Abstract
Severe acquired respiratory syndrome coronavirus 2 (SARS-CoV-2) rapidly spread worldwide and acquired multiple mutations in its genome. Orf3a, an accessory protein encoded by the genome of SARS-CoV-2, plays a significant role in viral infection and pathogenesis. In the present in-silico study, 15,928 sequences of Orf3a reported worldwide were compared to identify variations in this protein. Our analysis revealed the occurrence of mutations at 173 residues of Orf3a protein. Subsequently, protein modelling was performed that revealed twelve mutations which can considerably affect the stability of Orf3a. Among the 12 mutations, three mutations (Y160H, D210Y and S171L) also lead to alterations in secondary structure and protein disorder parameters of the Orf3a protein. Further, we used predictive tools to identify five promising epitopes of B-cells, which resides in the mutated regions of Orf3a. Altogether, our study sheds light on the variations occurring in Orf3a that might contribute to alteration in protein structure and function.
Keywords: SARS-CoV-2, Mutations, Orf3a, Protein stability, Protein disorder, B-cell epitopes
Highlights
-
•
The Open reading frame 3a (Orf3a) protein acquired mutations as it spread to new locations.
-
•
The Orf3a protein harbours 173 mutations.
-
•
These mutation alters secondary structure, protein disorder parameters and also affects Orf3a protein dynamicity.
-
•
The high rank B-cell epitopes resides in the mutated regions of Orf3a that might help the virus to evade immune system.
1. Introduction
The severe acquired respiratory syndrome coronavirus-2 (SARS-CoV-2), the etiological agent of coronavirus disease 19 (COVID-19), is an RNA virus that induces mild to severe respiratory distress in infected individuals [[1], [2], [3]]. The disease, started from wet seafood market area of Wuhan province (China), has now affected 218 countries leading to a global pandemic threat with severe implications on healthcare system worldwide [4]. As of January 15, 2021, the SARS-CoV-2 have already infected more than 90 million people worldwide and caused about two million deaths.
The genome of SARS-CoV-2 is comprised of a single-stranded positive sense RNA, about 30 kb in length [5]. It contains 29 open reading frames (Orfs) that encode four structural, sixteen non-structural and nine accessory proteins [6]. Orf3a is the largest accessory protein of 275 amino acids in SARS-CoV-2 [7] which is involved in critical steps of viral infection cycle and is required for viral replication, and assembly that determines virulence of SARS-CoV-2 [8]. Structurally, this protein is a multi-pass membrane protein that forms a homotetrameric viroporin with TRAF, ion channel and caveolin binding domain [8]. Functionally, Orf3a has been demonstrated to impact host immune system by activating pro-IL-1β gene expression as well as IL-1β secretion that eventually activates NF-kB signalling and NLRP3 inflammasome and contributes to the generation of cytokine storm [9,10]. A recent analysis of human protein interactome revealed that Orf3a interacts with TRIM59 (an E3 ubiquitin ligase) to regulate antiviral innate immune signalling [11]. Altogether, Orf3a is directly involved in pathogenesis of SARS coronaviruses and also acts as an important immune modulator.
The global sequencing efforts of the SARS-CoV-2 genome from different countries revealed that its genome is rapidly evolving by acquiring mutations [[12], [13], [14]]. As the Orf3a protein plays a very crucial role in virus infection and pathogenesis, it is quite intriguing to understand the structural and functional implications of Orf3a mutations. Present in-silico study was conducted to identify and characterize mutations in Orf3a protein. We compared a total of 15,928 sequences of Orf3a protein, reported till September 14, 2020 worldwide with the first reported sequence from Wuhan, China. Our study revealed 173 mutations in Orf3a protein. The probable implications of these mutations on the structure and function of Orf3a were discussed.
2. Materials and methods
2.1. Orf3a sequence retrieval
The Orf3a sequences were retrieved from the NCBI-virus-database that has 15,928 sequences of Orf3a deposited till September 14, 2020. All these sequences were downloaded from the database (listed in Supplementary Table 1). The amino acid sequences of the Orf3a were exported in the FASTA format. The polypeptide sequences with characters other than standard amino acid sequences such as ‘X’ represent sequencing errors were excluded from the analysis. Jalview visualization tool was used to identify and remove the redundant sequences from the analysis. After considering these exclusion criteria, the remaining Orf3a polypeptide sequences were used for mutational analyses. The reference or wild-type sequence used in this study (accession ID: YP_009724391)was the first reported sequence of SARS-CoV-2 from Wuhan, China [5].
