Abstract
As a widespread industrial chemical, formaldehyde carcinogenicity has been highly controversial. Meanwhile, formaldehyde is an essential metabolite in all living cells. Previously, we have demonstrated exogenous formaldehyde causes DNA adducts in a nonlinear manner between 0.7 ppm to 15.2 ppm using [13CD2]-formaldehyde for exposure coupled with the use of sensitive mass spectrometry. However, the responses from exposure to low doses of formaldehyde are still unknown. In this study, rats were exposed to 1, 30, and 300 ppb [13CD2]-formaldehyde for 28 days (6 hours/day) by nose-only inhalation, followed by measuring DNA mono-adduct (N2-HOMe-dG) and DNA-protein crosslinks (dG-Me-Cys) as formaldehyde specific biomarkers. Both exogenous and endogenous DNA mono-adducts and dG-Me-Cys were examined with ultrasensitive nano-liquid chromatography-tandem mass spectrometry. Our data clearly show that endogenous adducts are present in all tissues analyzed, but exogenous adducts were not detectable in any tissue samples, including the most susceptible nasal epithelium. Moreover, formaldehyde exposure at 1, 30 and 300 ppb did not alter the levels of endogenous formaldehyde induced DNA adducts or DNA-protein crosslinks. The novel findings from this study provide new data for risk assessment of exposure to low doses of formaldehyde.
Keywords: Formaldehyde, DNA damage, DNA adducts, DNA-protein crosslinks, mass spectrometry
Introduction
Formaldehyde (FA) is classified as a known carcinogen by the International Agency for Research on Cancer due to the resulting nasal squamous cell carcinomas in rats and nasopharyngeal cancers in humans (Swenberg et al. 1980; IARC 2006; Zhang et al. 2010; Nielsen et al. 2013 and 2017). FA is a widespread chemical, which mainly arises from exhaust gas emission and a variety of consumer products such as building materials, tobacco smoke, as well as through metabolism of foods and drugs. Thus, FA exposure poses a potential risk to public health owing to its carcinogenicity and extensive distribution. However, it has been highly controversial about formaldehyde carcinogenicity, especially on inducing leukemia (Langevin et al. 2011; Garaycoechea et al. 2012, 2014).
FA shows high reactivity with DNA and protein without the requirement of metabolic activation. The aldehyde moiety tends to react with nucleophilic sites in proteins and DNA, resulting in a number of DNA and protein adducts such as N2-hydroxymethyl-deoxyguanosine (N2-HOMe-dG) (Moeller et al. 2011; Lu et al. 2011) and N6-formyllysine (Edrissi et al. 2013; Edrissi et al. 2017). On the other hand, the FA-induced Schiff bases have the potential to react with other ambient nucleophilic sites, therefore generating DNA-DNA crosslinks (Lu et al. 2009, 2010a, 2010b) and DNA-protein crosslinks (DPCs) (Lu et al. 2009, 2010b; Magana-Schwencke et al. 1978; McGhee et al. 1977; Lai et al. 2016). The resulting dG-Me-Cys hinders normal DNA-protein interactions during DNA replication and transcription, which may lead to accelerated aging and cancer development (Stingele et al. 2017; Ide et al. 2011). Therefore, it would be essential to determine the formation and repair associated with DPCs. Our recent studies have revealed dG-Me-Cys to be an abundant and stable for measurement as formaldehyde-induced DPCs, as a biomarker of FA exposure (Liu et al. 2018).
Beyond exogenous exposure, endogenous FA is an essential metabolic intermediate in all living cells (Swenberg et al. 2011). Endogenous aldehydes cause DNA damage and increase the risk of cancers in mice deficient in the Fanconi anemia DNA repair pathway (Garaycoechea et al. 2012, 2014; Ren et al. 2013; Pontel et al. 2015). Previously, using inhaled [13CD2]-FA for exposure in rats and sensitive LC-MS/MS, we successfully interrogated the fate of exogenous FA in the presence of a substantial background of endogenous FA (Lu et al. 2011; Swenberg et al. 2013; Yu et al. 2015). It has been demonstrated that exogenous N2-HOMe-dG was formed in a highly nonlinear manner in rats exposed to 0.7 to 15 ppm formaldehyde (Lu et al. 2011). Examination of the ratio of exogenous versus endogenous FA induced DNA adducts clearly demonstrated that endogenous DNA adducts predominate at all doses used. The exogenous N2-HOMe-dG readily formed in the nasal tissues but was undetectable in tissues distant to the site of exogenous exposure. The data generated in previous studies provide clear scientific evidence for the assessment of risk resulting from FA exposure through inhalation. However, the formation of DNA adducts caused by low doses of exogenous FA exposure (<0.7 ppm) is still unclear. To address this, in this study, both exogenous and endogenous DNA mono-adduct (N2-HOMe-dG) and DNA-protein crosslinks (dG-Me-Cys) were measured to assess the formation of DNA adducts arising from the inhalation to 1, 30, 300 ppb [13CD2]-FA in rats for 28 days using ultrasensitive nano liquid chromatography-tandem mass spectrometry.
Methods
Chemicals and Materials
Unless otherwise specified, all reagents and chemicals used in this study were purchased from Sigma Aldrich (St. Louis, MO). Methanol, acetonitrile, high-performance liquid chromatography (HPLC) grade water, and optima LC-MS grade water were all purchased from Thermo Fisher Scientific (Pittsburgh, Pennsylvania). High-purity LC-MS grade water and acetonitrile were purchased from Burdick and Jackson Honeywell (Muskegon, Michigan). [15N5]-deoxyguanosine and [13CD2]-FA solution (20% w/w in D2O) were obtained from Cambridge Isotope Laboratories, Inc. (Tewksbury, MA). DNAzol and Turbo DNase were obtained from Lift Technologies. Proteinase K was obtained from FivePrime (San Francisco, CA). Nanosep Centrifugal Devices (MWCO 3K) were purchased from Pall Lift Sciences. All solvents used were at least HPLC-grade.
