Abstract
Cellular condensation of intrinsically disordered proteins (IDPs) through liquid–liquid phase separation (LLPS) allows dynamic compartmentalization and regulation of biological processes. The IDP tau, which promotes the assembly of microtubules and is hyperphosphorylated in Alzheimer's disease, undergoes LLPS in solution and on the surface of microtubules. Little is known, however, about the influence of tau phosphorylation on its ability to nucleate microtubule bundles in conditions of tau LLPS. Herein, we show that unmodified tau as well as tau phosphorylated at disease‐associated epitopes condense into liquid‐like droplets. Although tubulin partitioned into and reached high concentrations inside all tau droplets, it was unable to grow into microtubules form the inside of droplets formed by tau phosphorylated at the AT180 epitope (T231/S235). In contrast, neither phosphorylation of tau in the repeat domain nor at its tyrosine residues inhibited the assembly of tubulin from tau droplets. Because LLPS of IDPs has been shown to promote different types of cytoskeletal assembly, our study suggests that IDP phosphorylation might be a broadly used mechanism for the modulation of condensate‐mediated cytoskeletal assembly.
Keywords: IDP, liquid-liquid phase separation, NMR spectroscopy, phosphorylation, tau
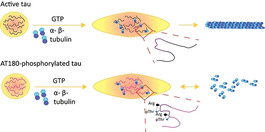
Active tau recruits tubulin into liquid‐like condensates and promotes microtubule assembly. Upon phosphorylation at the disease‐associated AT180‐epitope intramolecular salt bridges are formed and the microtubule‐assembly activity is lost.
Cells contain compartments that are linked to a diverse set of biochemical functions and pathological processes but are not surrounded by membranes. [1] Many of these membrane‐less organelles form by LLPS of IDPs. Condensation of IDPs into membrane‐less organelles strongly increases their concentration inside the condensates. In addition, enzymes and other factors are recruited to membrane‐less organelles, changing both the basic biophysical properties of membrane‐less organelles and allowing for their energy‐dependent regulation. [2]
The IDP tau promotes the assembly and bundling of microtubules (MTs), protects MTs from depolymerization, [3] drives neurite outgrowth, [4] and is critical for cell polarity. [5] In addition, tau can locally nucleate MT assembly from non‐centrosome sites in cells. [6] The ability to locally nucleate MT assembly has been linked to LLPS of tau: [7] tubulin efficiently partitions into tau droplets and thus drives nucleation of MTs. [8] In addition, tau forms liquid‐like condensates on the surface of MTs, regulating the movement of MT‐associated motors and fending off enzymes that would cut up tubulin strands. [9] Little is known, however, about the influence of post‐translational modifications on tau's ability to nucleate MT bundles from condensed phases. Using a combination of LLPS experiments, tubulin polymerization assays, site‐directed mutagenesis, proline‐directed phosphorylation and NMR spectroscopy we herein show that disease‐associated phosphorylation of tau at T231 perturbs the binding of tau's proline‐rich region P2 to tubulin and blocks tubulin assembly within tau condensates.
Microscopy showed that the 441‐residue isoform of tau (hTau40) formed spherical droplets in the presence of 10 % dextran (Figure S1a). The droplets were sensitive to ionic strength and the aliphatic alcohol 1,6‐hexanediol (Figure S1b), that is, they fulfill characteristics of LLPS. In addition, the shorter 352‐residue isoform of tau (hTau23) underwent LLPS in the presence of dextran (Figure S1c). Next, we incubated hTau40 with catalytic amounts of the cyclin‐dependent kinase 2 (Cdk2), which phosphorylates tau at several residues (Figure 1 a, S2a,b), including T231/S235, an epitope recognized by the monoclonal antibody AT180 in insoluble tau deposits in patient brains. [10] pTau(Cdk2) formed spherical condensates in the presence of dextran (Figure 1 b and S2c). The pTau(Cdk2) droplets fused and dissolved at increasing ionic strength or with hexanediol (Figure 1 b,c and Figure S2d,e). The intra‐droplet diffusion of Cdk2‐phosphorylated hTau40 was similar to that of the unmodified protein (Figure 1 d). Because Cdk2 not only phosphorylates T231 and S235, but also other sites (Figure 1 a, S2b), we prepared a mutant tau, in which T231 and S235 were replaced by glutamate to mimic AT180‐specific phosphorylation. The mutant protein hTau23(T231E, S235E) formed spherical condensates, which were sensitive to addition of NaCl and dissolved in the presence of 5–10 % 1,6‐hexanediol, which confirmed that they display characteristics of LLPS (Figure 1 e).
