Summary
Background
Fatty liver disease (FLD) is the most common cause of liver dysfunction in developed countries. There is great interest in developing clinically valid and minimally invasive biomarkers to enhance early diagnosis of FLD.
Aim
To investigate the potential of circulatory microRNAs (miRNAs) as biomarkers of FLD at the population level.
Methods
Plasma levels of 2083 miRNAs were measured by RNA sequencing in 1999 participants from the prospective population‐based Rotterdam Study cohort. The Hounsfield Unit (HU) attenuation of liver was measured using non‐enhanced computed tomography (CT) scan. Logistic and linear regression models adjusting for potential confounders were used to examine the association of circulatory miRNAs with liver enzymes (n = 1991) and CT‐based FLD (n = 954). Moreover, the association of miRNAs with hepatic steatosis and liver fibrosis was assessed longitudinally in individuals who underwent abdominal ultrasound (n = 1211) and transient elastography (n = 777) after a median follow‐up of >6 years.
Results
Cross‐sectional analysis showed 61 miRNAs significantly associated with serum gamma‐glutamyl transferase and/or alkaline phosphatase levels (Bonferroni‐corrected P < 8.46 × 10−5). Moreover, 17 miRNAs were significantly associated with CT‐based FLD (P < 8.46 × 10−5); 14 were among miRNAs associated with liver enzymes. Longitudinal analysis showed that 4 of these 14 miRNAs (miR‐193a‐5p, miR‐122‐5p, miR‐378d and miR‐187‐3p) were significantly associated with hepatic steatosis (P < 3.57 × 10−3) and three (miR‐193a‐5p, miR‐122‐5p and miR‐193b‐3p) were nominally associated with liver fibrosis (P < 0.05). Nine of the 14 identified miRNAs were involved in pathways underlying liver diseases.
Conclusions
Plasma levels of several miRNAs can be used as biomarkers of FLD, laying the groundwork for future clinical applications.
1. INTRODUCTION
Fatty liver disease (FLD), is the most common cause of liver dysfunction in developed countries, that is also increasing in developing countries, 1 is defined as an excess accumulation of fat in hepatocytes. 2 Specifically, non‐alcoholic fatty liver disease (NAFLD) is characterised by fat accumulation in hepatocytes not due to excess alcohol consumption. 3 The disorder covers a broad spectrum of underlying conditions, ranging from simple fatty liver to inflammation, which can progress to fibrosis, cirrhosis and even liver cancer. 4 FLD is strongly associated with obesity, hypertension, dyslipidaemia and insulin resistance, regarded as hepatic manifestation of the metabolic syndrome. 5 Currently, liver biopsy is the gold standard for diagnosing and staging of FLD, but its application is limited by the invasive nature, risk of complications and high cost. 6 Various imaging modalities, such as computed tomography (CT) scan and ultrasound, have also been used for detecting the presence or quantifying the severity of liver fat noninvasively. 7 However, the limited diagnostic accuracy of detecting mild degree hepatic steatosis with CT and ultrasound is an issue that should be taken into consideration. 7 Transient elastography is non‐invasive technique that uses both ultrasound and low‐frequency elastic waves to qualify liver fibrosis. However, recent research suggests that steatosis may influence its diagnostic performance. 8 Controlled attenuation parameter (CAP) is an ultrasound‐based diagnostic method and added to transient elastography enables simultaneous assessment of steatosis and fibrosis, 9 but the clinical application of CAP is limited by influences of covariates. 10 Therefore, the development of clinically valid and minimally invasive methods is required to enhance early diagnosis of FLD.
MicroRNAs (miRNAs) are small non‐coding RNA molecules of 20‐25 nucleotides in length that regulate gene expression at the post‐transcriptional level. 11 Recently, the interest in miRNAs has increased tremendously because they offer new insights into disease mechanisms and have a great potential to be used in the clinic as diagnostic biomarkers and/or even therapeutic targets. 12 , 13 In line with this, numerous studies have reported increased levels of circulating miR‐122 in liver diseases with different aetiologies and suggested this miRNA as a potential biomarker and target of therapy in liver dysfunction. 14 , 15 Although extensive research has explored the role of miRNAs in the pathophysiology of liver diseases, little is known about the potential of circulatory miRNAs as FLD biomarker in the population level. Moreover, the available studies have mainly used qPCR‐based methods and limited to small number of miRNAs and sample sizes. 16 , 17
The aim of this study was to systematically investigate the association of circulating miRNAs in plasma with FLD in a population‐based setting. To achieve this aim, we conducted regression models to identify miRNAs that are associated with FLD and liver enzymes at the baseline in the Rotterdam Study (RS) cohort. Moreover, we performed subsequent analyses to check whether the identified miRNAs are linked to the risk of hepatic steatosis or liver fibrosis after follow‐up and are involved in the known pathways underlying liver diseases.
2. MATERIALS AND METHODS
2.1. Study population
This study was embedded within the framework of the RS, a prospective cohort study of individuals aged ≥45 years living in the Ommoord district of Rotterdam, the Netherlands. The objectives and design of the RS have been described in detail elsewhere. 18 In 1989, the first cohort of study participants (RS‐I) comprised 7983 persons aged 55 years or over. In 2000, the second cohort (RS‐II) was extended to include an additional 3011 participants who moved into the study district or had become 55 years of age. A further extension of the RS cohort (RS‐III) formed in 2006 and include 3932 participants living in the research area and aged 45 years and older. Follow‐up examinations were scheduled periodically, approximately every 3‐5 years. All participants in the study provided written inform consent to participate and to obtain information from their treating physicians.
For this study, we used the expression profiles of circulating miRNA in plasma, collected between 2002 and 2005, from a random subset (n = 1000) of the fourth visit of the first cohort (RS‐I‐4) and a random subset (n = 999) of the second visit of the second cohort (RS‐II‐2). Among them, 1991 participants had serum gamma‐glutamyl transferase (GGT) and Alkaline phosphatase (ALP) levels available at baseline that were included to investigate the association of miRNAs with liver enzymes. Moreover, 954 participants who underwent CT‐scan from June 2003 to February 2006 were included for investigating the associations of miRNAs with FLD in a cross‐sectional setting (Figure 1).
Figure 1.