Preparation of Standards
The internal standard of mono-adduct, was synthesized following the previously described procedure (Lu et al. 2010a). Briefly, 10 mM [13C1015N5]-dG solution was treated by 100 mM FA in phosphate buffer (pH = 7.2) overnight at 37°C. The reaction mixture was separated by HPLC using a 150×2.5 mm C18 T3 analytical column. N2-HOMe-dG was collected and incubated with 50 mM NaCNBH3 (pH 7.1) overnight at 37°C, followed by further separation using HPLC. The synthesized dG-Me-Cys standards were prepared by enzymatic digestion of dG-Me-glutathione. Briefly, glutathione (GSH) was incubated with FA in sodium phosphate buffer at 37°C for 3 hours. Then, deoxyguanosine (dG) was added for crosslinking reaction at 37°C for 14 hours to produce dG-Me-GSH. The dG-Me-GSH was digested by carboxypeptidase Y and leucine aminopeptidase M in sodium phosphate buffer with the presence of MgCl2 and CaCl2 at room temperature for 15 hours in order to release dG-Me-Cys. The same method was applied to synthesize [15N5]-dG-[13CD2]-Me-Cys using [15N5]-dG and [13CD2]-FA as starting materials. The dG-Me-Cys and [15N5]-dG-[13CD2]-Me-Cys were further purified and quantified as described previously (Yu et al. 2015).
Formaldehyde Exposure
The exposure was done at the Lovelace Respiratory Research Institute (Albuquerque, NM, USA) according to the approved protocols for the use of vertebrate animals in experiments. Animal study was conducted in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals. Fischer 344 rats (6-week old, male) were served as the research cohort and exposed to [13CD2]-FA vapor by single-exposure, nose-only inhalation with the final target exposure concentration of 0 (air control), 1, 30, 300 ppb for 28 days (6 h/day). Rats that served as the positive control samples were exposed to 10 ppm [13CD2]-FA for 5 days (6 h/day). The formation of exogenous formaldehyde-induced DNA adducts in these 10 ppm exposed samples have been well validated in our previous study (Lu et al. 2010a). The concentration of the exposure chamber was monitored by collection of Waters XpoSure Aldehyde Sampler cartridges every 10 min continuously throughout the exposure. Animals were euthanized with a pentobarbital-based euthanasia solution to induce surgical level anesthesia and pneumothorax was introduced. Tissue samples were harvested, wrapped in aluminum foil, and immediately snap frozen in liquid nitrogen. They were stored at −80 °C pending analyses.
Extraction and Enzymatic Digestion for DNA mono-adduct analysis
The experimental procedures for the extraction and enzymatic hydrolysis of genomic DNA, and HPLC enrichment of DNA adducts were similar to those described previously (Yu et al. 2015; Liu et al. 2018). For DNA mono-adduct analysis, DNA was isolated from the tissues using a NucleoBond DNA isolation kit (Macherey-Nagel, Bethlehem, Pennsylvania), as instructed by the manufacturer with small modifications. The resultant DNA was then quantified by Nanodrop 2000 (Thermo Scientific). To reduce endogenous and exogenous N2-HOMe-dG to N2-Me-dG, DNA was thawed and incubated with NaCNBH3 (50 mM) and sodium phosphate buffer (100 mM, pH 7.2) for 6 h at 37 °C. Following reduction, DNA was frozen at −80 °C until digestion. Reduced DNA was thawed, and 200 μL of sodium phosphate/MgCl2 buffer (50/20 mM final concentration, pH 7.2) along with 2 fmol of the internal standard, [13C1015N5]-N2-Me-dG, were added. DNA was digested with DNAse I (200 units), alkaline phosphatase (5 units), and phosphodiesterase (0.005 units) for 1 h at 37 °C.
Following digestion, hydrolyzed DNA was filtered with the Pall NanoSep 3 kDa filter (Port Washington, New York) at 8000 rpm for 50 min to remove enzymes prior to HPLC purification. Hydrolyzed DNA was then injected onto an Agilent 1200 HPLC fraction collection system equipped with a DAD (Agilent Technologies, Santa Clara, California). Analytes were separated by reversed-phase liquid chromatography using an Atlantis C18 T3 column (150×4.6 mm, 3 μm). The column temperature was kept at 30 °C. The mobile phases were water with 0.1% acetic acid (A) and acetonitrile with 0.1% acetic acid (B). The flow rate was 1.0 mL/min with a starting condition of 2% B, which was held for 5 min, followed by a linear gradient of 4% B at 20 min, 10% B at 30 min, followed by 6 min at 80% B, then re-equilibrated to the starting conditions for 20 min. The dG and N2-Me-dG eluted with retention times of 13.1 and 25.5 min, respectively. The amount of digested dG in each sample was quantitated by the UV peak area (λ = 254 nm) based on each freshly prepared calibration curve to estimate the dG amount in each sample loaded on column.
Extraction and Enzymatic Digestion for DPC analysis
The DNA-protein crosslinks were isolated from the tissues using DNAzol (invitrogen). The samples were dissolved in 1 mL DNAzol reagent by pipetting before adding proteinase K for overnight digestion at room temperature. DPCs were precipitated by 100% ethanol at −20 °C for 2 h, and then centrifuged at 14,000 × g at 4 °C for 5 min. The pellets (DPCs) were washed with 75% ethanol before being reconstituted with 500 μL pre-digestion buffer (40 mmol/L ammonium acetate (pH 6.0), CaCl2 (10 mmol/L), and pronase (0.4 mg/mL)) for overnight digestion at room temperature. After digestion, the supernatant was collected by centrifuged at 12,000 × g at 4 °C for 5 min. Then, DPCs were precipitated at −20 °C for 2 h using 10% volume sodium acetate (3 M) and 2.5 fold volume of 100% ethanol. DPCs were pelleted by centrifugation at 12,000 × g at 4 °C for 5 min, and then washed with 75% ethanol. Pellets were reconstituted with 450 μL digestion buffer (40 mmol/L ammonium acetate (pH 6.0), MgCl2 (10 mmol/L), CaCl2 (10 mmol/L), DNase I (44.4 U/mL), alkaline phosphatase (3.3 U/mL), phosphodiesterase I (0.0067 U/mL), prolidase (3.3 U/mL), carboxypeptidase Y (16.67 μg/mL), and aminopeptidase M (0.083 U/mL)) for overnight digestion at room temperature. The enzymatic reaction was terminated with the addition of 10 μL of 30% acetic acid. After adding 8 fmol of internal standard ([15N5]-dG-[13CD2]-Me-Cys), the final reaction mixture was subjected to centrifugation at 12,000 × g at 4 °C for 40 minutes in the Pall NanoSep 3 kDa filter (Port Washington, New York) to remove enzymes prior to HPLC purification.
