Summary
Chromosome rearrangements and the way that they impact genetic differentiation and speciation have long raised questions from evolutionary biologists. They are also a major concern for breeders because of their bearing on chromosome recombination. Banana is a major crop that derives from inter(sub)specific hybridizations between various once geographically isolated Musa species and subspecies. We sequenced 155 accessions, including banana cultivars and representatives of Musa diversity, and genotyped‐by‐sequencing 1059 individuals from 11 progenies. We precisely characterized six large reciprocal translocations and showed that they emerged in different (sub)species of Musa acuminata, the main contributor to currently cultivated bananas. Most diploid and triploid cultivars analyzed were structurally heterozygous for 1 to 4 M. acuminata translocations, highlighting their complex origin. We showed that all translocations induced a recombination reduction of variable intensity and extent depending on the translocations, involving only the breakpoint regions, a chromosome arm, or an entire chromosome. The translocated chromosomes were found preferentially transmitted in many cases. We explore and discuss the possible mechanisms involved in this preferential transmission and its impact on translocation colonization.
Keywords: chromosome segregation, genome evolution, Musa, reciprocal translocation, recombination
Significance Statement
Chromosome rearrangements, the way that they emerged and their potential impact on speciation and breeding raise important questions. We report on the characterization of several large reciprocal translocations in banana, Musa acuminata, identify the genetic group in which they emerged, characterize their impacts on chromosome recombination, and explore possible mechanisms allowing their colonization.
Introduction
Chromosomal rearrangements and their possible link with lineage diversification and speciation have fascinated evolutionary biologists for a long time (Faria and Navarro, 2010; Kirkpatrick, 2010; Huang and Rieseberg, 2020). Many researchers have claimed that they play a role in speciation by contributing to reproductive isolation (White, 1978; Grant, 1981; Baker and Bickham, 1986; King, 1993; Ostevik et al., 2020). Indeed, structurally heterozygous hybrids are often unviable or have reduced fertility (Lai et al., 2005; Stathos and Fishman, 2014; Quach et al., 2016). Studies on inversions have shown, in various species, that suppressed recombination induced by inversion facilitated divergence and speciation. In the case of Robertsonian fusions, evidence of fixation in small island isolated populations through genetic drift was reported (Britton‐Davidian et al., 2000). Finally, conflicting data and unresolved questions persist despite over 100 years of research on the chromosome speciation model, which led Carracedo et al. (2000) to propose that various mechanisms may be involved depending on the evolutionary history of each species. To date, most evolutionary studies have been on inversions. Other types of rearrangement have been explored to a lesser extent.
Structural variations also complicate breeding because they generally lead to reduced or no fertility and to a recombination reduction (Tadmor et al., 1987; Quillet et al., 1995; Ostberg et al., 2013), which hamper combining targeted characters. Many crops are derived from or improved through inter(sub)specific hybridizations (Mcfadden and Sears, 1946; Simmonds, 1962; Zamir, 2001; Wu et al., 2014; Garsmeur et al., 2018; Flowers et al., 2019; Santos et al., 2019) between (sub)species potentially differing for structural variations.
Banana is a major crop derived from inter(sub)specific hybridization between various species and subspecies of the Musa genus. Musa acuminata (2n = 22, x = 500 Mb, A genome) is involved in all cultivars and Musa balbisiana (2n = 22, B genome), Musa schizocarpa (2n = 22, S genome) and Musa textilis (2n = 20, T genome) in some of them (Simmonds and Shepherd, 1955; Carreel et al., 1994; Němečková et al., 2018). Musa acuminata has been divided into several subspecies, namely banksii, malaccensis, zebrina, burmannica, burmannicoides, siamea, microcarpa, truncata, and errans (Simmonds, 1962; Perrier et al., 2009). The subdivision of burmannica, burmannicoides, and siamea has been questioned recently because they clustered together in molecular diversity studies (Perrier et al., 2009; Martin et al., 2017; Dupouy et al., 2019; Martin et al., 2020). They thus are referred to here as a single burmannica subspecies. The subspecies status of ssp. errans, truncata and microcarpa has been questioned by molecular genetic studies (Carreel et al., 1994; Perrier et al., 2009; Sardos et al., 2016a; Christelová et al., 2017; Dupouy et al., 2019; Martin et al., 2020).
Musa species and subspecies diverged following geographical isolation in distinct Southeast Asian continental regions and islands (Janssens et al., 2016). The current domestication scenario for bananas suggests that humans transported plant material during their migrations, which led to contacts between these subspecies, probably during the Holocene (Perrier et al., 2011). This resulted in the emergence of inter(sub)specific hybrids with reduced fertility (Dodds and Simmonds, 1948; Fauré, Bakry, et al., 1993; Shepherd, 1999). Early farmers would then have selected parthenocarpic diploid and triploid hybrids producing fruit with high flesh and low seed content. Cultivars were classified, based on morphology and ploidy, into genomic groups (‘AA’, ‘AAA’, ‘AB’, ‘AAB’, ‘ABB’, ‘AS’, ‘AT’, and ‘AAT’) to reflect the main species contributing to their genomes (Simmonds and Shepherd, 1955). Current cultivars have been vegetatively propagated for centuries or millennia, resulting in subgroups of phenotypic somaclonal variants.
Recent analysis of the chromosome contribution of these species and subspecies to several Musa cultivars revealed complex inter(sub)specific chromosome mosaics (Baurens et al., 2019; Cenci et al., 2020; Martin et al., 2020). This suggests that the origin of cultivars is more complex than previously assumed, with more hybridization steps and more ancestral contributors. These results also revealed a contribution from an unidentified ancestral genetic group to cultivated bananas (Martin et al., 2020).
Several large chromosomal rearrangements were suspected between M.acuminata subspecies. These suspicions were based on cytogenetic studies showing that chromosomal pairing at meiosis in M.acuminata is generally regular in bivalents within subspecies, but irregular with some multivalents and univalents in inter(sub)specific hybrids (Dodds, 1943; Dodds and Simmonds, 1948; Dessauw, 1987; Fauré, Bakry, et al., 1993; Shepherd, 1999). Similar suspicions were based on segregation distortions and chromosomal co‐segregation in genetic maps (Fauré, Noyer, et al., 1993; Hippolyte et al., 2010; D’Hont et al., 2012; Mbanjo et al., 2012). Recently, three large structural rearrangements in the form of reciprocal translocations were precisely characterized within M.acuminata (Martin et al., 2017; Dupouy et al., 2019). In addition, a reciprocal translocation and an inversion were characterized between M.acuminata and M.balbisiana (Baurens et al., 2019; Šimoníková et al., 2019).
Here, we report on the precise characterization of three reciprocal translocations in Musa acuminata that added to the three we reported previously. We analyzed the distribution of the six reciprocal translocations in 155 accessions representative of wild Musa diversity and cultivated bananas and identified in which (sub)species each rearrangement emerged. We analyzed chromosome segregation in 1059 individuals from 11 progenies from structurally heterozygous parents and revealed that translocation induced a reduction of recombination of extent and intensity that vary depending on rearrangements. In addition, in many cases, we observed that the translocated structures were preferentially transmitted. We discuss these results in terms of genome evolution and breeding prospects.
Results
Characterization of the reciprocal translocations
Eleven progenies involving 18 parental accessions were genotyped‐by‐sequencing (Table 1). Genetic linkage between single nucleotide polymorphism (SNP) markers from each parent was calculated and projected on the M.acuminata reference genome sequence (D’Hont et al., 2012; Martin et al., 2016). Chromosomes from this reference genome produced with an accession of ssp. malaccencis have been proposed to correspond to the ancestral chromosome architecture of M.acuminata (Shepherd, 1999; Martin et al., 2017; Dupouy et al., 2019) and thus are referred to here as reference chromosomes.
Table 1.
Statistics on studied banana populations
| Population ID | Female parent | Male parent | Structure α | Population size | Reduction of recombination β | Number of triploids individuals | Number of aneuploid individuals | Aneuploid on translocated chromosomes | Alternate structure proportion in progeny γ | Aneuploid on translocated chromosomes | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Segmental aneuploid | True aneuploid | Complex aneuploid δ | ||||||||||
| Borli* | Borneo | Pisang Lilin° | 1/4 | 82 | T1 & bp | 0 | 3 | 2 (2.4%) | 71% | 0 | 0 | 2 |
| AFPTBA00267** | PT‐BA‐00267° | PT‐BA‐00267° | 1/4 | 177 | T1 & bp | 0 | 6 | 5 (2.8%) | 72% | 5 | 0 | 0 |
| PBC | PB1° (PT‐BA‐00267 X Banksii) | Calcutta4 F09 | 1/4 | 133 | T1 & bp | 1 | 5 | 4 (3.0%) | 98% | 1 | 1 | 1 + 1' |
| AFMadu | Pisang Madu° | Pisang Madu° | 1/7 | 29 | Chr1 | 0 | 1 | 1 (3.4%) | 49% | 0 | 1 | 0 |
| Madga | Pisang Madu° | Galeo | 1/7 | 154 | Chr1 | 0 | 6 | 6 (3.9%) | 59% | 0 | 5 | 1' |
| BCM | BC1° (Banksii X Calcutta4) | Malaccensis nain | 1/9, 2/8 | 75 | bp | 0 | 16 | 16 (21.3%) | 72ζ–61% ε | 0 ζ –15 ε | 0 ζ –1 ε | 0 |
| PCMo | PC1° (PT‐BA‐00267 X Calcutta4) | Monyet P09 | 1/9, 2/8 | 74 | bp | 0 | 17 | 17 (23.0%) | 68ζ–65% ε | 0 ζ –11 ε | 1 ζ –6 ε | 0 |
| PCZ | PC1° (PT‐BA‐00267 X Calcutta4) | Zebrina R07 | 1/9, 2/8 | 99 | bp | 0 | 23 | 23 (23.2%) | 60ζ–46% ε | 0 ζ –20 ε | 1 ζ –2 ε | 0 |
| PMK | PM1° (PT‐BA‐00267 X Maia'Oa) | Khae Phrae N8 | 3/8 | 90 | T8 | 0 | 1 | 0 (0.0%) | 57% | 0 | 0 | 0 |
| PMP | PM2° (PT‐BA‐00267 X Maia'Oa) | Pa rayong | 3/8 | 70 | T8 | 1 | 0 | 0 (0.0%) | 44% | 0 | 0 | 0 |
| Pakid | Paka° T | IDN 110° | 1/4/ 1/7 | 76 | Chr1 | NA | 18 | 16 (21.1%) | NA | 9 | 6 | 1' |
NA, not applicable.