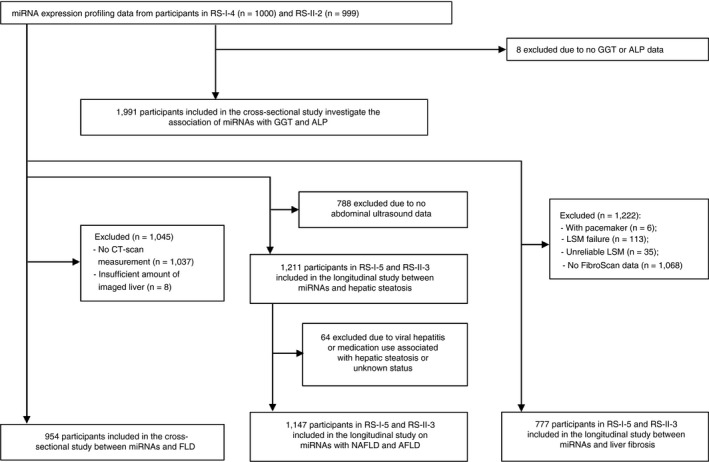
An overview of the study to identify circulatory miRNAs associated with FLD. Cross‐sectional studies at baseline were performed in participants from RS‐I‐4 and RS‐II‐2 with liver enzymes and CT‐scan data available. Longitudinal studies were performed in participants from RS‐I‐5 and RS‐II‐3 who underwent abdominal ultrasound or transient elastography. Abbreviations: AFLD, alcoholic fatty liver disease; ALP, Alkaline phosphatase; CT, computed tomography; FLD, fatty liver disease; GGT, gamma‐glutamyl transferase; LSM, liver stiffness measurement.; miRNAs, microRNAs; NAFLD, non‐alcoholic fatty liver disease; RS, Rotterdam Study; RS‐I‐4, the fourth visit of the first cohort; RS‐I‐5, the fifth visit of the first cohort; RS‐II‐2, the second visit of the second cohort; RS‐II‐3, the third visit of the second cohort
For the longitudinal analysis, 1999 participants at the baseline were followed up >6 years, until the fifth visit of the first cohort (RS‐I‐5) and the third visit of the second cohort (RS‐II‐3). Among these, 1211 participants who underwent abdominal ultrasound between January 2009 and June 2014 were included to investigate the association of miRNAs with hepatic steatosis (424 cases). Of these, 1147 participants were included to investigate the association of miRNAs with NAFLD (321 cases) and alcoholic FLD (76 cases). Moreover, out of the 1999 individuals, 777 participants who underwent transient elastography were included to investigate the association of miRNAs with liver fibrosis (33 cases). A more detailed flow chart for the selection of study participants is shown in Figure 1.
2.2. MiRNA expression profiling
Plasma levels of cell‐free miRNAs were determined using the HTG EdgeSeq miRNA Whole Transcriptome Assay (WTA), which measures the expression of 2083 human mature miRNAs. The WTA characterises miRNA expression patterns, and measures the expression of 13 housekeeping genes, that allows flexibility in data normalisation and analysis. Plasma samples, for two re‐measurements that generally is sufficient to obtain a valid result for all samples, were sent to HTG Molecular Diagnostics for sequencing. Each sample was tagged individually with molecular barcodes, tagged samples were pooled and sequenced on an Illumina NextSeq 500 sequencer (Illumina). Quantification of miRNA expression was based on counts per million (CPM). The log2 transformation of CPM was used as standardisation and adjustment for total reads within each sample. The miRNAs with log2 CPM < 1.0 were considered as not expressed in the samples. Of the 2083 miRNAs, 591 miRNAs were expressed at good levels in plasma. These 591 well‐expressed miRNAs are those with >50% values above Lower Limit of Quantification (LLOQ). The LLOQ level is based on a monotonic decreasing spline curve fit between the means and standard deviations of all miRNAs.
2.3. Assessment of liver fat with CT scan
As part of a larger project on the assessment of vascular calcification, ECG‐gated, cardiac, non‐enhanced CT scanning on a 16‐slice (n = 251) or 64‐slice (n = 703) multi‐detector CT scanner (Somatom Sensation 16 or 64, Siemens). Imaging parameters of the scans are described in detail elsewhere. 19
Using these cardiac scans, we evaluated the liver density (attenuation) using a standardised procedure. First, we placed three circular regions of interest (ROIs) in the liver and calculated the mean liver attenuation (LA) within these regions. 20 These ROIs are delineated throughout the imaged liver tissue (including both the left and right liver lobes) are carefully chosen to include only liver tissue, and avoiding the large blood vessels, cysts or focal lesions. Next, we calculated the mean Hounsfield unit (HU) value from these three measurements as a marker of total amount of liver fat, which is a reliable proxy for the mean LA value of the whole liver. 20 All measurements were done using Philips iSite Enterprise software (Royal Philips Electronics NV 2006) and described in detail elsewhere. 21
The CT diagnosis of liver fat is made by measuring mean LA in HU or the difference between the liver and spleen. 20 As the amount of liver fat increases, the measured LA decreases, that means low LA was equal to high risk of fatty liver. However, in the present study FLD was defined as mean LA <40 HU. 22
2.4. Assessment of hepatic steatosis and liver fibrosis
Hepatic steatosis was assessed by using abdominal ultrasound, which was carried out by a certified and skilled technician (Pavel Taimr) on Hitachi HI VISION 900. 23 Images were stored digitally and re‐assessed by a single hepatologist with more than 10 years of experience in ultrasonography. Diagnosis of steatosis was determined dichotomously as presence of a hyperechogenic liver parenchyma according to the protocol by Hamaguchi et al 24
Moreover, liver fibrosis was assessed using transient elastography (FibroScan®, EchoSens). Applied implementation of this examination has been described in detail previously. 25 Liver stiffness measurement (LSM) was performed by a single certified and experienced operator, who obtained 10 serial measurements using either the M or XL‐probe dependent on the thickness of the subcutaneous fat layer. 23 Moreover, LSM interquartile range/median LSM > 0.3 kilopascals (kPa) and LSM ≥ 7.1 kPa were regarded as poorly reliable. 26 In the present study, LSM ≥ 9.0 kPa was used as a cut‐off suggesting clinically relevant liver fibrosis. 8
2.5. Assessment of covariates and liver enzymes
Information on smoking behaviour, medication use and blood sampling, was obtained during home interviews. 18 Height and weight were measured, and the body mass index (BMI) [(weight in kg)/(height in m)2] was calculated. Waist circumferences were measured at the level midway between the lower rib margin and the iliac crest with the participant in a standing position. Smoking status was categorised into never, current or former. These data were further classified to two groups including never‐smokers and ever‐smokers (current and former smokers combined). Alcohol consumption was assessed in grams of ethanol per day and was classified (yes/no). Excessive alcohol consumption was defined as alcohol intake >30 g/day for men and >20 g/day for women. 23 Hypertension was defined as a systolic blood pressure (BP) ≥140 mm Hg or a diastolic BP ≥ 90 mm Hg or the use of BP‐lowering drugs prescribed for hypertension. 27 Diabetes mellitus was defined according to recent WHO guidelines 28 as fasting blood glucose ≥7.0 mmol/L or non‐fasting blood glucose between ≥11.1 mmol/L or the use of antidiabetic medication. From the blood samples, concentrations high‐density lipoprotein (HDL) cholesterol were determined using enzymatic procedures. 29 Serum GGT and ALP levels were determined within 2 weeks using a Merck Diagnostica kit on an Elan Autoanalyzer (Merc). According to local cut‐offs, elevation of GGT was defined as >34 U/L for women and >49 U/L for men, and elevation of ALP was defined as >97 U/L for women and >114 U/L for men. 30
2.6. Statistical analysis
Continuous variables are reported as mean ± standard deviation (SD) unless stated otherwise and categorical variables were presented as sample sizes and percentages. To obtain a normal distribution, skewed variables (serum HDL cholesterol, GGT and ALP) were log transformed. In addition, the amount of liver fat (A) had a left skewed and we used exponential transformed values (B) using the formula [B = A 3.5/10 000]. 21
Multivariable linear and logistic regression models were used to investigate the association between miRNA levels and CT‐based FLD (with the HU continues and dichotomous data). The multivariable linear regression models were used to check the association of miRNA levels with serum GGT and ALP levels. Beta, standard error (SE), P value were reported. The Bonferroni‐corrected p value threshold was calculated based on the number of tested miRNAs (0.05/591 = 8.46 × 10−5). In basic model (model 1), we adjusted the analysis for age and sex. The multivariable model (model 2), was additionally adjusted for waist circumference, ever smoking, alcohol consumption, hypertension, diabetes mellitus and serum HDL cholesterol. Because the missing values were likely to be missing at random and for avoidance of loss in efficiency, missing values on covariates (ranging from 0.1% to 1.7%) were imputed using a multiple imputation technique (N = 5 imputations). All analyses were done using SPSS statistical software (SPSS, version 25; IBM Corp) and R software version 3.5.2 (The R Foundation for Statistical Computing).
