Abstract
Zika virus (ZIKV) is a mosquito-borne pathogen that caused an epidemic in 2015–2016. ZIKV-specific T cell responses are functional in animal infection models, and helper CD4 T cells promote avid antibodies in the vaccine context. The small volumes of blood available from field research limit the determination of T cell epitopes for complex microbes such as ZIKV. The goal of this project was efficient determination of human ZIKV CD4 T cell epitopes at whole proteome scale, including validation of reactivity to whole pathogen, using small blood samples from convalescent time points when T cell response magnitude may have waned. Polyclonal enrichment of candidate ZIKV-specific CD4 T cells used cell-associated virus, documenting that T cells in downstream peptide analyses also recognize whole virus after antigen processing. Sequential query of bulk ZIKV-reactive CD4 T cells with pooled/single ZIKV peptides and molecularly defined antigen presenting cells (APC) allowed precision epitope and HLA restriction assignments across the ZIKV proteome and enabled discovery of numerous novel ZIKV CD4 T cell epitopes. The research workflow is useful for the study of emerging infectious diseases with very limited blood sample availability.
Keywords: Zika virus, CD4 T cells, epitope
Introduction
Acquired, specific T-cell immunity is typically required to clear self-limited infections, to maintain an equilibrium with chronic infections, and to enhance functional antibody responses in the vaccination context. Inclusion of microbial antigens that strongly stimulate T-cell immunity, and in particular specific CD4 T cells, is required to trigger lymph node (LN) germinal center (GC) reactions in which antigen-specific B cells undergo immunoglobulin (Ig) class switching and affinity maturation. T cells typically recognize discrete microbial peptides termed epitopes. Epitopes are also essential for analytic methods that use peptide-HLA oligomeric reagents such as tetramers to analyze pathogen-specific T cells ex vivo. Finally, knowledge of T cell epitopes facilitates therapy using virus-specific T cells that are isolated for adoptive immunotherapy or for the sequencing of their T cell receptors (TCR), which can then be used for transgenic TCR therapy.
Identification of viral T cell epitopes typically uses peripheral blood mononuclear cells (PBMC) as a source of T cells. Blood contains only a small minority of the total T cells in a human (1), such that T cells that function as T follicular helper (TFH) cells in LN GC reactions (2), and tissue resident memory (TRM) cells that control infections in barrier tissues (3), might be considered ideal for epitope discovery. These sites are difficult to access, especially in field research conditions during emerging epidemics. It is now recognized, using TCR hypervariable complementarity determining region 3 (CDR3) sequencing as barcodes for T cell clonotypes, that blood can serve as a surrogate for actual sampling of the T cell repertoire at tissue sites. In the case of ZIKV, infected cells circulate in blood (4), such that T cells may also exert a direct antiviral function in the circulation.
Commonly, T cell epitope discovery methods test peptides from the microbial proteome and use PBMCs for readout. One challenge is that as pathogen genomes increase is size, more and more blood is needed. If T cells recognize diverse epitopes and the integrated size of the response is low, reactivity to individual peptides may fall below the lower limit of detection of direct ex vivo assays. For example, we found that for the large-genome virus HSV-1, only a subset of proven CD4 and CD8 T cell epitopes recognized by blood T cells could be detected by ex vivo IFNγ ELISPOT (5, 6). Peptide pools can partially mitigate low blood availability, but solubility and solvent toxicity can still be limiting. Follow-up assays, and thus more blood, is required to definitively show reactivity to single peptides within reactive pools. In addition, techniques such as intracellular cytokine staining (ICS), enzyme-linked spot assay (ELISPOT), and mRNA detection are cell-destructive. Thus, important follow-up work after initial epitope discovery, such as determination of minimal epitopes via truncations, testing for cross-reactivity with peptides from phylogenetically related organisms or strain variants, measurement of functional avidity from peptide dose-response assays, and definition of TCR sequences of reactive T cells and determination of HLA restriction all require additional blood.
To overcome these obstacles, several groups use T-cell surface activation induced markers (AIM), or surrogates for activation such as fluorescent dye dilution, to sort live peptide-reactive T cells. After expansion, enriched, live peptide-reactive cells can be used for downstream studies (7). AIM enrichment using peptide stimulation of PBMC does not, however, document T cell reactivity with whole pathogen. Because T cell cross-reactivity to diverse sequence-related and even disparate microbial peptides is ubiquitous (8, 9), it is important to include tests of recognition of the microbial pathogen in the process of T cell epitope determination.
The recent ZIKV epidemic presents an urgent need for vaccine development. Several lines of evidence from animal models suggest that T cells are a functionally important component of the host response to both vaccination and infection (10). We sought to query the ~10,800 nucleotide ZIKV RNA genome encoding a predicted 3,423 amino acid (AA) polyprotein using one aliquot of approximately 10–15 × 106 PBMC, from approximately 10 ml of blood. The AIM workflow builds in recognition of whole ZIKV antigen and provides ample T cells for downstream analyses. Using these methods, we have discovered many novel ZIKV CD4 T cell epitopes. The results indicate that broad CD4 recognition of ZIKV in the context of not just HLA-DR, but also frequently of HLA-DQ and -DP alleles, consistent with antigen presentation by professional APC in vivo. The response is diverse, and includes several proteins in addition to the envelope (E) protein important for neutralizing antibody (11). The methods are relevant to field studies for novel and emerging pathogens that are conducted under conditions where large blood specimens are impractical.
Materials and Methods
Cells and viruses.
Vero cells (CCL-81) were obtained from ATCC (Manassas, VA). The Fortaleza strain of ZIKV was originally provided by Dr. Michael Diamond (Washington University, St. Louis, MO). ZIKV was expanded on Vero cells to titers of 107 plaque forming units/ml. PBMC were obtained in Puerto Rico or Florida, USA from consented adult blood donors identified by screening with serologic/PCR tests for ZIKV infection. Venous blood was processed to PBMC using standard Ficoll centrifugation and cells maintained in liquid nitrogen until use. Artificial APC (aAPC) were cell lines transduced to express single human HLA class II heterodimers (Supplementary Table 1). aAPC based on RM3 human lymphoma cells (12) were maintained in RPMI 1640 with 10% FCS, 25 mM HEPES, 1% penicillin-streptomycin, 1% non-essential amino acids and 1 mM sodium pyruvate. Selection for transductants was maintained with 700 μg/ml G418 and 12 μg/ml blasticidin (all ThermoFisher, Waltham, MA). For selected aAPC, HLA expression was induced with 10 μg/ml sodium butyrate (ThermoFisher) for 24–48 hours prior to testing. aAPC and non-transfected controls based on murine adherent DAP.3 cells were cultured in the same media with 200 μg/ml G418 for transductants. HLA surface expression on transduced cell lines were checked after trypsinizing cells if necessary and staining first with unconjugated anti-pan-HLA DR, DP, or DQ mAbs (13), or no first antibody negative control, followed by washing and staining with PE-conjugated secondary goat anti-mouse IgG (Biolegend, San Diego, CA) and analyzed by flow cytometry. Some cell lines were checked with PE-Cy7-conjugated anti-HLA DR clone L243 or relevant isotype control (Biolegend). Only aAPC with HLA expression were used (Supplementary Table 2). Cell lines were tested for Mycoplasma (MycoAlert, Lonza, Walkersville, MD). Positive cell lines were treated for Mycoplasma positivity with ciprofloxacin (10 μg/ml, Hospira, Forest, IL) for 3–4 weeks and re-tested to ensure clearance. Epstein-Barr virus-transformed lymphocyte continuous lines (EBV-LCL) were cultured (14) from ~2.5 × 105 donor thawed PBMC for use as autologous APC.
