though the cations were present in the crystallization buffers. It has previously been noted (37) that in cases where highly polar and naturally occurring inositol polyphosphates such as InsP6 have been captured in crystal complexes with non-catalytic, ligand binding proteins, no coordinating cations are present. Thus, it can be considered unsurprising that the putative process of ligand capture by electrotrostatic steering also does not appear to involve Mg2+. It follows that the catalytic requirement for Mg2+ coordination with the α- and β-phosphates of PP-InsPs must be satisfied at a later stage of the reaction. Perhaps subtle differences in pKa values and dihedral angles of the natural pyrophosphate moiety vis-a-vis the PCP group (38-40) are sufficient to reduce the latter’s complexation with Mg2+ and/or impair the analogues’ transfer into the catalytic pocket.
We mutated to Ala two of the residues which we hypothesize to specifically participate in substrate capture (His42, Arg45), which resulted in modest (2 to 5-fold) decreases in binding affinities for natural substrates (Table 1, 3; Supplementary Figure S4). These individual effects were more than additive in the His42Ala plus Arg45Ala double mutant (Table 1,3; Supplementary Figure S4). The Arg41Ala DIPP1 mutant also exhibited decreased substrate binding affinity, consistent with our structural data that indicate this residue has roles in both substrate capture and catalysis (see above).
Capture of multiple product states in crystallo.
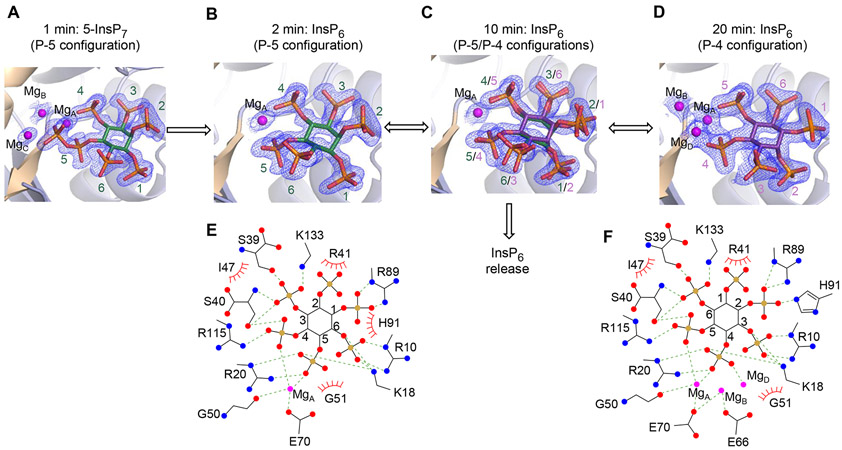
Having identified pre-reactions state (see above), in our final experiments in this study we investigated the characteristics of the final product-bound stages. To accomplish this, we transferred DIPP1/5-InsP7 crystals into F−-free buffer, and then performed a time-resolved analysis by flash-freezing samples at various intervals (41).
The buffer transfer led to a very rapid depletion of fluoride from within the catalytic site (Figure 7A, B), and consequently the 5-phosphoric anhydride was hydrolysed. There was an accompanying loss of electron density from two of the three catalytic Mg2+ ions, presumably reflecting their increased mobility. The InsP6 configuration in these crystals is designated as P-5, reflecting the retention of the position of this monophosphate relative to the 5-α-phosphate of 5-InsP7. We performed longer times of fluoride depletion, from a few minutes to several hours, and crystal complexes were observed in which the InsP6 product was observed in two configurations: in addition to P-5, we observed a configuration designated as P-4, representing a 180° ring flip across the 3-6 axis, relative to P-5 (Figure 7C). These data confirm the earlier study by Thorsell et al (18), although the relative positions of our Mg2+ ions are slightly different. Presumably, the P-5 InsP6 must at least partly exit the catalytic site to undergo the ring-flip, and there may be a critical point at which there are two competing outcomes: either InsP6 exit from the protein (as proposed in Figure 7C), or rebinding in the P-4 configuration. .
