Abstract
Coronavirus disease 2019 (COVID-19) has emerged from China and globally affected the entire population through the human-to-human transmission of a newly emerged virus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The genome of SARS-CoV-2 encodes several proteins that are essential for multiplication and pathogenesis. The main protease (Mpro or 3CLpro) of SARS-CoV-2 plays a central role in its pathogenesis and thus is considered as an attractive drug target for the drug design and development of small-molecule inhibitors. We have employed an extensive structure-based high-throughput virtual screening to discover potential natural compounds from the ZINC database which could inhibit the Mpro of SARS-CoV-2. Initially, the hits were selected on the basis of their physicochemical and drug-like properties. Subsequently, the PAINS filter, estimation of binding affinities using molecular docking, and interaction analyses were performed to find safe and potential inhibitors of SARS-CoV-2 Mpro. We have identified ZINC02123811 (1-(3-(2,5,9-trimethyl-7-oxo-3-phenyl-7H-furo[3,2-g]chromen-6-yl)propanoyl)piperidine-4-carboxamide), a natural compound bearing appreciable affinity, efficiency, and specificity towards the binding pocket of SARS-CoV-2 Mpro. The identified compound showed a set of drug-like properties and preferentially binds to the active site of SARS-CoV-2 Mpro. All-atom molecular dynamics (MD) simulations were performed to evaluate the conformational dynamics, stability and interaction mechanism of Mpro with ZINC02123811. MD simulation results indicated that Mpro with ZINC02123811 forms a stable complex throughout the trajectory of 100 ns. These findings suggest that ZINC02123811 may be further exploited as a promising scaffold for the development of potential inhibitors of SARS-CoV-2 Mpro to address COVID-19.
Keywords: SARS-CoV-2 main protease, Natural compounds, Drug discovery, Virtual high-throughput screening, Molecular dynamics simulation, Small molecule inhibitors
1. Introduction
A newly emerged pneumonia outbreak of coronavirus disease 2019 (COVID-19) spread through severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Asrani et al., 2020b, Huang et al., 2020), now globally affected over 100 million individuals and accounts for over two million deaths worldwide (https://www.worldometers.info/coronavirus/). There are several diagnostic and therapeutic approaches have emerged to handle COVID-19; however, still, no effective therapy has been developed (Asrani et al., 2020a, Graham, 2020). The use of Lopinavir and Ritonavir (Nutho et al., 2020), antiviral drugs (Grein et al., 2020), chloroquine and hydroxychloroquine (Jakhar and Kaur, 2020), convalescent plasma (Chen et al., 2020), stem cell therapy, etc., have shown some positive outcomes on admitted patients of COVID-19 (Fatima et al., 2020, Kumari et al., 2020). Hence, there is an emergent need to discover potential therapeutic agents to control the pathogenesis of SARS-CoV-2 (Corey et al., 2020).
The genome profiling of SARS-CoV-2 leads to the identification of a few drug targets including, the SARS-CoV-2 main protease (Mpro, 3CLpro), that mediates viral replication and transcription together and can be considered as a potential target for therapeutic development (Naqvi et al., 2020, Zhang and Holmes, 2020). Due to its critical role in pathogenesis, combined with the absence of closely related homologues in humans, Mpro is acting as an effective target for drug design and development (Zhang et al., 2020). Mpro cleaves polyproteins to generate Non-Structural Proteins (NSPs) that form a replicase-transcriptase complex (RTC). Mpro exists in NSP5 which releases the majority of NSPs from the polyproteins, is vital for the life cycle of SARS-CoV-2 (V’kovski et al., 2020). The structure of Mpro advances the field of modern drug discovery which results in the development of potential lead molecules that block Mpro function in cell-based assays (Zhang et al., 2020). SARS-CoV-2 Mpro is consists of 306 amino acid residues with a cysteine-histidine catalytic dyad including Cys145 and His41 (Dai et al., 2020; Jin et al., 2020). Structural analysis suggests that these residues including a few others found in the active site pocket of SARS-CoV-2 Mpro can be served as a platform for the discovery of its selective inhibitors in the therapeutic management of COVID-19 (Zhang et al., 2020).
In many attempts of ongoing research, new leads are being identified by utilizing advanced computational approaches to screen large chemical libraries (Jin et al., 2020; Shamsi et al., 2020). For the quick development of effective therapeutics, the experimental screening approach alone may not improve lead productivity (Padhi and Tripathi, 2020). Bioinformatics and computational biology play a crucial role in the drug discovery process while employing a structure-based drug design approach such as molecular docking-based virtual high-throughput screening (vHTS) which is currently largely implemented in the modern drug-discovery pipeline to find potential lead molecules among various chemical libraries (Mohammad et al., 2020b, Naqvi et al., 2018, Naqvi and Hassan, 2017). There are many chemical repositories on the web such as the ZINC database which contains the structural coordinates of millions of chemical compounds that can be screened to identify potential leads against predefined targets (Mohammad et al., 2020a, Mohammad et al., 2019a, Mohammad et al., 2019b, Sterling and Irwin, 2015).
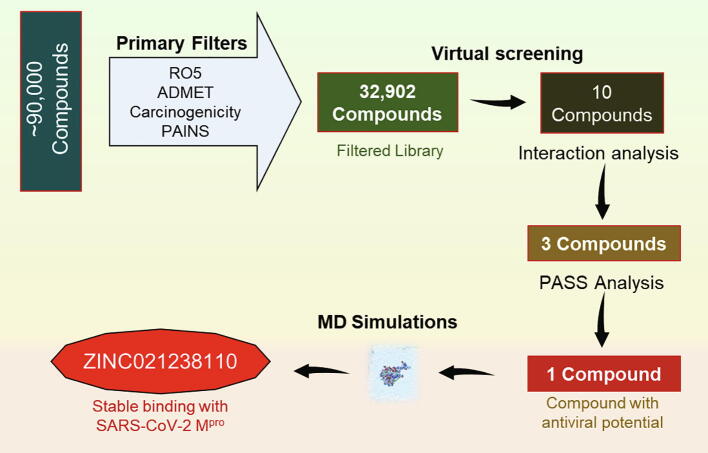
Here, we have employed a structure-based drug design approach to find natural leads that can act as potential inhibitors of SARS-CoV-2 Mpro and used in the drug development of effective COVID-19 therapy. Structure-based vHTS of natural products from the ZINC database was performed in search of high-affinity binding partners of SARS-CoV-2 Mpro. First, the library of ~90,000 natural compounds was filtered out by applying Rule of five, ADMET, carcinogenicity, and PAINS filters. Then, we estimated the binding affinities of filtered compounds with SARS-CoV-2 Mpro using the molecular docking approach, and subsequent interaction analysis was carried out to find better hits. Based on the specific interaction, we identified three compounds bearing appreciable affinity and specific interaction towards the binding site of SARS-CoV-2 Mpro. The identified compounds were further subjected to PASS analysis where we have selected one compound bearing antiviral potential (Lagunin et al., 2000). We further performed all-atom molecular dynamics (MD) simulation for 100 ns to see conformational changes in SARS-CoV-2 Mpro without and with the identified compound. A systematic approach of vHTS used in this study is described in Fig. 1.