2.2. Multiple sequence alignments (MSAs)
The MSAs were performed using Clustal Omega tool [15], and the first reported sequence Orf3a (accession ID: YP_009724391) from Wuhan, China was used as a reference sequence for comparison. First, the Orf3a fasta sequences were uploaded into the Clustal Omega webserver as an input to run the programe that utilizes HMM and pairwise alignment to generate the MSA data. The variations were recorded carefully and used for further analysis.
2.3. Secondary structure prediction
In order to understand the implications of mutation on the secondary structure of Orf3a, the secondary structure prediction tool CFSSP was used. The CFSSP programe was developed by Ashok et al. [16] which predicts the secondary structure from the input polypeptide sequences. To run this webserver, we uploaded the wild type and the corresponding Orf3a sequence containing the identified mutations as an input. The predicted secondary structure from wild type and mutant sequences were obtained as an output. We analysed the secondary structure between wild type and mutants and the differences, if any, were marked.
2.4. Protein disorder prediction
PONDR-VSL2 webserver was used to calculate the per-residue disorder distribution in the query sequences as described elsewhere [17]. The PONDR-VSL2 provides the per-residue disorder predisposition scores on the scale from 0 to 1. The value 0 represents fully ordered residues while 1 depicts fully disordered residues. The value of 0.5 is threshold above which residues are considered disordered. Residues are considered highly and moderately flexible if the disorder score ranges from 0.25 to 0.5 and 0.1 to 0.25 respectively.
2.5. Protein modelling studies
The protein modelling studies were performed to understand the impact of mutation on the stability of the Orf3a protein. This analysis was conducted using DynaMut programe [18]. The solved structure of Orf3a, RCSB ID: 6XDC [19] was used for protein modelling studies. The effect of mutations on protein was shown in terms of difference in free energy (ΔΔG). The positive value of ΔΔG indicates stabilizing mutation; however, negative value represents destabilizing mutation. The DynaMut webserver can only predict ΔΔG for those regions of protein whose structure have been solved. The three regions, (1–39, 175–180 and 239–275) appeared as unmodeled regions of Orf3a [19], therefore, the mutations residing in these areas have not been used for stability prediction.
2.6. Epitope predictions
B-cell epitope predictions were performed as described by Jesperson et al. [20] using IDEB analysis resource. The parameters such as hydrophilicity, flexibility, accessibility, turns, exposed surface, polarity and antigenic propensity of polypeptide chains have been correlated with the location of epitopes. This webserver uses these properties to predict epitopes from the provided input sequence. All prediction calculations are based on propensity scales for each of the 20 amino acids.
3. Results
3.1. Identification of mutations in Orf3a of SARS-CoV-2