Figure 7.
Identification and analysis of post-reaction DIPP1/InsP6 crystal complexes. A through D, DIPP1/5-InsP7/F− crystal complexes were placed in F−-free buffer and then snap frozen at the following times: 1 min (A), 2 min (B), 10 min (C) and 20 min (D). Data similar to those in panel C were obtained at 3 min and 7 min; data similar to those in panel D were obtained at 1 hr, 2 hr and 15 hr. The 5-InsP7 and InsP6 are shown in stick models, in which oxygen is red, phosphorous is orange, and inositol ring configurations are named P-5 (green) or P-4 (purple), based on the which phosphate has the closest relative position to that of the 5-α-phosphate of 5-InsP7. Magnesium is colored magenta. The 2Fo-Fc electron density maps are contoured at 1.0 σ. E and F are the ligplots that correspond to the cartoons shown in panels B and D, respectively. Polar contacts are denoted as broken lines; Van der Waals contacts are shown as “eyelash” graphics.
The P-4 configuration is sustained by more polar contacts compared to those for P-5 (Figure 7E, F); in the P-4 configuration the 2-phosphate has polar contact with His91, two additional Mg atoms form stabilizing interactions, and Glu66 makes also a contribution to ligand binding (Figure 7E, F). Thus, we hypothesize that the P-4 state may be the more long-lived product-bound inhibitory condition. Moreover, we have determined the Kd for InsP6 binding to DIPP1 in the presence and absence of Mg2+. The values are within the range of those for individual PP-InsPs (Table 1). Consistent with the structural data (Figure 7A, B, C, D), Mg2+ substantially increases the potency of InsP6 binding (Table 1).
Concluding comments
We have gained considerable structure-based insight into the reaction cycle for DIPP1-catalyzed PP-InsP hydrolysis, through our atomic-level analysis of a comprehensive range of protein crystals in complex with a variety of PP-InsP and PCP-InsP ligands. For example, we have substantially improved our knowledge of the determinants of substrate recognition and the catalytic mechanism (Figures 3-5), including our identification of the proton donor necessary to complete phosphate hydrolysis. Additionally, the DIPP1/PCP-InsP crystal complexes (Figure 6) have offered insight into putative pre-reaction states which have led us to propose an important role for several amino-acid residues in electrostatic steering for substrate capture. Some of these particular residues form ridges that overhang the substrate-binding pocket; these structural elements, that have not been noted in previous work, could also be important physical constraints for trapping bound substrate. Our new cryo-trapping data also reveal two separate DIPP1/InsP6 post-reaction states (Figure 7), one of which may serve as a hitherto-unsuspected, and relatively long-lived inhibitory condition. Taken together, our studies describe a reaction cycle of surprising complexity for such a relatively small catalytic core structure (<16 kDa).
Another significant observation is that our crystallization conditions captured DIPP1 in complex with 1-InsP7, but not 3-InsP7 (see Methods Section). This observation is relevant to a recent study by Dollins et al (20), who described DIPP1 crystals in complex with the reaction product of InsP6 phosphorylation by the Saccharomyces cerevisiae Vip1 (the PPIP5K ortholog); their X-ray crystallographic data diagnosed the reaction product as 1-InsP7. However, Dollins et al (20) cautiously noted that X-ray crystallographic data alone is not sufficient to exclude 3-InsP7 from being an additional reaction product. Our data underscore this point by showing that the absence of 3-InsP7 in DIPP1 could have reflected the intractable nature of such crystal complexes, rather than 3-InsP7 not being a significant Vip1 reaction product. Hence it is valuable that other experimental approaches consistently demonstrate that Vip1/PPIP5Ks synthesize 1-InsP7 and 1,5-InsP8 (20, 21).