Fig. 1.
The workflow illustrates the process of virtual high-throughput screening used in this study. RO5, Lipinski's rule of five; ADMET, Absorption, Distribution, Metabolism, Excretion, and Toxicity; PAINS, Pan-assay interference compounds.
2. Material and methods
2.1. Computational tools and web-servers
A well-defined computational pipeline of drug-design and discovery using different bioinformatics software, such as MGL AutoDock Tools (Jacob et al., 2012), AutoDock Vina (Trott and Olson, 2010), Discovery Studio Visualizer (Biovia, 2015) and GROMACS was used for vHTS and MD simulations. Online resources such as RCSB-Protein Data Bank (PDB), the ZINC database (Sterling and Irwin, 2015), SwissADME (Daina et al., 2017), CarcinoPred-EL (Zhang et al., 2017), VMD (Humphrey et al., 1996), QtGrace (Turner, 2005), etc., were used in retrieval, evaluation, and analysis. The atomic coordinates of SARS-CoV-2 Mpro were downloaded from PDB (PDB ID: 6LU7) (Jin et al., 2020). All co-crystallized hetero molecules including N3 inhibitor were removed from the parent structure. Finally, the protein structure was prepared for vHTS in MGL tools by adding hydrogen to polar atoms and assigning appropriate atom types. A library of ~90,000 natural products was downloaded from the ZINC database in processed form.
2.2. Filtration of compounds
All compounds from the ZINC library were filtered out based on their physicochemical and ADMET properties through SwissADME and Discovery Studio Visualizer. First, we have selected compounds based on their physicochemical and drug-like properties satisfying the Lipinski’s rule of five (Lipinski, 2000). We further applied Pan-assay interference compounds (PAINS) filter to avoid compounds with specific patterns with a higher tendency to bind towards multiple biological targets. We further screened the compounds for their carcinogenic patterns and ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) properties. Here, only compounds having well ADMET properties and non-carcinogenic patterns were selected for further docking-based vHTS studies.
2.3. Molecular docking-based vHTS
We performed a molecular docking-based vHTS to filter compounds based on their binding affinities towards SARS-CoV-2 Mpro. The docking was performed using AutoDock Vina with a grid size of a blind search space as 54, 68 and 62 Å, centralized at −26.34, 12.60 and 58.91 for X, Y and Z coordinates, respectively. The grid spacing was set to 1.00 Å with default docking parameters. The docking results were screened for high binding affinity and then all possible docked conformations were splitted for each compound which were further analyzed using PyMOL and Discovery Studio Visualizer for their possible interaction towards Mpro. The polar contacts depicted in PyMOL within 3.5 Å of distance were mapped as close interactions between the compounds and Mpro. Discovery Studio Visualizer was utilized to explore detailed interactions formed between the selected compounds with the SARS-CoV-2 Mpro. Interaction analysis resulted in the identification of three compounds interacting with the critical residues of the binding pocket of SARS-CoV-2 Mpro. Here, we have selected only those compounds which were specifically interacting with the binding-site residues including Cys145 of SARS-CoV-2 Mpro.
2.4. PASS analysis: biological activity predictions
The potential biological properties of the selected compounds were investigated through the PASS web server (Lagunin et al., 2000). The PASS tool allows us to explore the possible biological properties of compounds, based on their chemical formula. It uses 2D molecular fragments known as multilevel neighbors of atoms (MNA) descriptors which suggest that the biological activity of a chemical compound is the function of its molecular structure. It gives the prediction score for biological properties on the ratio of ‘probability to be active (Pa)’ and ‘probability to be inactive (Pi)’. A higher Pa means the biological property is having more probability for a compound.
2.5. MD simulations
All-atom MD simulations were performed on two systems, SARS-CoV-2 Mpro in the free state and SARS-CoV-2 Mpro-ZINC02123811 complex at 300 K at the molecular mechanics level using GROMOS 54A7 force-field in GROMACS v5.1.2 simulation package (Abraham et al., 2015). Gromacs topology parameters for ZINC02123811 were generated from the PRODRG server and merged to the protein topology to make the Mpro-ZINC02123811 complex. Both, Mpro and Mpro-ZINC02123811 systems were solvated in a cubic box with the SPC (spc216) water model for aqueous simulations. Energy minimization for 1500 steps of the steepest descent method was performed for 1000 ps. The temperature of both systems was consequently increased from 0 to 300 K during the equilibration period. The equilibration period was performed for 100 ps at constant volume under periodic boundary conditions with a stable pressure of 1 bar. The final MD run was performed for 100 ns for both systems, and resulting trajectories were analyzed using the inbuilt utilities of GROMACS as described (Gulzar et al., 2019, Khan et al., 2016, Khan et al., 2017b, Shahbaaz et al., 2018).
2.6. Principal component and free energy landscape analyses
To explore the conformational sampling, atomic motions and structural stability of Mpro and Mpro-ZINC02123811 complex, principal component (PC) and free energy landscape (FEL) studies were performed by the essential dynamics approach which employs the calculation of the covariance matrix (Altis et al., 2008, Fatima et al., 2019, Gupta et al., 2019, Mohammad et al., 2019b). The following formula was used to calculate the covariance matrix:
where xi/xj is the coordinate of the ith/jth atom of the system, and <−> is the ensemble average.
The FELs of Mpro and Mpro-ZINC02123811 complex were attained using the conformational sampling approach which allows exploring the protein conformations near the native state (Papaleo et al., 2009). FELs were generated to investigate the stability and native states of SARS-CoV-2 Mpro, before and after ZINC02123811 binding. The FELs were generated utilizing the following formula:
where KB is the Boltzmann constant, T is the temperature of simulation, and P(X) is the probability distribution of the system along with the PCs.