Sensitivity analyses were performed by adjusting for more variables. Model 3 was built by adding GGT and ALP to model 2. In model 4, we further adjusted for potential intermediator factors including in the model 2, use of lipid‐lowering medication, use of bile and liver medications. In model 5, we adjusted for all potential intermediator factors (including model 2, GGT, ALP, use of lipid‐lowering medication, use of bile and liver medications).
Furthermore, multivariable logistic regression models were used to investigate longitudinally the association of the plasma levels of the identified miRNAs with prevalence of hepatic steatosis and liver fibrosis after a median follow‐up of 6.4 years [interquartile range (IQR): 5.9‐7.0 years]. The Bonferroni correction was used to set the significance threshold.
Two databases, the Human miRNA tissue atlas (https://ccb‐web.cs.uni‐saarland.de/tissueatlas) 31 and Human miRNA expression profiles (https://guanfiles.dcmb.med.umich.edu/mirmine/index.html), were used to check whether the identified miRNAs are expressed in the liver. We also searched the literature 32 , 33 and several web tools (eg miR2Disease and GWAS catalogue) to see whether the identified miRNAs are associated with liver function and diseases.
3. RESULTS
At baseline, 954 participants who had miRNA expression data and CT‐based liver fat measurement were included to test the association of miRNAs and FLD. The mean age of the study population was 68.8 ± 6.7 years, and 46.6% were male. The mean LA in the population was 61.6 HU (IQR: 55.4‐65.6 HU). Among the study participants, 14.8% were diagnosed with cancer, but none of them was diagnosed with liver cancer. At follow‐up, 1211 participants who had underwent abdominal ultrasound were included to test the association of miRNAs and hepatic steatosis. The mean age of the study population was 76.3 ± 6.5 years, and 42.4% were male. Lifestyle, clinical and biochemical characteristics of all study participants are presented in Table 1. Comparison of characteristics between healthy controls and FLD patients based on CT‐scan and ultrasound data (in baseline and follow‐up study) are shown in Table S1. The participants with FLD have significantly higher BMI, waist circumference and alcohol consumption than healthy controls. In addition, compare to healthy controls, individuals with FLD have significantly lower serum HDL cholesterol.
Table 1.
Characteristics of the study population
| Characteristic | Baseline (n = 954) with CT‐scan data | Follow‐up (n = 1211) with ultrasound data |
|---|---|---|
| Age, years | 68.8 ± 6.7 | 76.3 ± 6.5 |
| Male, n (%) | 445 (46.6) | 513 (42.4) |
| Body mass index (kg/m2) | 27.9 ± 4.0 | 27.5 ± 4.1 |
| Waist circumference (cm) | 94.4 ± 11.7 | 93.2 ± 12.0 |
| Hypertension, n (%) | 709 (74.3) | 1047 (86.5) |
| Blood pressure‐lowering medication, n (%) | 385 (40.4) | 644 (53.2) |
| Smoking status, n (%) | ||
| Ever | 677 (71.0) | 791 (65.3) |
| Current | 130 (13.6) | 116 (9.6) |
| Former | 547 (57.4) | 675 (55.7) |
| Diabetes mellitus, n (%) | 123 (12.9) | 156 (12.9) |
| Use of lipid‐lowering medication, n (%) | 246 (25.8) | 372 (30.7) |
| Alcohol intake (g/day) | 8.6 (1.4‐20.0) | 8.6 (1.6‐8.6) |
| Mean liver attenuation (HU) | 61.6 (55.4‐65.6) | − |
| Serum HDL cholesterol (mmol/L) | 1.4 (1.2‐1.7) | 1.4 (1.2‐1.7) |
| GGT level (U/L) | 26.0 (18.0‐39.0) | 24.0 (17.0‐34.2) |
| ALP level (U/L) | 77.0 (66.0‐91.0) | 68.0 (57.0‐80.0) |
| Cancer, n (%) | 141 (14.8) | − |
| Liver cancer, n (%) | 0 (0) | − |
| Fatty liver, n (%) | 47 (4.93) | 424 (35) |
The table shows characteristics of 954 participants with CT‐scan data at baseline and 1211 participants with ultrasound data at follow‐up. Values are represented as mean (±standard deviation), sample sizes (%), or median (inter‐quartile range) for characteristics with skewed distributions.
Abbreviations: ALP, Alkaline phosphatase; BP, blood pressure; GGT, Gamma‐glutamyl transferase; HDL, high‐density lipoprotein; HU, Hounsfield unit.
In the linear regression analysis with liver enzymes, 37 miRNAs were significantly associated with serum GGT levels and 29 miRNAs with serum ALP levels, at the Bonferroni‐corrected P < 8.46 × 10−5 (0.05/591 well‐expressed miRNAs) (Tables S2 and S3 respectively). Volcano plot showing differently expressed miRNAs in relation to GGT and ALP levels is depicted in Figure 2A,B.
Figure 2.