The endogenous and exogenous dG-Me-Cys, and their internal standard, were purified from the filtrate using an Agilent 1200 Series UV HPLC System with C18 reverse-phase column (Waters Atlantis T3, 3 μm, 15 cm × 4.6 mm). The detection wavelength and column temperature were set at 254 nm and 15 °C, respectively. The mobile phases consisted of 0.05% acetic acid in water (A) and CH3CN (B). The flow rate was 0.45 mL/min, and elution gradient conditions were set as follows: 0 min, 2% B; 3 min, 2% B; 42 min, 4.2% B; 43 min, 4.2% B; 43.5 min, 80% B; 46 minutes, 80% B; 47.5 min, 2% B; 55 min, 2% B. The target analytes were eluted at a retention time range between 37 and 41 min. The amount of digested dG in each sample was quantitated by the UV peak area (λ = 254 nm) based on each freshly prepared calibration curve to estimate the dG amount in each sample loaded on column.
nLC-nESI-MS/MS Analyses
nLC-nESI-MS/MS analyses of mono-adduct, N2-HOMe-dG, and DPCs, dG-Me-Cys, were performed on two mass spectrometry platforms. First, the sensitive nano-LC-MS-MS instrument, AB SCIEX Triple Quad 6500 mass spectrometer (Foster City, California) operated in SRM mode, was responsible for the detection of N2-HOMe-dG. The mass spectrometer was interfaced with an Eksigent nanoLC Ultra 2D system (Dublin, California). A 150×0.075 mm Eksigent ChromXP 3C18-CL-120 analytical column (3 mm particle size) was used with loading volume of 1 μL. The column was kept at room temperature during each run. Mobile phases were comprised of water with 0.1% formic acid (A) or acetonitrile with 0.1% formic acid (B). Analytes were injected into the analytical column with an initial starting condition of 5% B at 0.3 mL/ min for 10 min followed by a linear gradient to 35% B over 12 min and to 60% B over 1 min. The flow was then held at 60% B for 6 min followed by re-equilibration for an additional 10 min. The analytes were introduced to the mass spectrometer using positive-mode nanospray ionization with a source voltage of 2400 V with collision energy of 15 eV. N2-HOMe-dG was quantified as N2-Me-dG after reduction using the transition of m/z 282.2 to m/z 166.1, and [13CD2]-N2-HOMe-dG was quantified as [13CD2]-N2-Me-dG with the transition of m/z 285.2 to m/z 169.1. Two additional transitions, including m/z 284.2 to m/z 168.1 and m/z 283.2 to m/z 167.1, were also monitored in case hydrogen-deuterium (H-D) exchange occurred. Similarly, the transition of m/z 297.2 to m/z 176.1 was chosen for the internal standard, [13C1015N5]-N2-Me-dG. The calibration curves were obtained using the integrated peak area and amount of injected analytical standard and internal standard.
On the other hand, the UltiMate 3000 RSLC nano system coupled to a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific) was responsible for the detection of dG-Me-Cys. Briefly, the DPCs sample of 6 μL was firstly loaded into a C18 trapping column (5 μm particle, 0.5 cm x 300 μm, Thermo Fisher Scientific) at a flow rate of 5 μL/min for 3 minutes using 0.1% formic acid in water as a loading solvent. A binary solvent system consisting of 0.1% formic acid in water (solvent A) and 0.1% formic acid in CH3CN (solvent B) was used for LC separation at a flow rate of 250 nL/min. dG-Me-Cys was then separated on a PepMap C18 analytical column (2 μm particle, 50 cm x 75 μm, Thermo Fisher Scientific). LC separation was performed with the gradient of holding at 1% B for 3 min, then from 1% to 20% B in 16 min, 20% to 90% B in 0.1 min, then maintaining at 90% B for 9.9 min, then from 90% to 1% B in 0.1 min, and finally holding at 4% B for 13.9 min for re-equilibration. The spectra were acquired by one full scan coupled with targeted PRM mode with an inclusion list comprised of m/z 401.12 (endogenous dG-Me-Cys), 404.14 (exogenous dG-[13CD2]-Me-Cys), and m/z 409.13 (internal standard [15N5]-dG-[13CD2]-Me-Cys). Precursors in targeted PRM mode were isolated with a window of 1.4 m/z and fragmented with HCD of normalized collision energy of 25. The raw data generated from Q Exactive HF instrument were analyzed by Xcalibur software (Thermo Fisher Scientific). The quantification analysis was performed using Skyline v3.7.0.11317. Endogenous dG-Me-Cys was quantified using the transition of m/z 401.12377 to m/z 164.05668, and exogenous dG-[13CD2]-Me-Cys was quantified with the transition of m/z 404.13948 to m/z 167.07238. Similarly, the transition of m/z 409.12465 to m/z 172.05756 was chosen for the internal standard, [15N5]-dG-[13CD2]-Me-Cys. The calibration curves were obtained using the integrated peak area and amount of injected analytical standard and internal standard.
Statistical analysis:
Analysis of variance (ANOVA) was used to compare the means of formaldehyde induced DNA adducts and DPCS across the control and exposure groups.
Results
The primary goal of this study is to examine DNA adducts after inhalation exposure to low doses of FA by measuring DNA mono-adduct, N2-HOMe-dG, and DPCs, dG-Me-Cys, as formaldehyde-specific biomarkers. Fischer F344 rats were exposed to [13CD2]-FA for 6 hours/day by nose-only inhalation for 28 consecutive days. This exposure caused no adverse clinical observations or gross pathology. Following completion of exposures, tissue samples were collected, immediately frozen, and stored at −80 °C until DNA isolation, reduction, digestion, HPLC purification, and adducts quantitation by nano-LC-MS-MS. These standard procedures have been extensively used in our previous studies (Lu et al. 2010a, 2011, 2012; Yu et al. 2015; Lai et al. 2016; Liu et al. 2018).
We first tested the sensitivity of our methods for both N2-HOMe-dG and dG-Me-Cys. The limit of detection for N2-HOMe-dG was 0.5 attomole on the column. The limit of detection for dG-Me-Cys was 5 attomole on the column. The sensitivity reported here are equivalent or better than our previously reported methods, which makes it feasible to detect adduct formation at low doses. In addition, these sensitive methods for the analysis of N2-HOMe-dG and dG-Me-Cys exhibited excellent intra-day or inter-day precision and accuracy. Satisfactory accuracy and precision of the assays were validated by adding known amounts of adducts analytical standards to 20 μg of rat DNA, as listed in Table 1. The CV for intra-day tests is typically within ~5%, while the CV for inter-day samples is within ~10%.