3. Results
3.1. Filtration of compounds
The physicochemical properties of all the natural compounds present in the library were calculated and analyzed through the SwissADME webserver and Discovery Studio Visualizer. Here we identified a set of 32,902 compounds based on Lipinski’s rule of five (Mol wt. ≤ 500 Da, log P ≤ 5, H-bond donor ≤ 5 and H- bond acceptor ≤ 10), bioavailability score, and PAINS pattern (Lipinski, 2000). The physicochemical properties of the finally selected three compounds satisfying the rule of five, along with the standard SARS-CoV-2 Mpro inhibitor N3 are shown in Table 1.
Table 1.
List of identified compounds and their physiochemical properties.
| S. No. | Compound ID | Mol wt. (Da) | Rotatable bond | H-bond acceptor | H-bond donor | LogP | Lipinski Violation |
|---|---|---|---|---|---|---|---|
| 1 | ZINC02123811 | 486.56 | 5 | 5 | 1 | 4.78 | 0 |
| 2 | ZINC02128147 | 488.49 | 6 | 7 | 3 | 4.01 | 0 |
| 3 | ZINC02161101 | 472.60 | 3 | 3 | 2 | 3.98 | 0 |
| 4 | N3-ILP | 680.79 | 17 | 9 | 5 | 2.08 | 2 |
The compounds were also screened out based on their ADMET properties and carcinogenic patterns to identify safe and non-carcinogenic compounds. ADMET properties of the finally selected three compounds and N3 are given in Table 2.
Table 2.
List of identified compounds and their ADMET properties.
| Compound ID | Absorption |
Distribution |
Metabolism |
Excretion |
Toxicity |
|
|---|---|---|---|---|---|---|
| GI absorption (%) | Water Solubility | BBB/CNS permeation | CYP2D6Inh/Subs | OCT2 substrate | AMES/ skin sens | |
| ZINC02123811 | 99.03 | Soluble | No | No | No | No |
| ZINC02128147 | 69.67 | Soluble | No | No | No | No |
| ZINC02161101 | 90.69 | Soluble | No | No | No | No |
| N3-ILP | 57.88 | Soluble | No | No | No | No |
3.2. Molecular docking-based vHTS
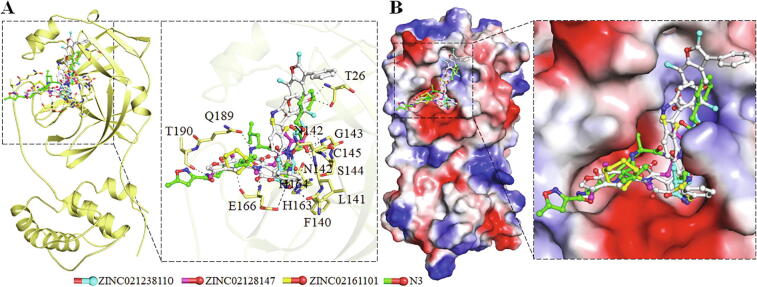
In this attempt of molecular docking-based vHTS, the compounds having appreciable binding affinities with SARS-CoV-2 Mpro were selected for further analysis. The vHTS analysis results in the identification of 10 compounds from a pool of 32,902 natural compounds showing a considerable binding score (−9.4 to −9.8 kcal/mol) towards Mpro (Table 3). We further calculated their mean affinity after performing redocking up to 10 different runs of AutoDock Vina with independent random seeds and found good consistency in the resultant output (Table 3). Further, detailed interaction analysis of the top 10 hits was carried out using the PyMOL and Discovery Studio Visualizer. Here, a total of 90 possible docked conformers were splitted from the out files of the selected hits. From the analysis of all possible docked conformers, we identified three natural compounds that have commonly interacted with the active-site ‘Cys145′ of SARS-CoV-2 Mpro. The binding pattern of the finally selected three compounds with SARS-CoV-2 Mpro is illustrating in Fig. 2.
Table 3.
Binding affinities of the selected compounds in 10 different runs of AutoDock Vina with independent random seeds. R1, R2, R3, …., R10 shows replicates of AutoDock Vina run.
| S. No. | Compound ID |
Affinity (kcal/mol) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 | Mean | ||
| 1 | ZINC02161101 | −9.8 | −9.4 | −8.9 | −9.4 | −9.9 | −9.8 | −9.4 | −9.5 | −9.9 | −9.4 | −9.5 |
| 2 | ZINC02113993 | −9.8 | −9.8 | −9.8 | −9.2 | −9.8 | −9.9 | −8.9 | −8.7 | −8.8 | −9.5 | −9.4 |
| 3 | ZINC02123811 | −9.7 | −9.8 | −9.6 | −9.6 | −9.7 | −9.4 | −9.6 | −9.7 | −9.7 | −9.7 | −9.6 |
| 4 | ZINC02125386 | −9.6 | −9.6 | −8.4 | −9.5 | −8.2 | −9.6 | −9.6 | −9.6 | −9.6 | −9.7 | −9.3 |
| 5 | ZINC02113878 | −9.6 | −9.5 | −9.6 | −9.6 | −9.6 | −9.5 | −9.6 | −9.6 | −9.5 | −9.5 | −9.6 |
| 6 | ZINC02110106 | −9.5 | −8.9 | −9.5 | −9.5 | −9.5 | −9.4 | −9.5 | −9.4 | −9.5 | −9.5 | −9.4 |
| 7 | ZINC02123668 | −9.5 | −8.6 | −9.5 | −9.5 | −9.4 | −9.5 | −8.5 | −9.5 | −9.5 | −9.5 | −9.3 |
| 8 | ZINC02128147 | −9.5 | −9.5 | −9.5 | −8.2 | −9.5 | −9.5 | −9.6 | −9.5 | −9.5 | −8.9 | −9.3 |
| 9 | ZINC02111094 | −9.4 | −9.1 | −9.4 | −9.2 | −9.3 | −9.1 | −9.3 | −9.4 | −9.4 | −9.1 | −9.3 |
| 10 | ZINC02112091 | −9.4 | −9.1 | −8.4 | −9.4 | −8.4 | −9.3 | −9.4 | −8.2 | −9.5 | −8.4 | −9.0 |
Fig. 2.