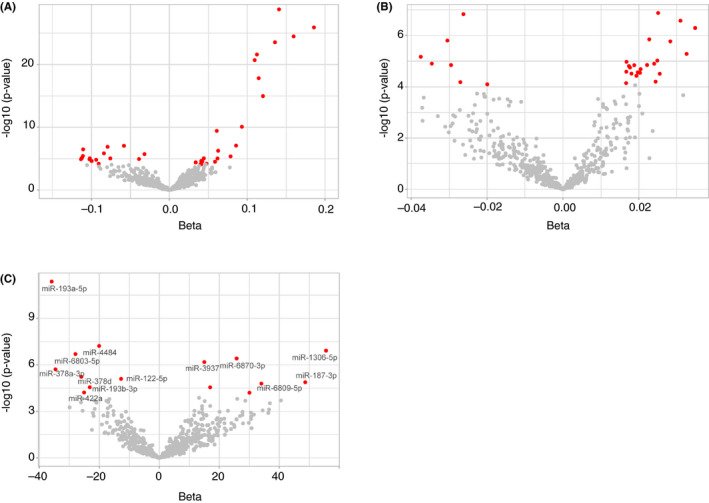
Volcano plots showing correlation between plasma levels of miRNAs and GGT (A), ALP (B), and continuous Hounsfield Unit values (C). The red dots indicate miRNAs significantly associated at Bonferroni‐corrected P < 8.46 × 10−5. The grey dots indicate miRNAs with no significant association. The name of miRNAs that were significantly associated with liver enzymes and the continuous Hounsfield Unit values are mentioned in (C). Abbreviations: ALP, Alkaline phosphatase; GGT, Gamma‐glutamyl transferase; miRNAs, microRNAs
Using a linear regression analysis of the continuous HU values, in the multivariable model 2, we found 15 miRNAs to be significantly associated at Bonferroni‐corrected P < 8.46 × 10−5 (Table S4). Volcano plot showing differently expressed miRNAs in relation to the continuous HU values is depicted in Figure 2C. In addition, in a logistic regression model testing the association of miRNA levels with the dichotomous HU values assessing FLD (mean LA ≤ or >40), six miRNAs were significantly associated (P < 8.46 × 10−5) (Figure 3). In model 3, further adjusting for GGT and ALP changed slightly the associations and with less miRNAs significant associated, but 2 of the 17 and 6 of 17 miRNAs remained significant using dichotomous and continuous Hounsfield Unit values respectively (P < 8.46 × 10−5) (Tables S5 and S6). Additional adjustment for use of lipid‐lowering medication, use of bile and liver medications (model 4) did not change the observed associations between miRNAs and FLD (Tables S5 and S6). In model 5, we added GGT, ALP, use of serum lipid reducing agents, use of bile and liver medications to model 2, the associations of miRNAs with FLD were similar to model 3, the same miRNAs remained significant (Tables S5 and S6).
Figure 3.
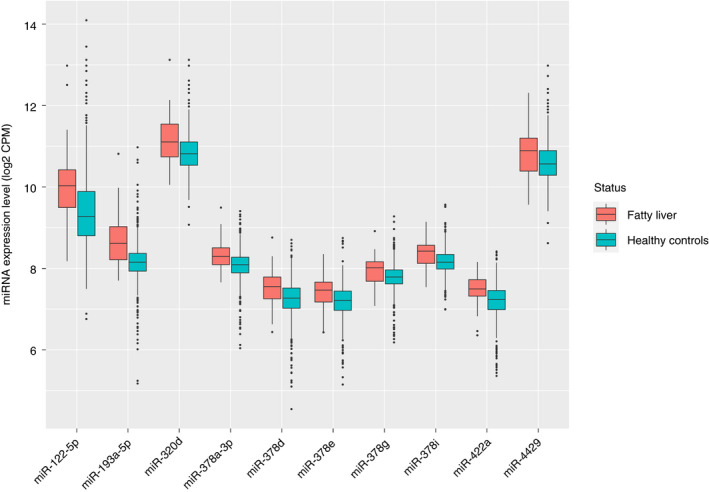
Comparison between expression levels of the top 10 miRNAs in patients with fatty liver disease and healthy controls. Of these, 6 miRNAs (miR‐193a‐5p, miR‐378a‐3p, miR‐422a, miR‐378d, miR‐320d and miR‐378e) were significantly associated with fatty liver disease at Bonferroni‐corrected P < 8.46 × 10−5. Abbreviations: CPM, counts per million; miRNAs, microRNAs
Collectively, our cross‐sectional studies with the baseline data revealed 61 unique miRNAs associated with liver enzymes (GGT and/or ALP) and 17 unique miRNAs associated with CT‐based FLD (continuous or dichotomous HU values). Of these, 14 miRNAs were common in both lists that were selected for further analyses (Table 2).
Table 2.
Circulatory miRNAs significantly associated with CT‐scan‐based fatty liver disease and serum liver enzymes
| miRNA ID | CT‐based FLD | Liver Enzymes | ||||||
|---|---|---|---|---|---|---|---|---|
| Dichotomous HU values | Continues HU values | GGT | ALP | |||||
| Beta | P value | Beta | P value | Beta | P value | Beta | P value | |
| miR‐193a‐5p | 1.86 | 2.02 × 10− 10a | −35.88 | 4.26 × 10−12a | 0.14 | 6.15 × 10−29a | 0.001 | 7.99 × 10−01 |
| miR‐4484 | 0.71 | 8.33 × 10−4 | −20.04 | 6.02 × 10−08a | 0.03 | 4.02 × 10−05a | 0.01 | 3.68 × 10−04 |
| miR‐1306‐5p | −0.60 | 3.15 × 10−01 | 55.59 | 1.21 × 10−07a | −0.09 | 6.45 × 10−05a | −0.02 | 1.12 × 10−01 |
| miR‐378a‐3p | 2.36 | 1.90 × 10−07 | −34.61 | 1.95 × 10−06a | 0.09 | 8.45 × 10−08a | −0.01 | 3.98 × 10−01 |
| miR‐6803‐5p | 0.70 | 2.68 × 10−02 | −27.96 | 2.01 × 10−07a | 0.05 | 6.31 × 10−05a | 0.01 | 1.03 × 10−02 |
| miR‐6870‐3p | −0.59 | 1.60 × 10−02 | 25.79 | 3.87 × 10−07a | −0.06 | 9.04 × 10−08a | −0.02 | 8.00 × 10−05a |
| miR‐3937 | −0.54 | 1.83 × 10−03 | 15.02 | 6.59 × 10−07 a | −0.03 | 1.90 × 10−06 a | −0.01 | 3.84 × 10−04 |
| miR‐122‐5p | 0.54 | 1.72 × 10−04 | −12.74 | 8.05 × 10−06 a | 0.14 | 2.39 × 10−116 a | 0.01 | 4.73 × 10−04 |
| miR‐422a | 2.01 | 9.87 × 10−06a | −25.07 | 6.09 × 10−05a | 0.06 | 9.71 × 10−06a | 0.002 | 7.37 × 10−01 |
| miR‐378d | 1.75 | 1.11 × 10−05a | −26.00 | 5.87 × 10−06a | 0.06 | 5.55 × 10−07a | −0.004 | 4.92 × 10−01 |
| miR‐187‐3p | −1.31 | 2.52 × 10−02 | 48.67 | 1.33 × 10−05a | −0.11 | 3.78 × 10−06a | −0.03 | 1.80 × 10−02 |
| miR‐6809‐5p | −0.73 | 5.51 × 10−02 | 34.03 | 1.61 × 10−05a | −0.08 | 1.45 × 10−06a | −0.01 | 1.11 × 10−01 |
| miR‐193b‐3p | 1.01 | 2.70 × 10−03 | −23.17 | 2.77 × 10−05 a | 0.11 | 2.01 × 10−21a | −0.001 | 7.85 × 10−01 |
| miR‐4713‐3p | −0.55 | 1.02 × 10−02 | 16.96 | 2.80 × 10−05a | −0.03 | 8.53 × 10−04 | ‐0.01 | 1.13 × 10−02 |
| miR‐320d | 1.16 | 2.87 × 10−05a | −14.30 | 4.54 × 10−03 | 0.02 | 1.72 × 10−01 | 0.004 | 4.10 × 10−01 |
| miR‐34b‐3p | −0.76 | 8.88 × 10−02 | 30.06 | 6.27 × 10−05a | −0.06 | 1.79 × 10−04 | −0.01 | 3.41 × 10−01 |
| miR‐378e | 1.64 | 6.88 × 10−05a | −21.54 | 6.86 × 10−04 | 0.06 | 3.13 × 10−05a | −0.01 | 4.42 × 10−01 |
Model 2: adjusted for age, sex, waist circumference, ever smoking, alcohol consumption, hypertension, diabetes mellitus and serum HDL cholesterol.