Table 1.
The precision and accuracy of assays for formaldehyde-induced mono-adduct and dG-Me-Cys.
| N2-Me-dG | Intra-day (n=3) | Inter-day (n=3) | ||||
| Added (fmol) | Measured | CV % | Accuracy % | Measured | CV % | Accuracy % |
| 2 | 2.11 | 5.18 | 105.5 | 1.93 | 8.82 | 96.5 |
| 5 | 5.23 | 4.20 | 104.6 | 5.19 | 7.65 | 103.8 |
| 10 | 9.27 | 5.57 | 92.7 | 10.81 | 9.16 | 108.1 |
| dG-Me-Cys | Intra-day (n=3) | Inter-day (n=3) | ||||
| Added (fmol) | Measured | CV % | Accuracy % | Measured | CV % | Accuracy % |
| 0.2 | 0.192 | 5.55 | 96.0 | 0.219 | 6.58 | 109.5 |
| 0.5 | 0.534 | 4.97 | 106.8 | 0.537 | 7.13 | 107.4 |
| 1 | 1.077 | 4.88 | 107.7 | 0.976 | 8.91 | 97.6 |
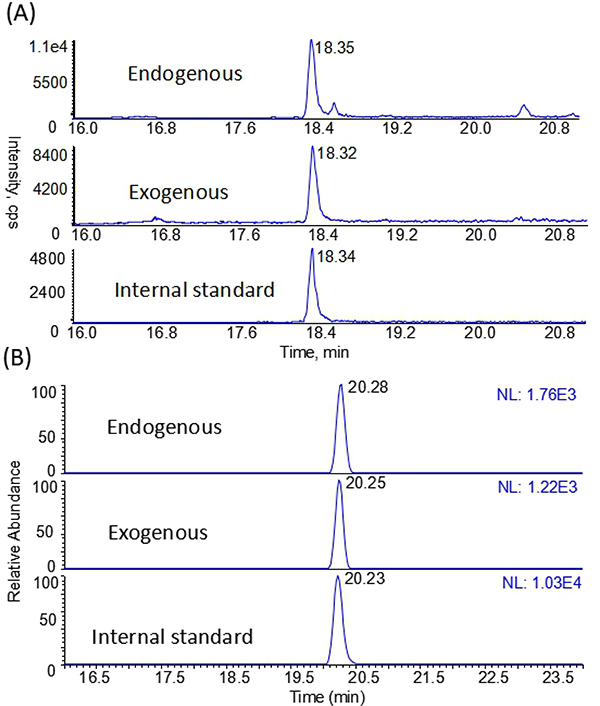
Next, we tested our methods using nasal epithelium samples of rats that were exposed to 10 ppm for 5 days (6h/day). The formation of exogenous formaldehyde induced DNA adducts has been well validated in our previous study (Lu et al. 2010a). As shown in Fig. 1A, the endogenous and exogenous N2-HOMe-dG were both detected and quantified with the amounts of 3.41 and 2.70 per 107 dG, respectively. Likewise, the endogenous and exogenous dG-Me-Cys were detected and quantified with the amounts of 3.67 and 2.47 per 108 dG, respectively, as shown in Fig. 1B. The amounts of N2-HOMe-dG and the ratios of endogenous and exogenous adducts were very consistent with previous reports (Lu et al. 2010a; Lai et al. 2016).
Fig. 1.
Typical nano-LC-MS-MS chromatograms from nasal tissues exposed to the [13C2H2]-FA of 10 ppm as positive controls for the detections of (A) N2-HOMe-dG and (B) dG-Me-Cys.
Taken together, these results have demonstrated that our methods are capable to detect both endogenous and exogenous formaldehyde-induced DNA adducts or DPCs. These methods are highly sensitive, accurate and precise as shown by our extensive validation.
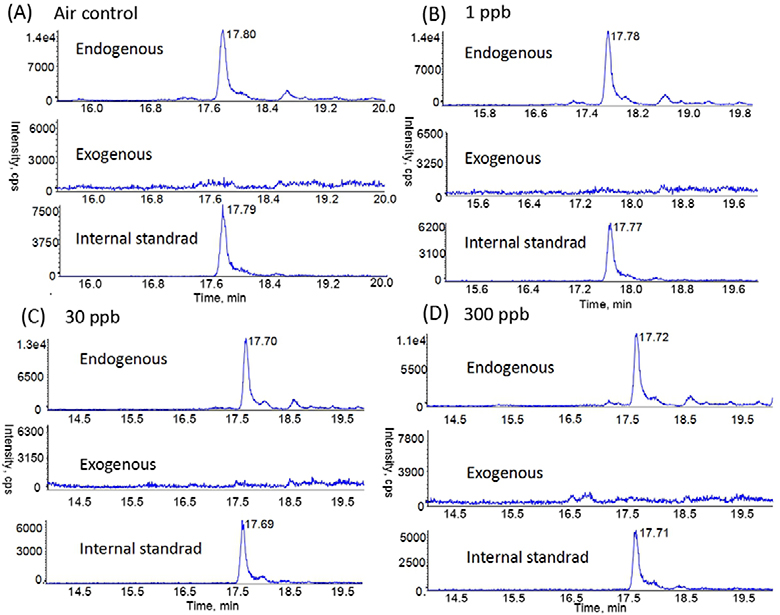
We next applied our methods to analyze the tissue samples of rats exposed to 1, 30 and 300 ppb FA. Fig. 2 shows the typical chromatograms of N2-HOMe-dG in the nasal DNA from the rats exposed to different concentrations of [13CD2]-FA. It can be seen that endogenous formaldehyde DNA adducts were all detected, but no exogenous formaldehyde DNA adducts could be found in any groups. Likewise, endogenous adducts were detectable in all other tissues distant to nasal epithelium, with the levels ranging from 2.35 to 5.25 adducts per 107 dG, depending on the specific tissue types as shown in Table 2. However, no exogenous formaldehyde-induced N2-HOMe-dG was detected in any tissues we analyzed.
Fig. 2.