Structural representation of docked compounds in the binding pocket of SARS-CoV-2 Mpro. (A) Cartoon representation of Mpro with the selected three compounds along with co-crystallized inhibitor N3. (B) Surface potential view of Mpro binding pocket occupied by the selected compounds and N3.
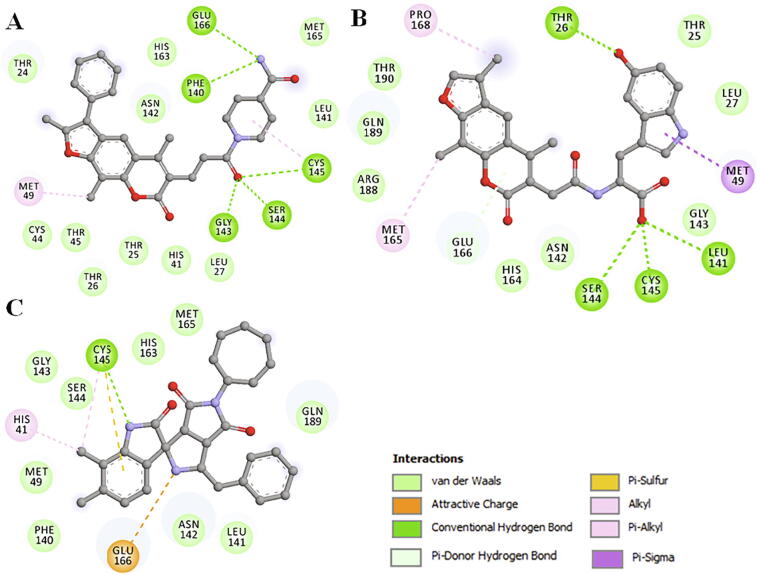
Compounds docked to the binding pocket of SARS-CoV-2 Mpro were checked for their interaction with the functionally important residues of the protein. Detailed interaction of the three selected compounds is shown in Fig. 3 where it is evident that all the selected compounds interact with Cys145 of SARS-CoV-2 Mpro. The docking poses of the identified compounds and N3 suggests that they could fit inside the substrate-binding pocket of Mpro. The compounds binding to critical residues of the substrate-binding pocket including Cys145, Met165, and Glu166 may hinder the substrate accessibility to Mpro thus its inhibition.
Fig. 3.
2D structural representation of SARS-CoV-2 Mpro residues interacting to the compound (A) ZINC0212381, (B) ZINC02128147, and (C) ZINC02161101.
3.3. PASS analysis: biological activity predictions
The exploration of biological activities of the selected compounds through the PASS analysis resulted in similar kinds of biological activities. The reference compound, N3 showed to have SARS-CoV-2 Mpro inhibitory potential, validating the results predicted. The compounds ZINC02123811, ZINC02128147 and ZINC02161101 have shown predictions for antithrombotic, antineoplastic, and antiviral potential, with Pa ranging from 0,340 to 0,552 when Pa > Pi. Table 4 shows the biological properties of all three compounds with higher Pa. Here, based on the specific interactions with SARS-CoV-2 Mpro and biological properties showing antiviral potential. Finally, we have selected one compound for MD simulation studies.
Table 4.
List of identified compounds and their biological properties calculated through PASS webserver.
| S. No. | Compound ID | Pa | Pi | Biological Activity |
|---|---|---|---|---|
| 1 | ZINC02123811 | 0,552 | 0,101 | CDP-glycerol glycerophosphotransferase inhibitor |
| 0,406 | 0,051 | Antithrombotic | ||
| 0,340 | 0,053 | Antiviral, HCV IRES inhibitor | ||
| 2 | ZINC02128147 | 0,543 | 0,105 | CDP-glycerol glycerophosphotransferase inhibitor |
| 0,494 | 0,113 | Phosphatase inhibitor | ||
| 0,354 | 0,073 | Antithrombotic | ||
| 3 | ZINC02161101 | 0,490 | 0,080 | Nicotinic alpha2beta2 receptor antagonist |
| 0,410 | 0,099 | Antineoplastic | ||
| 0,385 | 0,167 | Nicotinic alpha4beta4 receptor agonist | ||
| 4 | N3-ILP | 0,477 | 0,003 | Antiviral, SARS-CoV-2 Mpro inhibitor |
3.4. MD simulations
The finally selected compound, ZINC02123811 in complex with SARS-CoV-2 Mpro along with the free state of SARS-CoV-2 Mpro were subjected to all-atom MD simulations for 100 ns. The various systematic and structural parameters were calculated to analyze the stability and dynamics of SARS-CoV-2 Mpro before and after ZINC02123811 binding.
3.4.1. Structural deviations and compactness
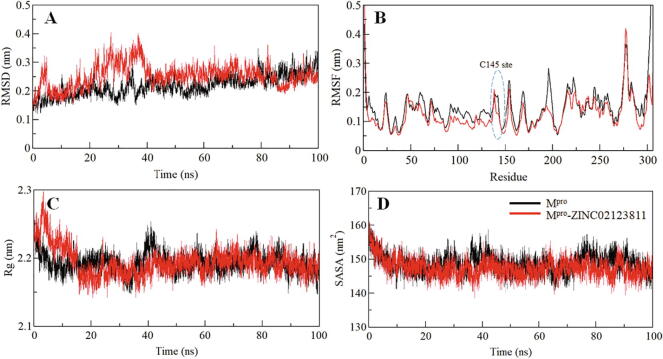
To investigate the structural deviations and dynamics of a protein structure, root-mean-square deviation (RMSD) has been utilized (Dahiya et al., 2019, Kuzmanic and Zagrovic, 2010). We also calculated the time- evolution of RMSDs for Mpro and Mpro-ZINC02123811 complex during the simulations and found average value as 0.22 nm and 0.25 nm, respectively. The RMSD plot shows both free Mpro and Mpro-ZINC02123811 complex are stable throughout the simulation (Fig. 4A). However, a minor increase in random fluctuations of up to 0.15 nm is observed between 0 and 40 ns in the case of complexed Mpro. But, after 40 ns, the plot is showing stable and equilibrated RMSD throughout the simulation. Variations in RMSD of the Mpro-ZINC02123811 complex reduces after 75 ns and stabilized throughout the trajectory as compared with free Mpro.