The table is sorted based on Bonferroni‐corrected P value association of miRNAs with dichotomous or continues Hounsfield Unit values in model 2.
The Bonferroni‐corrected significance threshold is P < 8.46 × 10−5 (0.05/591 miRNAs). The P values surpassing the significance threshold are “a”.
Abbreviations: ALP, Alkaline phosphatase; FLD, fatty liver disease; GGT, Gamma‐glutamyl transferase; HU, Hounsfield Unit; miRNA, microRNA.
The longitudinal analysis was performed for the 14 miRNAs by using ultrasound and FibroScan data. Using a logistic regression in the multivariable model 2, we found significant association between 4 of the 14 miRNAs (miR‐193a‐5p, miR‐122‐5p, miR‐378d and miR‐187‐3p) with hepatic steatosis at Bonferroni‐corrected P < 3.57 × 10−3 (0.05/14 miRNAs) (Table 3). Moreover, we found significant association of miR‐122‐5p and miR‐187‐3p with NAFLD (P < 3.57 × 10−3), and miR‐3937 was nominally (P < 0.05) associated with alcoholic FLD (Table S7). Using FibroScan data and in a multivariable logistic regression model, we also found miR‐193a‐5p (P = 5.58 × 10−3, β = 1.11), miR‐122‐5p (P = 0.0147, β = 0.45) and miR‐193b‐3p (P = 0.0102, β = 1.19) to be nominally associated with liver fibrosis (Table 3).
Table 3.
Longitudinal study of the 14 identified miRNAs with hepatic steatosis and liver fibrosis
| miRNA ID | Hepatic Steatosis | Liver Fibrosis | |||||
|---|---|---|---|---|---|---|---|
| Beta | SE | P value | Beta | SE | P value | ||
| miR‐193a‐5p | 0.54 | 0.16 | 2.02 × 10−10a | 1.11 | 0.40 | 5.58 × 10−03b | |
| miR‐4484 | 0.21 | 0.10 | 4.65 × 10−02b | 0.41 | 0.28 | 1.37 × 10−01 | |
| miR‐1306‐5p | −0.33 | 0.28 | 2.36 × 10−01 | −1.16 | −0.73 | 1.14 × 10−01 | |
| miR‐378a‐3p | 0.37 | 0.21 | 8.19 × 10−02 | 1.23 | 0.64 | 5.67 × 10−02 | |
| miR‐6803‐5p | 0.13 | 0.15 | 3.79 × 10−01 | 0.70 | 0.41 | 8.42 × 10−02 | |
| miR‐6870‐3p | −0.22 | 0.14 | 1.18 × 10−01 | 0.07 | 0.45 | 8.80 × 10−01 | |
| miR‐3937 | −0.13 | 0.09 | 1.44 × 10−01 | −0.18 | 0.26 | 4.87 × 10−01 | |
| miR‐122‐5p | 0.33 | 0.08 | 6.06 × 10−05a | 0.45 | 0.18 | 1.47 × 10−02b | |
| miR‐422a | 0.37 | 0.18 | 4.12 × 10−02b | 0.96 | 0.54 | 7.61 × 10−02 | |
| miR‐378d | 0.54 | 0.17 | 1.65 × 10−03a | 0.73 | 0.53 | 1.69 × 10−01 | |
| miR‐187‐3p | −1.01 | 0.34 | 2.76 × 10−03a | −0.42 | 0.98 | 6.68 × 10−01 | |
| miR‐6809‐5p | −0.55 | 0.24 | 2.49 × 10−02b | −0.66 | −0.76 | 3.83 × 10−01 | |
| miR‐193b‐3p | 0.22 | 0.15 | 1.42 × 10−01 | 1.19 | 0.46 | 1.02 × 10−02b | |
| miR‐378e | 0.50 | 0.18 | 7.25 × 10−03b | 0.94 | 0.60 | 1.14 × 10−01 | |
Model 2: adjusted for age, sex, waist circumference, ever smoking, alcohol consumption, hypertension, diabetes mellitus and serum HDL cholesterol. The table is sorted based on the association of 14 miRNAs with the continues Hounsfield Unit values in the cross‐sectional study. The P values surpassing the Bonferroni‐corrected threshold of P < 3.57 × 10−3 (0.05/14 miRNAs) are marked with “a” and nominal associations with P < 0.05 are marked with “b”. Abbreviations: miRNA, microRNA; SE, standard error.
Additionally, we searched the Human miRNA tissue atlas and the miRmine database to see whether the 14 miRNAs associated with FLD in plasma are expressed in the liver that are shown in Table S8. Among them, miR‐122‐5p is a specifically expressed miRNA with the tissue specificity index (TSI) of 0.97 and highly expressed in the liver. Then, we sought to find whether the 14 identified miRNAs are reported in previous studies to be associated with liver function or/and diseases. A summary of evidence for associations between nine of these miRNAs and liver diseases are shown in Table S9. Finally, we extracted SNPs annotated to the 16 identified miRNAs and checked their associations with FLD and liver enzymes using summary statistics data from pervious GWAS. 34 , 35 There were 63 SNPs related to miR‐193a‐5p, miR‐378d and miR‐193b‐3p, none of them showed significant association after correcting the P value for multiple testing based on the number of tested SNPs.