Typical nano-LC-MS/MS SRM chromatograms of endogenous and exogenous N2-HOMe-dG in rat nasal respiratory samples from (A) air control group, (B) 1 ppb group, (C) 30 ppb group, and (D) 300 ppb group.
Table 2.
Levels of endogenous and exogenous N2-HOMe-dG (adducts/107 dG) in rat tissues exposed to [13CD2]-formaldehyde (1, 30, 300 ppb) for 28 days.
| Tissues | Air control | 1 ppb | 30 ppb | 300 ppb | na | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endogenous | Exogenous | Endogenous | Exogenous | Endogenous | Exogenous | Endogenous | Exogenous | ||
| Nasal Mucosa | 3.23±0.85 | ndb | 3.59±0.90 | nd | 3.27±0.76 | nd | 3.48±0.83 | nd | 8 |
| Bone Marrow | 4.83±1.54 | nd | 4.32±1.21 | nd | 5.03±1.71 | nd | 4.42±0.69 | nd | 8 |
| PBMC | 2.64±1.03 | nd | 2.72±0.73 | nd | 2.80±1.11 | nd | 2.94±1.15 | nd | 8 |
| Trachea | 3.14±0.61 | nd | 3.23±1.02 | nd | 3.34±0.75 | nd | 3.23 ±0.47 | nd | 6 |
| Liver | 2.48±0.21 | nd | 2.57±0.31 | nd | 2.44±0.34 | nd | 2.60±0.76 | nd | 6 |
| Hippo campus | 2.35±0.56 | nd | 2.62±0.74 | nd | 2.52±0.82 | nd | 2.86±0.76 | nd | 5 |
| Olfactory Bulbs | 2.51±0.62 | nd | 2.74±1.05 | nd | 2.84±0.45 | nd | 2.59±0.38 | nd | 5 |
| Cerebellum | 2.45±0.76 | nd | 2.62±0.67 | nd | 2.46±0.43 | nd | 2.35±0.57 | nd | 5 |
| Lung | 5.25±3.23 | nd | 3.72±2.20 | nd | 4.79±3.22 | nd | 5.06±2.51 | nd | 7 |
na same sample number with each group.
nd: not detected.
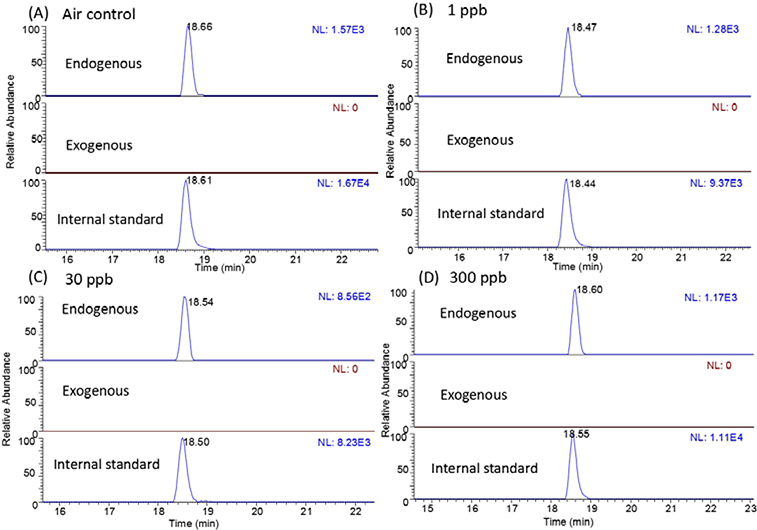
Fig 3 illustrates the chromatogram of dG-Me-Cys in the nasal epithelium samples of rats exposed to 1, 30 and 300 ppb FA for 28 days. Again, no exogenous formaldehyde DPC was found in nasal epithelium of rats exposed to formaldehyde, while endogenous formaldehyde adduct was detected in all samples. Besides, endogenous formaldehyde-induced dG-Me-Cys was also detected in all tissues distant to nasal epithilium, with 1.52 to 8.03 adducts per 108 dG across different tissues (Table 3). However, no exogenous formaldehyde-induced dG-Me-Cys was detected in any analyzed tissues.
Fig. 3.
Typical nano-LC-MS-MS PRM chromatograms of endogenous and exogenous dG-Me-Cys in rat nasal respiratory samples from (A) air control group, (B) 1 ppb group, (C) 30 ppb group, and (D) 300 ppb group. NL, normalized spectrum to largest peak in particular chromatogram.
Table 3.
Levels of endogenous and exogenous dG-Me-Cys (adducts/108 dG) in rat tissues exposed to [13CD2]-formaldehyde (1, 30, 300 ppb) for 28 days.
| Tissues | Air control | 1 ppb | 30 ppb | 300 ppb | na | ||||
|---|---|---|---|---|---|---|---|---|---|
| Endogenous | Exogenous | Endogenous | Exogenous | Endogenous | Exogenous | Endogenous | Exogenous | ||
| Nasal Mucosa | 2.66±0.54 | ndb | 2.77±0.61 | nd | 3.01±0.85 | nd | 2.85±0.74 | nd | 8 |
| Bone Marrow | 2.19±0.46 | nd | 2.28±0.55 | nd | 1.98±0.42 | nd | 2.45±0.48 | nd | 8 |
| PBMC | 1.96±0.66 | nd | 2.08±0.56 | nd | 1.88±0.64 | nd | 1.93±0.85 | nd | 8 |
| Trachea | 1.52±0.70 | nd | 2.30±1.03 | nd | 2.41±0.83 | nd | 1.99±0.57 | nd | 8 |
| Liver | 7.27±1.66 | nd | 8.03±1.46 | nd | 7.93±1.58 | nd | 7.13±1.58 | nd | 8 |
| Hippo campus | 1.81±0.46 | nd | 1.87±0.41 | nd | 1.63±0.51 | nd | 1.94±0.39 | nd | 5 |
| Olfactory Bulbs | 1.69±0.37 | nd | 2.55±0.40 | nd | 1.89±0.34 | nd | 2.04±0.42 | nd | 5 |
| Cerebellum | 2.71±0.87 | nd | 2.37±0.68 | nd | 2.39±1.60 | nd | 2.33±0.73 | nd | 5 |
| Lung | 4.07±1.11 | nd | 3.99±0.61 | nd | 3.34±0.67 | nd | 3.48±0.65 | nd | 8 |
na : same sample number with each group.
nd: not detected.
