Abstract
Cancer stem cells (CSCs) are self-renewing cells that facilitate tumor initiation, promote metastasis, and enhance cancer therapy resistance. Transcriptomic analyses across many cancer types have revealed a prominent association between stemness and immune signatures, potentially implying a biological interaction between such hallmark features of cancer. Emerging experimental evidence has substantiated the influence of CSCs on immune cells, including tumor-associated macrophages, myeloid-derived suppressor cells, and T cells, in the tumor microenvironment and, reciprocally, the importance of such immune cells in sustaining CSC stemness and its survival niche. This review covers the cellular and molecular mechanisms underlying the symbiotic interactions between CSCs and immune cells and how such heterotypic signaling maintains a tumor-promoting ecosystem and informs therapeutic strategies intercepting this co-dependency.
INTRODUCTION
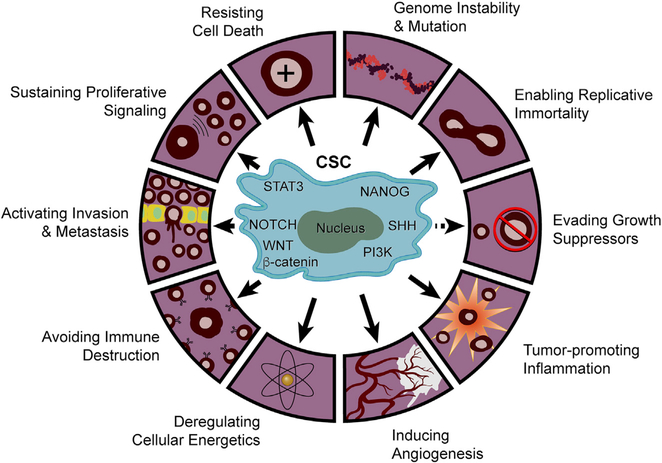
The cancer stem cell (CSC) paradigm emerged from the study of acute myeloid leukemia (AML), which identified a subpopulation of less-differentiated CD34+/CD38− cells possessing stem-cell-like renewal capacity and robust tumor-initiating capacity (Lapidot et al., 1994). Cancer cells with these biological properties have since been detected in virtually all solid tumors, including melanoma and cancers of the brain, breast, colon, thyroid, pancreas, prostate, liver, lung, ovary, head and neck, and stomach (Turdo et al., 2019). The clinical and biological significance of CSCs has been reinforced by a positive correlation between stem cell signatures and poor survival (Ben-Porath et al., 2008). Although CSCs share properties and surface markers with normal stem cells (Turdo et al., 2019), they maintain renewal capacity via specific altered signaling pathways with common and unique patterns across many tumor types (Figure 1). For instance, breast cancer CSCs show CD44 standard splice isoform (CD44s)-activated platelet-derived growth factor receptor β (PDGFRβ)/signal transducer and activator of transcription 3 (STAT3), forkhead box C1 (FOXC1)-activated sonic hedgehog (SHH), and sphingosine-1-phosphate (S1P)/S1PR3-activated NOTCH pathways (Han et al., 2015; Hirata et al., 2014; Zhang et al., 2019b). In contrast, CSC stemness in other cancer types, such as glioma and colon, gastric, and prostate cancers, is maintained via CD133-mediated phosphatidylinositol-3-kinase (PI3K)/protein kinase B (AKT, leucine-rich G-protein-coupled receptor 5 (LGR5)-mediated WNT/β-catenin and speckle-type POZ protein (SPOP)-mediated NANOG pathways (Morgan et al., 2018; Wang et al., 2010b, 2019a; Wei et al., 2013; Zhang et al., 2019c). Such CSC-associated patterns belie a high degree of biological complexity and tumor type specificity.
Figure 1. The Mechanism Underlying CSC Stemness Regulation and the Contribution of CSCs to Cancer Hallmarks.
CSC stemness is regulated by several key signaling pathways, including STAT3, SHH, NOTCH, PI3K, WNT/β-catenin, and NANOG. CSCs promote tumorigenesis and progression by contributing to at least nine out ten hallmarks of cancer. Whether CSCs contribute to the tenth hallmark of cancer, evading growth suppressors, remains unknown.
The hallmark traits of CSCs are well established and include self-renewal, clonal tumor initiation capacity, clonal long-term repopulation potential, and plasticity between stem and non-stem states (Plaks et al., 2015). This plasticity is particularly relevant because it enables CSCs to adapt and survive in the face of therapeutic perturbations as well as the ever-changing biological stresses of the tumor microenvironment (TME) throughout tumor evolution (Agliano et al., 2017; Hatina, 2012; Müller et al., 2020; Plaks et al., 2015). Mechanistically, the role of CSCs in tumor initiation, metastasis, and therapy resistance has been shown to be driven by interactions between cancer cells and host cells in the TME (Ayob and Ramasamy, 2018; Plaks et al., 2015), where the molecules and pathways driving CSC biology often power multiple cancer hallmarks (Figure 1). For example, the tumor-initiating capacity of CSCs relates to their stemness driven by the transcription factor sex-determining region Y-box 2 (SOX2), which also upregulates genes governing the cancer hallmarks of proliferation, survival, and invasion (Boumahdi et al., 2014; Zhou et al., 2009). In the case of the metastasis hallmark, stem cell signatures correlate positively with enhanced metastatic propensity (Ayob and Ramasamy, 2018); moreover, diverse CSC pathways and associated biological processes contribute to each step of the metastatic process—from dissemination to metastatic niche formation to distant organ growth—by inducing epithelial-mesenchymal transition (EMT), stimulating exosome production from myeloid cells and upregulating niche-derived factors, such as insulin-like growth factor-1 (IGF-1) and interleukin (IL)-6, respectively (Agliano et al., 2017; Ayob and Ramasamy, 2018; Shiozawa et al., 2013). Indeed, experimental and clinical evidence demonstrates that CSCs in primary tumors disseminate and colonize to distal sites (de Sousa e Melo et al., 2017) and that their location at the invasive front correlates negatively with patient survival (Kodama et al., 2017). Finally, with respect to therapy resistance, CSC pathways alter signaling molecules governing drug metabolism (e.g., high expression of ATP-binding cassette transporter proteins that increase drug efflux rate), EMT (e.g., increased SOX2, octamer-binding transcription factor 4 [OCT4], and NANOG expression), and metabolic reprogramming (e.g., enhanced glucose transporter 1, oxidative phosphorylation, and reactive oxygen species activity; Ayob and Ramasamy, 2018).