ZIKV antigens.
Vero cells were infected at MOI ~ 0.1. At 72 hours, moderate cytopathic effect was visible. ZIKV- or mock-infected Vero cells were scraped from plastic 75 cm2 culture flasks and collected by centrifugation at 400 × g for 10 minutes. Supernatant was collected, aliquoted into 100 μL droplets, and UV-C irradiated for 30 minutes at 10 cm from a GT15T8 bulb for 30 minutes. Peptides (Supplementary Table 2) covered the ZIKV strain Fortaleza proteome (Genbank KX811222.1). These were synthesized as 20 AA long, overlapping by 10 AA for ZIKV proteins NS (non-structural protein) 1, NS3, NS5, and E (envelope) as reported (15, 16) (GL Biochem, Shanghai, China). Similar peptides were obtained for ZIKV proteins ancC (anchored capsid protein, also termed C for capsid), preM (glycosylated precursor of M), NS2A, NS2B, NS4A, NS4B, and 2K (Genscript, Piscataway, NJ). Peptides were dissolved in DMSO (ThermoFisher) at 20 mg/ml. Pool stocks of ≤ 20 peptides containing 1 mg/ml each peptide (detailed in Supplementary Table 2) were tested at final concentrations of 1 μg/ml each. Single peptides were tested at specified concentrations.
ZIKV-reactive T cell lines.
We modified AIM-based sorting (6, 8, 17–19) to enrich ZIKV-reactive cells. Thawed PBMC were cultured at 2–4 × 106/well in 24-well plates in 2 ml/well T-cell medium (TCM, RPMI 1640 with 25mM HEPES, 1% penicillin-streptomycin, 2 mM L-glutamine, 5% FCS (ThermoFisher), and 5% human serum (Valley Biomedical, Winchester, VA). Mock or ZIKV antigens were added at 1:60 dilution for 18 hours in humidified, 37°C, 5% CO2 conditions. Cells were stained with a cocktail of anti-CD3-ECD (clone UCHT1, Beckman Coulter, Brea, CA), anti-CD4-PE (clone A161A1, Biolegend), anti-CD8-FITC (clone MHCD0801–4, ThermoFisher) and anti-CD137-APC (clone 4B4–1, BD, San Jose, CA) and 7-AAD viability stain (BD). Live CD3+CD8−CD4+CD137high cells were sorted (FacsAria II, BD). The threshold for CD137 positivity was set using PBMC from separate persons seropositive for HSV-1 and stimulation with UV-treated HSV-1 antigen as described (6). CD137high cells were expanded polyclonally once with phytohemagglutinin (PHA) as mitogen, and further expanded with anti-CD3 mAb as mitogen (19).
T cell functional assays.
Bulk responders were tested in 3H thymidine proliferation assays. For a peptide pool or single peptide to be considered reactive, the response measured as counts per minute were required to be at least twice that of DMSO negative control, and single peptides were required to be reactive in at least three separate assays at 1 μg/ml or lower concentration (below). Triplicate 96-well wells contained 5 × 104 autologous EBV-LCL, 5–10 × 104 responder cells, and antigen in 200 μl TCM. On day 3, 0.5 μCi/well 3H thymidine (Perkin Elmer, Waltham, MA) was added and 18 hours later, cells were harvested and counted (TopCount, Perkin Elmer). After identification of active 20 AA peptides, triplicate HLA restricting locus/dose-response assays used 0.01, 0.1, and 1 μg/ml peptide in the absence or presence of blocking anti-HLA-DR, -DP, or –DQ murine mAb or media control. The in-house, cell culture supernatant-based mAb preparations were used at a final concentration of 25% (13). HLA restricting alleles were then studied in triplicate using aAPC (above) and secreted IFNγ ELISA readout. For RM3-based aAPC, 5 × 104 cells/well were used in 96-well U-bottom plates with responders and peptide as above. For DAP.3-based aAPC, 3 × 104 cells were seeded the day before use into 96-well flat bottom plates and responders and peptides added as above. Selected DAP.3 aAPC were treated with 10 μg/ml sodium butyrate diluted from a 1 mg/ml stock in sterile PBS after seeding, with PBS wash prior to adding 1 × 105 responder cells. aAPC-based assays used TCM with selective antibiotic discontinuation after T cell addition. After 24 hours, supernatants were removed and IFNγ measured (20).
HLA typing.
Dried blood spots were provided to Scisco (Seattle, WA) where DNA was extracted and HLA type determined by sequencing (21).
Database submission.
Epitope-level information epitope and assay identification codes are deposited at IEDB (22) as submission 1000842.
Results
ZIKV-infected donors.
Subjects with PCR-proven ZIKV infection diagnosed in Puerto Rico or Florida in Fall, 2016 were enrolled in the Recipient Epidemiology and Donor Evaluation Study (REDS-III). Characteristics of the REDS-III study design, specimen collection during and after the ZIKV outbreak, and ZIKV PCR and antibody assays have been published (23, 24). Each subject was seropositive for dengue IgG antibodies at their initial visit. Six subjects ranging in age from 21 to 70 years old (4 women, 2 men) at the time of ZIKV diagnosis were studied between 82 and 100 days after initial diagnosis (Table 1). At this time, all subjects were ZIKV IgG seropositive. Each had been IgM positive at one or more time points, but three had reverted to negative and two were equivocal.
Table 1.
Participants and specimens used in this study.
| PBMC specimen1 | HLA class II | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| donor | specimen ID | sex | age2 | day | ZIKV IgG | ZIKV IgM | DRB1 | DRB3/4/5 | DQB1 | DPB1 |
| 4 | ZVPR8726 | F | 21 | 100 | pos | pos | *04:11 *13:03 |
3*01:01 4*01:03 |
*03:01 *04:02 |
*04:01 *14:01 |
| 10 | ZVPR2339 | M | 54 | 100 | pos | equiv | *11:04 *16:01 |
3*02:02 5*01:01 |
*03:01 *05:02 |
*01:01 *02:01 |
| 12 | ZVPR6617 | F | 36 | 90 | pos | equiv | *13:03 *16:01 |
3*01:01 5*02:02 |
*03:01 *05:02 |
*01:01 *10:01 |
| 17 | ZVPR6947 | F | 56 | 88 | pos | neg | *04:07 *13:01 |
3*02:02 4*01:03 |
*03:02 *06:02 |
*04:02 *01:05 |
| 24 | ZVFL4418 | F | 70 | 95 | pos | neg | *01:01 *15:03 |
5*01:01 | *05:01 *06:02 |
*01:01 *30:01 |
| 28 | ZVPR5399 | M | 52 | 82 | pos | neg | *01:02 *04:11 |
4*01:03 | *03:02 *05:01 |
*04:02 *01:24 |
Day of PBMC specimen after initial diagnosis of ZIKV infection and seroreactivity to ZIKV antigen on day of PBMC collection. pos = positive, neg = negative, equiv = equivocal.