Observations of interest to the wider Nudix field include our description of the functional significance of G1 (i.e., Gly51) in the Nudix motif, GX5EX5UXREX2EEXGU. This residue helps maintain the protein fold by clamping the β−3 sheet to the antiparallel β−1 sheet through backbone-to-backbone hydrogen bonds with Ala21 (Figure 3F). Additionally, the absence of a side chain at Gly51 permits the close approach of the substrate’s 5-β-phosphate and its coordination with Mg and Gly52 (Figure 3F). Nevertheless, another residue, Gly50, plays a more direct role in catalysis by coordination of MgA (Figure 4A). In the context that the sequence motif is recognized to create a versatile scaffold for metal binding and basic hydrolase chemistry (4), we propose the motif criteria be relaxed beyond its typical 23-residue restriction, so as to include DIPP’s Gly50 (Supplementary Figure S1). Such a strategy has wider applicability. For example, a metal-ion binding Gly residue also lies one residue N-terminal to the Nudix consensus sequence in both the ADP-ribose pyrophosphatases from Methanococcus jannaschii (42) and Enterobacterial GDP-Mannose Glycosyl Hydrolase (43).
Our data are also pertinent to previous biochemical assays that led to the conclusion dephosphorylation of 1,5-InsP8 by human DIPPs predominantly proceeds by hydrolysis of the 5-phosphoanhydride group (31). For example, we discovered that hydrolysis of 1,5-InsP8 in crystallo yields 1-InsP7. Moreover, the configuration of 1,5-PCP-InsP8 in our crystal complex is shown to orient the 5-diphosphate group towards the active site; the plausibility that this is indeed a catalytically-relevant substrate conformation is enhanced by our demonstration it is superimposable upon the orientation of 5-PCP-InsP7, which by definition must have the 5-β-phosphate directed into the active site.
The conclusion that 1,5-InsP8 predominantly undergoes 5-β-phosphate hydrolysis is a centerpiece of the proposed cyclical pathway of 1,5-InsP8 synthesis and metabolism, which we have proposed to be of significance to enzymatic control of PP-InsP signaling (Figure 1 and (7)). Fungi and plants reinforce this selective pathway of 1,5-InsP8 metabolism by expressing a separate, 5-β-phosphate specific PP-InsP phosphatase (23, 44). A human ortholog of that phosphatase has not been found; it is intriguing to speculate that the selectivity of DIPPs towards the 5-β-phosphate may have contributed to loss of a separate 1,5-InsP8 5-phosphatase activity during animal evolution.
Supplementary Material
Acknowledgements:
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, and the German Research Foundation (DFG) under Germany’s Excellence Strategy (CIBSS – EXC-2189 – Project ID 390939984). H. Jessen acknowledges financial support from the German Research Foundation (DFG, Grant JE 572/4-1). S. Hostachy acknowledges financial support from the Swiss National Foundation Sinergia Grant CRSII5_170925. We are grateful to the NIEHS Collaborative crystallography group, and the Advanced Photon Source (APS) SE Regional Collaborative Access Team (SER-CAT) 22-ID and 22-BM beam lines, for assistance with crystallographic data collection. All authors declare there is no conflict of interest.
Non-standard Abbreviations:
The literature in this field has a divergent usage of abbreviations to denote individual inositol pyrophosphates and their analogues, accompanied by dueling opinions as to which system is the more accessible to a multidisciplinary audience; one is systematic in nature, whereas the other is more colloquial. The latter is the approach taken in the text of the current study, except at first usage, whereupon the corresponding systematic abbreviation follows in parentheses, i.e.,
- 1-InsP7 (1-PP-InsP5)
1-diphosphoinositol 2,3,4,5,6-pentakisphosphate
- 2-InsP7 (2-PP-InsP5)
2-diphosphoinositol 1,3,4,5,6-pentakisphosphate
- 3-InsP7 (3-PP-InsP5)
3-diphosphoinositol 1,2,4,5,6-pentakisphosphate
- 4-InsP7 (4-PP-InsP5)
4-diphosphoinositol 1,2,3,5,6-pentakisphosphate
- 5-InsP7 (5-PP-InsP5)
5-diphosphoinositol 1,2,3,4,6-pentakisphosphate
- 6-InsP7 (6-PP-InsP5)
6-diphosphoinositol 1,2,3,4,5-pentakisphosphate
- 1,5-InsP8 (1,5-[PP]2-InsP4)
1,5-bis-diphosphoinositol 2,3,4,6-tetrakisphosphate
- 5-PP-InsP4 (also 5-PP-InsP4)
5-diphosphoinositol 1,3,4,6-tetrakisphosphate
- 5-PCP-InsP7 (5-PCP-InsP5)
5-methylene-diphosphonate inositol 1,2,3,4,6-pentakisphosphate
- 1,5-PCP-InsP8 (1,5-[PCP]2-InsP4)
1,5-bismethylene-diphosphonate inositol 2,3,4,6-tetrakisphosphate.