The phenotypic plasticity of CSCs can contribute to additional cancer hallmarks via their capacity to transdifferentiate into pericytes, endothelial cells, and fibroblasts, thus contributing to tumor angiogenesis, stem cell niche development, and inflammation (Figure 1; Cheng et al., 2013; Dongre and Weinberg, 2019; Hu et al., 2016; Nair et al., 2017; Ricci-Vitiani et al., 2010; Wang et al., 2010a). This plasticity is also reflected in the capacity of “differentiated” cancer cells to re-adopt an immature CSC state, a dedifferentiation process that can be stimulated by signals emanating from the TME, including tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs), T cells, cancer-associated fibroblasts (CAFs), and other immune cells (Plaks et al., 2015). Most notably, the strong CSC-immune cell connection was evidenced by unbiased profiling studies, showing a strong negative correlation between cancer cell stemness and anti-tumor immunity signatures across 21 types of solid tumors (Miranda et al., 2019). Specifically, increased stemness was associated with reduced anti-cancer immune cells, including CD8+ T cells, natural killer (NK) cells and B cells, and enhanced polarization of infiltrating macrophages (Miranda et al., 2019). Similarly, the cancer genome atlas (TCGA) and tissue microarray analyses have revealed that cancer cell stemness correlates negatively with activated CD4+ and CD8+ T cells in solid tumors (Hou et al., 2019; Malta et al., 2018).
These in silico findings in human cancer align well with emerging experimental findings from studies of various mouse models of human cancer. It has been shown that the proportion of CSCs in melanoma is dependent on the specific immune-compromised mouse strain employed, suggesting an important role of immune system in regulation of CSCs (Quintana et al., 2008). On the other hand, CSCs can shape a specific TME by their regulation of immune cells. For instance, the expression of CSC marker and regulator doublecortin-like kinase 1 (Westphalen et al., 2014) correlates positively with abundance of TAMs and regulatory T cells (T-reg cells) and elevated expression of factors that inhibit CD8+ T cell activity (Wu et al., 2020). CKLF-like MARVEL transmembrane-domain-containing 6, which is expressed on cancer cell plasma membranes, can enhance CSC stemness via the WNT/β-catenin pathway, suppress anti-tumor immunity via programmed death-ligand 1 (PD-L1) upregulation, and reduce CD8+ and CD4+ T cells in many types of cancer, including squamous cell carcinoma of the head and neck (SCCHN) (Chen et al., 2020a), melanoma, and breast cancer (Burr et al., 2017; Mezzadra et al., 2017). Fat-mass- and obesity-associated protein (FTO) is an m6A demethylase and overexpressed in AML. Inhibition of FTO impairs leukemia stem cell stemness and reprograms immune response by suppressing immune checkpoint genes, such as LILRB4 (leukocyte immunoglobulin-like receptor subfamily B member 4), thus sensitizing AML cells to T-cell-mediated cytotoxicity (Su et al., 2020). Similarly, single-cell RNA sequencing (scRNA-seq) analyses of AML revealed a subpopulation of stem-like AML cells that co-express stemness-related and myeloid-priming genes (van Galen et al., 2019). In addition, CSC-derived exosomes, which transfer cargo between cells (Mathieu et al., 2019), can enhance the survival of suppressive neutrophils to promote colon cancer growth (Hwang et al., 2019). This shared stemness and immune transcriptional profile aligns with the recent finding that lack of natural killer group 2 member D ligands, which defines leukemia stem cells, contributes to their selective escape from NK-cell-mediated immune surveillance (Paczulla et al., 2019). In addition to these immune cells, CAFs and their interactions with CSCs are also important for tumorigenesis and therapy resistance (Chan et al., 2019). Together, these findings highlight an intimate link between the molecules and mechanisms governing CSC biology and tumor immunity across many tumor types.
In summary, mounting translational and experimental evidence underscores the myriad interactions and intertwined tumor biological roles of CSCs and immune cells, particularly myeloid cells (TAMs and MDSCs) and T cells. This review summarizes the current knowledge of the molecular crosstalk and functional impact of these symbiotic interactions on the hallmarks of cancer. With respect to myeloid cells, we highlight TAM and MDSC individually, although they share the same cell of origin and similar function to suppress T-cell-mediated anti-tumor immunity (Engblom et al., 2016). These emerging insights provide a roadmap for the development of novel anti-cancer therapeutic strategies that disrupt this dynamic circuit in specific tumor types.