Age at time of ZIKV diagnosis.
Enrichment of ZIKV-reactive CD4+ T cells.
Exposure of PBMC from most ZIKV-immune individuals to cell-associated ZIKV antigen led to a small but detectable increment in live CD3+CD4+CD137high lymphocytes at 18 hours (representative donor, Fig. 1, summarized in Table 2). For some donors, the proportion of CD4 T cells expressing CD137 was < 0.01% and there was no mathematical difference from mock stimulation, but cells were collected regardless. Previously, we used PHA as a functional viability control, leading to CD137 expression by 5 to 50% of T cells (8, 19). We omitted this to use as many cells as possible for ZIKV stimulation. A small number of activated T cells, ranging from 83 to 354 cells per donor, were sorted (Table 2). CD137high cells were expanded polyclonally to > 108 cells/donor and frozen as multiple identical aliquots.
Figure 1.

Gating scheme of AIM enrichment of polyclonal ZIKV-reactive CD4 T cell lines. Left four panels illustrate sequential gating scheme of single cells followed by live CD3 T cells and then CD4+CD8- cells. At right, a representative donor shows in the upper panel low CD137 expression 18 hours after stimulation with negative control Vero cell preparation, and in contrast specific activation of rare CD4 T cell after stimulation with cell-associated, UV-treated ZIKV cell lysate.
Table 2.
ZIKV activation-induced marker-sorted CD4 T cells
| donor | specimen ID | mock1 | ZIKV1 | CD137high cells2 |
|---|---|---|---|---|
| 4 | ZVPR8726 | <0.01 | <0.01 | 83 |
| 10 | ZVPR2339 | 0 | 0.1 | 122 |
| 12 | ZVPR6617 | <0.01 | <0.01 | 175 |
| 17 | ZVPR6947 | 0.01 | 0.02 | 161 |
| 24 | ZVFL4418 | 0.04 | 0.06 | 301 |
| 28 | ZVPR5399 | 0.08 | 0.10 | 354 |
Percent of live, lymphocyte gated, CD3+CD4+CD8- cells expressing CD137 in response to mock or ZIKV antigen.
Cell sorter count of live CD3+CD4+CD8-CD137high cells sorted in cells with ZIKV antigen.
Detection of ZIKV epitope-specific CD4 T cells.
We detected ZIKV peptide-reactive lymphocytes in five of six CD137high-origin T cell lines. Representative results for subject 24 (Table 1) show several positive peptide pools, confirmed in follow up assays of single peptides (example, Fig. 2). In this case, two adjacent peptides, NS4B AA 13–32 LSHLMGRREEGATIGFSMDI and NS4B AA 22–41 EGATIGFSMDIDLRPASAWA (overlapping region AA 22–32 underlined; peptides detailed in Supplementary Table 2) were strongly positive, suggesting the presence of T cells recognizing AA 22–32. Reactivity was also noted for the C-terminal adjacent peptide NS4B AA 31–50, indicating that this particular donor recognized at least two discrete epitopes in the N-terminal region of NS4B. In addition, the polyclonal ZIKV-reactive CD4 T cell line also reacted to NS4A AA 73–92 within this pool.
Figure 2.

Definition of ZIKV peptides recognized by polyclonal ZIKV-reactive CD4 T cell lines enriched from ZIKV-seropositive donors. At left, the AIM-enriched bulk T cells from donor 24 were queried with pooled proteome-covering ZIKV-peptides using autologous EBV-LCL as APC. At right, single peptides within pool 17 were tested. Assays are triplicate’; stdev= standard deviation of the mean.
Epitope confirmation and HLA restriction.
HLA studies typically began at the locus level, with inclusion of blocking murine mAb capable of inhibiting antigen presentation by all HLA-DR, -DP, or -DQ allelic variants. To improve discrimination, peptides were titrated from 1.0 to 0.01 μg/ml. As representative examples, for subject 24, reactivity to NS4B AA 13–32 and AA 22–41 was nearly completely inhibited by anti-HLA-DQ at each peptide concentration. There was slight inhibition for anti-HLA-DR and no inhibition for anti-HLA-DP (Fig. 3). To determine the HLA restricting allele, we tested a panel of aAPC expressing subject-specific HLA DQ as well as DR molecules. Only aAPC expressing HLA-DQB1*06:02 were able to trigger T-cell reactivity, again with identical findings for both overlapping NS4B peptides (Fig. 4). Concordant with the identification of this candidate HLA restricting allele, both longer NS4B peptides and the shared AA 22–32 region were predicted to tightly bind HLA-DQB1*06:02 using computational algorithms (22).
Figure 3.

Example of determination of HLA restricting locus and allele. Left two panels show reactivity of polyclonal ZIKV-reactive CD4 T cells from donor 24 to the indicated peptides in proliferation assays using autologous EBV-LCL as APC. Peptide identities and titrations from 1 to 0.01 μg/ml (1000 to 10 ng/ml) are indicated on the X axis. Anti-HLA class II mAbs or no antibody control used to test inhibition are indicated at top. Anti-HLA DQ inhibited the response to both peptides. Right panel shows IFNγ secretion by the same effector cells in response to 1 μg/ml peptide NS4A AA 13–32 (black bars) and AA 22–41 (hatched bars) using defined aAPC expressing donor-specific HLA alleles. Only RM3 cells expressing DQB1*06:02 were active.
Figure 4.