4. DISCUSSION
In this study, we investigated the association between circulating miRNAs and liver enzymes in a population‐based setting and found 61 unique miRNAs to be associated with serum GGT or ALP levels. Moreover, we found plasma levels of 17 miRNAs to be associated with CT‐based FLD, 14 of these were also associated with the liver enzymes. Higher plasma levels of three and lower plasma level of 1 of the 14 miRNAs were significantly associated with hepatic steatosis after >6 years follow‐up. These findings suggest that plasma levels of miRNAs can be considered as potential biomarkers of FLD and hepatic steatosis in the general population.
Several studies have demonstrated the potential of miRNAs to be used as biomarkers for liver diseases. 36 , 37 However, previous studies have conducted for subset of miRNAs, using qPCR‐based methods, or on the modest sample sizes. While our study is embedded within the RS with much larger sample size, based on RNA‐sequencing method, conducted genome‐wide profiling of almost all important cell‐free miRNAs, and adjusted for a broad range of potential confounders, such as waist circumference, smoking status, alcohol consumption, hypertension and diabetes mellitus, which have been overlooked in most of previous studies. 36 , 39 Such a large‐scale population‐based study with long‐term follow‐up data provided a more statistical power to detect multiple significant associations. Compared to microarray or qPCR‐based profiling techniques, the cell‐free RNA‐seq analysis can provide higher sensitivity to measure miRNAs expression levels over a wide dynamic range and with ability to identify novel miRNAs. 40 Additionally, due to the high stability of cell‐free miRNAs in body fluids and accessibility of plasma compared with the target tissue, the identified miRNAs can be considered as potential easy‐to‐use biomarkers in clinical routine. 41
Previous studies on human or mouse model have demonstrated particularly miR‐122‐5p as potential biomarkers and therapeutic target for liver diseases. 39 , 42 , 43 In line with previous studies, we found that the higher plasma miR‐122‐5p level is significantly associated with FLD and liver enzymes also in a population‐based setting. In addition to the well‐established liver‐associated miR‐122, we found evidence in previous studies for eight of the other identified miRNAs in our study to be associated with liver diseases, indicating the importance of these miRNAs in pathways underlying liver function and diseases. In particular, the expression of miR‐193a‐5p, which is one of our top miRNAs associated with FLD and hepatic steatosis, is reported to be upregulated in HCC tissues, 44 whereas miR‐193a‐5p can distinguish HCC from other non‐HCC individuals 43 and inhibited HCC development through targeting SPOCK1. 45 Similarly, miR‐422a was related to NAFLD, 46 miR‐378d 47 miR‐187‐3p, 48 miR‐6809‐5p 49 and miR‐4484 50 were associated with HCC. Moreover, miR‐193b‐3p which were significantly associated with FLD and nominally associated with liver fibrosis in our study, have been verified previously to be involved in the pathogenesis of liver fibrosis in vitro. 51 Finally, the members of miR‐378 family, in particular miR‐378a‐3p that has been also proposed to have a therapeutic potential for liver fibrosis. 52
Our results showed a minimal of the use of serum lipid reducing agents, use of bile and liver medications on the observed associations between miRNAs and FLD. We observed slightly change in the associations between miRNAs and FLD by adding GGT and ALP to the model. This difference may indicate that liver enzymes have more stronger links to FLD compared to medication use. Also, we did not find significant association between SNPs related to the identified miRNAs and FLD in the summary statistics from previous GWAS, but we need to take into consideration the sample sizes of available GWAS of liver diseases. To date, the GWAS on liver diseases is mainly from two studies, 34 , 35 Nakamura et al conducted a study to identify susceptibility loci for primary biliary cirrhosis, a GWAS in 963 Japanese individuals and in a subsequent replication study including 1402 other Japanese individuals. The sample sizes of this study are limited and the analysis conducted in Asia population, while our study was conducted in European population, and miRNAs expression might exhibits population differences. 53 In addition, Namjou et al conducted a GWAS using both adult and paediatric participants (1106 NAFLD cases and 8571 controls) from electronic medical records to identify genetic contributions to NAFLD. As the cohorts in that GWAS study represent many geographic area in US, other ancestry groups are under‐represented in the electronic medical records. Thus, it is possible that future trans‐ethnic GWAS with larger samples sizes find association between some of the identified miRNAs and FLD.
Our study has some limitations that should be considered. First, in the cross‐sectional observational study the ability to assess causality or temporality is limited. We therefore assessed additionally the associations of the identified miRNAs as biomarkers for diagnosis hepatic steatosis after follow‐up. Future studies are still needed to confirm our findings in longitudinal settings considering the incidence date, longer follow‐up time and in different age groups. Second, we defined the mean LA< 40 HU as FLD and found 47 cases out of 954 individuals (5%), which is lower than the excepted prevalence of FLD in the general population. A liver‐to‐spleen ratio < 1.0 is comparable to using a mean LA cut‐off ≤ 51 HU for diagnosis of mild liver fat. 54 However, previous studies have demonstrated different cut‐offs, mainly a cut‐off value of 40 HU on non‐enhanced CT as the most clinically indicator for moderate‐to‐severe steatosis. 20 , 22 In our study, the cut‐off value is 40 HU for FLD, it is relatively strict than using the mean LA ≤ 51 HU, which increases the certainty of identifying participants with true FLD and also results in a lower prevalence. Therefore, we performed cross‐sectional analysis at baseline with the continuous HU values and liver enzymes as well. The majority (14 of 17) of the identified miRNAs with CT‐scan data showed significant association with liver enzyme, indicating the robustness of our results. Yet, compare to the liver biopsy as the gold standard, but invasive method, to measure FLD, CT‐based liver fat has limited diagnostic accuracy of detecting mild degree hepatic steatosis. Therefore, there might be some known or unknown causes for low density of the liver on CT scan. Also, there might be some inconsistency between CT‐scans and ultrasound data for diagnosing FLD and hepatic steatosis. In an optimal setting, one should use the repeated measurement of liver fat by a similar diagnostic method for longitudinal analysis.
In conclusion, we found that plasma levels of several miRNAs were significantly associated with FLD that can be considered as plasma biomarkers of the disease in the population level. Future research need to be conducted even with more sample sizes and longer follow‐up times in order to confirm the potential of the identified miRNAs as biomarkers for early diagnosis and progression of FLD and also to uncover underlying molecular mechanisms by which these miRNAs may control liver fat.