Figure 1.
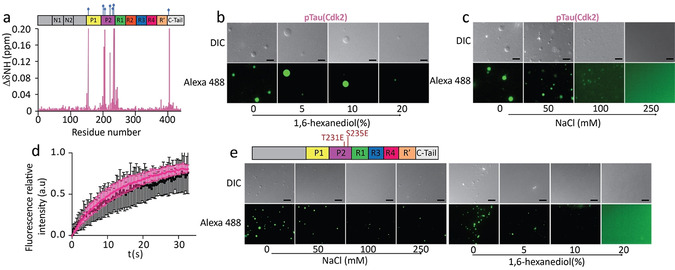
LLPS of tau phosphorylated at disease‐associated epitopes. a) 1H‐15N chemical shift perturbation of hTau40 upon Cdk2 phosphorylation identifying sites of phosphorylation. b,c) pTau(Cdk2) (25 μm) forms liquid‐like droplets that dissolve upon addition of 1,6‐hexanediol (b) or with increasing ionic strength (c). Scale bars, 10 μm. d) FRAP of 50 μm pTau(Cdk2) (pink) and hTau40 (black). FRAP curves are averaged from four individual measurements; error bars represent standard deviation. e) /S235E‐mutant hTau23 (50 μm) forms liquid‐like droplets that dissolve with increasing ionic strength (left), or the addition of 1,6‐hexanediol (right). Scale bars, 10 μm. Top panel, domain organization of hTau23 mutated at T231E and S235E.
Tau lowers the critical concentration for tubulin polymerization.[ 3 , 11 ] In agreement with previous studies, which showed that MT growth does not occur spontaneously at 25 °C but requires nucleation, [11] we did not observe polymerization of tubulin when mixed at room temperature with GTP and hTau40 (Figure S3a). Thus, under these solution conditions the concentrations of tubulin and tau are not sufficient for MT assembly. Because a key property of LLPS is the formation of a highly concentrated phase, we repeated the experiment, but first forming droplets of hTau40 through addition of dextran and then adding tubulin and GTP. Fluorescence microscopy showed that the added tubulin concentrated inside the hTau40 droplets (Figure 2 a and Figure S3b). In addition, the droplets deformed and small MT filaments started to grow from the droplets. Indeed, within 30 minutes long MT bundles were observed under the microscope (Figure 2 a). In the absence of hTau40, no MTs formed when 10 % dextran was present (Figure S3c). The observations are consistent with the previously reported partitioning of tubulin into tau droplets and the subsequent polymerization of tubulin from the tau‐dense phase. [8] The data show that partitioning of tubulin into a tau condensate provides an efficient mechanism for MT nucleation.
Figure 2.
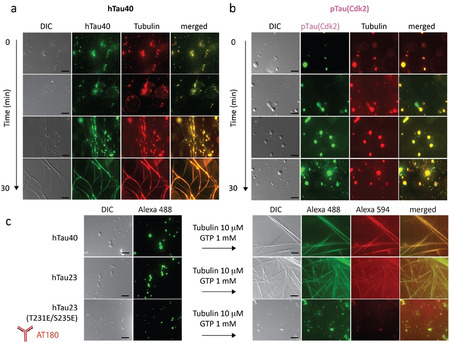
AT180‐phosphorylation blocks nucleation of MT filaments inside tau droplets. a) DIC and fluorescence microscopy of the growth of MTs from performed hTau40 (25 μm) droplets upon addition of tubulin (10 μm) and GTP; scale bars, 10 μm. b) Microscopy of pTau(Cdk2) droplets into which tubulin partitions but no MTs are formed in the presence of GTP; scale bars, 10 μm. c) LLPS of hTau40, hTau23 and hTau23 (T231E/S235E) and the assembly of tubulin into MTs in the case of the wild‐type proteins but not for the hTau23 mutant that mimics AT180‐phosphorylation. Scale bars, 10 μm.