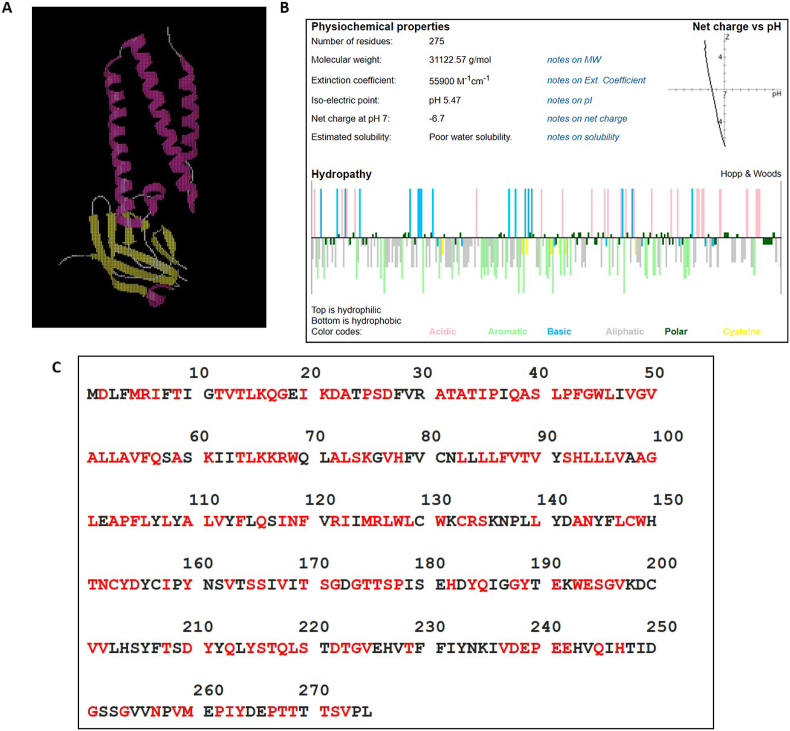
Recently, the structure of Orf3a has been solved [19] as represented by the cartoon (Fig. 1A). It is mainly comprised of helical regions, and forms a channel like structure in the membrane. A standalone Innovagen's peptide calculator (https://pepcalc.com/) was used to understand the overall physiochemical properties of Orf3a. It derives calculations and estimations on physiochemical properties of input molecule that includes peptide molecular weight, peptide extinction coefficient, peptide net charge at neutral pH, peptide iso-electric point and peptide water solubility. The colour coded display of amino acid classification and peptide hydropathy plot of Orf3a have been shown in the Fig. 1B. In order to identify the variations among Orf3a proteins, Clustal Omega mediated multiple sequence alignments (MSA) were performed between the Orf3a protein sequences among SARS-CoV-2 reported till September 14, 2020. The analysis revealed as many as 173 point mutations as highlighted in red font (Fig. 1C) and details of each mutation have been mentioned in Table 1.
Fig. 1.
A) The cartoon structure of Orf3a. The red color represents helical structure while yellow color represents beta sheets. B) The various properties of residues of Orf3a. The data were obtained from Innovagen's peptide calculator (https://pepcalc.com/). The colour coded display of amino acid classification and peptide hydropathy plot are shown. C) The sequence of SARS-CoV-2 Orf3a protein. The mutated amino acids are shown in red color. The number denotes the position of residues in Orf3a. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
List of Orf3a mutations identified in this study. The sequence of SARS-CoV-2 Orf3a protein reported till 14th Sept 2020 was aligned with the sequence from Wuhan (wet sea food market) SARS-CoV-2. The mutations were recognized by amino acid sequence alignment by CLUSTAL Omega.
| S. No. | Mutation | S. No. | Mutation | S. No. | Mutation |
|---|---|---|---|---|---|
| 1 | D2G, D2Y | 59 | L83F | 117 | S171L |
| 2 | M5V | 60 | L85F | 118 | G172V, G172C |
| 3 | R6T | 61 | L86W | 119 | G174D |
| 4 | I7T | 62 | F87L | 120 | T175K, T175I |
| 5 | T9K, T9I | 63 | V88L, V88A | 121 | T176I |
| 6 | T12N | 64 | T89I | 122 | S177I |
| 7 | V13L, V13A, V13I | 65 | V90F, V90I | 123 | P178S |
| 8 | T14I | 66 | S92L | 124 | H182Y |
| 9 | L15F | 67 | H93Y | 125 | Y184H |
| 10 | K16N | 68 | L94P, L94I, L94F | 126 | Q185H |
| 11 | Q17R | 69 | L95F | 127 | G188C |
| 12 | G18S, G18C, G18V, G18D | 70 | L96F | 128 | Y189C |
| 13 | I20T | 71 | V97A, V97F | 129 | E191K |
| 14 | K21Q, K21N | 72 | A99T, A99S, A99V | 130 | W193C, W193R |
| 15 | D22Y | 73 | G100L, G100C, G100F, G100V | 131 | E194Q |
| 16 | A23S | 74 | L101F | 132 | S195Y |
| 17 | P25L, P25S | 75 | A103S, A103V | 133 | G196V, G196R |
| 18 | S26L, S26P | 76 | P104L, P104S | 134 | V197L, V197I |
| 19 | D27Y, D27H | 77 | F105L | 135 | V201I |
| 20 | A31T | 78 | L106F | 136 | V202L |
| 21 | T32I | 79 | L108F | 137 | T208A |