CSC-TAM Crosstalk
Impact of CSCs on Macrophage Biology
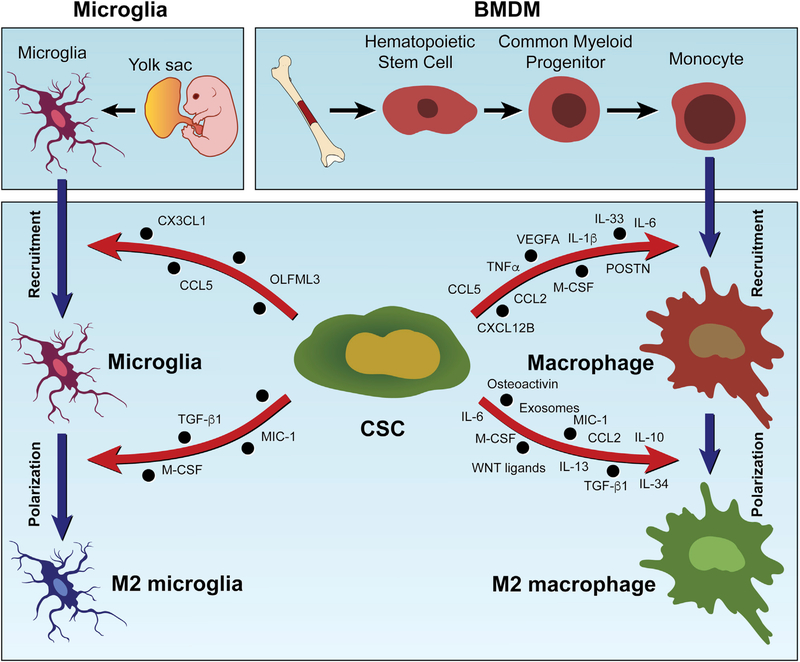
Factors secreted by various cell types in the TME, including CSCs, are known to recruit and polarize TAMs (Chen et al., 2017, 2019a, 2020b; Colegio et al., 2014). These TAMs are sourced from bone-marrow-derived macrophages (BMDMs) and local tissue-resident macrophages (e.g., microglia in the brain, Kupffer cells in the liver, and alveolar macrophages in the lung), which originate from hematopoietic stem cells and progenitors seeding in embryonic tissues (e.g., the yolk sac for microglia and the fetal liver for Kupffer cells and alveolar macrophages), respectively (Figure 2; Pathria et al., 2019). TAM recruitment is driven by a variety of chemokines, including C-C motif chemokine ligand 2 (CCL2), CCL3, C-X-C motif chemokine ligand 14 (CXCL14), and lysyl oxidase (LOX), which are secreted by cancer cells, macrophages, and other stromal cells in the TME (Chen et al., 2019a; Pathria et al., 2019; Wei et al., 2020). Increasing evidence shows that CSCs also contribute to the infiltration of macrophages and microglia via distinct molecules and mechanisms in various cancers (Figure 2; Table 1). Of note, some of these chemokines are specifically produced by CSCs, which highlight a unique role of CSCs in regulation of TAM infiltration. For example, periostin (POSTN) is preferentially expressed and secreted by CSCs in glioblastoma (GBM) and cholangiocarcinoma (CCA), which in turn recruits BMDMs through binding with αvβ3 integrin (Zeng et al., 2018; Zhou et al., 2015).
Figure 2. Origins of Macrophages and Microglia and the Mechanism of CSC-Driven TAM Infiltration and Polarization.
Macrophages in tumors have two distinct origins: BMDMs and tissue-resident macrophages, such as microglia in the brain. BMDMs originate from bone marrow progenitor cells that differentiate into monocytes. Monocytes traffic to other sites upon stimulation and then differentiate into macrophages. Microglia originate from microglial progenitors seeded from the embryonic yolk sac. CSCs contribute to the infiltration and polarization of BMDMs and microglia via secretion of a variety of chemokines and factors (as indicated).
Table 1.
Roles and Mechanisms of CSCs in Macrophage Biology
| CSCs Contribute to Macrophage Recruitment | |||
| Cancer Type | CSC-Derived Chemokines | Chemokine Receptors | References |
| Glioma | OLFML3 | not shown | Chen et al., 2020b |
| POSTN | αvβ3 integrin | Zhou et al., 2015 | |
| CXCL12B | CXCR4 | Chia et al., 2018 | |
| CCL5 and CX3CL1 | CCL5R and CX3CR1 | Guo et al., 2019b | |
| Cholangiocarcinoma | POSTN | Αvβ3 integrin | Zeng et al., 2018 |
| Lung cancer | TNF-α, IL-1β, and IL-6 | IL-6R | Lu et al., 2018 |
| Bladder cancer | IL-6 and CCL2 | IL-6R and CCR2 | Kobatake et al., 2020 |
| CRC | IL-33 | ST2 | Fang et al., 2017 |
| Liver cancer | CCL2 and M-CSF | CCR2 and CSF1R | Guo et al., 2017 |
| Breast cancer | M-CSF, CCL2, CCL5, and vascular endothelial growth factor A | not shown | Valeta-Magara et al., 2019 |
| CSCs Contribute to Macrophage Polarization | |||
| Cancer Type | CSC-Derived Chemokines | Mechanisms | References |
| Glioma | IL-6, IL-10, M-CSF, TGF-b1, and MIC-1 | STAT3 pathway | Wu et al., 2010; Yao et al., 2016 |
| WISP1 | α6β1 integrin/AKT pathway | Tao et al., 2020 | |
| exosomes | STAT3 pathway | Gabrusiewicz et al., 2018 | |
| Ovarian cancer | IL-10 and WNT | NF-κB pathway | Raghavan et al., 2019 |
| Cholangiocarcinoma | IL-13, IL-34, and osteoactivin | not shown | Raggi et al., 2017 |
| Bladder cancer | IL-6 and CCL2 | STAT3 pathway | Kobatake et al., 2020 |
| Breast cancer | IL-6 | STAT3 pathway | Weng et al., 2019 |
Genetic and epigenetic changes in CSCs regulate chemokine production. For example, PTEN deficiency or AKT overexpression in GSCs and neural stem cells upregulates LOX and CXCL12B, which recruits TAMs through β1 integrin (Chen et al., 2019a) and C-X-C motif chemokine receptor 4 (CXCR4) (Chia et al., 2018), respectively. Optic GSCs isolated from the Nf1flox/neo;GFAP-Cre low-grade glioma mouse model secrete CX3CL1 and CCL5 to recruit microglia, and this effect is further amplified by loss of Pten (Guo et al., 2019b). Similarly, NF1 deficiency in human GSCs can promote the infiltration of macrophages and microglia, although the NF1-regulated chemokines are not known (Wang et al., 2017). In many tumor types, epidermal growth factor receptor (EGFR) amplification and mutation can promote CSC stemness and macrophage recruitment (An et al., 2018; McCann et al., 2018; Rutkowska et al., 2019). In liver cancer, EGFR/AKT activation activates the yes-associated protein (YAP)/TEA domain family member (TEAD) transcription factor complex in CSCs, which in turn upregulates macrophage recruitment factors CCL2 and macrophage colony stimulating factor (M-CSF) (Guo et al., 2017). In non-small cell lung cancer (NSCLC), increased ubiquitin-specific protease 17, a deubiquitinase required for trafficking and oncogenic activity of mutant EGFR (McCann et al., 2018), increases cancer cell stemness, which in turn upregulates macrophage infiltration by augmented production of cytokines, including tumor necrosis factor (TNF)-α, IL-1β, and IL-6 (Lu et al., 2018). In bladder cancer, loss-of-function mutations of the histone modifier gene lysine (K)-specific demethylase 6A (KDM6A) promote CSC stemness and secretion of IL-6 and CCL2, which in turn increase macrophage recruitment (Kobatake et al., 2020). In GBM, circadian locomotor output cycles kaput (CLOCK), an epigenetic and circadian regulator amplified in 5% of cases, enhances GSC stemness and secretion of the chemokine olfactomedin-like protein 3 (OLFML3), which recruits microglia into the TME (Chen et al., 2020b). Finally, caveat emptor, although many studies have established the involvement of CSC-derived factors in macrophage infiltration, the converse has also been observed due to the specific CSC genotype and its unique TME. For example, TP53-mutated and cisplatin-resistant CSCs from lung cancer inhibit macrophage infiltration into the TME (Xu et al., 2019).
In addition to stimulating TAM recruitment, CSCs can influence the biological state of these macrophages. Macrophages are known to exhibit a spectrum of phenotypes, ranging from an anti-tumor to pro-tumor phenotype (formerly referred to as M1 and M2; Pathria et al., 2019). Once macrophages infiltrate into tumors, they typically undergo polarization toward a pro-tumor phenotype, a process driven by chemokines (e.g., IL-4 and IL-13) and metabolites (e.g., lactate), which are derived from both cancer cells and host cells in the TME (Chen et al., 2017; Colegio et al., 2014; Qian and Pollard, 2010). Several lines of evidence demonstrate that CSCs can further provoke anti- to pro-tumor polarization of macrophages. First, upon co-culture with CSCs, pro-tumor macrophage markers (e.g., CD206, IL-10, and arginase 1) are upregulated, whereas anti-tumor macrophage markers (e.g., TNF-α, nitric oxide synthase 2 [NOS2], and CD86) are downregulated (Deng et al., 2015). Second, CSCs can secrete various soluble factors known to induce polarization toward a pro-tumor phenotype (Figure 2; Table 1). For example, Wnt-induced signaling protein 1 (WISP1) is preferentially secreted by GSCs in GBM, which promotes the survival of pro-tumor TAMs through activation of the α6β1 integrin/AKT pathway on macrophages (Tao et al., 2020). Similarly, CSC-derived IL-6 and IL-10 can skew TAMs toward a pro-tumor phenotype in ovarian cancer (Raghavan et al., 2019), bladder cancer (Kobatake et al., 2020), GBM (Wu et al., 2010; Yao et al., 2016), and breast cancer (Weng et al., 2019). In addition to secreted factors, GSCs release exosomes containing eukaryotic initiation factor 2, mammalian target of rapamycin (mTOR), and ephrin B signaling pathways that home to the monocyte membrane and promote macrophage pro-tumor polarization (Gabrusiewicz et al., 2018). Finally, in the context of tumor necrosis in GBM, GSC-derived particles, defined as “autoschizis-like products,” can be engulfed by TAMs, which in turn upregulate IL-12 to polarize these TAMs toward an anti-tumor phenotype (Tabu et al., 2020). Thus, CSCs secrete a variety of products that encourage macrophage polarization.
In multiple cancer types, macrophage and microglia pathways and/or factors involved in pro-tumor polarization include STAT3, nuclear factor (NF)-κB, and PI3Kγ pathways (Kaneda et al., 2016; Qian and Pollard, 2010). Among these, the STAT3 transcription factor plays a prominent role. Macrophage STAT3 is activated by CSC-derived IL-6, IL-10, and/or exosome cargo, resulting in the upregulation of genes promoting pro-tumor programming and inhibition of genes encoding anti-tumor cytokines (Malyshev and Malyshev, 2015). Moreover, STAT3 inhibition abolishes CSC-induced pro-tumor macrophage polarization in GBM (Gabrusiewicz et al., 2018; Wu et al., 2010; Yao et al., 2016), bladder cancer (Kobatake et al., 2020), and breast cancer (Weng et al., 2019). The NF-κB pathway is essential for CSC-induced pro-tumor macrophage polarization in ovarian cancer (Deng et al., 2015). In summary, the study of CSC-induced macrophage polarization has identified prominent and therapeutically actionable macrophage pathways in specific cancers (Table 1).