Previous studies have shown that phosphorylation of tau at the AT180‐epitope can interfere with tau‐promoted tubulin polymerization. [10c] Because the concentration of tubulin in hTau40 droplets is increased (Figure S3b), [8] we asked if partitioning of tubulin into pTau(Cdk2) condensates overcomes this inhibitory effect. We therefore added tubulin and GTP to a solution with preformed pTau(Cdk2) droplets. Fluorescence microscopy showed that tubulin efficiently partitioned into the pTau(Cdk2) droplets (Figure 2 b, S3b). Notably, the concentration of pTau(Cdk2) inside the droplets was higher than for the unmodified protein, in agreement with an increased LLPS‐propensity of tau upon phosphorylation.[ 7a , 7b , 8 ] Strikingly, however, tubulin did not grow into MTs from pTau(Cdk2) droplets (Figure 2 b). We then repeated the experiments with twice the pTau(Cdk2) concentration (Figure S3d), but no MT assembly was observed (Figure S3e). In addition, Cdk2‐phosphorylation of hTau23 inhibited tubulin polymerization from droplets (Figure S3f,g). Consistent with these results, tubulin partitioned into preformed hTau23(T231E, S235E)‐droplets, but did not polymerize into MTs (Figure 2 c). The experiments show that even high concentrations of tubulin inside of tau condensates are not sufficient for MT formation when tau is phosphorylated at the AT180 epitope.
We then asked whether the impaired MT nucleation inside tau droplets is specific to AT180‐phosphorylation. To this end, we phosphorylated hTau40 with the kinase MARK2 (Figure S4a–c). In agreement with previous studies,[ 7a , 7b , 8 ] pTau(MARK2) formed spherical droplets in the presence of 10 % dextran (Figure S4d, left panel). Upon addition of tubulin and GTP to the solution, MT filaments formed (Figure S4d, right panel). However, these were less abundant/regular when compared to unmodified hTau40, potentially due to the attenuated binding of MARK2‐phosphorylated tau to tubulin (Figure S5a). [12] Next we phosphorylated hTau40 with the tyrosine kinase C‐Abl (Figure S4e–g, S6). [13] Binding of pTau(C‐Abl) to MTs was indistinguishable from that of the unmodified protein by NMR spectroscopy (Figure S5b). In addition, pTau(C‐Abl) underwent LLPS and—upon addition of tubulin and GTP—polymerized into MTs (Figure S4h).
To characterize the interaction of tau with tubulin at high resolution, we performed NMR experiments with tau in the presence of increasing concentrations of tubulin. To decrease NMR signal broadening, [7b] the experiments were first recorded in the absence of molecular crowding agents, that is, in conditions in which hTau40 does not phase separate and tubulin does not polymerize into MTs. In agreement with previous results, [14] we observed progressive changes in NMR signal intensity and position of individual hTau40 residues at increasing tubulin concentrations (Figure 3 a). Particularly strong signal broadening was observed for residues ≈224–237 in the P2 region, ≈245–253 in repeat R1, ≈274–287 at the boundary between R1/R2, and ≈300–318 at R2/R3 (Figure 3 b). Additional tubulin‐induced broadening was present in P1 and R′, indicating that residues in these two domains further contribute to the tau/tubulin interaction. The importance of the repeat domain of tau for binding to unpolymerized tubulin is supported by biochemical studies. [15]
Figure 3.
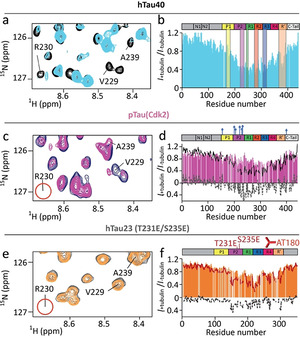
AT180‐phosphorylation perturbs the interaction of tau's P2 region with tubulin. a,c,e) Superposition of a selected region of the 1H‐15N HSQC of hTau40 (a), pTau(Cdk2) (c) and hTau23 (T231E/S235E) (e) in its free state (a: black; c: blue; e: grey) and in presence of tubulin (1:0.5 tau:tubulin molar ratio; a: light blue; c: pink; e: orange). Upon phosphorylation by Cdk2 or T231E/S235E mutation the cross peak of R230 disappears (marked by red circle). b,d,f) Signal attenuation of 1H‐15N signals of hTau40 (b), pTau(Cdk2) (d) and hTau23 (T231E/S235E) (f) (each 10 μm) upon addition of 5 μm unpolymerized tubulin. Previously identified hot spots of the tau/tubulin interaction [14a] are marked in (b). The profiles of hTau40 (black) and hTau23 (red) are shown for comparison. Residue‐specific difference are shown as dashed line.