Glossary
Other abbreviations:
- PP-InsPs
diphosphoinositol polyphosphates
- Ins(1, 3, 4,5, 6)P5
inositol 1,3,4,5,6-pentakisphosphate
- InsP6
inositol 1,2,3,4,5,6-hexakisphosphate
- InsP5
inositol 1,3,4,5,6-pentakisphosphate.
References
- 1.Kramer S, and McLennan AG (2019) The complex enzymology of mRNA decapping: Enzymes of four classes cleave pyrophosphate bonds. Wiley Interdiscip Rev RNA 10, e1511. [DOI] [PubMed] [Google Scholar]
- 2.Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang LW, and Amzel LM (2005) Structures and mechanisms of Nudix hydrolases. Arch. Biochem Biophys 433, 129–143 [DOI] [PubMed] [Google Scholar]
- 3.Bessman MJ, Frick DN, and O’Handley SF (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. jbc 271, 25059–25062 [DOI] [PubMed] [Google Scholar]
- 4.Gabelli SB, Bianchet MA, Bessman MJ, and Amzel LM (2001) The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nature Structural Biology 8, 467–472 [DOI] [PubMed] [Google Scholar]
- 5.Srouji JR, Xu A, Park A, Kirsch JF, and Brenner SE (2017) The evolution of function within the Nudix homology clan. Proteins 85, 775–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Safrany ST, Caffrey JJ, Yang X, Bembenek ME, Moyer MB, Burkhart WA, and Shears SB (1998) A novel context for the “MutT” module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J 17, 6599–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shears SB (2018) Intimate Connections: Inositol Pyrophosphates at the Interface of Metabolic Regulation and Cell-Signaling. J. Cell Physiol 233, 1897–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caffrey JJ, Safrany ST, Yang X, and Shears SB (2000) Discovery of Molecular and Catalytic Diversity Among Human Diphosphoinositol Polyphosphate Phosphohydrolases: An Expanding NUDT Family. jbc 275, 12730–12736 [DOI] [PubMed] [Google Scholar]
- 9.Hua LV, Hidaka K, Pesesse X, Barnes LD, and Shears SB (2003) Paralogous murine Nudt10 and Nudt11 genes have differential expression patterns but encode identical proteins that are physiologically competent diphosphoinositol polyphosphate phosphohydrolases. Biochem J 373, 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leslie NR, McLennan AG, and Safrany ST (2002) Cloning and characterization of hAps1 and hAps2, human diadenosine polyphosphate-metabolizing Nudix hydrolases. BMC Biochemistry 3, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winward L, Kilari R, and Safrany S (2020) Characterizing Enzymes of the Diphosphoinositol Polyphosphate Phosphohydrolase (DIPP) Family. Vol. 2091 pp. 75–82 [DOI] [PubMed] [Google Scholar]
- 12.Williams MJ, Eriksson A, Shaik M, Voisin S, Yamskova O, Paulsson J, Thombare K, Fredriksson R, and Schiöth HB (2015) The Obesity-Linked Gene Nudt3 Drosophila Homolog Aps Is Associated With Insulin Signaling. Molecular Endocrinology 29, 1303–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winward L, Whitfield WG, McLennan AG, and Safrany ST (2010) Oxidation of the diphosphoinositol polyphosphate phosphohydrolase-like Nudix hydrolase Aps from Drosophila melanogaster induces thermolability--A possible regulatory switch? Int J Biochem Cell Biol 42, 1174–1181 [DOI] [PubMed] [Google Scholar]