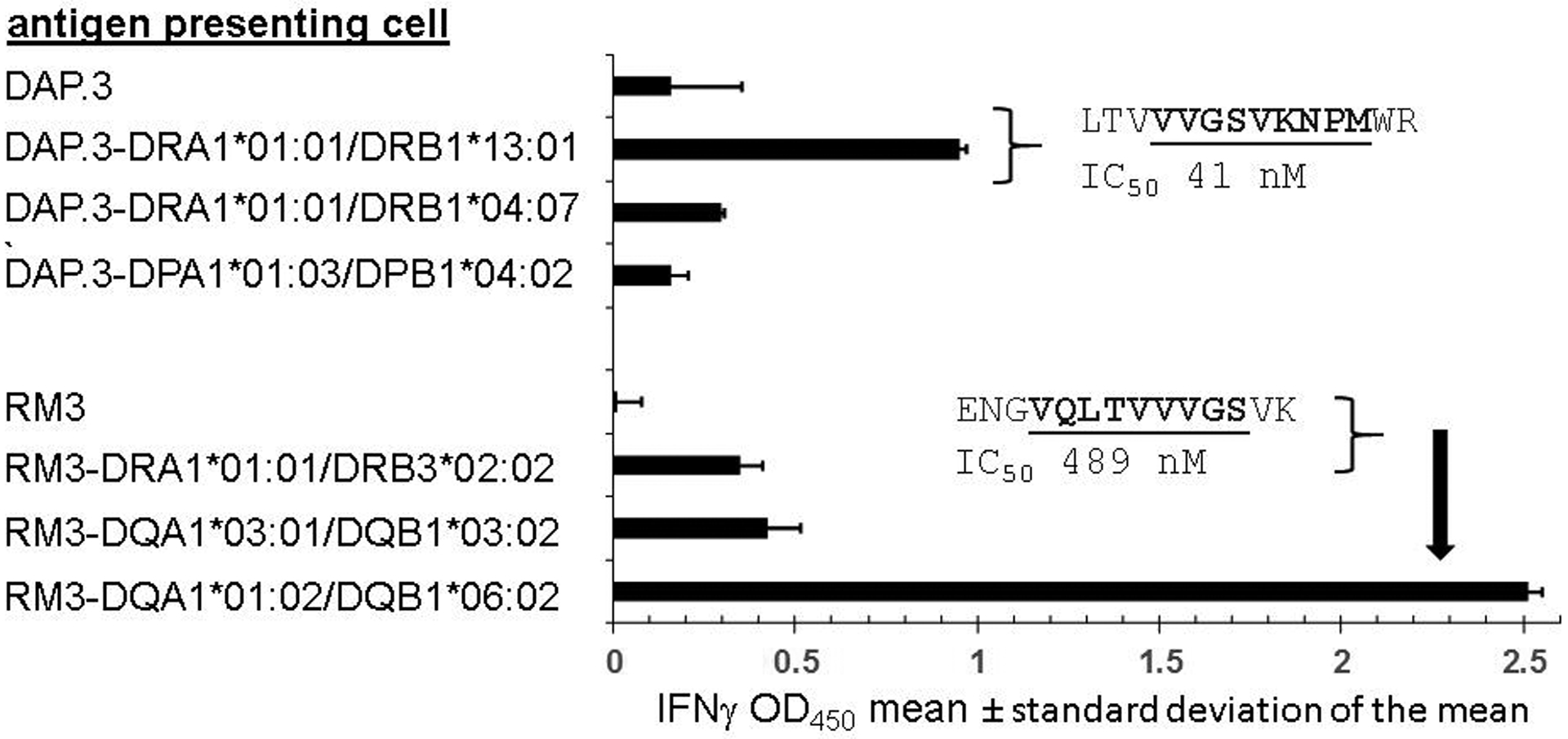
CD4 T cell recognition of ZIKV peptide NS1 81–100 is restricted by two distinct HLA alleles. APC expressing defined HLA class II heterodimers or non-expressing parental cells DAP.3 or RM3 (at left) were incubated with 20-mer peptide NS 81–100, washed, and co-cultured with expanded polyclonal CD4 T cells enriched using CD137 expression after contact with whole UV-treated ZIKV viral lysate. Data from triplicate assays are mean and standard deviation of IFNγ secretion at 24 hours as measured by ELISA. Insets are 14 to 15 AA peptides internal to NS1 81–100 and their predicted binding to the indicated HLA heterodimers, with core HLA binding motifs bold and underlined. The vertical arrow in the lower portion connects the peptide sequence with HLA DQA1*01:02/DQB1*06:02, the heterodimer used to predict that IC50 value in the figure.
Integrated CD4 T cell reactivity to ZIKV.
The conservative criteria outlined above were used to assign ZIKV epitopes for 5 participants (Table 3). For peptide-participant pairings with peptides that were reactive at least twice at 1 μg/ml, but for which HLA restriction could not be determined, epitopes are still considered confirmed. In some cases, no substantial inhibition was noted in anti-HLA blocking mAb experiments. In other cases, restricting HLA loci could be discerned using mAb, but specific alleles could not be demonstrated given lack of suitable aAPC.
Table 3.
T cell epitopes in ZIKV detected using bulk ZIKV viral lysate-reactive CD4 T cell lines.
| donor | protein and AA | sequence | HLA loci1 | HLA restricting allele(s)2 |
|---|---|---|---|---|
| 4 | ENV [301–320] | KGVSYSLCTAAFTFTKIPAE | DP | HLA-DPB1*04:01 |
| 4 | NS1 [91–110] | GSVKNPMWRGPQRLPVPVNE | DP | HLA-DPB1*04:01 |
| 4 | NS1 [101–120] | PQRLPVPVNELPHGWKAWGK | DP | HLA-DPB1*04:01 |
| 4 | NS2B[36–55] | GLLIVSYVVSGKSVDMYIER | DR | HLA-DRB1*04:11 |
| 10 | ancC [10–29] | GFRIVNMLKRGVARVSPFGG | HLA-DPB1*02:01 | |
| 10 | ancC [82–101] | KKFKKDLAAMLRIINARKEK | ||
| 10 | ENV [101–120] | WGNGCGLFGKGSLVTCAKFA | HLA-DQB1*03:01 | |
| 10 | ENV [111–130] | GSLVTCAKFACSKKMTGKSI | HLA-DRB3*02:02 | |
| 10 | ENV [131–150] | QPENLEYRIMLSVHGSQHSG | HLA-DPB1*02:01 | |
| 10 | NS1 [141–160] | KECPLKHRAWNSFLVEDHGF | ||
| 12 | NS1 [31–50] | RYKYHPDSPRRLAAAVKQAW | DR | |
| 12 | NS1 [91–110] | GSVKNPMWRGPQRLPVPVNE | DP | |
| 17 | ancC [10–29] | GFRIVNMLKRGVARVSPFGG | DR | HLA-DRB1*13:01 |
| 17 | ancC [37–56] | LLLGHGPIRMVLAILAFLRF | DR | |
| 17 | ancC [46–65] | MVLAILAFLRFTAIKPSLGL | DR/DP | HLA-DRB1*13:01 |
| 17 | ENV [61–80] | YEASISDMASDSRCPTQGEA | DQ | HLA-DQB1*03:02 |
| 17 | ENV [391–410] | VGEKKITHHWHRSGSTIGKA | DR | HLA-DRB1*04:07 |
| 17 | NS1 [81–100] | ENGVQLTVVVGSVKNPMWRG | DQ | HLA-DRB1*13:01, HLA-DQB1*06:02 |
| 17 | NS1 [151–170] | NSFLVEDHGFGVFHTSVWLK | DP | HLA-DPB1*04:02, HLA-DQB1*03:02 |
| 17 | NS1 [161–180] | GVFHTSVWLKVREDYSLECD | DP | HLA-DPB1*04:02, HLA-DQB1*03:02 |
| 17 | NS1 [201–220] | WIESEKNDTWRLKRAHLIEM | HLA-DPB1*04:02 | |
| 17 | NS5 [221–240] | WVSGAKSNTIKSVSTTSQLL | DR | HLA-DRB1*04:07, HLA-DRB1*13:01 |
| 17 | PreM [64–83] | DVDCWCNTTSTWVVYGTCHH | DQ | HLA-DQB1*06:02 |
| 17 | PreM [73–92] | STWVVYGTCHHKKGEARRSR | DQ | HLA-DQB1*06:02 |
| 24 | ENV [131–150] | QPENLEYRIMLSVHGSQHSG | DR | HLA-DRB1*01:01 |
| 24 | NS4A [73–92] | MRNKGIGKMGFGMVTLGASA | DQ | HLA-DQB1*06:02 |
| 24 | NS4B [13–32] | LSHLMGRREEGATIGFSMDI | DQ | HLA-DQB1*06:02 |
| 24 | NS4B [22–41] | EGATIGFSMDIDLRPASAWA | DQ | HLA-DQB1*06:02 |
| 24 | NS4B [31–50] | DIDLRPASAWAIYAALTTFI | ||
| 24 | NS5 [41–60] | RRALKDGVATGGHAVSRGSA | DR | HLA-DRB1*01:01, HLA-DRB5*01:01 |
Specificity of HLA locus-specific mAb or mAbs causing substantial inhibition of proliferative responses by bulk ZIKV-reactive CD4 T cell lines in 3H thymidine incorporation assays. In some instances, inhibition was not noted with any anti-HLA mAb while in others, more than one mAb led to inhibition.