Chromosome structures found at heterozygous state in parent indicated by (°).
bp means breakpoint; T1 and T8 refer to translocated fragments of 1/4 or 2/8, respectively.
Calculated excluding aneuploid on translocated chromosome individuals (i.e. calculated on green cells of Table S2).
1' refers to individuals for which segmental aneuploidy at the end of chromosome 1 is observed but could not be explained.
Statistics calculated for the 1/9 translocation.
Statistics calculated for the 2/8 translocation.
Population described in Hippolyte et al. (2010).
Population described in Martin et al. (2017).
Refers to tetraploid parent.
For four accessions (Monyet, Zebrina and hybrids PM1 and PM2), genetic linkage between markers of reference chromosomes 3 and 8 was observed (Figure S1a,b,c and d) suggesting the presence of a reciprocal translocation involving one arm of reference chromosome 3 (T3) and one arm of reference chromosome 8 (T8). In accession Monyet (Figure 1a and 1b), the linkage breaks around the centromeric regions indicated that the accession is homozygous for this 3/8 translocation. The linkage involved markers from pericentromeric part of reference chromosome 8 and a distal part of reference chromosome 3. This showed that this reciprocal translocation is associated with the inversion of segment T3. The position of the linkage breaks allowed us to locate the translocation breakpoint between 15.4 and 17.3 Mb of reference chromosome 3 and between 16.1 and 23.5 Mb of reference chromosome 8. The linkage break position estimates were wide because they were located in centromeric regions that have a high number of repetitive sequences and, consequently, a low number of SNPs suitable for genetic analysis. The second accession Zebrina, although largely homozygous and thus displaying a limited number of segregating markers, showed a genetic linkage pattern suggesting that it was homozygous for the 3/8 translocation (Figure S1b). Analysis of its marker linkage pattern enabled us to refine the position of the translocation breakpoint of chromosome 8 between 16.1 and 17.6 Mb. Close linkage between reference chromosomes 3 and 8 was also observed in the two PM1 and PM2 hybrids, but without any linkage breaks, indicating that PM1 and PM2 were structurally heterozygous for the 3/8 reciprocal translocation (Figure S1c and S1d). Chromosomes resulting from this 3/8 translocation were named 3T8 and 8T3 (Figure 1b). Translocation 3/8 was also detected in the Khi Maeo accession through bacterial artificial chromosome‐fluorescence in situ hybridization (BAC‐FISH), with BACs originating from reference chromosomes 3 and 8 found together on one chromosome, whereas the other signals were found on two distinct chromosomes (Figure 2a).
Figure 1.
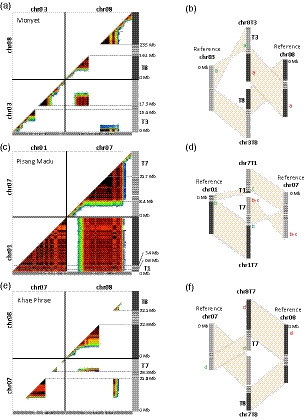
Characterization of reciprocal translocations through genetic analysis. (a, c, e) Dot‐plots with pairwise marker genetic linkage in the analyzed accession along the 11 Musa acuminata reference chromosomes (V2). Marker linkage is represented by a color gradient from red (strong) to dark blue (weak). Gray boxed arrows at the bottom represent scaffolds from the V2M. acuminatareference sequence. (a) Monyet accession, PCMo population. (c) Pisang Madu accession, Magda population. (e) Khae Phrae, PMK population. (b, d, f) Schematic representation of the inferred chromosome structures. Gray hatched boxes indicate the translocation breakpoint regions. Different green and red lowercase letters refer to the position and color of detection of the bacterial artificial chromosomes (BACs) used for BAC‐fluorescence in situ hybridization in Figure 2.
Figure 2.
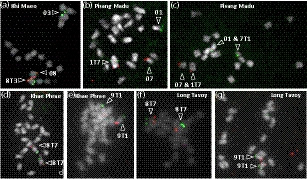
Characterization of reciprocal translocations through cytogenetic analysis. Bacterial artificial chromosome (BAC)‐fluorescence in situ hybridization on chromosomes at metaphase. Accession names are indicated on the pictures: (a) Khi Maeo, (b and c) Pisang Madu, (d and e) Khae Phrae, (f and g) Long Tavoy. Chromosomes were counterstained using 4′‐6‐diamidino‐2‐phenylindole (shown in gray). Locations of the BAC on Musa acuminata reference and translocated chromosome structures are indicated in Figure 1; their names and precise positions are provided in Table S3. Arrows point to the detected reference and translocated chromosome structures.
In the Pisang Madu accession, genetic linkage was observed between all markers of reference chromosome 1 and part of the markers of reference chromosome 7 (Figure 1c, Figure S1e and S1f) suggesting the presence of a rearrangement involving reference chromosomes 1 and 7 at the heterozygous state in this accession. FISH experiments with BACs originating from reference chromosomes 1 and 7 revealed a reciprocal translocation with translocation breakpoints located between 0.9 and 3.4 Mb on chromosome 1 and in the pericentromeric region for chromosome 7 (Figure 2b,c; Figure 1d). Chromosomes resulting from this 1/7 translocation were named 1T7 and 7T1.
In the Khae Phrae accession, genetic linkage patterns suggested the presence of a reciprocal translocation at the homozygous state involving one arm of reference chromosome 7 (T7) and one arm of reference chromosome 8 (T8) (Figure 1e and f, Figure S1g). The linkage break positions allowed us to locate the translocation breakpoint between 21.8 and 26.3 Mb of reference chromosome 7 and between 22.6 and 32.1 Mb of reference chromosome 8. In the Khae Phrae (Figure 2d) and Long Tavoy accessions (Figure 2f), BACs from reference chromosomes 7 and 8 were found on the same chromosome through BAC‐FISH, demonstrating that both accessions were homozygous for this 7/8 translocation. Chromosomes resulting from this 7/8 translocation were named 7T8 and 8T7. The Khae Phrae and Long Tavoy accessions were previously characterized as structurally homozygous for the 2/8 translocation (Dupouy et al., 2019). As a result of its strong homozygosity in some regions, including those around the 2/8 translocation, this rearrangement could not be detected with genetic markers from Khae Phrae (Figure S1g). The position of the 7/8 translocation breakpoint on chromosome 8 is much more proximal than that of the 2/8 translocation breakpoint (37.7 Mb) (Dupouy et al., 2019). This suggests that Khae Phrae and Long Tavoy have both translocated structures with chromosomes 2T8, 8T7, and 7T8T2 (Figure S2).
Three translocations that we previously described, one involving chromosomes 1 and 4 (Martin et al., 2017), one involving chromosomes 2 and 8 (Dupouy et al., 2019), and one involving chromosomes 1 and 9 (Dupouy et al., 2019), were also found in some parents of the studied progenies. For four accessions (Pisang Lilin, PT‐BA‐00267, Malaccensis nain, and PB1 hybrid), the genetic linkage patterns observed between reference chromosomes 1 and 4 (Figure S1h, i, j and k) confirmed the presence of the 1/4 reciprocal translocation at homozygous state in Malaccensis nain (Martin et al., 2017) and heterozygous state in Pisang Lilin, PT‐BA‐00267 (Martin et al., 2017) and also showed that PB1 hybrid was structurally heterozygous. Chromosomes with the translocated structure were named 1T4 and 4T1.
For four accessions (Calcutta 4, Pa Rayong, and hybrids PC1 and BC1), the genetic linkage patterns observed between reference chromosomes 1 and 9 and 2 and 8 (Figure S1l, m, n, o and p) showed the presence of the 1/9 and 2/8 reciprocal translocations at homozygous state in Pa Rayong and Calcutta 4 accessions and heterozygous state in PC1 and BC1 hybrids. In addition, BAC‐FISH experiments showed the presence of the 1/9 translocation at homozygous state in Khae Phrae and Long Tavoy (Figure 2e and 2g). Chromosomes with the translocated structure were named 1T9, 9T1, 2T8, and 8T2.
For the IDN 110 accession, the genetic linkage pattern observed between reference chromosomes 1, 4, and 7 suggested that this accession is structurally heterozygous for both the 1/4 and 1/7 translocations (Figure S1q).