- 14.Singh AN, and Gasman B (2020) Disentangling the genetics of sarcopenia: prioritization of NUDT3 and KLF5 as genes for lean mass & HLA-DQB1-AS1 for hand grip strength with the associated enhancing SNPs & a scoring system. BMC Medical Genetics 21, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.York SJ, Armbruster BN, Greenwell P, Petes TD, and York JD (2005) Inositol diphosphate signaling regulates telomere length. J. Biol. Chem 280, 4264–4269 [DOI] [PubMed] [Google Scholar]
- 16.Saiardi A, Resnick AC, Snowman AM, Wendland B, and Snyder SH (2005) Inositol pyrophosphates regulate cell death and telomere length via PI3K-related protein kinases. pnas 102, 1911–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Randall TA, Gu C, Li X, Wang H, and Shears SB (2020) A two-way switch for inositol pyrophosphate signaling: Evolutionary history and biological significance of a unique, bifunctional kinase/phosphatase. Adv Biol Regul 75, 100674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorsell AG, Persson C, Graslund S, Hammarstrom M, Busam RD, and Hallberg BM (2009) Crystal structure of human diphosphoinositol phosphatase 1. Proteins 77, 242–246 [DOI] [PubMed] [Google Scholar]
- 19.Thomas MP, and Potter BVL (2013) The enzymes of human diphosphoinositol metabolism. FEBS Journal 281, 14–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dollins DE, Bai W, Fridy PC, Otto JC, Neubauer JL, Gattis SG, Mehta KPM, and York JD (2020) Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling. Proc Natl Acad Sci U S A 117, 9356–9364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Falck JR, Hall TM, and Shears SB (2012) Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol 8, 111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu M, Chong LS, Capolicchio S, Jessen HJ, Resnick AC, and Fiedler D (2014) Elucidating Diphosphoinositol Polyphosphate Function with Nonhydrolyzable Analogues. Angew. Chem. Int. Ed Engl 53, 9508–9511 [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Gu C, Rolfes RJ, Jessen HJ, and Shears SB (2018) Structural and biochemical characterization of Siw14: A protein-tyrosine phosphatase fold that metabolizes inositol pyrophosphates. J Biol Chem 293, 6905–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capolicchio S, Wang H, Thakor DT, Shears SB, and Jessen HJ (2014) Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation. Angew. Chem. Int. Ed Engl 53, 9508–9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capolicchio S, Thakor DT, Linden A, and Jessen HJ (2013) Synthesis of Unsymmetric Diphospho-Inositol Polyphosphates. Angew. Chem. Int. Ed Engl 52, 6912–6916 [DOI] [PubMed] [Google Scholar]
- 26.Gerasimaite R, Pavlovic I, Capolicchio S, Hofer A, Schmidt A, Jessen HJ, and Mayer A (2017) Inositol pyrophosphate specificity of the SPX-dependent polyphosphate polymerase VTC. ACS Chem. Biol 12, 648–653 [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Godage HY, Riley AM, Weaver JD, Shears SB, and Potter BVL (2014) Synthetic Inositol Phosphate Analogs Reveal that PPIP5K2 Has a Surface-Mounted Substrate Capture Site that Is a Target for Drug Discovery. Chemistry & Biology 21, 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlovic I, Thakor DT, and Jessen HJ (2016) Synthesis of 2-diphospho-myo-inositol 1,3,4,5,6-pentakisphosphate and a photocaged analogue. Org. Biomol. Chem 14, 5559–5562 [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, and Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30.Tong J, Pei J, Otwinowski Z, and Grishin NV (2015) Refinement by shifting secondary structure elements improves sequence alignments. Proteins 83, 411–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilari RS, Weaver JD, Shears SB, and Safrany ST (2013) Understanding inositol pyrophosphate metabolism and function: Kinetic characterization of the DIPPs. FEBS Lett 587, 3464–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laussmann T, Pikzack C, Thiel U, Mayr GW, and Vogel G (2000) Diphospho-myo-inositol phosphates during the life cycle of Dictyostelium and Polysphondylium. ejb 267, 2447–2451 [DOI] [PubMed] [Google Scholar]
- 33.Yang X, Safrany ST, and Shears SB (1999) Site-directed mutagenesis of DIPP, a dual specificity MutT/Nudix-type hydrolase that attacks diadenosine polyphosphates and diphosphoinositol polyphosphates. jbc 274, 35434–35440 [DOI] [PubMed] [Google Scholar]
- 34.Wilcox RA, Safrany ST, Lampe D, Mills SJ, Nahorski SR, and Potter BVL (1994) Modification at C2 of myo-inositol 1,4,5-trisphosphate produces inositol trisphosphates and tetrakisphosphates with potent biological activities. Eur. J. Biochem 223, 115–124 [DOI] [PubMed] [Google Scholar]
- 35.Riley AM, Unterlass J, Konieczny V, Taylor CW, Helleday T, and Potter BVL (2018) A synthetic diphosphoinositol phosphate analogue of inositol trisphosphate. MedChemComm 9, 1105–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wade RC, Gabdoulline RR, Ludemann SK, and Lounnas V (1998) Electrostatic steering and ionic tethering in enzyme-ligand binding: insights from simulations. Proc. Natl. Acad. Sci. U. S. A 95, 5942–5949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson MS, Jessen HJ, and Saiardi A (2019) The inositol hexakisphosphate kinases IP6K1 and −2 regulate human cellular phosphate homeostasis, including XPR1-mediated phosphate export. J Biol Chem 294, 11597–11608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elliott TS, Slowey A, Ye Y, and Conway SJ (2012) The use of phosphate bioisosteres in medicinal chemistry and chemical biology. Med. Chem. Commun 3, 735–751 [Google Scholar]
- 39.Engel R (1977) Phosphonates as analogues of natural phosphates. Chemical Reviews 77, 349–367 [Google Scholar]
- 40.Flohr A, Aemissegger A, and Hilvert D (1999) alpha-Functionalized phosphonylphosphinates: synthesis and evaluation as transcarbamoylase inhibitors. J Med Chem 42, 2633–2640 [DOI] [PubMed] [Google Scholar]
- 41.Gao Y, and Yang W (2016) Capture of a third Mg(2)(+) is essential for catalyzing DNA synthesis. Science 352, 1334–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabelli SB, Bianchet MA, Ohnishi Y, Ichikawa Y, Bessman MJ, and Amzel LM (2002) Mechanism of the Escherichia coli ADP-ribose pyrophosphatase, a Nudix hydrolase. Biochemistry 41, 9279–9285 [DOI] [PubMed] [Google Scholar]
- 43.Gabelli SB, Bianchet MA, Azurmendi HF, Xia Z, Sarawat V, Mildvan AS, and Amzel LM (2004) Structure and Mechanism of GDP-Mannose Glycosyl Hydrolase, a Nudix Enzyme that Cleaves at Carbon Instead of Phosphorus. Structure 12, 927–935 [DOI] [PubMed] [Google Scholar]
- 44.Steidle EA, Chong LS, Wu M, Crooke E, Fiedler D, Resnick AC, and Rolfes RJ (2016) A novel inositol pyrophosphate phosphatase in Saccharomyces cerevisiae: Siw14 selectively cleaves the beta-phosphate from 5-diphosphoinositol pentakisphosphate (5PP-IP5). J. Biol. Chem 291, 6772–6783 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.