Fig. 4.
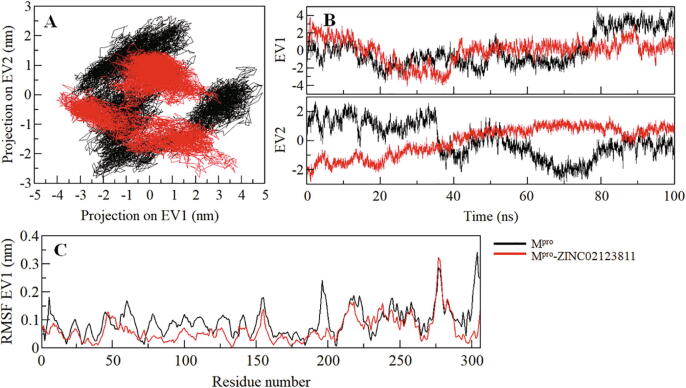
Structural dynamics and compactness of SARS-CoV-2 Mpro upon ZINC02123811 binding as a function of time. (A) RMSD plot of Mpro in complexed with ZINC02123811. (B) Residual fluctuations (RMSF) plot of Mpro before and after ZINC02123811 binding. (C) Time evolution of radius of gyration. (D) SASA plot of Mpro as a function of time.
To explore the residual flexibility in Mpro in the free-state and upon ZINC02123811 binding, the average fluctuation of all residues was considered and plotted as root-mean-square fluctuation (RMSF). The RMSF plot showed several residual fluctuations in Mpro in different regions. These fluctuations were found to be stable and minimized upon ZINC02123811 binding with the progression of simulation at region spanning from N- to C- termini (Fig. 4B).
The conformational stability of Mpro before and after ZINC02123811 binding was also evaluated by calculating the radius of gyration (Rg) of both systems. The average Rg values for Mpro before and after ZINC02123811 binding were estimated to be the same as calculated 2.19 nm. Rg plot suggested no major changes in the packing of Mpro when bound with ZINC02123811. An initial fluctuation until 15 ns of MD trajectories might occur due to packing adjustment of Mpro, but thereafter, the Rg became stable and equilibrated throughout the simulation suggesting complex stability (Fig. 4C).
The average solvent-accessible surface area (SASA) values for Mpro and Mpro-ZINC02123811 complexes were found to be 148.38 nm2, and 147.46 nm2, respectively. The SASA plot is showed to have a similar pattern of equilibration in the case of both the systems. A slight decrease in the average SASA might be owing to tighter packing of the Mpro upon ZINC02123811 binding (Fig. 4D).
3.4.2. Dynamics of hydrogen bonds
To further examine the stability of Mpro before and after ZINC02123811 binding, the time-evolution of hydrogen bonds (H-bonds) formed within 0.35 nm during the simulation was explored. In Mpro, the average number of intramolecular H-bonds before and after ZINC02123811 binding was estimated to be 216 and 214, respectively (Fig. 5A). We also plotted the probability distribution function (PDF) of the H-bonds for both systems (Fig. 5B).
Fig. 5.
(A) Time evolution and stability of Hydrogen bonds formed within 0.35 nm Intra-Mpro, and (B) The probability distribution function (PDF) of the H-Bonds for both the systems.
3.5. Principal component and free energy landscape analyses
Principal components analysis (PCA) is a useful approach to extract the dominant modes in a protein motion. It helps to identify the configurational space of the protein that contains a few degrees of freedom while the motion occurs. We have performed PCA to explore the conformational sampling of the Mpro and Mpro-ZINC02123811 complex via studying their collective motions through the essential dynamics approach (Amadei et al., 1993). The conformational sampling of Mpro and Mpro-ZINC02123811 complex in the essential subspace is portrayed in Fig. 6A. The projection shows the conformational sampling of Mpro along with the EV-1 and EV-2 projected by the protein Cα atoms. We found that the Mpro-ZINC02123811 complex occupied the same conformational subspace as Mpro in the free-state. A little decrease was observed at both EVs in the case of the complex with no overall shift of the motion (Fig. 6B).
Fig. 6.
Principal component analysis. (A) 2D projections of trajectories on eigenvectors (EVs) showing conformational projections of SARS-CoV-2 Mpro and Mpro-ZINC02123811 (B) The time-evolution of projections of trajectories on both EVs (C) Residual fluctuations of Mpro on EV1.
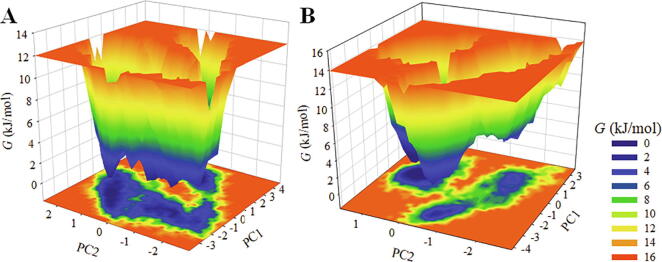
The FEL analysis provides an atomic resolution of a protein–ligand bound system, possible binding transition states, and metastable states, which can be useful in designing inhibitor. To study the conformational stability and native states of Mpro and Mpro-ZINC02123811 complex, the FELs were generated using the first two PCs. The contoured FELs of Mpro and Mpro-ZINC02123811 complex are illustrated in Fig. 7. While exploring the plots, a deeper blue is portentous to the conformational states with lower energy near to native states. We observed that Mpro is having only a single global minimum confined within three local basins. Similarly, Mpro in presence of ZINC02123811 acquires different states with multiple minima showing three local basins with different conformational motions (Fig. 7B).
Fig. 7.
The Gibbs energy landscapes for (A) free Mpro (B) Mpro- ZINC02123811.
Overall, the drug-like properties including physicochemical and ADMET, higher and specific binding towards the SARS-CoV-2 Mpro binding site, and stability during MD simulation studies suggest that ZINC02123811 can act as a potential lead in drug development against SARS-CoV-2 infection. The compound is showing antiviral potential with improved pharmacological properties and considerably high affinity and stability with SARS-CoV-2 Mpro hence could be implemented in effective therapeutic development against COVID-19 after required validation.