Finally, for Borneo and Galeo accessions, no genetic linkage was observed between markers from distinct reference chromosomes despite the overall good distribution of markers along the chromosomes (Figure S1r and s). This suggested that these accessions were structurally homozygous with the reference chromosome structure.
Translocated chromosome distribution in Musa germplasm
To be able to trace the distinct translocated chromosome structures in Musa germplasm, we developed a methodology tailored to exploit SNP data from the 11 studied progenies for identifying alleles specific to each chromosome structures (see Experimental procedures). These diagnostic alleles were then searched in whole‐genome sequencing data from 155 accessions representative of banana germplasm.
Among the 13 Musa and one Ensete species tested, the six targeted translocated structures were only found in M.acuminata and hybrids involving this species (Table S1).
We performed a factorial analysis with the 92M. acuminata diploid accessions using a subset of 3979 SNP markers derived from whole‐genome sequencing data to highlight the distribution of each translocated chromosome in the M.acuminata diversity and to explore the subspecies or genetic group from which it originated. The first two axes explained 26 and 23%, respectively, of the diversity, allowing us to separate the wild representatives of the four main M.acuminata subspecies (i.e. banksii, zebrina, malaccensis, and burmannica) (Figure 3, Figure S3). The cultivars were in an intermediate or central position between these subspecies, as expected from their intersubspecific hybrid origin.
Figure 3.
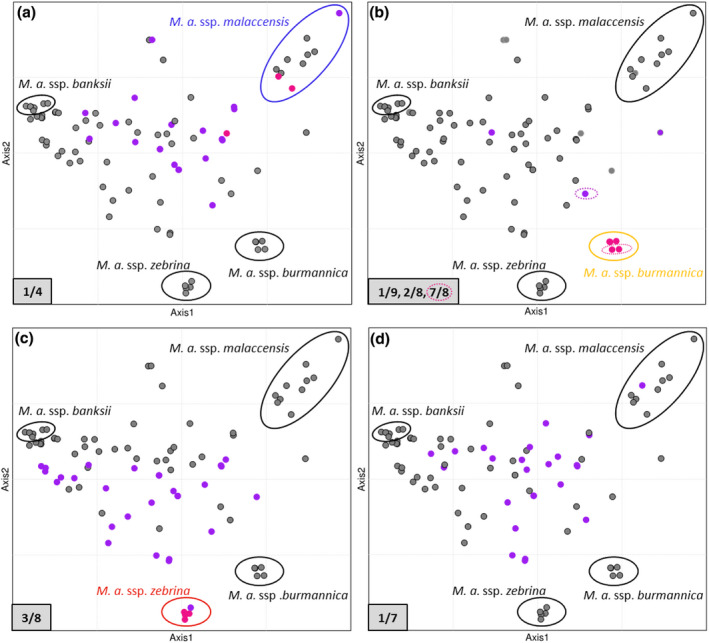
Translocation distributions in diploid Musa acuminata germplasm. Factorial analysis was performed on 34 wild M. acuminata accessions with projection of 58 diploid cultivars and hybrids along the synthetic axes (the first two axes are represented). Accessions homozygous or heterozygous for a translocation are represented by pink and purple dots, respectively. (a) Translocation 1/4. (b) Translocations 1/9 and 2/8. Translocation 7/8 is indicated by hashed circles. (c) Translocation 3/8. (d) Translocation 1/7. Note that some dots are superposed; for more details, see Figure S3.
Among the three wild accessions identified as homozygous for the 1/4 translocation, two were found within the genetic group corresponding to ssp. malaccensis (Figure 3a), with the last one (Ambihy) being in a more intermediate position toward the center.
The five wild accessions identified as homozygous for translocations 7/8 and/or 2/8 and 1/9 were found within the genetic group corresponding to ssp. burmannica, whereas the four wild accessions identified as homozygous for the 3/8 translocation were found within the genetic group corresponding to ssp. zebrina (Figure 3b and 3c, Table S1, Figure S3b and S3c). Accessions heterozygous for one or more of all these translocations were mainly found among cultivars or hybrids in central positions in the factorial analysis.
No homozygous accessions were identified for the 1/7 translocation and most accessions heterozygous for this 1/7 translocation were cultivars or hybrids that were found in a central position in the factorial analysis (Figure 3d, Table S1, Figure S3d). One exception is the seedy Pisang Serun 400 accession that clustered with ssp. malaccensis, but had introgressions from other Musa origins.
Among the 52 M. acuminata diploid cultivars tested, 10 were structurally homozygous with reference chromosome structure and the remaining were structurally heterozygous for 1 to 4 translocations, with 34, 43, and 43% having the 1/4, 1/7, and 3/8 translocations, respectively, and 2, 4, and 2% having the 1/9, 2/8, and 7/8 translocations, respectively (Table S1). The M.acuminata triploid cultivars tested were structurally heterozygous for 1 to 3 translocations. Among these 17 triploid cultivars, eight had the 1/4 translocation, 13 had the 1/7 translocation, and 14 had the 3/8 translocation, but none had 1/9, 2/8, or 7/8 translocations (Table S1).
Among the 21 interspecific hybrids tested involving M.acuminata and M.balbisiana, the 1/4 translocation was found in one of the two ‘AB’ cultivars, the 1/4, 1/7, and 3/8 translocations were found in five, one, and five of the 12 ‘AAB’ cultivars, respectively and the 1/4 translocation in one of the seven ‘ABB’ cultivars (Table S1).
Chromosomal recombination in structurally heterozygous accessions
The recombination rate along chromosomes was calculated for each diploid parent based on marker segregation in progeny individuals, excluding aneuploid individuals (Figure 4, Figure S4) (see below for aneuploid identification).
Figure 4.
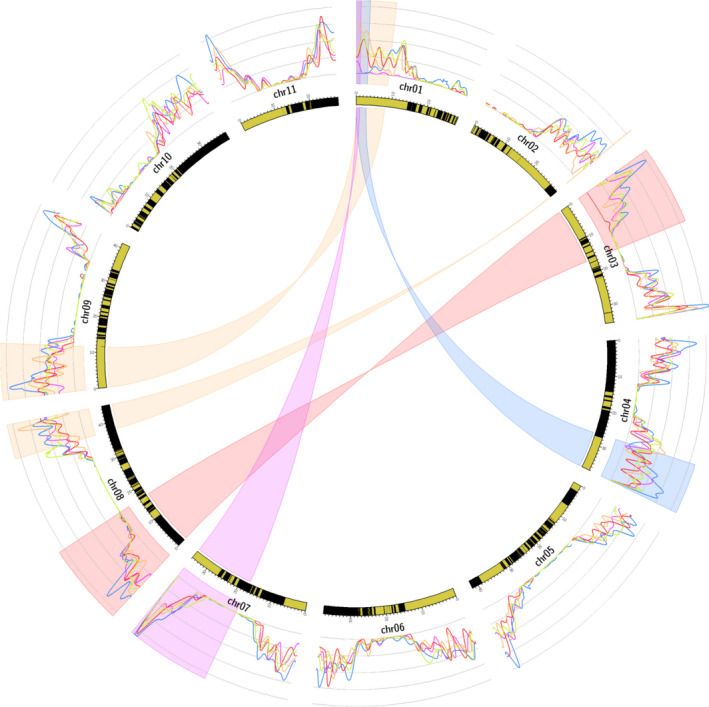
Circos representing the impact of translocations on recombination in structurally heterozygous accessions. Highlighted regions indicate the translocated chromosome segments for the 3/8 (red), 1/7 (purple), 1/4 (blue), and 1/9 and 2/8 (orange) translocations. The curves represent the average recombination rate for heterozygous parents for the 3/8 translocation (red), 1/7 translocation (purple), 1/4 translocation (blue), and 1/9 and 2/8 translocations (orange). The green curve represents structurally homozygous parents. The average recombination rate, calculated as the number of observed recombinations in 1‐Mb windows divided by the number of individuals with at least two markers in these regions, was obtained from the markers corrected matrix and by grouping all parents structurally heterozygous for the same structure. The inner circle represents theM.acuminatareference genome sequence V2, with black and yellow boxes representing the successive scaffolds.
For accessions structurally heterozygous for the 1/4 translocation, no recombination was observed in the T1 fragment and the recombination rate was reduced in a region of 4–5 Mb after the translocation breakpoint of chromosome 1 (Figure 4, blue curve, Figure S4). Recombination was also reduced around the recombination breakpoint of chromosome 4.
For accessions structurally heterozygous for the 1/9 and 2/8 translocations, no or very few recombinations were observed in the translocated T2 segment, which was very small in size (240 kb). A recombination reduction was observed around translocation breakpoints of chromosomes 1, 8, and 9 (Figure 4, orange curve, Figure S4). Note that recombination was suppressed in a region 1 Mb from the translocation breakpoint of chromosome 2.
For accessions structurally heterozygous for the 3/8 translocation, recombination was highly reduced in the inverted translocated segment T3 (Figure 4, red curve, Figure S4), with only one individual with a double recombination of 850 kb in the 158 individuals studied. However, no recombination reduction was observed on chromosome 8.
For accessions structurally heterozygous for the 1/7 translocation, the recombination rate was highly reduced for the entire chromosome 1 relative to structurally homozygous accessions. This recombination reduction was less pronounced in translocated segment T1 in which rare recombinations were observed (Figure 4, purple curve, Figure S4). No recombination reduction was observed on chromosome 7.