AUTHORSHIP
Guarantor of the article: Mohsen Ghanbari.
Author contributions: MG and MAI designed the research; XZ conducted the analyses; MM and YA helped in the miRNA analyses; MG, DB, SDM and RJK provided consultation regarding the fatty liver data analyses and interpretation of the data. All authors critically reviewed and approved the final manuscript.
Supporting information
Table S1‐S9
ACKNOWLEDGEMENTS
All authors are grateful to the Rotterdam Study participants, the staff involved with the Rotterdam Study, and the participating general practitioners and pharmacists. We thank the members of the VERI/O laboratory, and bioinformatics teams of HTG Molecular Diagnostics, Tucson, Arizona for performing the miRNA expression analyses by HTG EdgeSeq WTA.
Declaration of personal interests: None.
Zhang X, Mens MMJ, Abozaid YJ, et al. Circulatory microRNAs as potential biomarkers for fatty liver disease: the Rotterdam Study. Aliment Pharmacol Ther. 2021;53:432–442. 10.1111/apt.16177
The Handling Editor for this article was Professor Stephen Ryder, and it was accepted for publication after full peer‐review.
Michelle MJ Mens and Yasir J. Abozaid denote equal contribution.
Funding
The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. MiRNA expression analyses by HTG EdgeSeq WTA were funded by Johnson & Johnson.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in the supplementary material of this article.
REFERENCES
- 1. Younossi Z, Tacke F, Arrese M, et al. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2019;69:2672‐2682. [DOI] [PubMed] [Google Scholar]
- 2. Fatiha Nassir R, Rector S, Hammoud GM, Ibdah JA. Pathogenesis and prevention of hepatic steatosis. Gastroenterol Hepatol. 2015;11:167‐175. [PMC free article] [PubMed] [Google Scholar]
- 3. VanWagner LB, Ning H, Allen NB, et al. Alcohol use and cardiovascular disease risk in patients with nonalcoholic fatty liver disease. Gastroenterology. 2017;153:1260‐1272.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DiStefano JK, Gerhard GS. Circulating microRNAs in nonalcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol. 2016;10:161‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eslam M, Sanyal AJ, George J International Consensus P . MAFLD: A consensus‐driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158:1999‐2014.e1. [DOI] [PubMed] [Google Scholar]
- 6. Musaddaq G, Shahzad N, Ashraf MA, Arshad MI. Circulating liver‐specific microRNAs as noninvasive diagnostic biomarkers of hepatic diseases in human. Biomarkers. 2019;24:103‐109. [DOI] [PubMed] [Google Scholar]
- 7. Lee DH. Imaging evaluation of non‐alcoholic fatty liver disease: focused on quantification. Clin Mol Hepatol. 2017;23:290‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eddowes PJ, Sasso M, Allison M, et al. Accuracy of FibroScan controlled attenuation parameter and liver stiffness measurement in assessing steatosis and fibrosis in patients with nonalcoholic fatty liver disease. Gastroenterology. 2019;156:1717‐1730. [DOI] [PubMed] [Google Scholar]
- 9. Karlas T, Petroff D, Sasso M, et al. Impact of controlled attenuation parameter on detecting fibrosis using liver stiffness measurement. Aliment Pharmacol Ther. 2018;47:989‐1000. [DOI] [PubMed] [Google Scholar]
- 10. Oeda S, Takahashi H, Imajo K, et al. Accuracy of liver stiffness measurement and controlled attenuation parameter using FibroScan((R)) M/XL probes to diagnose liver fibrosis and steatosis in patients with nonalcoholic fatty liver disease: a multicenter prospective study. J Gastroenterol. 2020;55:428‐440. [DOI] [PubMed] [Google Scholar]
- 11. Otsuka M, Kishikawa T, Yoshikawa T, et al. MicroRNAs and liver disease. J Hum Genet. 2016;62:75. [DOI] [PubMed] [Google Scholar]
- 12. Schueller F, Roy S, Vucur M, Trautwein C, Luedde T, Roderburg C. The role of miRNAs in the pathophysiology of liver diseases and toxicity. Int J Mol Sci. 2018;19:261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pirola CJ, Fernández Gianotti T, Castaño GO, et al. Circulating microRNA signature in non‐alcoholic fatty liver disease: from serum non‐coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800‐812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roderburg C, Luedde T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J Hepatol. 2014;61:1434‐1437. [DOI] [PubMed] [Google Scholar]
- 15. Liu C‐H, Ampuero J, Gil‐Gómez A, et al. miRNAs in patients with non‐alcoholic fatty liver disease: a systematic review and meta‐analysis. J Hepatol. 2018;69:1335‐1348. [DOI] [PubMed] [Google Scholar]
- 16. Mehta R, Otgonsuren M, Younoszai Z, Allawi H, Raybuck B, Younossi Z. Circulating miRNA in patients with non‐alcoholic fatty liver disease and coronary artery disease. BMJ Open Gastroenterol. 2016;3:e000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zarrinpar A, Gupta S, Maurya MR, Subramaniam S, Loomba R. Serum microRNAs explain discordance of non‐alcoholic fatty liver disease in monozygotic and dizygotic twins: a prospective study. Gut. 2016;65:1546‐1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ikram MA, Brusselle G, Ghanbari M, et al. Objectives, design and main findings until 2020 from the Rotterdam Study. Eur J Epidemiol. 2020;35:483‐517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Odink AE, van der Lugt A, Hofman A, et al. Association between calcification in the coronary arteries, aortic arch and carotid arteries: the Rotterdam study. Atherosclerosis. 2007;193:408‐413. [DOI] [PubMed] [Google Scholar]
- 20. Kodama Y, Ng CS, Wu TT, et al. Comparison of CT methods for determining the fat content of the liver. Am J Roentgenol. 2007;188:1307‐1312. [DOI] [PubMed] [Google Scholar]
- 21. Wolff L, Bos D, Murad SD, et al. Liver fat is related to cardiovascular risk factors and subclinical vascular disease: the Rotterdam Study. Eur Heart J Cardiovasc Imaging. 2016;17:1361‐1367. [DOI] [PubMed] [Google Scholar]
- 22. Park SH, Kim PN, Kim KW, et al. Macrovesicular hepatic steatosis in living liver donors: Use of CT for quantitative and qualitative assessment. Radiology. 2006;239:105‐112. [DOI] [PubMed] [Google Scholar]
- 23. Alferink LJM, Trajanoska K, Erler NS, et al. Nonalcoholic fatty liver disease in The Rotterdam Study: about muscle mass, sarcopenia, fat mass, and fat distribution. J Bone Miner Res. 2019;34:1254‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamaguchi M, Kojima T, Itoh Y, et al. The severity of ultrasonographic findings in nonalcoholic fatty liver disease reflects the metabolic syndrome and visceral fat accumulation. Am J Gastroenterol. 2007;102:2708‐2715. [DOI] [PubMed] [Google Scholar]
- 25. Koehler EM, Plompen EPC, Schouten JNL, et al. Presence of diabetes mellitus and steatosis is associated with liver stiffness in a general population: The Rotterdam study. Hepatology. 2016;63:138‐147. [DOI] [PubMed] [Google Scholar]
- 26. Boursier J, Zarski J‐P, de Ledinghen V, et al. Determination of reliability criteria for liver stiffness evaluation by transient elastography. Hepatology. 2013;57:1182‐1191. [DOI] [PubMed] [Google Scholar]
- 27. Williams B, Mancia G, Spiering W, et al. 2018 ESC/ESH Guidelines for the management of arterial hypertension. Eur Heart J. 2018;39:3021‐3104. [DOI] [PubMed] [Google Scholar]
- 28. World Health Organization and International Diabetes Federation . Definition and diagnosis of diabetes mellitus and intermediate hyperglycaemia. ISBN: 9789241594936, 2016.