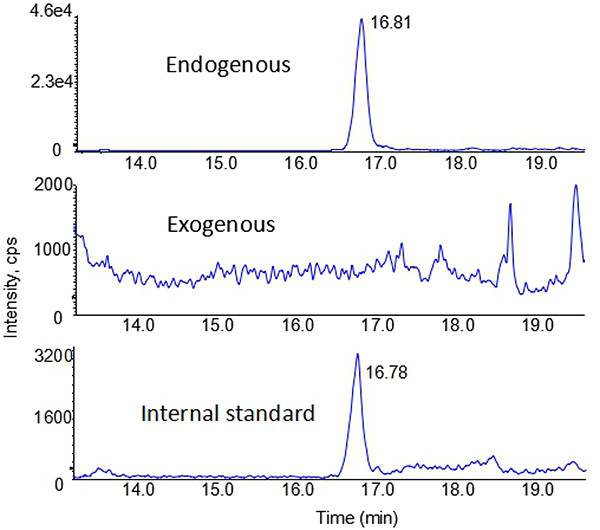
Since no N2-HOMe-dG and dG-Me-Cys caused by exogenous FA was detected in any tissues, even at the highest exposure level of 300 ppb in this study, it indicates that the levels of exogenous DNA adducts were below the limit of detection. Typically, DNA amount from each rat nasal epithelium is small (10~20 μg), which could limit our capability of detecting these extremely low abundant DNA adduct following a low dose exposure. To address this, we pooled five nasal samples exposed to 300 ppb FA together in this study, as shown in Fig. 4. However, we still found the exogenous N2-HOMe-dG to be absent (the middle panel, Fig 4). This further illustrated that the levels of exogenous N2-HOMe-dG were below the limit of detection when exposed to low dose of FA at 300 ppb or lower.
Fig. 4.
Typical nano-LC-MS-MS chromatograms of N2-HOMe-dG from pooled five nasal samples exposed to 300 ppb [13CD2]-FA for 28 days.
Table 2 and 3 summarize the amounts of formaldehyde induced DNA mono-adducts and DPCs in 9 different tissue samples, including nasal epithelium, bone marrow, trachea, liver, brain and lung. They show the levels of endogenous DNA adducts and DPCs across different tissues, but no exogenous formaldehyde DNA adducts were detected.
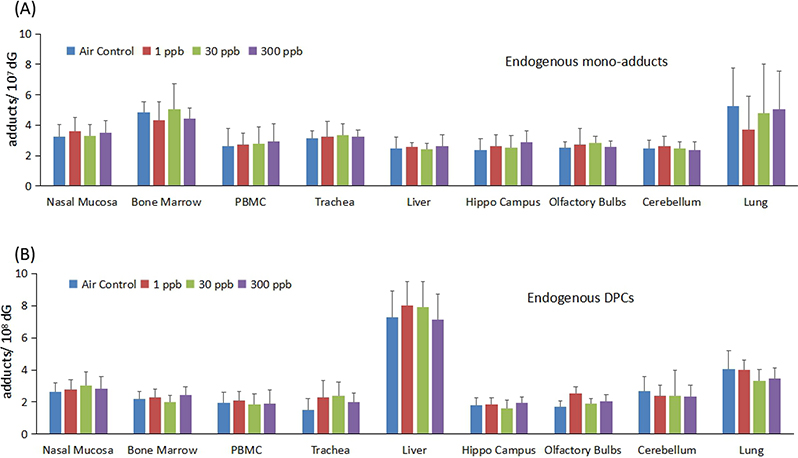
We next examined whether low doses of formaldehyde exposure altered the endogenous levels of DNA adducts in tissues. As shown in Fig 5, no statistical significant difference was found for either mono-adduct or DPCs in any tissues, indicating that exogenous FA did not alter the endogenous adducts levels at 300 ppb or lower. This is consistent with results in previous studies of higher FA doses (Lu et al. 2010a; Yu et al. 2015). The concentrations of dG-Me-Cys in the liver were relatively higher than the other tissues examined. In addition, endogenous levels of N2-HOMe-dG and dG-Me-Cys reported herein were consistent with previous results (Lu et al. 2010a; Yu et al. 2015; Lai et al. 2016), which again indicates the robustness and reliability of our methods used in this study.
Fig. 5.
Exogenous formaldehyde at 300 ppb or lower did not alter the levels of endogenous formaldehyde induced (A) mono-adduct, N2-HOMe-dG, and (B) DPC, dG-Me-Cys.
Discussion
Formaldehyde-induced DNA adducts have been extensively used as biomarkers to assess its exposure and associated risks. However, the fact that formaldehyde is ubiquitously present in all cells makes it challenging to evaluate the contribution of exogenous formaldehyde exposure. To address this, we have used stable isotope labeled formaldehyde for exposure, which allowed us to differentiate endogenous and exogenous formaldehyde-induced DNA adducts in numerous previous studies that involved higher formaldehyde doses for exposure (Lu et al. 2010a, 2011, 2012; Yu et al. 2015; Lai et al. 2016; Liu et al. 2018). Here, we applied sensitive mass spectrometry methods to analyze samples of rats exposed to 1, 30, 300 ppb [13CD2]-FA for 28 days (6h/day). No exogenous formaldehyde induced DNA adducts or DPCs was detected, although endogenous formaldehyde adducts were detected in all tissue we analyzed. This finding again demonstrates that endogenous formaldehyde-induced DNA damage predominates under the exposure scheme of this study.
It is well know that endogenous FA originates from numerous sources including one-carbon pool metabolism, amino acid metabolism, methanol metabolism, lipid peroxidation, cytochrome P450–catalyzed demethylation, and histone demethylation reactions (Dhareshwar and Stella 2008; Liu et al. 2013; Casanova et al. 1988; Shi et al. 2004). Due to its water solubility, endogenous FA is present in all aqueous body fluids and cellular nuclei, secondary to demethylation of histone (Shi et al. 2004). FA is released in close proximity to DNA, which provides an important source for FA induced endogenous DNA adducts and DPCs. Consistent with this, endogenous formaldehyde induced DNA damage is always present, as shown in this study and elsewhere (Lu et al. 2010a, 2011; Yu et al. 2015; Lai et al. 2016). Therefore, the risk assessment of formaldehyde exposure should not ignore the effects of such abundant endogenous formaldehyde.