Aneuploidy in progeny from structurally heterozygous accessions
Analysis based on regular marker phase shifts and sequence coverage (see Experimental procedures) revealed 96 aneuploids out of the 1059 progeny individuals studied (Table 1). Two triploids resulting from diploid parents were also observed.
Among the aneuploid individuals, 78 were found in diploid populations from which 74 (95%) involved chromosomes contributing to structural heterozygosity in the parents. The presence and proportion of aneuploid individuals varied depending on the translocation involved. No aneuploid was observed in diploid progeny from a parent heterozygous for the 3/8 translocation and very few (between 0 and 3.9%) from a parent heterozygous for the 1/4, 1/7, and 1/9 translocations. In progenies from diploid parents heterozygous for the 2/8 translocation, aneuploids represented 21–23% (Table 1, Table S2).
Analysis of these 74 aneuploids found in diploid populations through sequence coverage and in silico parental chromosome painting showed that they could be classified in three categories (Table 1): (i) 52 aneuploids were segmental aneuploids with a euploid chromosome number but a chromosome segment in one or three copies instead of two. This aneuploidy resulted from a gamete that combined a translocated structure and a reference structure for only one of the chromosomes involved in a reciprocal translocation. One exception in this category concerned a particular individual that lacked a region from its Calcutta 4 homozygous parent at the start of chromosome 1; (ii) 17 aneuploid individuals had an additional chromosome and one individual cumulated one additional chromosome and a segmental aneuploidy; (iii) Five aneuploids had a complex structure involving chromosome 1. Finally, the triploid progeny (Pakid) from a diploid parent structurally heterozygous for the 1/4 and 1/7 translocations and a tetraploid parent structurally heterozygous for the 1/4 translocation showed a high proportion of aneuploids (21%).
Bias in chromosomal segregation in progeny from structurally heterozygous accessions
Segregation distortion along chromosomes was calculated as the average proportion of euploid individuals deviating from the expected segregation in each progeny. Depending on parental accessions, distinct chromosomes were involved at various orders of magnitude (Figure S4). The distorted segregations concerned structurally heterozygous chromosomes, but not exclusively, and these chromosomes were not always those that showed the highest segregation distortion.
The transmission ratio of the translocated chromosomes versus the reference chromosomes was then calculated in each progeny, excluding aneuploids involving translocated chromosomes. Translocated chromosomes were found to be preferentially transmitted relative to the reference chromosomes in 10 out of the 13 progenies analyzed (Table 1 and Table S2). For the 1/4 translocation, translocated chromosomes were preferentially transmitted in proportions of 71–98% in the three populations tested. For the 1/9 translocation, this proportion was in the range 60–72% in the three populations tested. For the 2/8 translocation, this preferential transmission concerned two populations (61 and 65%) out of the three tested. For the 1/7 translocation, preferential transmission (59%) was observed in one of the two diploid populations tested. However, the other population was very small (29 individuals). For the 3/8 translocation, translocated chromosomes were preferentially transmitted in one of the two populations tested (57%).
Discussion
Translocations emerged in distinct wild Musa subspecies with different geographical distributions
We characterized three large reciprocal translocations in the M.acuminata species in addition to the three that we reported previously (Martin et al., 2017; Dupouy et al., 2019). Musa balbisiana has another reciprocal translocation involving chromosomes 1 and 3 and a large inversion compared to the M.acuminata reference structure (Baurens et al., 2019). No large structural variation was found in the M.schizocarpa genome assembly compared to the M.acuminata reference structure (Belser et al., 2018) (Figure 5).
Figure 5.
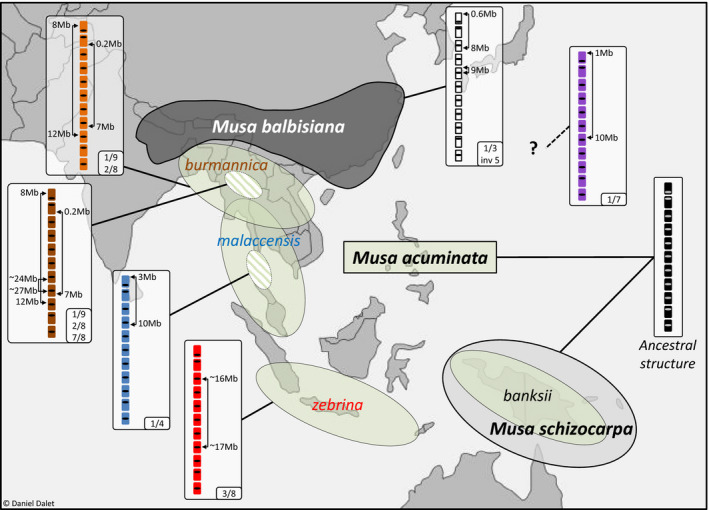
Geographical distribution of the main Musa species and subspecies involved in cultivated bananas and the associated translocated chromosome structures. Areas indicate the geographical distribution of the main Musa species and subspecies according to Perrier et al. (2011). Boxes represent structural rearrangements found in these species and subspecies. Hatched circles indicate that translocations were identified in part of the subspecies but do not point to specific regions. The outline map was modified from http://www.histgeo.ac‐aix‐marseille.fr/ancien_site/carto.
The distribution of the six M. acuminata translocations in 155 accessions representative of banana diversity, including 53 diploid wild representatives and 102 diploid and triploid cultivars or hybrids, revealed interesting features. The distribution showed that the six translocations were present only in M. acuminata and not in the other Musa species tested, suggesting that they had emerged in this species. These observations confirmed that the M. acuminata reference sequence, which was generated from a M.acuminata ssp. malaccensis accession, represents the ancestral chromosome structure of M.acuminata, as suggested by Dupouy et al. (2019). No large translocations were found in ssp. banksii and most accessions from ssp. malaccensis, suggesting that they had the ancestral structure. In addition, each translocation was found to be limited to a M.acuminata subspecies, supporting the hypothesis that translocation 1/4 emerged in ssp. malaccensis (Martin et al., 2017) and translocations 2/8 and 1/9 in ssp. burmannica (Dupouy et al., 2019). In addition, our results suggested that translocation 7/8 also emerged in ssp. burmannica after the 1/9 and 2/8 translocation and that translocation 3/8 emerged in ssp. zebrina. Most of these translocations corresponded to independent events, except for translocation 7/8 that involved the translocated 8T2 chromosome. The geographical distributions of each translocation likely correspond to the distributions of these subspecies that were each reported to be limited to one continental region or island of Southeast Asia (Figure 5) (Perrier et al., 2009).
The origin of translocation 1/7 could not be defined because it was found only at the heterozygous state and only in cultivars or hybrids. The sample we analyzed contained representatives of all species and subspecies previously indicated as being involved in banana cultivars, which suggested that this chromosome translocation emerged in a so far uncharacterized Musa gene pool that contributed to many M.acuminata cultivars. The distribution of this translocation in the cultivars analyzed (including Pisang Madu and Cavendish) suggested that this gene pool corresponds to the cryptic wild gene pool hypothesized by Martin et al. (2020).
Shepherd (1999), in an elegant in‐depth synthesis of chromosome pairing configurations analyses in intersubspecific hybrid accessions (Dodds, 1943; Dodds and Simmonds, 1948; Dessauw, 1987; Fauré, Bakry, et al., 1993; Shepherd, 1999), suggested the presence of nine to ten translocations in M.acuminata distributed in seven to eight translocation groups differing by one to four translocations (Figure S5). Our results confirmed five of these translocation groups and allowed us to determine the chromosomes involved and the architectures of these translocations while specifying their distributions. The 1/4 translocation corresponded to the ‘Northern Malayan’ group of Shepherd (1999), the 1/9 and 2/8 to the ‘Northern 1’ group, the 7/8 to the ‘Northern 2’ group and the 3/8 to the ‘Javanese’ group. The ancestral ‘Standard’ group of Shepherd corresponded to our ancestral structure. Interestingly, chromosomal pairing observations (Shepherd, 1999) combined with the new knowledge regarding the chromosomes involved, suggested that the remaining ‘East Africa’ group has a translocation involving chromosome 9 and one of the four chromosomes 5, 6, 10, or 11 (Appendix S1). A similar reasoning suggested that the ‘Malayan Highland’ group has a reciprocal translocation involving chromosome 1 and/or 4 and another chromosome. Meanwhile, the 1/7 translocation may correspond to the eighth group suggested by Shepherd; however, this hypothetical group was based on only one structurally heterozygous cultivar (Appendix S1).
Translocation distribution sheds light on banana cultivar origins
Most cultivars analyzed here (70.5%) were structurally heterozygous for 1 to 4M. acuminata translocations. Translocations 1/4 and 3/8, which reflect the presence of chromosomes from ssp. malaccensis and zebrina, were found frequently in cultivars. This supports previous hypotheses regarding the important contribution of both subspecies to cultivars (Carreel et al., 2002; Perrier et al., 2009; Perrier et al., 2011; Christelová et al., 2017). Conversely, translocations 1/9, 2/8, or 7/8, which reflect the presence of chromosomes from ssp. burmannica, were rarely found in cultivars, in accordance with the suspected low contribution of this genetic group to cultivars (Carreel et al., 1994; Perrier et al., 2011). The 1/7 translocated structure was found in many cultivated bananas, including Cavendish banana which represents 50% of world banana production. These data suggested that the so far uncharacterized wild Musa gene pool hypothesized by Martin et al. (2020), from which this translocation probably emerged, has contributed to many cultivars. This stresses the need for new collecting campaigns to identify the corresponding wild gene pool so that it could be used in banana breeding programs.