| 22 | A33E, A33S | 80 | A110S, A110V | 138 | D210Y |
| 23 | T34A | 81 | L111S | 139 | Y211C |
| 24 | I35T | 82 | V112F, V112L | 140 | Q213H |
| 25 | P36L | 83 | F114C | 141 | Y215H |
| 26 | Q38E, Q38P | 84 | Q116H | 142 | S216P |
| 27 | A39T | 85 | I118V | 143 | T217A |
| 28 | S40P, S40L | 86 | N119H | 144 | Q218R |
| 29 | L41I, L41H, L41F | 87 | F120L | 145 | L219F, L219S, L219V |
| 30 | P42S, P42L, P42R | 88 | R122K, R122I | 146 | S220N |
| 31 | F43Y | 89 | I123V | 147 | D222G |
| 32 | G44V | 90 | M125I | 148 | T223I |
| 33 | W45L, W45R | 91 | R126 M, R126S | 149 | G224C, G224V |
| 34 | L46F | 92 | L127F, L127I | 150 | V225L, V225F |
| 35 | V48F | 93 | W128C, W128L | 151 | T229I |
| 36 | G49D, G49S, G49V | 94 | L129F | 152 | V237A, V237F |
| 37 | V50A, V50I | 95 | W131S, W131R, W131L, W131V | 153 | D238N, D238E |
| 38 | A51S | 96 | C133F | 154 | E239D, E239G |
| 39 | L52F, L52I | 97 | R134H, R134L, R134C | 155 | P240L, P240S |
| 40 | L53F, L53H | 98 | S135P | 156 | E241A |
| 41 | A54T, A54S, A54V | 99 | L140F, L140I | 157 | E242A |
| 42 | V55G, V55F | 100 | A143S, A143V | 158 | Q245L |
| 43 | F56C | 101 | N144Y | 159 | H247Y |
| 44 | Q57Y, Q57H | 102 | L147F | 160 | G251C, G251V |
| 45 | A59V | 103 | C148Y, C148S | 161 | G254R |
| 46 | K61 N | 104 | W149L, W149C | 162 | N257S |
| 47 | T64I | 105 | T151I | 163 | V259L, V259E |
| 48 | L65F | 106 | N152S, N152I | 164 | M260K, M260I |
| 49 | K66N | 107 | C153Y | 165 | P262L, P262S |
| 50 | K67N, K67R | 108 | Y154C | 166 | I263M |
| 51 | R68I | 109 | D155Y | 167 | Y264C |
| 52 | W69C, W69L, W69R | 110 | I158V | 168 | P267S, P267L |
| 53 | A72S | 111 | Y160H | 169 | T268K, T268M |
| 54 | L73F | 112 | V163L | 170 | T269M |
| 55 | S74F, S74P | 113 | S165F, S165I | 171 | T271I |
| 56 | K75R, K75E | 114 | S166L | 172 | S272I |
| 57 | V77I, V77F | 115 | V168I | 173 | V273L |
| 58 | H78Q, H78Y | 116 | T170S |
3.2. Analysis of the effect of mutations on Orf3a stability
To assess the impact of mutations on Orf3a, protein modelling studies were performed using DynaMut webserver [18]. This webserver calculates the change in free energy (ΔΔG) due to the mutation induced variation in the target protein. The positive ΔΔG represents increase in stability while the negative ΔΔG represents decrease in stability. Our analysis revealed various mutations that alter stability of the protein as shown in Supplementary Table 1. Our analysis revealed that the mutations caused destabilization as well as stabilisation in Orf3a protein structure. Top twelve mutations have been shown in the Table 2. The maximum positive ΔΔG (1.7 kcal/mol) was obtained for G49V mutation, leading to increase in stability. Similarly, R126S mutation caused maximum negative ΔΔG (−2.02 kcal/mol), leading to decrease in the stability of Orf3a.
Table 2.
Calculations of ΔΔG between wild-type and mutant Orf3a. The top rank mutations are listed in the table. DynaMut webserver was used to calculate the predicted ΔΔG. The negative values indicate the destabilization of protein upon mutation.