HLA loci expressed by aAPC leading to specific secretion of IFNγ by bulk ZIKV-reactive CD4 T cell lines. In some instances, reactive aAPC were not detected.
A total of 27 distinct peptides were reactive, including 6 pairs of adjacent overlapping peptides and one triad of adjacent, overlapping peptides. Peptides NS1 AA 91–110, ancC AA 10–29, and ENV AA 131–150 were found to be antigenic in two donors each. The later two peptides drove distinct CD4 T cell responses, as the use of aAPC defined restriction by both DPB1*04:01 and DRB1*13:01 for ancC AA 10–29, and by both DPB1*02:01 and DRB1*01:01 for ENV AA 1313–150, Taken together, these data implies CD4 T cell responses to between 23 and 29 ZIKV epitopes. The presence of more than one HLA restricting locus and allele for some individual subject/peptides combinations (Table 3) likely indicates a polyclonal response. For example, for subject 17, polyclonal CD4 T cells reacted to peptide NS1 81–100 ENGVQLTVVVGSVKNPMWRG in the context of autologous EBV-LCL used as APC. Single antigen line (SAL) analyses showed that both DRA1*01:01/DRB1*13:01 and DQA1*01:02/DQB1*06:02 heterodimers were able to present this 20 AA peptide to the expanded CD137high CD4 T cell population (Fig. 4). Dual recognition of some 20 AA-long peptides, presumably by distinct T cell clonotypes, was supported by the presence of peptides with high predicted binding avidity for each implicated peptide. Using the NetMHCPan3.2 predictive algorithm (25) hosted at IEDB, for this example, NS1 81–100 contained internal peptides predicted to tightly bind to a peptide internal to both implicated HLA restricting alleles (Fig. 4). Without conducting truncation analyses within overlapping antigenic peptides combined with query with aAPC, including creation of CD4 T cell clones, the complexity of responses contained with these polyclonal T cell lines can only be estimated.
Given these levels of complexity, epitope counts and descriptions of antigen immunodominance and epitope diversity remain somewhat ambiguous. The number of reactive peptides detected ranged from two (donor 12) to 12 (donor 17) per participant (Table 3). Overall, reactivity was present to each structural protein derived by proteolysis of the ZIKV polyprotein: AncC, preM, and E, and in five nonstructural proteins: NS1, NS2B, NS4A, NS4B, and NS5. Amongst these, E and NS1 were the most frequently recognized. While the specific HLA restricting alleles reflected the subjects studied, there was fairly even representation of HLA-DR, -DP, and -DQ loci, which restricted responses to 9, 8, and 10 peptides, respectively.
Discussion
ZIKV caused globally widespread infections in regions hospitable to insect vectors in 2015–2016. During this crisis, children of thousands of pregnant women suffered profound neurodevelopmental damage related to ZIKV crossing the placenta and infecting embryonic brain cells (26). This has led to great interest in vaccine development with a principle goal of high titer neutralizing antibody (11). Recognizing the essential role of helper CD4 T cells in the development of long-lasting and highly avid antibodies, it is generally agreed that vaccines should at a minimum elicit substantial CD4 T cell responses. In addition, T cells can have important antiviral effector functions in animal models of ZIKV and other flavivirus infections (27). Therefore, there has been intensive research into the antigenic targets, phenotype, and Flaviviridae cross-reactivity of human ZIKV-specific T cells in both the infection and vaccine contexts (24, 28).
ZIKV is representative of epidemic and pandemic viral infections that periodically cause great morbidity, mortality, and economic damage. Once a pathogen’s nucleic acid sequence is identified, rapid determination of immunodominant T cell epitopes is essential for study of the immune correlates of infection severity and possible immunopathology, and as a benchmark for candidate vaccines. While blood sampling is accepted for medical purposes, health conditions such as anemia and cultural standards may limit blood donations for research (29). Even if generous research blood is possible, some important pathogens such as M. tuberculosis or Herpesviridae have quite large genomes, such that direct ex vivo study of the entire predicted proteome is impractical and expensive. Strategies to mitigate this include preliminary epitope mapping using large PBMC harvests from consented, immune donors and creation of large peptide pools (“megapools”) from accreted, documented peptide epitopes (30, 31). Related approaches use predictive algorithms to define peptides that can bind a set of population-prevalent HLA allelic variants and assemble megapools across the pathogen’s proteome (32). These pathways, while yielding much data, are limited to the HLA types included in the epitope discovery or prediction pipelines that contribute to pool creation, and do not provide confirmation of T cell recognition of the microbe.
In addition to these practical and medical considerations, the magnitude of the pathogen-specific T cell response, especially at memory time points, can be very low, providing an additional challenge to direct ex vivo approaches. For example, the integrated abundance of HHV-6B-reactive CD4 T cells in PBMC is on the order of 0.05% (19). Using cells expressing multiple AIM molecules (CD137, CD69, and CD154), we were able to reliably enrich virus-reactive cells to levels of up to 50% (19). This ZIKV project featured both low immune responses and very limited blood volumes. After addition of ZIKV antigen, both the absolute magnitude, and increment above negative control, for CD137 expression by CD4 T cells was minuscule or too small to detect (Fig. 1, Table 2).
Lack of PBMC to use as APC to query expanded, CD137high polyclonal CD4 T cell lines, a quality control step used for several pathogens (6, 8, 19), necessitated direct progression to ZIKV peptide screens without this reassurance. Peptides covering the predicted proteome were obtained for this purpose. Culture of autologous EBV-LCL from the equivalent of 0.25 ml of blood, while requiring a few weeks, provided an ongoing source of APC for tests of fine specificity, and is a simple procedure. Overall, despite the low levels of memory CD4 T cell responses, we identified up to 30 CD4 T cell epitopes from about 10 ml of blood per donor from 5 donors. The essentially limitless supply of responder cells and autologous aAPC, HLA typing, and a bank of aAPC expressing defined HLA class II heterodimers further allowed us to assign definitive HLA restriction to the majority of epitopes.
Supernatant-derived ZIKV antigen was able to re-stimulate rare CD4 T cells from PBMC that recognize not only abundant structural ZIKV antigens, such as E and ancC, but also non-structural proteins not thought to be present in virions. Overall, epitopes detected in non-structural proteins outnumbered those detected from classic structural proteins. It is possible that small amounts of viral proteins categorized as non-structural in fact were contained within virions. Survey of the literature reveals very little proteomic study of ZIKV virions. Non-structural proteins could also be shed into the supernatant. It is also interesting that prominent restriction by HLA-DQ and -DP alleles, in addition to HLA-DR, was documented in our small specimen set. This may correlate with in vivo infection of monocytes, as these cells constitutively express each of these HLA class II gene products (33). While definition of CD4 T cell epitopes in ZIKV is relatively mature, the precise HLA restriction of many epitopes has not been demonstrated experimentally.