TAMs Promote Cancer Cell Stemness and the CSC Niche
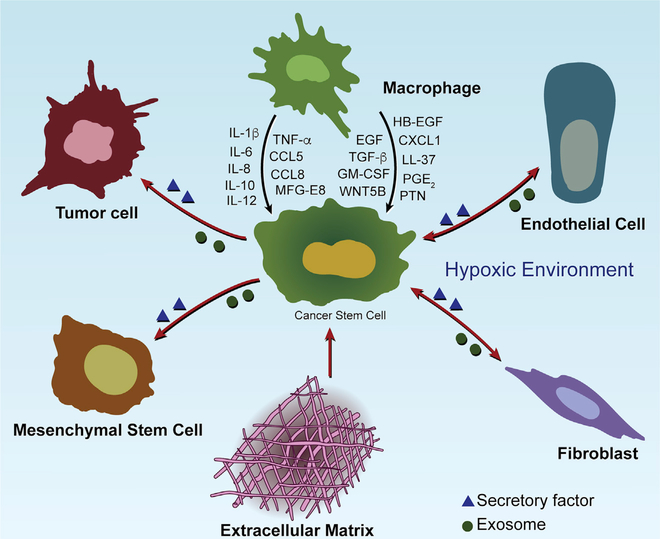
Mirroring CSC actions, TAMs can support CSC stemness and the CSC niche (Figure 3). The niche is particularly important in the maintenance of CSC self-renewal, repopulation potential, and tumor initiation. This supportive microenvironment is composed of cancer cells, immune cells, mesenchymal stem cells (MSCs), fibroblasts, endothelial cells, and extracellular matrix (ECM) components (Figure 3; Plaks et al., 2015). Paracrine factors derived from these diverse stromal cell types play prominent roles in promoting CSC stemness in the niche. Specifically, in breast and colon cancers, MSCs contribute to CSC niche formation by secreting prostaglandin E2 (PGE2), IL-6, IL-8, and CXCL1 (Li et al., 2012). Fibroblasts can induce a metastatic niche for breast cancer CSCs via secretion of POSTN (Malanchi et al., 2011). TAMs produce factors to enable “differentiated” cancer cells to acquire CSC-like features and to maintain CSC stemness in breast cancer (Lu et al., 2014), oral squamous cell carcinoma (Li et al., 2019), renal cell carcinoma (Yang et al., 2016), hepatocellular carcinoma (HCC) (Wang et al., 2016), and pancreatic cancer (Mitchem et al., 2013). Correspondingly, depletion of TAMs via inhibition of colony-stimulating factor 1 receptor (CSF1R) and C-C motif chemokine receptor 2 (CCR2) diminishes the tumor-initiating properties of CSCs in mouse models (Mitchem et al., 2013). In GBM, TAM support of the CSC niche depends on its pro-tumor phenotype, and reprogramming to an anti-tumor phenotype (using amphotericin B or vitamin B3) attenuates cancer cell stemness and tumorigenicity in vitro and in vivo and sensitizes these tumors to chemotherapy (Sarkar et al., 2014, 2020). These findings highlight the therapeutic potential of disrupting CSC niche via reprogramming TAMs toward an anti-tumor phenotype.
Figure 3. CSC Niche Components and the Role of Macrophages in CSC Stemness Maintenance.
CSC niches are composed of several types of cells (including macrophages, tumor cells, mesenchymal stem cells, fibroblasts, and endothelial cells) and their cytokine/growth factor networks, ECM components, and hypoxic environment. Macrophages represent one of these niches, which promote CSC stemness through producing a large number of soluble factors as indicated.
The important of TAMs in CSC biology is reinforced by a growing list of TAM-derived factors implicated in the maintenance of CSC stemness. Table 2 summarizes such factors and their purported mechanisms in different cancer models. EMT is an important process that enables cancer cells to acquire CSC-like features and maintain CSC stemness (Biddle and Mackenzie, 2012). In breast cancer cells, EMT is associated with upregulation of CD90 and EphA4, which mediate physical interactions between CSCs and TAMs (Lu et al., 2014). As a result, TAMs can further accelerate breast cancer cell EMT, thus inducing a positive feedback loop to reinforce CSC stemness via secreting a panel of CSC-supporting cytokines, such as IL-6, IL-8, and IL-1b (Guo et al., 2019a; Lu et al., 2014; Valeta-Magara et al., 2019). Similarly, accumulating evidence shows that TAMs in GBM (Hide et al., 2018), HCC (Fan et al., 2014; Wan et al., 2014), pancreatic cancer (Nomura et al., 2018; Sainz et al., 2015; Zhang et al., 2019a), and ovarian cancer (Raghavan et al., 2019) can promote cancer cell EMT and/or secrete a variety of CSC-supporting cytokines (including IL-1β, IL-6, and TGF- β), thus promoting CSC stemness, tumor progression, and therapy resistance. In addition to cytokines, TAMs can specifically produce unique factors to support CSC stemness. For example, CCL5, pleiotrophin (PTN), globule-epidermal growth factor-VIII (MFG-E8), and CCL8 are preferentially expressed and secreted by TAMs in prostate cancer (Huang et al., 2020), lymphoma (Wei et al., 2019b), colorectal cancer (CRC) (Jinushi et al., 2011), and GBM (Zhang et al., 2020), respectively, where they promote CSC stemness and tumor progression. Finally, in addition to soluble factors, TAMs can promote CSC stemness via direct interactions. Specifically, in breast cancer, liver, and lymph node sinusoidal endothelial cell C-type, lectin is a transmembrane protein highly expressed on TAMs that interacts with butyrophilin subfamily 3 member A3 receptor on cancer cells to enhance stemness (Liu et al., 2019).