We then performed the same NMR analysis for pTau(Cdk2) (Figure 3 c). Regions of pTau(Cdk2) downstream of repeat R2, that is, R3, R4 and R′, displayed a similar tubulin‐binding profile as unmodified tau (Figure 3 d). In contrast, residues located in the P2 region and repeat R1 were less broadened upon addition of tubulin (Figure 3 d). Furthermore, substitution of T231 and S235 by glutamate predominantly reduced signal broadening in P2 and R1 (Figure 3 e,f). The data demonstrate that phosphorylation at the AT180‐epitope perturbs the interaction of tau's P2 region with tubulin. This perturbation, however, does not decrease the overall affinity of tau for binding to tubulin. [10c]
Next we used NMR to probe the tau/tubulin‐interaction in crowded conditions. The experiments are challenging, because in crowded conditions rapid tubulin assembly occurs and LLPS enhances NMR line broadening. To minimize these problems, we decreased the temperature to 5 °C and reduced the NMR measurement time. Microscopy showed that at 5 °C both hTau40 and pTau(Cdk2) form droplets (Figure S7a,b). Subsequently, we recorded NMR spectra for hTau40 and pTau(Cdk2) in the absence and presence of tubulin. Detailed analysis showed that the overall NMR signal of hTau40 in the presence of tubulin and dextran was decreased to 30–40 % (Figure S7c). In the case of pTau(Cdk2), the decrease was much less (Figure S7c), suggesting that the strong decrease in case of hTau40 is due to tubulin assembly. At the same time, the residue‐specific analysis showed that both hTau40 and pTau(Cdk2) bind through their repeat region to tubulin in crowded conditions (Figure S7c,d).
Phosphorylation of tau's P2 region changes the overall electrostatic properties of the tau/tubulin interaction. In addition, the attached phosphate groups can engage in salt bridges. To provide experimental support for the formation of intramolecular salt bridges in phosphorylated tau, we recorded 1H‐15N HSQC spectra optimized for the arginine guanidinium side‐chain groups. For unmodified hTau40, we observed one large cluster of unresolved signals (1H shifts of 7.3–7.4 ppm) together with a broad, slightly‐downfield shifted signal at circa 7.45 ppm (Figure 4). The small dispersion and low intensity of these NMR signals is caused by rapid exchange of labile side‐chain guanidinium protons with bulk solvent. In contrast, NMR signals sharpened up and new cross‐peaks appeared in the spectrum of pTau(Cdk2) (Figure 4). To determine the identity of the pTau(Cdk2)‐specific cross peaks, we recorded the same spectral region for a tau peptide comprising residues 225–246 and phosphorylated at the AT180‐epitope (T231 and S235; termed Tau(225–246)‐AT180). Tau(225–246)‐AT180 contains two arginine residues, namely R230 and R242. Superposition of the spectrum of Tau(225–246)‐AT180 with that of pTau(Cdk2) showed that one arginine cross peak is located in the broad cluster of resonances originating from labile guanidinium protons. The other signal, however, overlaps with the cross peak at approximately 7.53 ppm (Figure 4), which is specific for the salt‐bridge forming, side‐chain guanidinium proton of R230. [16] The analysis suggests that Cdk2‐phosphorylation results in the formation of an intramolecular salt bridge between R230 and phosphorylated T231.
Figure 4.
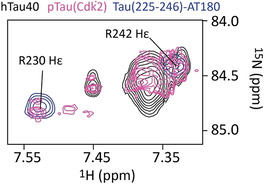
Detection of an intramolecular salt‐bridge in pTau(Cdk2). Superposition of the guanidinium region of the 2D 1H‐15N HSQC spectra of hTau40 (black), pTau(Cdk2) (pink) and Tau(225–246)‐AT180 (blue).