| S. No | Wild type residue | Residue position | Mutant residue | ΔΔG DynaMut (kcal/mol) |
|---|---|---|---|---|
| 1 | G | 49 | V | 1.74 |
| 2 | V | 88 | L | 1.507 |
| 3 | V | 90 | F | 1.512 |
| 4 | V | 112 | F | 1.30 |
| 5 | R | 126 | S | −2.024 |
| 6 | C | 148 | S | −1.741 |
| 7 | I | 158 | V | −1.583 |
| 8 | Y | 160 | H | −1.527 |
| 9 | S | 171 | L | 1.619 |
| 10 | D | 210 | Y | 1.442 |
| 11 | G | 224 | V | −1.528 |
| 12 | G | 224 | C | −1.717 |
3.3. Secondary structure and protein disorder predictions due to mutations in Orf3a
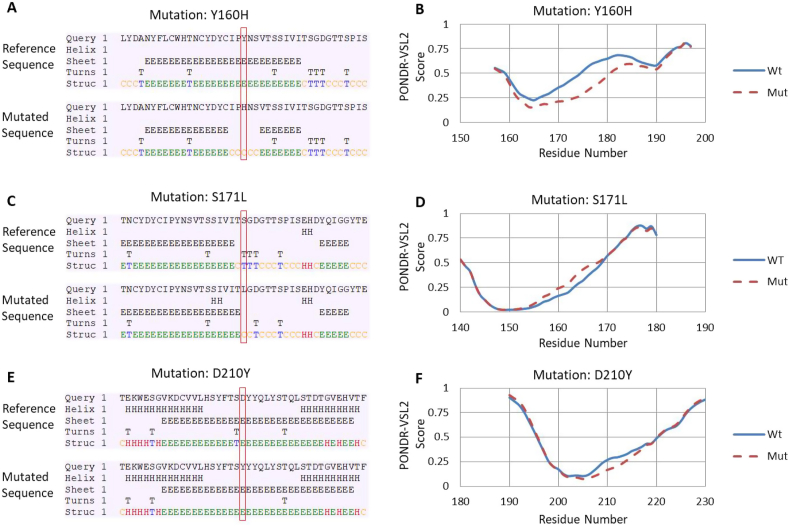
Subsequently, the twelve mutations were characterised that exhibited maximum variation in ΔΔG by predicting their effect on the secondary structure of the Orf3a protein. The CFSSP webserver was used to analyse the variations in secondary structure where these mutations reside. The data revealed that out of twelve mutations, only three positions led to change in the secondary structure (Fig. 2A, C and E). Rest of the nine locations exhibited no alteration in secondary structure (data not shown). The detailed analysis revealed that Y160H mutation has led to shift of beta-sheet to coiled-coil structure (Fig. 2A). The turn structure is replaced by coiled coil at S171L (Fig. 2C) mutation while D210Y mutation leads to replacement of turn structure by beta-sheet (Fig. 2E).
Fig. 2.
Analysis of the secondary structure and intrinsic disorder predisposition of the unique mutations of SARS-CoV2 Orf3a in comparison with the reference Orf3a protein (YP_009724391) from China, Wuhan. (A, C and E) Secondary structure predictions, the amino acid sequences near the mutation site were uploaded on CFSSP web tool that predict secondary structure. Each panel (A, C and E) shows the secondary structure of the wild type and mutated input sequences. The panel (i) represents the wild type or Wuhan sequence while panel (ii) represents the mutated Indian sequence. The mutation site is highlighted in the rectangular box. (B,D and F) protein disorder prediction, the analysis was conducted using PONDR-VSL2 algorithm. A disorder threshold is depicted at a score of 0.5). Residues/regions with the disorder scores >0.5 are considered as disordered.
The impact of these three mutations on protein disorder parameters was further analysed. The PONDR-VSL2 webserver was used to measure the protein disorder contributed by these three mutations. Our analysis revealed that Y160H (Fig. 2B) and D210Y (Fig. 2F) decreased the protein disorder while S171L (Fig. 2D) increased the protein disorder. Altogether, both secondary structure and protein disorder were altered due to the mutation in Orf3a.