Table 2.
Roles and Mechanisms of TAMs in CSC Stemness
| Cancer Type | TAM-Derived Factors | Mechanisms | References |
| Breast cancer | IL-6, IL-8, and GM-CSF | SRC and NF-κB pathways | Lu et al., 2014 |
| IL-6, IL-1β, TNFα, and IL-10 | STAT3/NF-κB pathway | Guo et al., 2019a | |
| IL-8 and CXCL1 | STAT3 pathway | Valeta-Magara et al., 2019 | |
| EGF | EGFR/STAT3/SOX2 pathway | Yang et al., 2013 | |
| GBM | HB-EGF and IL-1β | not shown | Hide et al., 2018 |
| IL-12 | not shown | Tabu et al., 2020 | |
| CCL8 | CCR1/CCR5/ERK1/2 pathway | Zhang et al., 2020 | |
| HCC | TNF-α | WNT/β-catenin pathway TNFR1/SRC/STAT3 pathway | Chen et al., 2019b; Li et al., 2017 |
| TGF-β | not shown | Fan et al., 2014 | |
| IL-6 | STAT3 pathway | Wan et al., 2014 | |
| Prostate cancer | CCL5 | β-catenin/STAT3 pathway | Huang et al., 2020 |
| Pancreatic cancer | IL-1β | NF-κB pathway | Nomura et al., 2018 |
| hCAP-18/LL-37 | Formyl peptide receptor 2 and P2X7R | Sainz et al., 2015 | |
| TGF-β | SMAD2/3/NANOG pathway | Zhang et al., 2019a | |
| CRC | PGE2 | not shown | Fang et al., 2017 |
| MFG-E8 | STAT3 and SHH pathways | Jinushi et al., 2011 | |
| Ovarian cancer | WNT5B and IL-6 | not shown | Raghavan et al., 2019 |
| Lymphoma | PTN | PTN receptor/β-catenin pathway | Wei et al., 2019b |
TAM-derived factors and TAM-cancer cell physical interactions activate several pathways in cancer cells that are pivotal to the maintenance of stemness. These key CSC pathways include STAT3, SHH, and NOTCH (Han et al., 2015; Hirata et al., 2014; Zhang et al., 2019b), as well as PI3K/AKT, WNT/ β-catenin, and NANOG (Morgan et al., 2018; Wang et al., 2010b, 2019a; Wei et al., 2013; Zhang et al., 2019c). Available evidence supports the view that TAM-derived factors activate these pathways to enhance or maintain CSC stemness (Table 2). Among them, STAT3 appears most important as a result of its potent upregulation of stemness-related genes and activation of stemness-promoting pathways, such as NF-κB (Galoczova et al., 2018). Accordingly, STAT3/NF-κB inhibition abolishes TAM-promoted stemness in breast cancer (Lu et al., 2014; Valeta-Magara et al., 2019; Yang et al., 2013), HCC (Li et al., 2017; Wan et al., 2014), prostate cancer (Huang et al., 2020), pancreatic cancer (Mitchem et al., 2013; Nomura et al., 2018), and CRC (Jinushi et al., 2011). The WNT/β-catenin and SHH pathways can also promote CSC stemness in some settings. Aberrant activation of WNT signaling, common in many tumor types, often defines the CSC state and maintenance of CSC biology (de Sousa E Melo and Vermeulen, 2016). Cell culture and mouse model systems demonstrate that TAMs activate the WNT/β-catenin pathway in CSCs and inhibition of this pathway impairs TAM-induced upregulation of CSC stemness in HCC (Chen et al., 2019b), prostate cancer (Huang et al., 2020), and lymphoma (Wei et al., 2019b). Similarly, the SHH pathway has been implicated in regulating CSC stemness either directly or through interaction with other stemness-related pathways, such as TGF-β (Takebe et al., 2015). With respect to TAM-supported CSC stemness, CRC relies on the SHH pathway (Jinushi et al., 2011), pancreatic cancer on the TGF-β1/SMAD2/3/NANOG pathway (Zhang et al., 2019a), HCC on the NOTCH pathway (Wang et al., 2016), breast cancer on the vsrc sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) (SRC) pathway (Lu et al., 2014), and glioma on the extracellular regulated kinase 1/2 (ERK1/2) pathway (Zhang et al., 2020). Collectively, these findings highlight STAT3/NF-κB and WNT/β-catenin as key pathways responsible for TAM-induced CSC stemness. However, the diversity of pathways across many cancers underscores the need to develop context-specific strategies to target them.