An important aspect of MT polymerization is the conformation which the tubulin monomers adopt. [17] While at the growing end of MTs curved αβ‐tubulin dimers are added, the heterodimers already incorporated into MTs adopt a straight conformation which gives stability to the structure. [17] Our observations in combination with previous results [8] suggest that non‐membrane bound compartments, in which tau is concentrated, might contribute to a non‐templated polymerization mechanism through stabilization of the αβ‐tubulin heterodimer. However, when tau is phosphorylated at T231 in the proline‐rich region P2, R230 engages in an intramolecular salt‐bridge with the T231 phosphate group. R230 is thus no longer able to engage in an intermolecular salt bridge with tubulin. This model is consistent with mutagenesis and biochemical experiments, which demonstrated that R230 is important for MT‐assembly. [18]
We showed that unmodified 3R and 4R‐tau as well as tau phosphorylated at disease‐associated epitopes condense into liquid‐like droplets. Although tubulin partitioned into and reached high concentrations inside all tau droplets, it was unable to grow into MTs form the inside of droplets formed by tau phosphorylated at the AT180 epitope (T231/S235). The blocked MT polymerization from droplets of AT180‐phosphorylated tau is in striking contrast to the highly efficient MT assembly inside droplets of unmodified tau. It is specific because neither phosphorylation of tau in the repeat domain nor at its tyrosine residues inhibited the assembly of tubulin from tau droplets. Notably, LLPS of IDPs has been shown to promote different types of cytoskeletal assembly. [19] Modulation of this process by IDP phosphorylation might therefore be a mechanism that is more broadly active in condensate‐mediated cytoskeletal assembly.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We thank Maria‐Sol Cima‐Omori for help with sample preparation. M.Z. was supported by the advanced grant “787679—LLPS‐NMR” of the ERC, and the DFG through SPP2191 (DFG ZW 71/9‐1) and SFB860 (project B2). E.M. was supported by the DFG through SPP2191 (DFG Ma 563/14‐1). Open access funding enabled and organized by Projekt DEAL.
A. Savastano, D. Flores, H. Kadavath, J. Biernat, E. Mandelkow, M. Zweckstetter, Angew. Chem. Int. Ed. 2021, 60, 726.
References
- 1.
- 1a. Alberti S., Dormann D., Annu. Rev. Genet. 2019, 53, 171–194; [DOI] [PubMed] [Google Scholar]
- 1b. Boeynaems S., Alberti S., Fawzi N. L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., Tompa P., Fuxreiter M., Trends Cell Biol. 2018, 28, 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.
- 2a. Nott T. J., Craggs T. D., Baldwin A. J., Nat. Chem. 2016, 8, 569–575; [DOI] [PubMed] [Google Scholar]
- 2b. Rai A. K., Chen J. X., Selbach M., Pelkmans L., Nature 2018, 559, 211–216. [DOI] [PubMed] [Google Scholar]
- 3.
- 3a. Cleveland D. W., Hwo S. Y., Kirschner M. W., J. Mol. Biol. 1977, 116, 207–225; [DOI] [PubMed] [Google Scholar]
- 3b. Weingarten M. D., Lockwood A. H., Hwo S. Y., Kirschner M. W., Proc. Natl. Acad. Sci. USA 1975, 72, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drubin D. G., Feinstein S. C., Shooter E. M., Kirschner M. W., J. Cell Biol. 1985, 101, 1799–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caceres A., Potrebic S., Kosik K. S., J. Neurosci. 1991, 11, 1515–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Drubin D. G., Kirschner M. W., J. Cell Biol. 1986, 103, 2739–2746; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b. Kanai Y., Chen J., Hirokawa N., EMBO J. 1992, 11, 3953–3961; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6c. Lewis S. A., Ivanov I. E., Lee G. H., Cowan N. J., Nature 1989, 342, 498–505; [DOI] [PubMed] [Google Scholar]
- 6d. Knops J., Kosik K. S., Lee G., Pardee J. D., Cohen-Gould L., McConlogue L., J. Cell Biol. 1991, 114, 725–733; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6e. Lee G., Rook S. L., J. Cell Sci. 1992, 102, 227–237; [DOI] [PubMed] [Google Scholar]
- 6f. Muroyama A., Lechler T., Development 2017, 144, 3012–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.