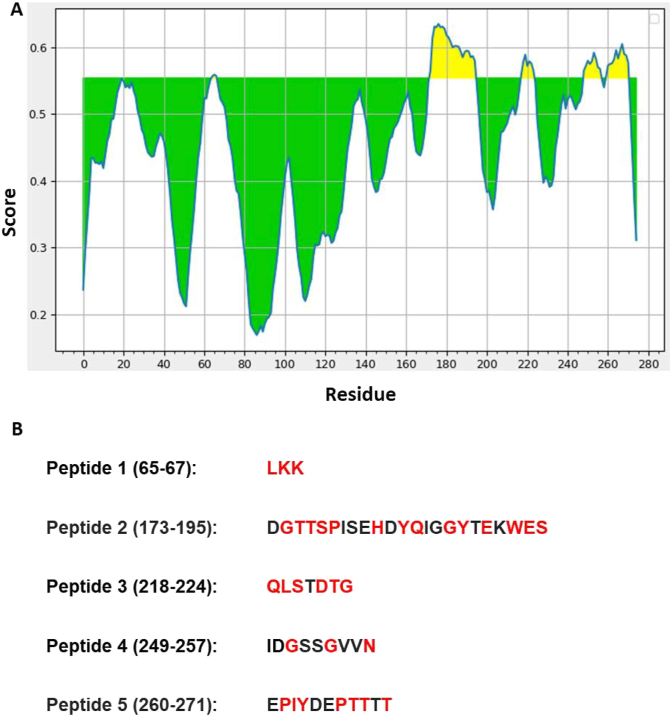
3.4. Effect on B cell epitopes due to Orf3a mutations
B cell epitopes were predicted using webserver as shown in IDEB analysis resource [20]. The data has been represented graphically (Fig. 3A). The yellow shaded area corresponds to the high score peptides that can act as potential B-cell epitopes. This tool provided five peptide sequences (B-cell epitopes) as shown in Fig. 3B. Subsequently, we compared these sequences with the mutations identified in this study. Our data revealed that peptide 1 was mutated at its all three positions while peptide 3 was also mutated at its all positions except one. Peptide 2 also has five mutations, out of nine. Similarly, peptide 4 and 5 were also found to harbour multiple mutations. It is plausible that due to these mutations the respective epitopes will change and they might help SARS-CoV-2 to evade immunogenic response of the host.
Fig. 3.
Prediction of B-cell epitopes. A) On the graphs, the Y-axis depicts for each residue the correspondent BepiPred score (averaged in the specified window); while the X-axis depicts the residue positions in the sequence. The larger score for the residues might be interpreted as that the residue might have a higher probability to be part of epitope (those residues are colored in yellow on the graphs). B) The top five peptides of Orf3a that showed the highest score. The sequences of all five peptides are shown. The number in parentheses represents the location of the peptide in the primary sequence of Orf3a. The red font shows the location of mutant residues of Orf3a. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussions
Due to the rapid spread of SARS-CoV-2 in various countries worldwide, WHO announced COVID-19 a global pandemic on March 11, 2020 [21]. With the spread of virus to new locations, it acquired mutations leading to evolution of SARS-CoV-2 variants that can potentially affect the rate of viral spread, its pathogenicity and interactions with host. In our study, 173 mutations in Orf3a were identified after analysing approximately 16,000 reported sequences of Orf3a. Our study also showed that there was a considerable alteration instability and dynamicity due to mutations at various positions that might alter Orf3a function. These data were further supported by the protein disorder analysis and secondary structure predictions (Fig. 2). Previous studies revealed that Orf3a, a widely expressed protein, triggered inflammatory responses in the host cells [22,23]. It is plausible that the mutations occurring in Orf3a can highly affect the function of this protein. To gain some insight into the altered function of Orf3a, in-silico analyses were performed to predict the possible B-cell epitopes generated by the peptides of this protein. Our data supports the fact that these mutations might help the virus to evade immune system of the host because of the loss of putative epitopes (Fig. 3).
The putative consequences of variations in Orf3a explained in our observation are in conformity with similar findings reported recently. In an analytical study, it has been observed that the accumulation of non-synonymous mutations in Orf3a of SARS-CoV-2 could be driving protein changes that might mediate immune evasion and thus favouring viral spread [24]. Occurrences of epitope loss due to mutation in SARS-CoV-2 has also been reported experimentally where six putative epitopes in wild type Orf3a are found to be replaced by five in mutant variants, and such loss of epitopes might allow the mutant to escape interaction with host immunity system [25]. Moreover, a novel missense mutation in the Orf3a gene has been found responsible for the global dissemination of SARS-CoV-2 [26]. Further, SARS-CoV-2 strain with Orf3a mutation often found to carry a mutation in its S (spike) gene, facilitating its interaction with ACE-2 receptors followed by viral entry in the host cells [27]. Majumdar and Niyogi [28] have also observed an appreciable association of Orf3a mutation in SARS-CoV-2 with higher infection and mortality rate.
In summary, structural variations and residue composition in the Orf3a protein might be related to rapid infection kinetics and spreading of SARS-CoV-2. Mutational analysis studies are, therefore, highly pertinent to determine the changes in the structure and function of viral proteins.