Among the main cultivars, only the cooking‐type banana cultivars, which are interspecific cultivars (‘AAB’ and ‘ABB’), have not inherited translocated chromosomes from M.acuminata. This is in line with the fact that their A genome is related to ssp. banksii, which has the ancestral structure (Hippolyte et al., 2012), and suggested that their origin is less complex than most other cultivars. In East‐African‐Banana ‘AAA’, which are considered to be mixed‐type (cooking and dessert), the presence of the 3/8 translocated chromosome in two copies, originating from ssp. zebrina, is in line with the suspected major contribution of ssp. zebrina to this subgroup (Hippolyte et al., 2012). Conversely, triploid cultivars with the most translocations (1/4, 1/7, and 3/8) are dessert‐type banana cultivars, including the world top ranking subgroup Cavendish and its predecessor Gros Michel. This translocation pattern agrees with the very complex origin of the Cavendish subgroup (Martin et al., 2020) and with its suspected parents (Raboin et al., 2005; Hippolyte et al., 2012). The Figue dessert‐type subgroup includes the sole intensively grown diploid cultivars, which are among the few diploid ‘AA’ cultivars that have three translocations. Finally, the combination of several chromosomal structures in many interspecific and inter(sub)specific cultivars (diploid and triploid) suggested that cultivated banana origins are more complex than previously assumed, with multiple hybridization steps as recently proposed (De Langhe et al., 2010; Baurens et al., 2019; Martin et al., 2020).
Impact of reciprocal translocations on genetic analysis and breeding
Our results showed that reciprocal translocations in structurally heterozygous parents generated (i) a reduction or absence of recombination; (ii) a bias in chromosome segregation in several cases; and (iii) genetic linkage between the four chromosome regions involved in the translocations. The reduction or absence of recombination would prevent precise localization of the genomic interval containing a targeted gene or quantitative trait locus (QTL), particularly when it concerns a large chromosome segment (case of translocations 1/4 and 3/8) or an entire chromosome (case of translocation 1/7). In addition, genetic linkage between the four chromosome regions involved in the reciprocal translocations would generate multiple QTL intervals on both chromosomes, again resulting in imprecise localization of the region involved in the targeted traits. These phenomena would also impact GWAS (i.e. genome‐wide association study) analysis by generating a strong population structure.
Information on the genetic determinism of traits in bananas is starting to be reported (Sardos et al., 2016b; Nyine et al., 2019) but is still lagging far behind other crops. Once known, this information could be combined with information regarding chromosome structure to help design breeding strategies. If recombination between alleles from genes located on the same chromosome is targeted, then parents structurally homozygous for these chromosomes or chromosome regions should be chosen or developed. Conversely, structurally heterozygous parents could be used or developed to fix genetic combinations on certain chromosomes.
In this context, the information that we have provided on 155 accessions regarding their chromosome structures (Table S1) will be very useful when designing genetic determinism studies for important agronomic traits and breeding strategies. Banana breeding programs are particularly hard to manage as a result of the low hybrid fertility rates, as well as the large size of banana plants, requiring large fields for selection trials. Improving their efficiency is of high priority considering the current global outbreak of Fusarium tropical race 4 (TR4) that is seriously endangering this crop (Dita et al., 2020) and given the massive use of pesticides to control other diseases (de Lapeyre de Bellaire et al., 2010).
Reciprocal translocations and Musa evolution
In M.acuminata, we characterized six large reciprocal translocations and detected no other types of large structural variations. This showed that reciprocal translocations are the most frequent type of large chromosomal rearrangements in this species.
In the diploid progenies that we studied, not all chromosomal combinations were found, suggesting a reduction of fitness of heterozygous individuals as a result of gametic and possibly zygotic selection. Indeed, because of translocations, some chromosome combinations lead to unbalanced gametes with a missing chromosome segment (Martin et al., 2017). This situation appears to be lethal for gametes in most cases, except when it involves segment T2 (e.g. in the case of translocation 2/8). This exception is likely a result of the very small size of this segment (240 kb) that encompass only 36 predicted protein coding gene models. In the sole triploid population that we analyzed, more aneuploids were observed because the absence of a chromosome or chromosome segment in a polyploid context may be offset by another copy, as previously proposed (Birchler, 2013; Baurens et al., 2019). It is important to note that embryo rescue has been applied in banana breeding and in the progenies that we analyzed. In the absence of this process, the number of descendants per progeny is very low, except for wild crosses within subspecies. Crosses between subspecies may induce endosperm formation problems, which would warrant investigation. Vegetative multiplication, which is a prime propagation mode in Musa species, may be a factor that could allow the colonization of rearrangements even if they induce a reduction of fertility in heterozygous individuals.
Most of these translocations were limited in their distribution in wild material to a subspecies or a subset within a subspecies (Figure 5). Some of them were found in all the accessions we tested, from a subspecies and may thus be fixed. Others were only found in some accessions within a subspecies, and so they are not fixed at the subspecies scale and may have emerged more recently. Because many M.acuminata subspecies are isolated from each other and may have been even more restricted in their distribution and number in past climatic cycles, genetic drift may be one factor that contributed to the fixation of these translocations.
Interestingly, in many progenies, we showed a chromosome transmission bias that favored the translocated chromosomes. This bias in translocated structure transmission was not observed in all progeny and its intensity was variable. This may be a result of the phylogenomic context of those translocations. To allow colonization of a translocation, it could be assumed that this bias would be efficient in the phylogenomic context in which the translocation emerged, but might not be efficient in other phylogenomic settings. In addition, the factors causing this bias may be selected during colonization of the translocation but may gradually disappear when the translocation is fixed in the population.
Various types of mechanisms favoring the colonization of structural rearrangements have been reported (Brown and O’Neill, 2010). Among those factors, the reduction of gene flow as a result of suppression of recombination could lead to the accumulation of incompatibility alleles or genes that confer adaptive advantages. This has been reported particularly in chromosome inversion cases (Kirkpatrick, 2010; Huang and Rieseberg, 2020). However, because major recombination reductions were only associated with some of the translocations we described, this may not be the main factor involved here. Rearrangements may also cause an increased expression of genes previously epigenetically silenced by a position effect near the centromere or other large blocks of heterochromatin (Dernburg et al., 1996). Interestingly, in amniotes, Larkin et al. (2009) showed that regions maintained collinear during evolution are enriched in genes involved in fundamental development processes and biological organization, and that regions often involved in the rearrangement breakpoints are enriched in genes associated with adaptive functions. This suggested that these breakpoint regions are hotspots of evolutionary activity where genes are created, amplified, and destroyed by a variety of molecular mechanisms.
Segregation distortions were observed in several progenies in regions not involved in structural variations, suggesting that allelic incompatibilities exist between Musa accessions. In populations with one parent in common, because these segregation distortions did not always involve the same regions, it could be hypothesized that they at least partly resulted from zygotic selection. This type of incompatibility, when located in these translocated regions, could contribute to the fixation of some translocations. Meiotic drive mechanisms (Buckler et al., 1999) could also be involved in the preferential transmission of translocated chromosomes and would be worthy of investigation.
Finally, it could be envisaged that translocations regularly occur in M.acuminata and that most of them are eliminated except those that are preferentially transmitted and could thus colonize the population.
Experimental procedures
Plant material and sequencing
Eleven progenies from 18 distinct parents, comprising 29 to 177 individuals and representing 1059 individuals in total, were analyzed (Table 1). Ten progenies were diploid and one was triploid. They were produced at the CIRAD research station in Guadeloupe (French West Indies). Borli and AFPTBA00267 populations have already been described in Hippolyte et al. (2010) and Martin et al. (2017), respectively. All populations, except AFPTBA00267, were genotyped‐by‐sequencing (GBS), with a library made at the GPTR platform (https://www.gptr‐lr‐genotypage.com) using PstI and MseI restriction enzymes and sequencing performed on the GeT‐PlaGe platform (https://get.genotoul.fr) or Genoscope (http://www.genoscope.cns.fr) using an Illumina HiSeq sequencer (Illumina, San Diego, CA, USA). Raw sequence reads were demultiplexed using gbsx, version 1.2 (Herten et al., 2015). Adaptors were removed and reads were quality trimmed using the cutadapt program (Martin, 2011).
In total, 155 accessions, most of which were M.acuminata accessions along with 21 from other species, obtained from the Musa CRB Plantes tropicales collection in Guadeloupe (http://crb‐tropicaux.com/Portail) or the International Transit Center (ITC) in Belgium (https://www.bioversityinternational.org/banana‐genebank), were analyzed (Table S1). Total DNA was sequenced using the Illumina HiSeq 4000 platform at Genoscope.
BAC‐FISH
Chromosome preparations and in situ hybridization were performed as described in D’Hont et al. (2000) with modifications. BAC clones (Table S3) from accession DH‐Pahang (D’Hont et al., 2012; http://banana‐genome.cirad.fr) were labeled by random priming with biotin‐14‐dUTP (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA) or Alexa 488‐5‐dUTP (Life Technologies; Thermo Fisher Scientific). Chromosome preparations were incubated in RNAse A (100 ng µl–1) and pepsin (100 mg ml–1) in 0.01 m HCl and fixed with paraformaldehyde (4%). Biotinylated probes were immunodetected by Texas Red avidin DCS (Vector Laboratories, Burlingame, CA, USA) and the signal was amplified with biotinylated antiavidin D (Vector Laboratories). Fluorescence images were captured using a cooled high‐resolution black and white CCD camera (ORCA; Hamamatsu, Hamamatsu City, Japan) fitted on a DMRXA2 fluorescence microscope (Leica Microsystems, Wetzlar, Germany) and analysed using volocity (Perkin Elmer, Waltham, MA, USA).