The use of a related, somewhat reversed workflow, starting with polyclonal enrichment and expansion of peptide-reactive cells, followed by readout with ZIKV-infected APC, has been successful for both flavivirus-immune and naïve blood donors. Effector CD4 T cell lines from dengue-immune, ZIKV-naïve donors, specific for cross-reactive peptides, showed brisk cytokine and granzyme B responses to HLA-appropriate, allogeneic EBV-LCL APC infected with an African strain of ZIKV (34). We plan to examine the cytotoxic and other effector potential of ZIKV-reactive CD4 T cells in response to ZIKV-infected APC in future studies. In other viral systems, we have been able to readily clone and expand T cell clones from AIM-enriched T cell lines, perhaps due to the strongly co-stimulatory effect of CD137 ligation with a mAb that is integral to initial AIM cell sorting (17, 35). Clones with fully defined specificity and HLA restriction should enable us to determine if physiologically relevant cells with limited HLA class II expression can be recognized by virus-specific CD4 T cells in the context of viral infection. Unfortunately, we were not able to correlate T cell responses with clinical outcomes because of our small sample size and because the cell donors were recruited from a blood donation program. Precisely-defined T cell epitopes are a necessary precursor for direct ex vivo studies with peptide-MHC staining reagents, an important method for establishing immune correlates of infection severity (36, 37).
Limited blood samples and logistics required us to cryopreserve PBMC. Some cells with the ability to present complex antigens, such as plasmacytoid dendritic cells (pDC), are sensitive to freeze-thaw and are studied fresh (38). However, pDC are generally thought to have primarily innate immunity functions, while other blood leukocytes mediate most antigen presentation to CD4 T cells. AIM work by our lab and others to enrich or measure virus-specific CD4 T cell responses to peptide or complex antigens generally uses cryopreserved PBMC (6, 8, 19, 32, 39, 40). This indicates that cells with APC function do survive freeze-thaw, while not ruling out a decrement. Limited research comparing cryopreserved with fresh PBMC for CD4 T cell recall to complex antigens is consistent with some decrease in magnitude of responses but retention of proportionality within the results (41, 42). While the use of cryopreserved PBMC is almost universal in human T cell research requiring complex cohorts, rare donors, and biocontainment, future research could compare responses between fresh and cryopreserved cells.
We were not able to include flavivirus-naïve individuals in our study. However, we feel it is unlikely that the epitope-specific responses we detected came from the naïve repertoire. In studies of replication-competent flavivirus vaccines such as yellow fever virus strain 17D and attenuated dengue that were conducted in flavivirus-naïve individuals, virus-reactive T cells were essentially undetectable at baseline (43, 44). We found that HSV- and CMV-seronegative persons had very low or undetectable virus-specific T cells in PBMC using direct ex vivo tests including CD137-based AIM (6, 38). Further research will be required to determine if the one-step re-stimulation used in this report was able to amplify naïve responses, which are reported to be present but very low abundance in virus-seronegative humans (45).
The parameters chosen for rare cell enrichment were tailored to both the use of complex viral antigen as the initial stimulus, and to the biology of CD137 up-regulation after stimulation of T cells. Typically, direct ex vivo assays to detect recall responses can short times, on the order of 6 to 8 hours, for peptides, while for complex antigens that require intracellular processing within APC, longer periods are used. Studies show that 18–24 hours or more is optimal for CD137 up-regulation regardless of whether peptide of complex antigens are used (7, 43, 46). These data inform the protocols used for CD137-based rare cell enrichment and quantitative assays (32, 39).
Our findings significantly extend previous communications concerning the specificity of ZIKV-reactive T cells. Koblishchke et al. used 15-mer OLP covering the ZIKV C, prM, and E proteins and detected IL-2 secretion in ex vivo ELISPOT from CD8-depleted PBMC. The donors were ZIKV-positive travelers returned from ZIKV-epidemic regions with prior flavivirus vaccination with live attenuated tick-born encephalitis (TBE) or yellow fever (YF); all were dengue-seronegative. Compared to flavivirus-naïve controls, positive responses to ZIKV peptides segregated with ZIKV immune status, and across the cohort, were detected for each ZIKV gene studied (47). Data integration with CD4 T cell epitopes in IEDB (22) identified several putative immunogenic regions including ENV 101–150, 301–320 and 391–410, NS1 151–180, and ancC 37–56 and 82–101 that overlap with epitopes regions described here. Previous studies summarized in the IEDB identified 89 different Zika-derived CD4 epitopes, about 30% (n=25) of which cluster (48) with the epitopes identified in the present study. However, exact MHC restriction has been defined for just 10 of the previously defined epitopes. By comparison, the present analysis has identified 27 different reactive peptide epitopes, of which 18 are novel, associated with 29 unique HLA Class II-epitope restrictions, none of which have been previously described. Thus the present study has contributed a 3-fold increase in the number of defined Zika CD4-epitope HLA class II restrictions that can subsequently be utilized for generation of multimeric reagents to further characterize Zika CD4 responses.
In conclusion, the level of ZIKV-reactive memory CD4 T cells expressing the activation marker CD137 in response to whole viral recall antigen in healthy, dengue-seropositive adults, several months after ZIKV infection, was quite low. Regardless, CD137 AIM-based virus-specific CD4 T cell enrichment led to readily expandable, polyclonal cell populations that contained epitope-specific, ZIKV-specific CD4 T cell recovery from the majority of donors. Importantly, very small blood samples, acceptable in diverse communities, were able to yield valuable viral T cell epitope information including precise HLA restriction information.
Supplementary Material
Importance.
Zika virus (ZIKV) caused an epidemic in 2015–2016 that resulted in impaired fetal brain growth. There is no licensed vaccine. An effective vaccine will require a specific immune response from T cells that express the CD4 molecule, which coordinate with other immune cells called B cells to produce antibodies. Because ZIKV is a complex virus with many proteins, discovery of the specific viral proteins that stimulate T cell responses is very challenging with the small blood samples typically obtained during field research. In this report, 8 ml blood samples were “stretched” to interrogate T cell responses to every ZIKV protein. Many small regions of ZIKV proteins recognized by T cells, termed epitopes, were discovered and validated. The workflow is suitable for emerging infectious pathogens when only limited blood specimens are available.
Abbreviations
Many abbreviations are introduced at first use, while some that are commonly used are not defined. Intermediate terms are listed here; the authors can adjust as needed after initial review.