- 7a. Wegmann S., Eftekharzadeh B., Tepper K., Zoltowska K. M., Bennett R. E., Dujardin S., Laskowski P. R., MacKenzie D., Kamath T., Commins C., Vanderburg C., Roe A. D., Fan Z., Molliex A. M., Hernandez-Vega A., Muller D., Hyman A. A., Mandelkow E., Taylor J. P., Hyman B. T., EMBO J. 2018, 37, 7; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7b. Ambadipudi S., Biernat J., Riedel D., Mandelkow E., Zweckstetter M., Nat. Commun. 2017, 8, 275; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7c. Zhang X., Lin Y., Eschmann N. A., Zhou H., Rauch J. N., Hernandez I., Guzman E., Kosik K. S., Han S., PLoS Biol. 2017, 15, e2002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B. T., Alberti S., Diez S., Hyman A. A., Cell Rep. 2017, 20, 2304–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a. Tan R., Lam A. J., Tan T., Han J., Nowakowski D. W., Vershinin M., Simo S., Ori-McKenney K. M., McKenney R. J., Nat. Cell Biol. 2019, 21, 1078–1085; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b. Siahaan V., Krattenmacher J., Hyman A. A., Diez S., Hernandez-Vega A., Lansky Z., Braun M., Nat. Cell Biol. 2019, 21, 1086–1092. [DOI] [PubMed] [Google Scholar]
- 10.
- 10a. Amniai L., Lippens G., Landrieu I., Biochem. Biophys. Res. Commun. 2011, 412, 743–746; [DOI] [PubMed] [Google Scholar]
- 10b. Baumann K., Mandelkow E. M., Biernat J., Piwnica-Worms H., Mandelkow E., FEBS Lett. 1993, 336, 417–424; [DOI] [PubMed] [Google Scholar]
- 10c. Amniai L., Barbier P., Sillen A., Wieruszeski J. M., Peyrot V., Lippens G., Landrieu I., FASEB J. 2009, 23, 1146–1152. [DOI] [PubMed] [Google Scholar]
- 11.
- 11a. Fygenson D. K., Braun E., Libchaber A., Phys. Rev. E 1994, 50, 1579–1588; [DOI] [PubMed] [Google Scholar]
- 11b. Johnson K. A., Borisy G. G., J. Mol. Biol. 1979, 133, 199–216. [DOI] [PubMed] [Google Scholar]
- 12. Biernat J., Gustke N., Drewes G., Mandelkow E. M., Mandelkow E., Neuron 1993, 11, 153–163. [DOI] [PubMed] [Google Scholar]
- 13. Derkinderen P., Scales T. M., Hanger D. P., Leung K. Y., Byers H. L., Ward M. A., Lenz C., Price C., Bird I. N., Perera T., Kellie S., Williamson R., Noble W., Van Etten R. A., Leroy K., Brion J. P., Reynolds C. H., Anderton B. H., J. Neurosci. 2005, 25, 6584–6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.
- 14a. Mukrasch M. D., Bibow S., Korukottu J., Jeganathan S., Biernat J., Griesinger C., Mandelkow E., Zweckstetter M., PLoS Biol. 2009, 7, e1000034; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b. Sillen A., Barbier P., Landrieu I., Lefebvre S., Wieruszeski J. M., Leroy A., Peyrot V., Lippens G., Biochemistry 2007, 46, 3055–3064. [DOI] [PubMed] [Google Scholar]
- 15.
- 15a. Gustke N., Trinczek B., Biernat J., Mandelkow E. M., Mandelkow E., Biochemistry 1994, 33, 9511–9522; [DOI] [PubMed] [Google Scholar]
- 15b. Butner K. A., Kirschner M. W., J. Cell Biol. 1991, 115, 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwalbe M., Kadavath H., Biernat J., Ozenne V., Blackledge M., Mandelkow E., Zweckstetter M., Structure 2015, 23, 1448–1458. [DOI] [PubMed] [Google Scholar]
- 17. Brouhard G. J., Rice L. M., Nat. Rev. Mol. Cell Biol. 2018, 19, 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goode B. L., Denis P. E., Panda D., Radeke M. J., Miller H. P., Wilson L., Feinstein S. C., Mol. Biol. Cell 1997, 8, 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. King M. R., Petry S., Nat. Commun. 2020, 11, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