4. Discussion
Computational approaches are commendable in lead discovery by screening large chemical libraries against predefined drug targets. Here in this study, a library of natural compounds was filtered out based on their physicochemical and ADMET properties to identify safe and effective compounds against SARS-CoV-2 Mpro. The finally selected compounds are showing well ADMET properties and drug-likeliness. The molecular docking identified that the selected natural compounds showing a considerable binding affinity towards SARS-CoV-2 Mpro. The interaction analysis suggests that all selected compounds occupy the same position where most of the co-crystalized ligands bind. The SARS-CoV-2 Mpro cysteine-histidine catalytic dyad including Cys145 and His41, located at the main catalytic center of the protein is responsible for the functional activity of Mpro. We found three natural compounds which are commonly interacting with the binding-site residue ‘Cys145′ of SARS-CoV-2 Mpro. PASS analysis was carried out to explore the biological properties of the compounds. Based on the PASS analysis, and specific interactions towards SARS-CoV-2 Mpro cysteine-histidine catalytic dyad, Cys145 and His41, we have selected ZINC02123811 as a potent compound against SARS-CoV-2 Mpro.
The binding of any small chemical compound can make significant conformational changes to a protein structure (Kalita et al., 2020). RMSD analysis shows that the binding of ZINC02123811 with Mpro showed to have equilibration in RMSD throughout the simulation time of 100 ns which suggesting stability of the docked complex. The RMSF fluctuations were taken during the simulation for each residue in the backbone of Mpro before and after ZINC02123811 binding. The fluctuations were found to be stable and minimized upon ZINC02123811 binding suggests significant stability of the protein–ligand complex.
The Rg of a protein is directly linked to its tertiary structure and thus it is one of the widely employed parameters to study the compactness of a protein structure (Gupta et al., 2020, Naqvi et al., 2018). The Rg plot suggested that Mpro was stably folded with ZINC02123811 and behaved like the Mpro free. The SASA of a macromolecule is the area that is accessible to its surrounding solvent (Mazola et al., 2015). It has been employed in exploring the folding behavior of proteins under solvent conditions. No switching in SASA was observed throughout the trajectory of 100 ns suggesting a stable complex of Mpro and ZINC02123811.
The intramolecular H-bonds within a protein plays a fundamental role in its stability (Hubbard and Kamran Haider, 2001, Khan et al., 2017a, Prakash et al., 2019, Prakash et al., 2018, Shahbaaz et al., 2019). The plot suggests that there is no major change in the number of H-Bonds formed intramolecular within Mpro and Mpro-ZINC02123811 complex. This analysis suggests that the Mpro-ZINC02123811 complex is quite stable throughout the simulation.
The structural dynamics and conformational sampling of a protein can be explored through its phase space performance (Naz et al., 2019, Naz et al., 2018, Papaleo et al., 2009). The projection of conformational sampling of Mpro-ZINC02123811 along with the EV-1 and EV-2 projected by the protein Cα atoms is overlapping the stable clusters with phase space of Mpro-apo. The PCA analysis including the EV1 RMSF indicates that Mpro and its complex with ZINC02123811 are pretty stable during the simulation course. FEL plots also suggest that the binding of ZINC02123811 to Mpro affects the size and the location of the sampled essential subspace but with a confined stable global minimum.
Overall analysis suggests that computational methods would play a significant role in the design and development of potential therapeutic molecules to address COVID-19. Mpro plays a significant role in mediating replication and transcription and subsequent pathogenesis of SARS-CoV-2. In a recent study, Wu et al. (2020) have demonstrated that Mpro of SARS-CoV-2 inhibits the IFN induction, which may reduce antiviral responses in infected cells and thus be considered as a novel target for potential therapeutic intervention on SARS-CoV-2 infection. In another study, using a combination of structure-based virtual and high-throughput screening, Jin et al. (2020) assayed a large number of compounds and identified potential Mpro inhibitors with half-maximal inhibitory concentration (IC50) values in the range of 0.67–21.4 μM. In a similar study, Douangamath et al. (2020) screened a larger set of the electrophile and non-covalent fragments using combined mass spectrometry and X-ray approach against the Mpro of SARS-CoV-2. Several of these compounds bind to the active site pocket of enzymes and offered promising antiviral activity. Our results demonstrate the efficacy of our in-silico screening strategy, which offers a rapid discovery of potential drug leads for COVID-19. The identified compounds are showing better drug-like properties and similar fashion of binding pattern as compared to the reported Mpro inhibitor N3, in-silico. The identified compound ZINC02123811 is also shown to have antiviral potential in PASS analysis making it a potent lead scaffold for drug development against COVID-19.
5. Conclusion
With the emergence of the COVID-19 pandemic, a quick drug development against SARS-CoV-2 is immediately needed. Targeting SARS-CoV-2 Mpro with natural compounds is an attractive strategy for antiviral therapy. We employed a structure-based drug discovery approach and identified a natural compound, ZINC02123811 (1-(3-(2,5,9-trimethyl-7-oxo-3-phenyl-7H-furo[3,2-g]chromen-6-yl)propanoyl)piperidine-4-carboxamide) which is showing antiviral potential with improved pharmacological properties and considerably high affinity and stability with SARS-CoV-2 Mpro. The MD simulation study suggests the formation of a highly stable complex of Mpro with ZINC02123811. Altogether, this study provides a strong indication that ZINC02123811 might be further employed as a lead to develop potent and selective inhibitors of SARS-CoV-2 Mpro for the therapeutic management of COVID-19 after required clinical validations.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
MIH acknowledges Council of Scientific and Industrial Research (Grant No. 27(0368)/20-EMRII) and Indian Council of Medical Research for financial support (Grant No. ISRM/12(22)/2020). Authors sincerely thank to the Department of Science and Technology, Government of India for the FIST support (FIST program No. SR/FST/LSI-541/2012). MFA and AH extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding of this research through the Research Group Project No. RGP-150. Authors sincerely acknowledge Natural Science Foundation of Sichuan Province of China (Grant No. 21GJHZ0266). DSJ sincerely acknowledge the support of CMMS, Arabian Gulf University, Kingdom of Bahrain (E006-PI-10/20).
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Mohamed F. Alajmi, Email: malajmii@ksu.edu.sa.