Variant calling and vcf filtration
Variant calling was performed using the M.acuminata reference sequence V2 (Martin et al., 2016). Two separate vcf files were obtained: one with all segregating population individuals and a second with the 155 banana diversity representatives. Variant calling was performed using the vcfhunter toolbox (https://github.com/SouthGreenPlatform/VcfHunter) (Garsmeur et al., 2018) as described in Baurens et al. (2019). Only bi‐allelic sites with no indels were selected for the analyses.
Segregating marker selection and phasing
For each population, a sub‐vcf was generated and filtered, according to the data point coverage, allele frequency, and missing data, using the ‘vcfFilter.1.0.py’ script of the vcfhunter toolbox, as described in Baurens et al. (2019). Segregating markers were selected according to their segregation ratio in the progeny (1:1 for bi‐parental cross or 1:2:1 for selfing) using the ‘vcf2popNew.1.0.py’ script of the vcfhunter toolbox. For each parent, the marker phase was determined using joinmap, version 4.1 (https://www.kyazma.nl/index.php/JoinMap). A new coding matrix was generated by arbitrarily reversing the marker coding of one phase (i.e. joinmap coding ‘nn’ and ‘np’ markers of Phase 1 were converted to ‘np’ and ‘nn’, respectively). This matrix manipulation allowed us to assign the same code to each parental haplotype and to easily visualize individual recombinations along reference chromosomes (Martin et al., 2016).
Marker linkage and segregation analysis
Selected markers from each parent of each population were used to calculate simple matching pairwise dissimilarities (corresponding to the observed recombination frequency between pairs of markers) and marker segregation distortions [calculated as –log10 (P‐value of chi‐squared test)]. These statistics were used to generate dot‐plots showing marker genetic linkage and segregation distortions along reference chromosomes. These analyses were performed using the ‘RecombCalculatorDDose.py’ and ‘Draw_dot_plot.py’ scripts of the vcfhunter toolbox.
Aneuploid identification
The presence of aneuploids was investigated for all progeny individuals and for each chromosome class via two complementary approaches:
Individuals showing unexpected parental marker distribution patterns along chromosomes were analyzed. Regions with both phases of one parent (i.e. the two haplotypes of a single parent) or lacking both haplotypes of the parent were identified. In these regions, all markers were heterozygous when both haplotypes of one parent were present and homozygous for all markers when both haplotypes of one parent were absent. Individuals showing these patterns were classified as aneuploid.
To detect aneuploids resulting from haplotype doubling (undetectable with the first approach), sequence coverage was calculated and plotted along the chromosomes using ‘vcf2cov.py’ (included in the vcfhunter toolbox) for each progeny individual. The obtained graphical sequence coverage representation allowed us to identify aneuploid individuals with complete or partial chromosome gain (increased sequence coverage) or loss (decreased sequence coverage).
Marker correction and chromosomal recombination rate
Missing data, sequencing errors, and aneuploidy generate false recombination breakpoints and lead to overestimation of the recombination rate. Aneuploid individuals were discarded and marker correction using the ‘GBS_corrector.py’ tool of scaffhunter (Martin et al., 2016) was applied following an iterative approach: (i) To prevent bias associated with distinct marker numbers in populations during correction, a subset of one marker every 100 kb was selected for each parental marker file. (ii) For each marker per individual, the central marker was corrected to the consensus of its two surrounding markers (i.e. no double recombination in a 200‐kb window). (iii) Then, correction was applied with the four surrounding markers (i.e. one or two markers could not generate double recombination in a 400‐kb window). (iv) A last polishing step with manual correction was applied to convert missing data at recombination breakpoints with the marker code of the preceding marker and to convert blocks of less than four markers considered as errors and corrected with the surrounding marker codes (i.e. no double recombinations allowed in a 400‐kb window).
The recombination rate was then calculated on corrected markers using the ‘CaReRa.py’ script (included in the vcfhunter toolbox). This script calculates, for each individual, the number and location of parental haplotype shifts corresponding to recombination events. These values were then used to calculate an average recombination number observed in the population in a 1‐Mb window corresponding to the recombination rate. The type of cross was taken into account for this calculation (i.e. in case of selfing, the observed recombination rate was divided by two because the observed recombination is the result of two independent meiosis of the same parent). The results were visualized with circos (Krzywinski et al., 2009).
Tracing chromosome structures in progenies and Musa diversity
Two distinct approaches were developed to trace the distinct chromosome structures in progenies and Musa germplasm.
First, taking advantage of the absence of recombination observed in some of the translocated regions, it was possible to identify haplotypes corresponding to these regions (i.e. segment T1 for translocations 1/4, most of chromosome 1 for translocations 1/7, segment T3 for translocations 3/8). Parental haplotypes for structurally heterozygous chromosomes were constructed using the ‘VcfAndCarto2haplo.py’ script of the vcfhunter toolbox, from the phased matrix of markers. Haplotypes in the non‐recombining regions were compared in two parental accessions that shared the same translocated chromosome structure (i.e. ‘PB1’ and ‘Pisang Lilin’ for the 1/4 translocation, ‘Pisang Madu’ and ‘IDN 110’ for the 1/7 translocation, ‘PM1’ and ‘PM2’ for the 3/8 translocation) aiming to identify haplotypes corresponding to the translocated chromosomes and then searched for corresponding specific alleles in individuals (Figure S6) using the ‘HaploProp.py’ script (added to vcfhunter toolbox).
This first approach could not be applied for 1/9 and 2/8 translocations because the involved chromosomes recombine. A second approach based on in silico chromosome painting was performed in the BCM, PCMo, and PCZ progenies heterozygous for 1/9 and 2/8 translocations transmitted by the ‘Calcutta 4’ parent (Table S2) using the ‘vcf2allPropAndCov.py’ script (Baurens et al., 2019) to identify the origin of chromosome segments surrounding breakpoints. Individuals of ‘Calcutta 4’ origin around translocation breakpoints would thus have the translocated chromosomes (Figure S7). No incongruences were observed when both approaches were applied to other populations (PMK, PMP, and PBC) (Table S2).
Specific alleles obtained from the first approach were used to trace 1/4, 1/7, and 3/8 chromosome structures in Musa diversity using a tailored methodology (see Methods S1, Figures S8 and S9). Similarly, the second approach was used to trace the 1/9, 2/8, and 7/8 chromosome structures in Musa diversity using a tailored methodology (see Methods S1, Figure S10, Table S4).
Distribution of distinct chromosome structures in Musa diversity
A subset of 3979 markers, corresponding to the 3092 polymorphous SNPs published in Dupouy et al. (2019) and including 144, 367, and 384 additional markers specific to 1/4, 1/7, and 3/8 translocated structures, respectively, was selected from the vcf obtained from the diversity dataset. This subset was used to calculate a dissimilarity matrix on M.acuminata diploid accessions using the ‘vcf2dis.1.0.py’ script (added to the vcfhunter toolbox) as described in Martin et al. (2017) and Dupouy et al. (2019). The dissimilarity index was calculated as the proportion of unmatching alleles. The dissimilarity matrix was used to perform a factorial analysis using r, version 3.2.4 (http://www.r‐project.org). Because the cultivar accessions originated from the wild banana genepools, a factorial analysis was performed with the 34 wild accessions. The 58 cultivar accessions were then projected along the synthetic axes.
AUTHOR CONTRIBUTIONS
GM, ADH, and NY designed the study; GM, FCB, ADH, FS, and SR designed the genetic crosses; FS, SR, and JMD developed the progenies; FCB, CH, and PM performed DNA extraction and constructed GBS libraries; CH performed the BAC‐FISH experiments. KL, AP, and AL produced the sequencing data; FCB, LB, and MD performed some segregation analysis; GM designed and performed the bioinformatic analysis and developed bioinformatic programs; GM, ADH, and FC interpreted the data; GM and ADH wrote the manuscript; NY, FC and FCB edited the manuscript. All authors read and approved the final manuscript submitted for publication.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Table S1. Distribution of the translocated structures in the studied Musa germplasm
Table S2. Chromosomal segregation in progeny from structurally heterozygous accessions.
Table S3. BAC used in the BAC‐FISH experiments.
Table S4. Accessions used to search for 1/9, 2/8, and 7/8 translocation in Musa germplasm.
Appendix S1. Inferences from earlier chromosome pairing configuration studies.
Methods S1. Distribution of distinct chromosome structures in Musa diversity.
Figure S1. Dot‐plots of pairwise marker genetic linkage in the diploid parents of the studied populations.
Figure S2. Schematic representation of the inferred 2/8 and 7/8 translocated chromosomes.
Figure S3. Translocation distributions in diploid Musa acuminata germplasm.
Figure S4. Circos with recombination rate and segregation distortion.
Figure S5. Inferences from earlier chromosome pairing configuration studies.
Figure S6. Example of determination of translocated structures using allele proportion.
Figure S7. Example of determination of translocated structures using in silico chromosome painting.
Figure S8. Detection of the translocated structures in the 155 tested accessions based on allele proportion.
Figure S9. Read proportion profiles for the detection of the 1/4, 1/7, and 3/8 translocations presence.