Previously, we have demonstrated that exogenous formaldehyde causes DNA adducts in a highly non-linear manner. For example, the number of exogenous N2-HOMe-dG adducts was 0.039±0.019, 0.19±0.08, 1.04±0.24, 2.03±0.43 and 11.15±3.01 adducts/107 dG for 0.7, 2.0, 5.8, 9.1 and 15.2 ppm [13CD2]-formaldehyde exposure for 6 hours, respectively (Lu et al. 2011). Although it was challenging to measure exogenous DNA adduct at low doses, such as 0.7 ppm for 1 day exposure in the previous study (Lu et al. 2011), we were able to detect and quantify exogenous formaldehyde induced DNA adducts after combining DNA from 4–6 rat samples. In contrast, this study used a more sensitive mass spectrometry method (limit of detection of 0.5 amol versus 20 amol) and a longer exposure period (28 day versus 1 day) that has been demonstrated to cause exogenous formaldehyde accumulation (Lu et al. 2010a; Yu et al. 2015; Lai et al. 2016). Using the limits of detection of this study, if the ratios of exogenous versus endogenous DNA adducts were above ~1.5/10000 or ~1.8/100 for N2-HOMe-dG and dG-Me-Cys, we would have detected them. In addition, we also pooled DNA from nasal epithelium of 5 rats for detection of DNA adducts. However, exogenous formaldehyde DNA adducts or DPC could not be detected in this study in rats exposed to 300 ppb [13CD2]-formaldehyde or lower.
It is somewhat surprising to observe that exogenous formaldehyde adducts were below the limit of detection of the assays in rats exposed to 300 ppb formaldehyde. However, these results may be consistent with formation mechanisms of exogenous formaldehyde DNA adducts. Previously, we have demonstrated that formaldehyde induced exogenous DNA adducts are formed via the degradation of exogenous formaldehyde induced DPCs (Lai et al. 2016). DPCs are critical intermediates for formaldehyde induced exogenous DNA adducts. Exogenous formaldehyde can first target neighboring proteins due to the high reactivity of lysine, followed by the formation of DPCs (Lu et al. 2009, 2010b). Protein binding with formaldehyde is a key step in the formation of exogenous formaldehyde-induced DPCs and its degradation products, N2-HOMe-dG adducts. Obviously, cross-linking between DNA and exogenous formaldehyde-adducted proteins occurs in the nuclei. However, if exogenous formaldehyde reacts with extracellular components and is not able to enter into the nuclei, or formaldehyde reacts with proteins and such modified proteins could not be transported into the nuclei, then no exogenous DPCs, as well as N2-HOMe-dG, will be produced. Previously, we have shown that the formation of exogenous DNA adducts in cells treated with formaldehyde depends on the culture medium used (Lu et al. 2012). At 250 or 500 μM [13CD2]-formaldehyde-treated cells, the numbers of exogenous N2-HOMe-dG adducts in cells exposed to formaldehyde in culture medium were at least 50% lower than those in cells exposed to formaldehyde in PBS. When formaldehyde concentration was further reduced to 125 μM, we could not detect exogenous dG adducts in cells exposed in culture medium, while we still could detect exogenous adducts in cells exposed to formaldehyde in PBS buffer. These results indicate that formaldehyde has high reactivity with proteins or amino acids present in culture medium, which significantly affects the formation of exogenous DNA adducts in cells. In rats exposed to low doses of formaldehyde at 300 ppb or below, if considerable amounts of exogenous [13CD2]-formaldehyde react or interact with extracellular components or proteins and thus have no means to enter into the nuclei, no exogenous DNA adducts or DPCs could be formed or detected, which may account for our inability to detect exogenous DNA adducts and DPCs in these low doses of formaldehyde exposed tissues.
In summary, highly sensitive and accurate nano-LC-MS/MS methods integrated with the use of stable isotope labeled FA enabled us to unambiguously examine the formation of DNA adducts induced by the low dose exposure of exogenous FA under a substantial endogenous background of FA. Examination of exogenous versus endogenous FA DNA adducts clearly showed that endogenous DNA adducts and DPCs are always present, while exogenous formaldehyde at 1, 30 and 300 ppb contributes undetectable amounts of exogenous DNA adducts. The data suggest that FA may also induce exogenous DNA adducts in a nonlinear manner at low doses. Exogenous formaldehyde binding with extracellular components or proteins may explain undetectable exogenous DNA adducts and DPCs. Regardless, exogenous DNA adducts and DPCs in rats exposed to 1, 30 and 300 ppb formaldehyde for 28 days are lower than the limit of detection of our assays, while endogenous formaldehyde DNA adducts and DPCs are ubiquitously present in all tissues.
Acknowledgements
We thank Dr. Leonard Collins for his assistance with HPLC purification and nano-UPLC-MS-MS. This project received funding from the American Chemistry Council and Formacare. The instrumentation was partially supported by funding from the NIEHS (R01ES024950 and P30ES010126). None of the funding agencies is involved in writing or have access to the manuscript before its submission.
Abbreviations
- FA
formaldehyde
- dG
deoxyguanosine
- DPCs
DNA-Protein Crosslinks
- HCD
higher-energy collisional dissociation
- MS/MS
tandem mass spectrometry
- PRM
Parallel reaction monitoring
- SRM
selected reaction monitoring
Footnotes
Conflict of interest
The authors declare they have no actual or potential competing financial interests.