Md. Imtaiyaz Hassan, Email: mihassan@jmi.ac.in.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- Altis A., Otten M., Nguyen P.H., Hegger R., Stock G. Construction of the free energy landscape of biomolecules via dihedral angle principal component analysis. J. Chem. Phys. 2008;128:06B620. doi: 10.1063/1.2945165. [DOI] [PubMed] [Google Scholar]
- Amadei A., Linssen A.B., Berendsen H.J. Essential dynamics of proteins. Prot.: Struct., Funct. Bioinf. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- Asrani P., Eapen M.S., Chia C., Haug G., Weber H.C., Hassan I., Sohal S.S. Diagnostic approaches in COVID-19: clinical updates. Exp. Rev. Respir. Med. 2020:1–16. doi: 10.1080/17476348.2021.1823833. [DOI] [PubMed] [Google Scholar]
- Asrani P., Hasan G.M., Sohal S.S., Hassan M.I. Molecular basis of pathogenesis of coronaviruses: a comparative genomics approach to planetary health to prevent zoonotic outbreaks in the 21st century. OMICS: J. Integr. Biol. 2020;24(11):634–644. doi: 10.1089/omi.2020.0131. [DOI] [PubMed] [Google Scholar]
- Biovia D.S. Dassault Systèmes; San Diego: 2015. Discovery Studio Modeling Environment. [Google Scholar]
- Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey L., Mascola J.R., Fauci A.S., Collins F.S. A strategic approach to COVID-19 vaccine R&D. Science. 2020;368:948–950. doi: 10.1126/science.abc5312. [DOI] [PubMed] [Google Scholar]
- Dahiya R., Mohammad T., Gupta P., Haque A., Alajmi M.F., Hussain A., Hassan M.I. Molecular interaction studies on ellagic acid for its anticancer potential targeting pyruvate dehydrogenase kinase 3. RSC Adv. 2019;9:23302–23315. doi: 10.1039/c9ra02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Ábrányi-Balogh P. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima S., Mohammad T., Jairajpuri D.S., Rehman M.T., Hussain A., Samim M., Ahmad F.J., Alajmi M.F., Hassan M.I. Identification and evaluation of glutathione conjugate gamma-l-glutamyl-l-cysteine for improved drug delivery to the brain. J. Biomol. Struct. Dyn. 2019;38(12):3610–3620. doi: 10.1080/07391102.2019.1664937. [DOI] [PubMed] [Google Scholar]
- Fatima U., Rizvi S.S.A., Fatima S., Hassan M.I. Impact of Hydroxychloroquine/chloroquine in COVID-19 therapy: two sides of the coin. J. Interferon Cytokine Res. 2020;40(10):469–471. doi: 10.1089/jir.2020.0105. [DOI] [PubMed] [Google Scholar]
- Graham B.S. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.X. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulzar M., Ali S., Khan F.I., Khan P., Taneja P., Hassan M.I. Binding mechanism of caffeic acid and simvastatin to the integrin linked kinase for therapeutic implications: a comparative docking and MD simulation studies. J. Biomol. Struct. Dyn. 2019;37:4327–4337. doi: 10.1080/07391102.2018.1546621. [DOI] [PubMed] [Google Scholar]
- Gupta P., Khan S., Fakhar Z., Hussain A., Rehman M., AlAjmi M.F., Islam A., Ahmad F., Hassan M. Identification of potential inhibitors of calcium/calmodulin-dependent protein kinase IV from bioactive phytoconstituents. Oxid. Med. Cell. Longev. 2020;2020 doi: 10.1155/2020/2094635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P., Mohammad T., Dahiya R., Roy S., Noman O.M.A., Alajmi M.F., Hussain A., Hassan M.I. Evaluation of binding and inhibition mechanism of dietary phytochemicals with sphingosine kinase 1: towards targeted anticancer therapy. Sci. Rep. 2019;9:1–15. doi: 10.1038/s41598-019-55199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard R.E., Kamran Haider M. eLS. (John Wiley & Sons; Ltd): 2001. Hydrogen bonds in proteins: role and strength. [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jacob R.B., Andersen T., McDougal O.M. Accessible high-throughput virtual screening molecular docking software for students and educators. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakhar D., Kaur I. Potential of chloroquine and hydroxychloroquine to treat COVID-19 causes fears of shortages among people with systemic lupus erythematosus. Nat. Med. 2020;26:632. doi: 10.1038/s41591-020-0853-0. [DOI] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Kalita P., Padhi A., Zhang K.Y., Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microbial Pathog. 2020 doi: 10.1016/j.micpath.2020.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F.I., Bisetty K., Gu K.-R., Singh S., Permaul K., Hassan M.I., Wei D.-Q. Molecular dynamics simulation of chitinase I from Thermomyces lanuginosus SSBP to ensure optimal activity. Mol. Simul. 2017;43:480–490. [Google Scholar]
- Khan P., Parkash A., Islam A., Ahmad F., Hassan M.I. Molecular basis of the structural stability of hemochromatosis factor E: a combined molecular dynamic simulation and GdmCl-induced denaturation study. Biopolymers. 2016;105:133–142. doi: 10.1002/bip.22760. [DOI] [PubMed] [Google Scholar]
- Khan P., Shandilya A., Jayaram B., Islam A., Ahmad F., Hassan M.I. Effect of pH on the stability of hemochromatosis factor E: a combined spectroscopic and molecular dynamics simulation-based study. J. Biomol. Struct. Dyn. 2017;35:1582–1598. doi: 10.1080/07391102.2016.1189359. [DOI] [PubMed] [Google Scholar]
- Kumari P., Singh A., Ngasainao M.R., Shakeel I., Kumar S., Lal S., Singhal A., Sohal S.S., Singh I.K., Hassan M.I. Potential diagnostics and therapeutic approaches in COVID-19. Clin. Chim. Acta. 2020;510:488–497. doi: 10.1016/j.cca.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmanic A., Zagrovic B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys. J. 2010;98:861–871. doi: 10.1016/j.bpj.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunin A., Stepanchikova A., Filimonov D., Poroikov V. PASS: prediction of activity spectra for biologically active substances. Bioinformatics. 2000;16:747–748. doi: 10.1093/bioinformatics/16.8.747. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods. 2000;44:235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Mazola Y., Guirola O., Palomares S., Chinea G., Menendez C., Hernandez L., Musacchio A. A comparative molecular dynamics study of thermophilic and mesophilic beta-fructosidase enzymes. J. Mol. Model. 2015;21:2772. doi: 10.1007/s00894-015-2772-4. [DOI] [PubMed] [Google Scholar]