Figure S10. Read proportion supporting 1/9, 2/8, and 7/8 translocations presence.
Acknowledgements
This work was supported by CIRAD, Genoscope, CEA, France Génomique (ANR‐10‐INBS‐09‐08) ‘DynaMo’ project, Agropolis Fondation (ID 1504‐006) ‘GenomeHarvest’ project through the French Investissements d’avenir programme (Labex Agro: ANR‐10‐LABX‐0001‐01), and the CGIAR Research Programme on Roots, Tubers and Bananas. We thank CRB Plantes Tropicales Antilles CIRAD‐INRA Guadeloupe France and the Bioversity International Transit Centre (https://www.bioversityinternational.org/banana‐genebank) for providing the plant materials. We are grateful to Franck Marius, Chantal Guiougou, and Jean‐Claude Efile from CIRAD Guadeloupe for their progeny development contributions. We thank Dr Eric Jenczewski for critically reading the manuscript. The CIRAD – UMR AGAP HPC Data Center of the South Green Bioinformatics platform (http://www.southgreen.fr) provided the computational resources.
Contributor Information
Guillaume Martin, Email: guillaume.martin@cirad.fr.
Angélique D’Hont, Email: angelique.d'hont@cirad.fr.
Data Availability Statement
The VCF files, specific allele list, and genetic mapping matrices are available in the download section of the Banana Genome Hub (http://banana‐genomehub.southgreen.fr).
The Illumina data of the 11 progenies are available in the SRA database under project PRJNA667853.
The raw whole genome sequencing illumina from the 155 accessions analyzed during the present study are not publicly available because they are part of a larger ongoing project, although they are available from the corresponding author on reasonable request.
References
- Baker, R.J. and Bickham, J.W. (1986) Speciation by monobrachial centric fusions. Proc. Natl. Acad. Sci. 83, 8245–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurens, F.‐C. , Martin, G. , Hervouet, C. et al (2019) Recombination and Large Structural Variations Shape Interspecific Edible Bananas Genomes. Mol. Biol. Evol. 36, 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser, C. , Istace, B. , Denis, E. et al (2018) Chromosome‐scale assemblies of plant genomes using nanopore long reads and optical maps. Nat. Plants, 4, 879. [DOI] [PubMed] [Google Scholar]
- Birchler, J.A. (2013) Aneuploidy in plants and flies: the origin of studies of genomic imbalance. Semin. Cell Dev. Biol. 24, 315–319. [DOI] [PubMed] [Google Scholar]
- Britton‐Davidian, J. , Catalan, J. , da Ramalhinho, M. , Ganem, G. , Auffray, J.‐C. , Capela, R. , Biscoito, M. , Searle, J.B. and da Luz Mathias, M. (2000) Rapid chromosomal evolution in island mice. Nature, 403, 158. [DOI] [PubMed] [Google Scholar]
- Brown, J.D. and O’Neill, R.J. (2010) Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu. Rev. Genomics Hum. Genet. 11, 291–316. [DOI] [PubMed] [Google Scholar]
- Buckler, E.S. , Phelps‐Durr, T.L. , Buckler, C.S.K. , Dawe, R.K. , Doebley, J.F. and Holtsford, T.P. (1999) Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics, 153, 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo, M.C. , Asenjo, A. and Casares, P. (2000) Location of Shfr, a new gene that rescues hybrid female viability in crosses between Drosophila simulans females and D. melanogaster males. Heredity, 84(Pt 6), 630–638. [DOI] [PubMed] [Google Scholar]
- Carreel, F. , Fauré, S. , De Leon, D.G. , Lagoda, P.J.L. , Perrier, X. , Bakry, F. , Du Montcel, H.T. , Lanaud, C. and Horry, J.P. (1994) Evaluation de la diversité génétique chez les bananiers diploïdes (Musa sp). Genet. Sel. Evol. 26, 125s–136s. [Google Scholar]
- Carreel, F. , de Leon, D.G. , Lagoda, P. , Lanaud, C. , Jenny, C. , Horry, J.P. and du Montcel, H.T. (2002) Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome, 45, 679–692. [DOI] [PubMed] [Google Scholar]
- Cenci, A. , Sardos, J. , Hueber, Y. , Martin, G. , Breton, C. , Roux, N. , Swennen, R. , Carpentier, S.C. and Rouard, M. (2020) Unravelling the complex story of intergenomic recombination in ABB allotriploid bananas. Ann. Bot Available at: https://academic.oup.com/aob/advance‐article/doi/10.1093/aob/mcaa032/5760888 [Accessed May 1, 2020]. [DOI] [PMC free article] [PubMed]
- Christelová, P. , De Langhe, E. , Hřibová, E. et al. (2017) Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers. Conserv. 26, 801–824. [Google Scholar]
- De Langhe, E. , Hřibová, E. , Carpentier, S. , Doležel, J. and Swennen, R. (2010) Did backcrossing contribute to the origin of hybrid edible bananas? Ann. Bot. 106, 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg, A.F. , Broman, K.W. , Fung, J.C. , Marshall, W.F. , Philips, J. , Agard, D.A. and Sedat, J.W. (1996) Perturbation of Nuclear Architecture by Long‐Distance Chromosome Interactions. Cell, 85, 745–759. [DOI] [PubMed] [Google Scholar]
- Dessauw, D. (1987) Etude des facteurs de la stérilité du bananier (Musa spp.) et des relations cytotaxinomiques entre M. acuminata Colla et M. balbisiana Colla. PhD thesis. Université de Paris‐sud. [Google Scholar]
- D’Hont, A. , Denoeud, F. , Aury, J.‐M. et al (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature, 488, 213–217. [DOI] [PubMed] [Google Scholar]
- Dita, M. , Teixeira, L.A.J. , O’Neill, W. et al (2020) Current state of Fusarium wilt of banana in the subtropics. In Acta Horticulturae International Society for Horticultural Science (ISHS), Leuven, Belgium, pp. 45–56.
- Dodds, K. and Simmonds, N. (1948) Sterility and parthenocarpy in diploid hybrids of musa. Heredity, 2, 101–117. [DOI] [PubMed] [Google Scholar]
- Dodds, K.S. (1943) Genetical and cytological studies of Musa. V. Certain edible diploids. J. Genet. 45, 113–138. [Google Scholar]
- Dupouy, M. , Baurens, F.‐C. , Derouault, P. et al (2019) Two large reciprocal translocations characterized in the disease resistance‐rich burmannica genetic group of Musa acuminata. Ann. Bot. 124, 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria, R. and Navarro, A. (2010) Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol. Evol. 25, 660–669. [DOI] [PubMed] [Google Scholar]
- Fauré, S. , Bakry, F. and González de Leon, D. (1993) Cytogenetic studies of diploid bananas Breeding Banana and Plantain for Resistance to Diseases and Pests. CIRAD‐FLHOR, Montpellier: Ganry J.,, pp. 77–92. [Google Scholar]
- Fauré, S. , Noyer, J.L. , Horry, J.P. , Bakry, F. , Lanaud, C. and de León, D.G. (1993) A molecular marker‐based linkage map of diploid bananas (Musa acuminata). Theor. Appl. Genet. 87, 517–526. [DOI] [PubMed] [Google Scholar]
- Flowers, J.M. , Hazzouri, K.M. , Gros‐Balthazard, M. et al (2019) Cross‐species hybridization and the origin of North African date palms. Proc. Natl. Acad. Sci. 116, 1651–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsmeur, O. , Droc, G. , Antonise, R. et al (2018) A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 9, 2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, V. (1981) Plant Speciation, 2nd edn. New York, NY, USA: Columbia University Press. [Google Scholar]
- Herten, K. , Hestand, M.S. , Vermeesch, J.R. and Van Houdt, J.K. (2015) GBSX: a toolkit for experimental design and demultiplexing genotyping by sequencing experiments. BMC Bioinformatics, 16, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippolyte, I. , Bakry, F. , Seguin, M. et al (2010) A saturated SSR/DArT linkage map of Musa acuminata addressing genome rearrangements among bananas. BMC Plant Biol. 10, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippolyte, I. , Jenny, C. , Gardes, L. et al (2012) Foundation characteristics of edible Musa triploids revealed from allelic distribution of SSR markers. Ann. Bot Available at: http://aob.oxfordjournals.org/content/early/2012/02/09/aob.mcs010.abstract. [DOI] [PMC free article] [PubMed]
- Hont, A.d.’. , Paget‐Goy, A. , Escoute, J. and Carreel, F. (2000) The interspecific genome structure of cultivated banana, Musa spp. revealed by genomic DNA in situ hybridisation. Theor. Appl. Genet. 100, 177–183. [Google Scholar]
- Huang, K. and Rieseberg, L.H. (2020) Frequency, Origins, and Evolutionary Role of Chromosomal Inversions in Plants. Front. Plant Sci, 11. Available at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00296/full [Accessed May 1, 2020]. [DOI] [PMC free article] [PubMed]
- Janssens, S.B. , Vandelook, F. , De Langhe, E. , Verstraete, B. , Smets, E. , Vandenhouwe, I. and Swennen, R. (2016) Evolutionary dynamics and biogeography of Musaceae reveal a correlation between the diversification of the banana family and the geological and climatic history of Southeast Asia. New Phytol., n/a‐n/a. [DOI] [PMC free article] [PubMed]
- King, M. (1993) Species evolution: The role of chromosome change. Cambridge: Cambridge University Press. [Google Scholar]
- Kirkpatrick, M. (2010) How and why chromosome inversions evolve. PLoS Biol. 8, e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski, M. , Schein, J. , Birol, İ. , Connors, J. , Gascoyne, R. , Horsman, D. , Jones, S.J. and Marra, M.A. (2009) Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Z. , Nakazato, T. , Salmaso, M. , Burke, J.M. , Tang, S. , Knapp, S.J. and Rieseberg, L.H. (2005) Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics, 171, 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bellaire, L.d.L. , Fouré, E. , Abadie, C. and Carlier, J. (2010) Black Leaf Streak Disease is challenging the banana industry. Fruits, 65, 327–342. [Google Scholar]
- Larkin, D.M. , Pape, G. , Donthu, R. , Auvil, L. , Welge, M. and Lewin, H.A. (2009) Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 19, 770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G. , Baurens, F.‐C. , Droc, G. et al (2016) Improvement of the banana “Musa acuminata” reference sequence using NGS data and semi‐automated bioinformatics methods. BMC Genom. 17, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G. , Cardi, C. , Sarah, G. et al (2020) Genome ancestry mosaics reveal multiple and cryptic contributors to cultivated banana. Plant J., Available at: https://onlinelibrary.wiley.com/doi/abs/10.1111/tpj.14683 [Accessed May 1, 2020] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G. , Carreel, F. , Coriton, O. et al (2017) Evolution of the Banana Genome (Musa acuminata) Is Impacted by Large Chromosomal Translocations. Mol. Biol. Evol. 34, 2140–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011) Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnetjournal Vol 17 No 1 Gener. Seq. Data Anal Available at: http://journal.embnet.org/index.php/embnetjournal/article/view/200/458.