CSC-MDSC Crosstalk
The Impact of CSCs on MDSC Biology
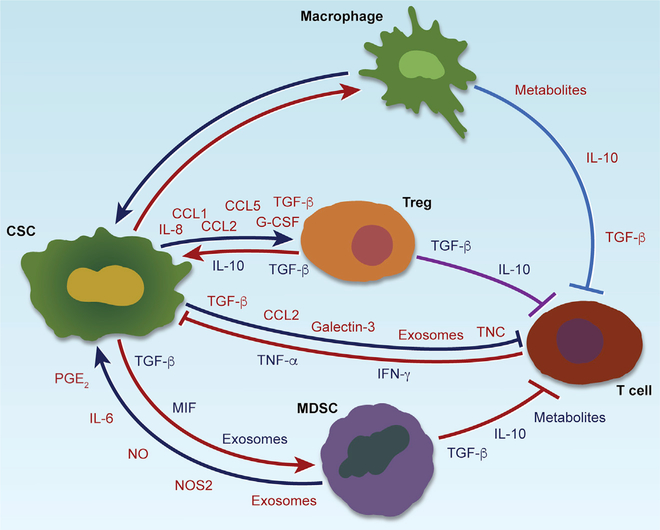
MDSCs are a heterogeneous population of myeloid cells that include granulocytic or polymorphonuclear (PMN-MDSC) and monocytic (M-MDSC) subgroups (Gabrilovich and Nagaraj, 2009). PMN-MDSCs account for more than 80% of all MDSCs, and M-MDSCs can differentiate into TAMs (Gabrilovich, 2017; Kumar et al., 2016). MDSCs are generated in the bone marrow and recruited into tumors by tumor-derived chemokines, such as CCL2 and CCL5. Similar to TAMs, MDSCs play an important role in regulation of tumor angiogenesis, growth, metastasis, and immune suppression (Gabrilovich, 2017; Kumar et al., 2016). Increasing evidence also has revealed symbiotic interactions between CSCs and MDSCs in the TME, where CSCs contribute to MDSC infiltration, expansion, and activation via secretion of soluble factors and exosomes in different cancer types (Figure 4; Table 3). In SCCHN, compared with CD44− cells, CD44+ CSCs secrete higher levels of IL-8, granulocyte-macrophage colony-stimulating factor (GM-CSF), and TGF-β and induce larger populations of MDSCs when they are co-cultured with peripheral blood mononuclear cells (PBMCs) (Chikamatsu et al., 2011). In GBM, GSCs not only promote the differentiation of PBMCs into M-MDSCs via secretion of exosomes (Domenis et al., 2017) but also activate MDSCs to suppress immune responses via secretion of macrophage migration inhibitory factor (MIF) (Otvos et al., 2016). In melanoma, the expression of miR-92 in CD133+ CSCs is reduced when compared to CD133− cells, which upregulates the expression of TGF-β via the α5 integrin/SMAD2 pathway, resulting in more PMN-MDSCs in the TME (Shidal et al., 2019).
Figure 4. CSC-Immune Cell Crosstalk and Interactions among Immune Cells in Cancer.
Different types of CSC-immune cell crosstalk (including CSC-TAM, CSC-MDSC, CSC-T cell, and CSC-T-reg cell) and the immunosuppressive function of TAMs, MDSCs, and T-reg cells on T cells in cancer. Crosstalk between two cell types is achieved by secretion of a variety of chemokines, cytokines, exosomes, or other factors as indicated.
Table 3.
CSC-MDSC Crosstalk
| Roles and Mechanisms of CSCs on MDSC Infiltration and Activation | |||
| Cancer Type | CSC-Derived Factors | Mechanisms | References |
| SCCHN | IL-8, GM-CSF, and TGF-β | not shown | Chikamatsu et al., 2011 |
| GBM | exosomes | directly on monocytes via internalization | Domenis et al., 2017 |
| MIF | Otvos et al., 2016 | ||
| Melanoma | TGF-β | not shown | Shidal et al., 2019 |
| Roles and Mechanisms of MDSCs on CSC Stemness | |||
| Cancer Type | MDSC-Derived Factors | Mechanisms | References |
| Ovarian cancer | unknown | miR-101/CtBP2 pathway | Cui et al., 2013 |
| unknown | GM-CSF/STAT3 pathway | Li et al., 2020 | |
| Ovarian, uterine cervical, and endometrial cancer | PGE2 | not shown |
Komura et al., 2020; Kuroda et al., 2018; Yokoi et al., 2019 |
| Multiple myeloma | unknown | piRNA-823/DNMT3B pathway | Ai et al., 2019 |
| Breast and pancreatic cancer | IL-6 | STAT3 pathway | Panni et al., 2014; Peng et al., 2016 |
| Breast cancer | nitric oxide | NOTCH pathway | Peng et al., 2016 |
| NOS2 | not shown | Ouzounova et al., 2017 | |
| CRC | exosomes | S100A9/STAT3/NF-κB pathway | Wang et al., 2019b |
MDSCs Promote CSC Stemness
Once MDSCs infiltrate into the TME, they can reciprocally promote CSC stemness via distinct mechanisms in a number of cancer types (Figure 4; Table 3). In ovarian cancer, MDSCs induce miR-101 and GM-CSF expression in cancer cells, which increases stemness via upregulation of the corepressor gene C-terminal binding protein-2 (CtBP2) (Cui et al., 2013) and activation of the STAT3 pathway (Li et al., 2020), respectively. In multiple myeloma, PMN-MDSCs trigger the expression of piwi-interacting RNA piRNA-823 in cancer cells, which promotes stemness via activation of DNA methyltransferase 3B (DNMT3B) to facilitate DNA methylation (Ai et al., 2019). Although these data highlight the essential role of MDSCs in promoting cancer cell stemness, the identities of the MDSC-derived factors in these cancer types are still emerging. Of note, STAT3 again appears to be one of the key pathways responsible for stemness. In breast and pancreatic cancers, MDSCs secrete IL-6 (in both cancer types) and nitric oxide (in breast cancer) to activate STAT3 and NOTCH pathways to promote stemness (Panni et al., 2014; Peng et al., 2016). In breast cancer, activated STAT3 promotes activation of NOTCH, which in turn facilitates persistent STAT3 activation, thus creating a feedback loop to reinforce stemness (Peng et al., 2016). In addition to stemness, M-MDSC-derived NOS2 can also promote EMT via activation of the STAT3 pathway, which drives cancer cell dissemination and metastasis (Ouzounova et al., 2017). In CRC, PMN-MDSCs can secrete exosomal S100A9 to promote cancer cell stemness via activation of the STAT3 and NF-κB pathways, which is further amplified under hypoxic conditions (Wang et al., 2019b), establishing NF-κB in MDSC-induced stemness. Indeed, MDSCs are the major source of PGE2 in several types of cancer (e.g., ovarian, cervical, and endometrial cancers; Komura et al., 2020; Kuroda et al., 2018; Yokoi et al., 2019), which can foster cancer cell stemness by activating NF-κB via E-type prostanoid receptor 4 (EP4)-PI3K and EP4-mitogen-activated protein kinase (MAPK) pathways (Wang et al., 2015). Thus, a number of factors and pathways underlie MDSC-CSC interactions, and STAT3 and NF-κB are again very prominent.