- Mohammad T., Arif K., Alajmi M.F., Hussain A., Islam A., Rehman M.T., Hassan I. Identification of high-affinity inhibitors of pyruvate dehydrogenase kinase-3: towards therapeutic management of cancer. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1711810. [DOI] [PubMed] [Google Scholar]
- Mohammad T., Batra S., Dahiya R., Baig M.H., Rather I.A., Dong J.-J., Hassan I. Identification of high-affinity inhibitors of cyclin-dependent kinase 2 towards anticancer therapy. Molecules. 2019;24:4589. doi: 10.3390/molecules24244589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad T., Khan F.I., Lobb K.A., Islam A., Ahmad F., Hassan M.I. Identification and evaluation of bioactive natural products as potential inhibitors of human microtubule affinity-regulating kinase 4 (MARK4) J. Biomol. Struct. Dyn. 2019;37:1813–1829. doi: 10.1080/07391102.2018.1468282. [DOI] [PubMed] [Google Scholar]
- Mohammad T., Siddiqui S., Shamsi A., Alajmi M.F., Hussain A., Islam A., Ahmad F., Hassan M. Virtual screening approach to identify high-affinity inhibitors of serum and glucocorticoid-regulated kinase 1 among bioactive natural products: combined molecular docking and simulation studies. Molecules. 2020;25:823. doi: 10.3390/molecules25040823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A.A., Mohammad T., Hasan G.M., Hassan M. Advancements in docking and molecular dynamics simulations towards ligand-receptor interactions and structure-function relationships. Curr. Top. Med. Chem. 2018;18:1755–1768. doi: 10.2174/1568026618666181025114157. [DOI] [PubMed] [Google Scholar]
- Naqvi, A.A.T., Fatima, K., Mohammad, T., Fatima, U., Singh, I.K., Singh, A., Atif, S.M., Hariprasad, G., Hasan, G.M., Hassan, M.I., 2020. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochimica et Biophysica Acta (BBA)-Mol. Basis Dis., 165878. [DOI] [PMC free article] [PubMed]
- Naqvi, A.A.T., Hassan, M.I. (2017). Methods for docking and drug designing. In: Oncology: Breakthroughs in Research and Practice, IGI Global, pp. 876–890.
- Naz H., Tarique M., Ahamad S., Alajmi M.F., Hussain A., Rehman M.T., Luqman S., Hassan M.I. Hesperidin-CAMKIV interaction and its impact on cell proliferation and apoptosis in the human hepatic carcinoma and neuroblastoma cells. J. Cell. Biochem. 2019;120:15119–15130. doi: 10.1002/jcb.28774. [DOI] [PubMed] [Google Scholar]
- Naz H., Tarique M., Khan P., Luqman S., Ahamad S., Islam A., Ahmad F., Hassan M.I. Evidence of vanillin binding to CAMKIV explains the anti-cancer mechanism in human hepatic carcinoma and neuroblastoma cells. Mol. Cell. Biochem. 2018;438:35–45. doi: 10.1007/s11010-017-3111-0. [DOI] [PubMed] [Google Scholar]
- Nutho B., Mahalapbutr P., Hengphasatporn K., Pattaranggoon N.C., Simanon N., Shigeta Y., Hannongbua S., Rungrotmongkol T. Why are lopinavir and ritonavir effective against the newly emerged Coronavirus 2019? Atomistic insights into the inhibitory mechanisms. Biochemistry. 2020;59:1769–1779. doi: 10.1021/acs.biochem.0c00160. [DOI] [PubMed] [Google Scholar]
- Padhi A.K., Tripathi T. Can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacol. Transl. Sci. 2020;3:1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaleo E., Mereghetti P., Fantucci P., Grandori R., De Gioia L. Free-energy landscape, principal component analysis, and structural clustering to identify representative conformations from molecular dynamics simulations: the myoglobin case. J. Mol. Graph. Model. 2009;27:889–899. doi: 10.1016/j.jmgm.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Prakash A., Kumar V., Meena N.K., Hassan M.I., Lynn A.M. Comparative analysis of thermal unfolding simulations of RNA recognition motifs (RRMs) of TAR DNA-binding protein 43 (TDP-43) J. Biomol. Struct. Dyn. 2019;37:178–194. doi: 10.1080/07391102.2017.1422026. [DOI] [PubMed] [Google Scholar]
- Prakash A., Kumar V., Pandey P., Bharti D.R., Vishwakarma P., Singh R., Hassan M.I., Lynn A.M. Solvent sensitivity of protein aggregation in Cu, Zn superoxide dismutase: a molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2018;36:2605–2617. doi: 10.1080/07391102.2017.1364670. [DOI] [PubMed] [Google Scholar]
- Shahbaaz M., Amir M., Rahman S., Mustafa Hasan G., Dohare R., Bisetty K., Ahmad F., Kim J., Hassan M.I. Structural insights into Rab21 GTPase activation mechanism by molecular dynamics simulations. Mol. Simul. 2018;44:179–189. [Google Scholar]
- Shahbaaz, M., Potemkin, V., Grishina, M., Bisetty, K., Hassan, I. (2019). The structural basis of acid resistance in Mycobacterium tuberculosis: insights from multiple pH regime molecular dynamics simulations. J. Biomol. Struct. Dyn., 38.15 (2020), 4483–4492. [DOI] [PubMed]
- Shamsi A., Mohammad T., Anwar S., AlAjmi M.F., Hussain A., Rehman M., Islam A., Hassan M. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci. Rep. 2020;40 doi: 10.1042/BSR20201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling T., Irwin J.J. ZINC 15–ligand discovery for everyone. J. Chem. Inf. Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, P. (2005). XMGRACE, Version 5.1. 19. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton, OR.
- V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020:1–16. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Ma L., Zhuang Z., Cai S., Zhao Z., Zhou L., Zhang J., Wang P.-H., Zhao J., Cui J. Main protease of SARS-CoV-2 serves as a bifunctional molecule in restricting type I interferon antiviral signaling. Signal Transd. Target. Therapy. 2020;5:221. doi: 10.1038/s41392-020-00332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ai H., Chen W., Yin Z., Hu H., Zhu J., Zhao J., Zhao Q., Liu H. CarcinoPred-EL: novel models for predicting the carcinogenicity of chemicals using molecular fingerprints and ensemble learning methods. Sci. Rep. 2017;7:2118. doi: 10.1038/s41598-017-02365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]