References
- Swenberg JA, Kerns WD, Mitchell RI, Gralla EJ, Pavkov KL (1980) Induction of squamous cell carcinomas of the rat nasal cavity by inhalation exposure to formaldehyde vapor. Cancer Res 40:3398–3402 [PubMed] [Google Scholar]
- (2006) International Agency for Research on Cancer. Formaldehyde, 2-butoxyethanol and 1-tert-butoxypropan-2-ol. IARC Monogr Eval Carcinog Risks Hum 88:1–478 [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tang X, Rothman N, Vermeulen R, Ji Z, Shen, M et al. (2010) Occupational exposure to formaldehyde, hematotoxicity, and leukemia-specific chromosome changes in cultured myeloid progenitor cells. Cancer Epidemiol Biomarkers Prev 19:80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen GD, Larsen ST, Wolkoff P (2013) Recent trend in risk assessment of formaldehyde exposures from indoor air. Arch Toxicol 87:73–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen GD, Larsen ST, Wolkoff P (2017) Re-evaluation of the WHO (2010) formaldehyde indoor air quality guideline for cancer risk assessment. Arch Toxicol 91:35–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ (2011) Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475:53–58 [DOI] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ (2012) Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature 489:571–575 [DOI] [PubMed] [Google Scholar]
- Garaycoechea JI, Patel KJ (2014) Why does the bone marrow fail in Fanconi anemia? Blood 123:26–34 [DOI] [PubMed] [Google Scholar]
- Moeller BC, Lu K, Doyle-Eisele M, McDonald J, Gigliotti A, Swenberg JA (2011) Determination of N2-hydroxymethyl-dG adducts in the nasal epithelium and bone marrow of nonhuman primates following 13CD2-formaldehyde inhalation exposure. Chem Res Toxicol 24:162–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edrissi B, Taghizadeh K, Moeller BC, Kracko D, Doyle-Eisele M, Swenberg JA, et al. (2013) Dosimetry of N6-formyllysine adducts following 13CD2-formaldehyde exposures in rats. Chem Res Toxicol 26:1421–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edrissi B, Taghizadeh K, Moeller BC, Yu R, Kracko D, Doyle-Eisele M, et al. (2017) N6-Formyllysine as a Biomarker of Formaldehyde Exposure: Formation and Loss of N6-Formyllysine in Nasal Epithelium in Long-Term, Low-Dose Inhalation Studies in Rats. Chem Res Toxicol 30:1572–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Ye W, Gold A, Ball LM, Swenberg JA (2009) Formation of S-[1-(N2-deoxyguanosinyl)methyl]glutathione between glutathione and DNA induced by formaldehyde. J Am Chem Soc 131:3414–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Collins LB, Ru H, Bermudez E, Swenberg JA (2010a) Distribution of DNA adducts caused by inhaled formaldehyde is consistent with induction of nasal carcinoma but not leukemia. Toxicol Sci 116:441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Ye W, Zhou L, Collins LB, Chen X, Gold A, et al. (2010b) Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J Am Chem Soc 132:3388–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Moeller B, Doyle-Eisele M, McDonald J, Swenberg JA (2011) Molecular dosimetry of N2-hydroxymethyl-dG DNA adducts in rats exposed to formaldehyde. Chem Res Toxicol 24:159–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Craft S, Nakamura J, Moeller BC, Swenberg JA (2012) Use of LC-MS/MS and stable isotopes to differentiate hydroxymethyl and methyl DNA adducts from formaldehyde and nitrosodimethylamine. Chem Res Toxicol 25:664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magana-Schwencke N, Ekert B (1978) Biochemical analysis of damage induced in yeast by formaldehyde. II. Induction of cross-links between DNA and protein. Mutat Res 51:11–19 [DOI] [PubMed] [Google Scholar]
- McGhee JD, von Hippel PH (1977) Formaldehyde as a probe of DNA structure. r. Mechanism of the initial reaction of Formaldehyde with DNA. Biochemistry 16:3276–3293 [DOI] [PubMed] [Google Scholar]
- Lai Y, Yu R, Hartwell HJ, Moeller BC, Bodnar WM, Swenberg JA (2016) Measurement of Endogenous versus Exogenous Formaldehyde-Induced DNA-Protein Crosslinks in Animal Tissues by Stable Isotope Labeling and Ultrasensitive Mass Spectrometry. Cancer Res 76:2652–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J, Bellelli R, Boulton SJ (2017) Mechanisms of DNA-protein crosslink repair. Nat Rev Mol Cell Biol 18:563–573 [DOI] [PubMed] [Google Scholar]
- Ide H, Shoulkamy MI, Nakano T, Miyamoto-Matsubara M, Salem AM (2011) Repair and biochemical effects of DNA-protein crosslinks. Mutat Res 711:113–122 [DOI] [PubMed] [Google Scholar]
- Liu CW, Tian X, Hartwell HJ, Leng J, Chi L, Lu K, et al. (2018) Accurate Measurement of Formaldehyde-Induced DNA-Protein Cross-Links by High-Resolution Orbitrap Mass Spectrometry. Chem Res Toxicol 31:350–357 [DOI] [PubMed] [Google Scholar]
- Swenberg JA, Lu K, Moeller BC, Gao L, Upton BP, Nakamura J, et al. (2011) Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol Sci 120 Suppl 1:S130–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Ji Z, McHale CM, Yuh J, Bersonda J, Tang M, et al. (2013) The impact of FANCD2 deficiency on formaldehyde-induced toxicity in human lymphoblastoid cell lines. Arch Toxicol 87:189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontel LB, Rosado IV, Burgos-Barragan G, Garaycoechea JI, Yu R, Arends MJ et al. (2015) Endogenous Formaldehyde Is a Hematopoietic Stem Cell Genotoxin and Metabolic Carcinogen. Mol Cell 60:177–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Lai Y, Hartwell HJ, Moeller BC, Doyle-Eisele M, Kracko D, et al. (2015) Formation, Accumulation, and Hydrolysis of Endogenous and Exogenous Formaldehyde-Induced DNA Damage. Toxicol Sci 146:170–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenberg JA, Moeller BC, Lu K, Rager JE, Fry RC, Starr TB (2013) Formaldehyde carcinogenicity research: 30 years and counting for mode of action, epidemiology, and cancer risk assessment. Toxicol Pathol 41:181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhareshwar SS, Stella VJ (2008) Your prodrug releases formaldehyde: should you be concerned? No! J Pharm Sci 97:4184–4193 [DOI] [PubMed] [Google Scholar]
- Liu J, Liu FY, Tong ZQ, Li ZH, Chen W, Luo WH et al. (2013) Lysine-Specific Demethylase 1 in Breast Cancer Cells Contributes to the Production of Endogenous Formaldehyde in the Metastatic Bone Cancer Pain Model of Rats. Plos one 8:e58957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova M, Heck HD, Everitt JI, Harrington WW, Popp JA (1988) Formaldehyde concentrations in the blood of rhesus monkeys after inhalation exposure. Food Chem Toxicol 26:715–716 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953 [DOI] [PubMed] [Google Scholar]
- Aydin S, Canpinar H, Undeger U, Guc D, Colakoglu M, Kars A, et al. (2013) Assessment of immunotoxicity and genotoxicity in workers exposed to low concentrations of formaldehyde. Arch Toxicol 87:145–153 [DOI] [PubMed] [Google Scholar]