- ELISPOT
enzyme-linked immunospot assay
- CD
cluster of differentiation
- IFNγ
Interferon gamma
- mAb
monoclonal antibody
Footnotes
Previous presentation: none
References
- 1.Morris SE, Farber DL, and Yates AJ. 2019. Tissue-Resident Memory T Cells in Mice and Humans: Towards a Quantitative Ecology. Journal of immunology 203: 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heit A, Schmitz F, Gerdts S, Flach B, Moore MS, Perkins JA, Robins HS, Aderem A, Spearman P, Tomaras GD, De Rosa SC, and McElrath MJ. 2017. Vaccination establishes clonal relatives of germinal center T cells in the blood of humans. The Journal of experimental medicine 214: 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gebhardt T, Whitney PG, Zaid A, Mackay LK, Brooks AG, Heath WR, Carbone FR, and Mueller SN. 2011. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature 477: 216–219. [DOI] [PubMed] [Google Scholar]
- 4.Foo SS, Chen W, Chan Y, Bowman JW, Chang LC, Choi Y, Yoo JS, Ge J, Cheng G, Bonnin A, Nielsen-Saines K, Brasil P, and Jung JU. 2017. Asian Zika virus strains target CD14(+) blood monocytes and induce M2-skewed immunosuppression during pregnancy. Nature microbiology 2: 1558–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jing L, Schiffer JT, Chong TM, Bruckner JJ, Davies DH, Felgner PL, Haas J, Wald A, Verjans GM, and Koelle DM. 2013. CD4 T-cell memory responses to viral infections of humans show pronounced immunodominance independent of duration or viral persistence. Journal of virology 87: 2617–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jing L, Haas J, Chong TM, Bruckner JJ, Dann GC, Dong L, Marshak JO, McClurkan CL, Yamamoto TN, Bailer SM, Laing KJ, Wald A, Verjans GMGM, Koelle DM 2012. Herpes simplex virus type 1 T-cells antigens in humans revealed by cross-presentation and genome-wide screening. Journal of Clinical Investigation 122: 654–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfl M, Kuball J, Ho WY, Nguyen H, Manley TJ, Bleakley M, and Greenberg PD. 2007. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood 110: 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nayak K, Jing L, Russell RM, Davies DH, Hermanson G, Molina DM, Liang X, Sherman DR, Kwok WW, Yang J, Kenneth J, Ahamed SF, Chandele A, Murali-Krishna K, and Koelle DM. 2015. Identification of novel Mycobacterium tuberculosis CD4 T-cell antigens via high throughput proteome screening. Tuberculosis 95: 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clute SC, Watkin LB, Cornberg M, Naumov YN, Sullivan JL, Luzuriaga K, Welsh RM, and Selin LK. 2005. Cross-reactive influenza virus-specific CD8+ T cells contribute to lymphoproliferation in Epstein-Barr virus-associated infectious mononucleosis. The Journal of clinical investigation 115: 3602–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grubor-Bauk B, Wijesundara DK, Masavuli M, Abbink P, Peterson RL, Prow NA, Larocca RA, Mekonnen ZA, Shrestha A, Eyre NS, Beard MR, Gummow J, Carr J, Robertson SA, Hayball JD, Barouch DH, and Gowans EJ. 2019. NS1 DNA vaccination protects against Zika infection through T cell-mediated immunity in immunocompetent mice. Science advances 5: eaax2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erasmus JH, Khandhar AP, Guderian J, Granger B, Archer J, Archer M, Gage E, Fuerte-Stone J, Larson E, Lin S, Kramer R, Coler RN, Fox CB, Stinchcomb DT, Reed SG, and Van Hoeven N. 2018. A Nanostructured Lipid Carrier for Delivery of a Replicating Viral RNA Provides Single, Low-Dose Protection against Zika. Molecular therapy : the journal of the American Society of Gene Therapy 26: 2507–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinney DM, Southwood S, Hinz D, Oseroff C, Arlehamn CS, Schulten V, Taplitz R, Broide D, Hanekom WA, Scriba TJ, Wood R, Alam R, Peters B, Sidney J, and Sette A. 2013. A strategy to determine HLA class II restriction broadly covering the DR, DP, and DQ allelic variants most commonly expressed in the general population. Immunogenetics 65: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koelle DM, Corey L, Burke RL, Eisenberg RJ, Cohen GH, Pichyangkura R, and Triezenberg SJ. 1994. Antigenic specificity of human CD4+ T cell clones recovered from recurrent genital HSV-2 lesions. Journal of virology 68: 2803–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tigges MA, Koelle DM, Hartog K, Sekulovich RE, Corey L, and Burke RL. 1992. Human CD8+ herpes simplex virus-specific cytotoxic T lymphocyte clones recognize diverse virion protein antigens. Journal of virology 66: 1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds CJ, Suleyman OM, Ortega-Prieto AM, Skelton JK, Bonnesoeur P, Blohm A, Carregaro V, Silva JS, James EA, Maillere B, Dorner M, Boyton RJ, and Altmann DM. 2018. T cell immunity to Zika virus targets immunodominant epitopes that show cross-reactivity with other Flaviviruses. Scientific reports 8: 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds CJ, Watber P, Santos CNO, Ribeiro DR, Alves JC, Fonseca ABL, Bispo AJB, Porto RLS, Bokea K, de Jesus AMR, de Almeida RP, Boyton RJ, and Altmann DM. 2020. Strong CD4 T Cell Responses to Zika Virus Antigens in a Cohort of Dengue Virus Immune Mothers of Congenital Zika Virus Syndrome Infants. Frontiers in immunology 11: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jing L, Laing KJ, Dong L, Russell RM, Barlow RS, Haas JG, Ramchandani MS, Johnston C, Buus S, Redwood AJ, White KD, Mallal SA, Phillips EJ, Posavad CM, Wald A, and Koelle DM. 2016. Extensive CD4 and CD8 T Cell Cross-Reactivity between Alphaherpesviruses. Journal of immunology 196: 2205–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramchandani MS, Jing L, Russell RM, Tran T, Laing KJ, Magaret AS, Selke S, Cheng A, Huang ML, Xie H, Strachan E, Greninger AL, Roychoudhury P, Jerome KR, Wald A, and Koelle DM. 2019. Viral Genetics Modulate Orolabial Herpes Simplex Virus Type 1 Shedding in Humans. The Journal of infectious diseases 219: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson DJ, Tsvetkova O, Rerolle GF, Greninger AL, Sette A, Jing L, Campbell VL, and Koelle DM. 2019. Genome-Wide Approach to the CD4 T-Cell Response to Human Herpesvirus 6B. Journal of virology 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koelle DM, Chen HB, Gavin MA, Wald A, Kwok WW, and Corey L. 2001. CD8 CTL from genital herpes simplex lesions: recognition of viral tegument and immediate early proteins and lysis of infected cutaneous cells. Journal of immunology 166: 4049–4058. [DOI] [PubMed] [Google Scholar]
- 21.Lind A, Akel O, Wallenius M, Ramelius A, Maziarz M, Zhao LP, Geraghty DE, Palm L, Lernmark Å, and Larsson HE. 2019. HLA high-resolution typing by next-generation sequencing in Pandemrix-induced narcolepsy. PLoS One 14: e0222882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, and Peters B. 2015. The immune epitope database (IEDB) 3.0. Nucleic acids research 43: D405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinman S, Busch MP, Murphy EL, Shan H, Ness P, and Glynn SA. 2014. The National Heart, Lung, and Blood Institute Recipient Epidemiology and Donor Evaluation Study (REDS-III): a research program striving to improve blood donor and transfusion recipient outcomes. Transfusion 54: 942–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grifoni A, Pham J, Sidney J, O’Rourke PH, Paul S, Peters B, Martini SR, de Silva AD, Ricciardi MJ, Magnani DM, Silveira CGT, Maestri A, Costa PR, de-Oliveira-Pinto LM, de Azeredo EL, Damasco PV, Phillips E, Mallal S, de Silva AM, Collins M, Durbin A, Diehl SA, Cerpas C, Balmaseda A, Kuan G, Coloma J, Harris E, Crowe JE Jr., Stone M, Norris PJ, Busch M, Vivanco-Cid H, Cox J, Graham BS, Ledgerwood JE, Turtle L, Solomon T, Kallas EG, Watkins DI, Weiskopf D, and Sette A. 2017. Prior Dengue Virus Exposure Shapes T Cell Immunity to Zika Virus in Humans. Journal of virology 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z, Sette A, Peters B, and Nielsen M. 2018. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154: 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rombi F, Bayliss R, Tuplin A, and Yeoh S. 2020. The journey of Zika to the developing brain. Molecular biology reports 47: 3097–3115. [DOI] [PubMed] [Google Scholar]