5. Conclusions
Altogether, this study identified several interesting mutations of Orf3a and characterised them showing their probable effects on immune evasion. However, the data obtained here warrants validation to better understand the implications of these mutations on the function of Orf3a.
CRediT authorship contribution statement
Gajendra Kumar Azad: Conceptualization, Supervision, Methodology, Validation, Visualization, Writing - original draft, and & editing. Parimal Kumar Khan: Validation, and Manuscript editing.
Declaration of competing interest
Authors declare no conflict of interests.
Acknowledgements
We would like to acknowledge the infrastructural support provided by the Department of Zoology, Patna University, Patna, Bihar (India) for this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.100933.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabi F.A., Al Zoubi M.S., Al-Nasser A.D., Kasasbeh G.A., Salameh D.M. Sars-cov-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020 doi: 10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bchetnia M., Girard C., Duchaine C., Laprise C. The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review of the current global status. J. Infect. Public Health. 2020 doi: 10.1016/j.jiph.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassan S.S., Choudhury P.P., Basu P., Jana S.S. Molecular conservation and differential mutation on ORF3a gene in Indian SARS-CoV2 genomes. Genomics. 2020 doi: 10.1016/j.ygeno.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyser J.M. Viroporins. 2015. [DOI]
- 9.Lu W., Xu K., Sun B. SARS accessory proteins ORF3a and 9b and their functional analysis. Mol. Biol. SARS-Coronavirus. 2010 doi: 10.1007/978-3-642-03683-5_11. [DOI] [Google Scholar]
- 10.Siu K.L., Yuen K.S., Castano-Rodriguez C., Ye Z.W., Yeung M.L., Fung S.Y., Yuan S., Chan C.P., Yuen K.Y., Enjuanes L., Jin D.Y. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. Faseb. J. 2019 doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y.F., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020 doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chand G.B., Banerjee A., Azad G.K. Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure. PeerJ. 2020;8 doi: 10.7717/peerj.9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chand G.B., Banerjee A., Azad G.K. Identification of twenty-five mutations in surface glycoprotein (Spike) of SARS-CoV-2 among Indian isolates and their impact on protein dynamics. Gene Rep. 2020;21:100891. doi: 10.1016/j.genrep.2020.100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashok Kumar T. CFSSP: chou and fasman secondary structure prediction server. Wide Spectr. 2013 doi: 10.5281/zenodo.50733. [DOI] [Google Scholar]
- 17.Obradovic Z., Peng K., Vucetic S., Radivojac P., Dunker A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins Struct. Funct. Genet. 2005 doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 18.Rodrigues C.H.M., Pires D.E.V., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kern D.M., Sorum B., Hoel C.M., Sridharan S., Remis J.P., Toso D.B., Brohawn S.G. Cryo-EM structure of the SARS-CoV-2 3a ion channel in lipid nanodiscs. 2020. BioRxiv Prepr. Serv. Biol. [DOI]
- 20.Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization . WHO Bull; 2020. Coronavirus Disease 2019 Situation Report 51. [Google Scholar]
- 22.Freundt E.C., Yu L., Goldsmith C.S., Welsh S., Cheng A., Yount B., Liu W., Frieman M.B., Buchholz U.J., Screaton G.R., Lippincott-Schwartz J., Zaki S.R., Xu X.-N., Baric R.S., Subbarao K., Lenardo M.J. The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 2010 doi: 10.1128/jvi.01662-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law P.T.W., Wong C.H., Au T.C.C., Chuck C.P., Kong S.K., Chan P.K.S., To K.F., Lo A.W.I., Chan J.Y.W., Suen Y.K., Chan E., Fung K.P., Waye M.M.Y., Sung J.J.Y., Lo Y.M.D., Tsui S.K.W. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005 doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- 24.Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020 doi: 10.1128/msystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta A.M., Chakrabarti J., Mandal S. Non-synonymous mutations of SARS-CoV-2 leads epitope loss and segregates its variants. Microb. Infect. 2020 doi: 10.1016/j.micinf.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M., Li M., Ren R., Li L., Chen E.-Q., Li W., Ying B. International expansion of a novel SARS-CoV-2 mutant. J. Virol. 2020 doi: 10.1128/jvi.00567-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majumdar P., Niyogi S. ORF3a mutation associated higher mortality rate in SARS-CoV-2 infection. Epidemiol. Infect. 2020 doi: 10.1017/S0950268820002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.