- Mbanjo, E. , Tchoumbougnang, F. , Mouelle, A. , Oben, J. , Nyine, M. , Dochez, C. , Ferguson, M. and Lorenzen, J. (2012) Molecular marker‐based genetic linkage map of a diploid banana population (Musa acuminata Colla). Euphytica, 188, 369–386. [Google Scholar]
- Mcfadden, E.S. and Sears, E.R. (1946) The origin of triticum spelta and its free‐threshing hexaploid relatives. J. Hered. 37, 81–89. [DOI] [PubMed] [Google Scholar]
- Němečková, A. , Christelová, P. , Čížková, J. et al (2018) Molecular and Cytogenetic Study of East African Highland Banana. Front. Plant Sci, 9. Available at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01371/full [Accessed November 13, 2018]. [DOI] [PMC free article] [PubMed]
- Nyine, M. , Uwimana, B. , Akech, V. , Brown, A. , Ortiz, R. , Doležel, J. , Lorenzen, J. and Swennen, R. (2019) Association genetics of bunch weight and its component traits in East African highland banana (Musa spp. AAA group). Theor. Appl. Genet Available at: 2019, 10.1007/s00122-019-03425-x [Accessed September 19. [DOI] [PMC free article] [PubMed]
- Ostberg, C.O. , Hauser, L. , Pritchard, V.L. , Garza, J.C. and Naish, K.A. (2013) Chromosome rearrangements, recombination suppression, and limited segregation distortion in hybrids between Yellowstone cutthroat trout (Oncorhynchus clarkii bouvieri) and rainbow trout (O. mykiss). BMC Genom. 14, 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostevik, K.L. , Samuk, K. and Rieseberg, L.H. (2020) Ancestral Reconstruction of Karyotypes Reveals an Exceptional Rate of Non‐random Chromosomal Evolution in Sunflower. Genetics, Available at: https://www.genetics.org/content/early/2020/02/07/genetics.120.303026 [Accessed March 4, 2020]. [DOI] [PMC free article] [PubMed]
- Perrier, X. , Bakry, F. , Carreel, F. , Jenny, C. , Horry, J.‐P. , Lebot, V. and Hippolyte, I. (2009) Combining Biological Approaches to Shed Light on the Evolution of Edible Bananas. Ethnobot. Res Appl., 7. Available at: http://lib‐ojs3.lib.sfu.ca:8114/index.php/era/article/view/362/231 [Accessed January 1, 2009].
- Perrier, X. , De Langhe, E. , Donohue, M. et al (2011) Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. 108, 11311–11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach, A.T. , Revay, T. , Villagomez, D.A.F. , Macedo, M.P. , Sullivan, A. , Maignel, L. , Wyss, S. , Sullivan, B. and King, W.A. (2016) Prevalence and consequences of chromosomal abnormalities in Canadian commercial swine herds. Genet. Sel. Evol. 48, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillet, M.C. , Madjidian, N. , Griveau, Y. , Serieys, H. , Tersac, M. , Lorieux, M. and Bervillé, A. (1995) Mapping genetic factors controlling pollen viability in an interspecific cross in Helianthus sect. Helianthus. Theor. Appl. Genet. 91, 1195–1202. [DOI] [PubMed] [Google Scholar]
- Raboin, L.M. , Carreel, F. , Noyer, J.‐L. et al (2005) Diploid ancestors of triploid export banana cultivars: molecular identification of 2n restitution gamete donors and n gamete donors. Mol. Breed. 16, 333–341. [Google Scholar]
- Santos, J.D. , Chebotarov, D. , McNally, K.L. , Bartholomé, J. , Droc, G. , Billot, C. and Glaszmann, J.C. (2019) Fine Scale Genomic Signals of Admixture and Alien Introgression among Asian Rice Landraces. Genome Biol. Evol. 11, 1358–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardos, J. , Perrier, X. , Doležel, J. , Hřibová, E. , Christelová, P. , Van den houwe, I. , Kilian, A. and Roux, N. (2016a) DArT whole genome profiling provides insights on the evolution and taxonomy of edible Banana (Musa spp.). Ann. Bot. 118, 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardos, J. , Rouard, M. , Hueber, Y. , Cenci, A. , Hyma, K.E. , van den Houwe, I. , Hribova, E. , Courtois, B. and Roux, N. (2016b) A Genome‐Wide Association Study on the Seedless Phenotype in Banana (Musa spp.) Reveals the Potential of a Selected Panel to Detect Candidate Genes in a Vegetatively Propagated Crop. PLoS One, 11, e0154448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, K. (1999) Cytogenetics of the genus Musa, IPGRI. [Google Scholar]
- Simmonds, N.W. (1962) The evolution of the bananas. London: Longmans. [Google Scholar]
- Simmonds, N.W. and Shepherd, K. (1955) The taxonomy and origins of the cultivated bananas. J. Linn. Soc. Lond. Bot. 55, 302–312. [Google Scholar]
- Šimoníková, D. , Němečková, A. , Karafiátová, M. , Uwimana, B. , Swennen, R. , Doležel, J. and Hřibová, E. (2019) Chromosome Painting Facilitates Anchoring Reference Genome Sequence to Chromosomes In Situ and Integrated Karyotyping in Banana (Musa Spp.). Front. Plant Sci, 10. Available at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01503/full [Accessed March 24, 2020]. [DOI] [PMC free article] [PubMed]
- Stathos, A. and Fishman, L. (2014) Chromosomal rearrangements directly cause underdominant F1 pollen sterility in Mimulus lewisii–Mimulus cardinalis hybrids. Evolution, 68, 3109–3119. [DOI] [PubMed] [Google Scholar]
- Tadmor, Y. , Zamir, D. and Ladizinsky, G. (1987) Genetic mapping of an ancient translocation in the genus Lens. Theor. Appl. Genet. 73, 883–892. [DOI] [PubMed] [Google Scholar]
- White, M.J. (1978) Modes of speciation. San Francisco: W.H. Freeman & Co. [Google Scholar]
- Wu, G.A. , Prochnik, S. , Jenkins, J. et al (2014) Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32, 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir, D. (2001) Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2, 983–989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Distribution of the translocated structures in the studied Musa germplasm
Table S2. Chromosomal segregation in progeny from structurally heterozygous accessions.
Table S3. BAC used in the BAC‐FISH experiments.
Table S4. Accessions used to search for 1/9, 2/8, and 7/8 translocation in Musa germplasm.
Appendix S1. Inferences from earlier chromosome pairing configuration studies.
Methods S1. Distribution of distinct chromosome structures in Musa diversity.
Figure S1. Dot‐plots of pairwise marker genetic linkage in the diploid parents of the studied populations.
Figure S2. Schematic representation of the inferred 2/8 and 7/8 translocated chromosomes.
Figure S3. Translocation distributions in diploid Musa acuminata germplasm.
Figure S4. Circos with recombination rate and segregation distortion.
Figure S5. Inferences from earlier chromosome pairing configuration studies.
Figure S6. Example of determination of translocated structures using allele proportion.
Figure S7. Example of determination of translocated structures using in silico chromosome painting.
Figure S8. Detection of the translocated structures in the 155 tested accessions based on allele proportion.
Figure S9. Read proportion profiles for the detection of the 1/4, 1/7, and 3/8 translocations presence.
Figure S10. Read proportion supporting 1/9, 2/8, and 7/8 translocations presence.
Data Availability Statement
The VCF files, specific allele list, and genetic mapping matrices are available in the download section of the Banana Genome Hub (http://banana‐genomehub.southgreen.fr).
The Illumina data of the 11 progenies are available in the SRA database under project PRJNA667853.
The raw whole genome sequencing illumina from the 155 accessions analyzed during the present study are not publicly available because they are part of a larger ongoing project, although they are available from the corresponding author on reasonable request.