- 27.Elong Ngono A, Young MP, Bunz M, Xu Z, Hattakam S, Vizcarra E, Regla-Nava JA, Tang WW, Yamabhai M, Wen J, and Shresta S. 2019. CD4+ T cells promote humoral immunity and viral control during Zika virus infection. PLoS pathogens 15: e1007474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tonnerre P, Melgaco JG, Torres-Cornejo A, Pinto MA, Yue C, Blumel J, de Sousa PSF, de Mello VDM, Moran J, de Filippis AMB, Wolski D, Grifoni A, Sette A, Barouch DH, Hoogeveen RC, Baylis SA, Lauer GM, and Lewis-Ximenez LL. 2020. Evolution of the innate and adaptive immune response in women with acute Zika virus infection. Nature microbiology 5: 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muthivhi TN, Olmsted MG, Park H, Sha M, Raju V, Mokoena T, Bloch EM, Murphy EL, and Reddy R. 2015. Motivators and deterrents to blood donation among Black South Africans: a qualitative analysis of focus group data. Transfusion medicine (Oxford, England) 25: 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tscharke DC, Karupiah G, Zhou J, Palmore T, Irvine KR, Haeryfar SM, Williams S, Sidney J, Sette A, Bennink JR, and Yewdell JW. 2005. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. The Journal of experimental medicine 201: 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, and Picker LJ. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. The Journal of experimental medicine 202: 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grifoni A, Weiskpof D, Ramirez SI, Mateus J, Dan JM, Rydyznski Moderbacker C, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin A, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A 2020. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed persons. Cell 181: 1489–1501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, Tam H, Adler L, Ilstad-Minnihan A, Macaubas C, and Mellins ED. 2017. The MHC class II antigen presentation pathway in human monocytes differs by subset and is regulated by cytokines. PLoS One 12: e0183594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanajiri R, Sani GM, Hanley PJ, Silveira CG, Kallas EG, Keller MD, and Bollard CM. 2019. Generation of Zika virus-specific T cells from seropositive and virus-naive donors for potential use as an autologous or “off-the-shelf” immunotherapeutic. Cytotherapy 21: 840–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones D, Como CN, Jing L, Blackmon A, Neff CP, Krueger O, Bubak AN, Palmer BE, Koelle DM, and Nagel MA. 2019. Varicella zoster virus productively infects human peripheral blood mononuclear cells to modulate expression of immunoinhibitory proteins and blocking PD-L1 enhances virus-specific CD8+ T cell effector function. PLoS pathogens 15: e1007650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James EA, DeVoti JA, Rosenthal DW, Hatam LJ, Steinberg BM, Abramson AL, Kwok WW, and Bonagura VR. 2011. Papillomavirus-specific CD4+ T cells exhibit reduced STAT-5 signaling and altered cytokine profiles in patients with recurrent respiratory papillomatosis. Journal of immunology (Baltimore, Md. : 1950) 186: 6633–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strickland N, Müller TL, Berkowitz N, Goliath R, Carrington MN, Wilkinson RJ, Burgers WA, and Riou C. 2017. Characterization of Mycobacterium tuberculosis-Specific Cells Using MHC Class II Tetramers Reveals Phenotypic Differences Related to HIV Infection and Tuberculosis Disease. Journal of immunology (Baltimore, Md. : 1950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moss NJ, Magaret A, Laing KJ, Kask AS, Wang M, Mark KE, Schiffer JT, Wald A, and Koelle DM. 2012. Peripheral blood CD4 T-cell and plasmacytoid dendritic cell (pDC) reactivity to herpes simplex virus 2 and pDC number do not correlate with the clinical or virologic severity of recurrent genital herpes. Journal of virology 86: 9952–9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, van den Akker JPC, Molenkamp R, Koopmans MPG, van Gorp ECM, Haagmans BL, de Swart RL, Sette A, and de Vries RD. 2020. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Science immunology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaunders JJ, Munier ML, Seddiki N, Pett S, Ip S, Bailey M, Xu Y, Brown K, Dyer WB, Kim M, de Rose R, Kent SJ, Jiang L, Breit SN, Emery S, Cunningham AL, Cooper DA, and Kelleher AD. 2009. High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40). Journal of immunology (Baltimore, Md. : 1950) 183: 2827–2836. [DOI] [PubMed] [Google Scholar]
- 41.Trück J, Mitchell R, Thompson AJ, Morales-Aza B, Clutterbuck EA, Kelly DF, Finn A, and Pollard AJ. 2014. Effect of cryopreservation of peripheral blood mononuclear cells (PBMCs) on the variability of an antigen-specific memory B cell ELISpot. Human vaccines & immunotherapeutics 10: 2490–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martikainen MV, and Roponen M. 2020. Cryopreservation affected the levels of immune responses of PBMCs and antigen-presenting cells. Toxicology in vitro : an international journal published in association with BIBRA 67: 104918. [DOI] [PubMed] [Google Scholar]
- 43.Blom K, Braun M, Ivarsson MA, Gonzalez VD, Falconer K, Moll M, Ljunggren HG, Michaëlsson J, and Sandberg JK. 2013. Temporal dynamics of the primary human T cell response to yellow fever virus 17D as it matures from an effector- to a memory-type response. Journal of immunology (Baltimore, Md. : 1950) 190: 2150–2158. [DOI] [PubMed] [Google Scholar]
- 44.Graham N, Eisenhauer P, Diehl SA, Pierce KK, Whitehead SS, Durbin AP, Kirkpatrick BD, Sette A, Weiskopf D, Boyson JE, and Botten JW. 2020. Rapid Induction and Maintenance of Virus-Specific CD8(+) T(EMRA) and CD4(+) T(EM) Cells Following Protective Vaccination Against Dengue Virus Challenge in Humans. Frontiers in immunology 11: 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geiger R, Duhen T, Lanzavecchia A, and Sallusto F. 2009. Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. The Journal of experimental medicine 206: 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfl M, Kuball J, Eyrich M, Schlegel PG, and Greenberg PD. 2008. Use of CD137 to study the full repertoire of CD8+ T cells without the need to know epitope specificities. Cytometry. Part A : the journal of the International Society for Analytical Cytology 73: 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koblischke M, Stiasny K, Aberle SW, Malafa S, Tsouchnikas G, Schwaiger J, Kundi M, Heinz FX, and Aberle JH. 2018. Structural Influence on the Dominance of Virus-Specific CD4 T Cell Epitopes in Zika Virus Infection. Frontiers in immunology 9: 1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dhanda SK, Vaughan K, Schulten V, Grifoni A, Weiskopf D, Sidney J, Peters B, and Sette A. 2018. Development of a novel clustering tool for linear peptide sequences. Immunology 155: 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