- 29. Braun KVE, Dhana K, de Vries PS, et al. Epigenome‐wide association study (EWAS) on lipids: the Rotterdam Study. Clin Epigenetics. 2017;9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koehler EM, Sanna D, Hansen BE, et al. Serum liver enzymes are associated with all‐cause mortality in an elderly population. Liver Int. 2014;34:296‐304. [DOI] [PubMed] [Google Scholar]
- 31. Ludwig N, Leidinger P, Becker K, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44:3865‐3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huan T, Rong J, Liu C, et al. Genome‐wide identification of microRNA expression quantitative trait loci. Nat Commun. 2015;6:6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nikpay M, Beehler K, Valsesia A, et al. Genome‐wide identification of circulating‐miRNA expression quantitative trait loci reveals the role of several miRNAs in the regulation ofcardiometabolic phenotypes. Eur Soc Cardiol. 2019;115:1629‐1645. [DOI] [PubMed] [Google Scholar]
- 34. Nakamura M, Nishida N, Kawashima M, et al. Genome‐wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am J Hum Genet. 2012;91:721‐728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Namjou B, Lingren T, Huang Y, et al. GWAS and enrichment analyses of non‐alcoholic fatty liver disease identify new trait‐associated genes and pathways across eMERGE Network. BMC Med. 2019;17:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salvoza NC, Klinzing DC, Gopez‐Cervantes J, Baclig MO. Association of circulating serum miR‐34a and miR‐122 with dyslipidemia among patients with non‐alcoholic fatty liver disease. PLoS One. 2016;11:e0153497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Auguet T, Aragonès G, Berlanga A, et al. miR33a/miR33b* and miR122 as possible contributors to hepatic lipid metabolism in obese women with nonalcoholic fatty liver disease. Int J Mol Sci. 2016;17:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raitoharju E, Seppälä I, Lyytikäinen L‐P, et al. Blood hsa‐miR‐122‐5p and hsa‐miR‐885‐5p levels associate with fatty liver and related lipoprotein metabolism‐The Young Finns Study. Sci Rep. 2016;6:38262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brandt S, Roos J, Inzaghi E, et al. Circulating levels of miR‐122 and nonalcoholic fatty liver disease in pre‐pubertal obese children. Pediatr Obes. 2018;13:175‐182. [DOI] [PubMed] [Google Scholar]
- 40. Eminaga S, Christodoulou DC, Vigneault F, Church GM, Seidman JG. Quantification of microRNA expression with next‐generation sequencing. Curr Protoc Mol Biol 2013;Chapter 4: Unit 4 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Szelenberger R, Kacprzak M, Saluk‐Bijak J, Zielinska M, Bijak M. Plasma MicroRNA as a novel diagnostic. Clin Chim Acta. 2019;499:98‐107. [DOI] [PubMed] [Google Scholar]
- 42. Cheng JL, Zhao H, Yang SG, Chen EM, Chen WQ, Li LJ. Plasma miRNA‐122‐5p and miRNA‐151a‐3p identified as potential biomarkers for liver injury among CHB patients with PNALT. Hepatol Int. 2018;12:277‐287. [DOI] [PubMed] [Google Scholar]
- 43. Jin YU, Wong YS, Goh BKP, et al. Circulating microRNAs as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Sci Rep. 2019;9:10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang JT, Wang ZH. Role of miR‐193a‐5p in the proliferation and apoptosis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:7233‐7239. [DOI] [PubMed] [Google Scholar]
- 45. Li P, Xiao Z, Luo J, Zhang Y, Lin L. MiR‐139‐5p, miR‐940 and miR‐193a‐5p inhibit the growth of hepatocellular carcinoma by targeting SPOCK1. J Cell Mol Med. 2019;23:2475‐2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Latorre J, Moreno‐Navarrete JM, Mercader JM, et al. Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. Int J Obes (Lond). 2017;41:620‐630. [DOI] [PubMed] [Google Scholar]
- 47. Gao L, Xiong D‐D, He R‐Q, et al. Identifying TF‐miRNA‐mRNA regulatory modules in nitidine chloride treated HCC xenograft of nude mice. Am J Translat Res. 2019;11:7503‐7522. [PMC free article] [PubMed] [Google Scholar]
- 48. Dou C, Liu Z, Xu M, et al. miR‐187‐3p inhibits the metastasis and epithelial‐mesenchymal transition of hepatocellular carcinoma by targeting S100A4. Cancer Lett. 2016;381:380‐390. [DOI] [PubMed] [Google Scholar]
- 49. Yang P‐W, Lu Z‐Y, Pan Q, et al. MicroRNA‐6809‐5p mediates luteolin‐induced anticancer effects against hepatoma by targeting flotillin 1. Phytomedicine. 2019;57:18‐29. [DOI] [PubMed] [Google Scholar]
- 50. Pascut D, Cavalletto L, Pratama MY, et al. Serum miRNA are promising biomarkers for the detection of early hepatocellular carcinoma after treatment with direct‐acting antivirals. Cancers (Basel). 2019;11:1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ju B, Nie Y, Yang X, et al. miR‐193a/b‐3p relieves hepatic fibrosis and restrains proliferation and activation of hepatic stellate cells. J Cell Mol Med. 2019;23:3824‐3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hyun J, Wang S, Kim J, et al. MicroRNA‐378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun. 2016;7:10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang RS, Gamazon ER, Ziliak D, et al. Population differences in microRNA expression and biological implications. RNA Biol. 2011;8:692‐701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zeb I, Li D, Nasir K, Katz R, Larijani VN, Budoff MJ. Computed tomography scans in the evaluation of fatty liver disease in a population based study: the multi‐ethnic study of atherosclerosis. Acad Radiol. 2012;19:811‐818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S9
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.