CSC-T Cell Crosstalk
Impact of CSCs on T Cell Biology
Computational analyses have also revealed correlations between cancer cell stemness and CD8+ T cells in a broad range of cancer types (Miranda et al., 2019). In SCCHN, GBM, and melanoma, high stemness correlates with low expression of cancer-associated antigens and immune stimulatory molecules (e.g., CD86, CD40, major histocompatibility complex [MHC] II, transporter associated with antigen processing, histocompatibility leukocyte antigen [HLA]-A2, melanoma antigen recognized by T cells 1, melanoma inhibitor of apoptosis, New York esophageal squamous cell carcinoma 1, and melanoma-associated antigen-A) and high expression of immune checkpoint inhibitors (e.g., PD-L1; Chikamatsu et al., 2011; Schatton et al., 2010; Wei et al., 2010). Consistent with these correlates, CSCs regulate the composition and function of T cells via several experimentally validated mechanisms (Figure 4; Table 4). First, in GBM, GSCs produce TGF-β, CCL2, and galectin-3, which suppress CD8+ and CD4+ T cell activation and proliferation (Wei et al., 2010). Second, GSC exosome tenascin-C (TNC) engages α5β1 and α5β6 integrins on T cells to downregulate AKT/mTOR signaling and inhibit T cell activation and proliferation (Domenis et al., 2017; Mirzaei et al., 2018). Finally, in prostate cancer, CSCs secrete TNC to inhibit the activation and proliferation of CD8+ and CD4+ T cells via interaction with α5β1 integrin on T cells (Jachetti et al., 2015).
Table 4.
CSC-T Cell Crosstalk
| Roles and Mechanisms of CSCs in T Cell Infiltration, Proliferation, and Activation | ||||
| Cancer Type | CSC-Derived Factors | T Cell Type | Mechanisms | References |
| GBM | TGF-β, CCL2, and galectin-3 | CD4+/CD8+ T cell and T-reg cell | STAT3 pathway | Wei et al., 2010 |
| exosomes | CD3+/CD4+ T cell | α5β1/αvβ6 integrin/mTOR pathway |
Domenis et al., 2017; Mirzaei et al., 2018 |
|
| Prostate cancer | TNC | CD4+/CD8+ T cell | α5β1 integrin pathway | Jachetti et al., 2015 |
| SCCHN | IL-8, G-CSF, and TGF-β | T cell and T-reg cell | not shown | Chikamatsu et al., 2011 |
| Ovarian cancer | CCL5 | T-reg cell | not shown | You et al., 2018 |
| Breast cancer | CCL1 | T-reg cell | CCL/CCR8 pathway | Xu et al., 2017a |
| Melanoma | unknown | T-reg cell | CD86-dependent pathway | Schatton et al., 2010 |
| TGF-β | T-reg cell | integrin-α/SMAD2 pathway | Shidal et al., 2019 | |
| Roles and Mechanisms of T Cells in CSC Stemness | ||||
| Cancer Type | T-Cell-Derived Factors | T Cell Type | Mechanisms | References |
| GBM | TNF-α and IFN-γ | T cell | not shown | Mirzaei et al., 2018 |
| NSCLC | IFN-γ | CD8+ T cell | PI3K/AKT/NOTCH1 pathway | Song et al., 2019 |
| Breast cancer | unknown | T-reg cell | SOX2/NANOG/OCT4 | Xu et al., 2017a |
| HCC | TGF-β | T-reg cell | EMT |
Shi et al., 2018a; Xu et al., 2009 |
| CRC | TGF-β | T-reg cell | driving dedifferentiation | Nakano et al., 2019 |
| AML | IL-10 | T-reg cell | PI3K/AKT/OCT4/NANOG pathway | Xu et al., 2017b |
| Gastric cancer | IL-17 | Th17 | STAT3 pathway | Jiang et al., 2017 |
| Ovarian and pancreatic cancer | IL-17 | Th17 and CD4+ T cells | p38 MAPK and NF-κB pathways |
Xiang et al., 2015; Zhang et al., 2018 |
T-reg cells are an immunosuppressive subset of CD4+ T cells characterized by expression of forkhead box P3 (FoxP3) and by tumor promotion via inhibition of effector T cells (Togashi et al., 2019). CSCs attract and activate T-reg cells via various soluble factors (Figure 4; Table 4), including, most notably, TGF-β, which controls T-reg cell recruitment and expansion. For example, CSCs can produce high levels of TGF-β in SCCHN (Chikamatsu et al., 2011), GBM (Wei et al., 2010), and melanoma (Shidal et al., 2019), which in turn promotes T-reg cell recruitment and expansion via activation of the α5 integrin/mothers against decapentaplegic homolog 2 (SMAD2) pathway, thus inducing T cell apoptosis and inhibiting T cell proliferation and activation. In addition, several C-C chemokine family members, such as CCL1 (Xu et al., 2017a), CCL2 (Wei et al., 2010), and CCL5 (You et al., 2018), have been shown to be highly produced by CSCs in distinct types of cancer, where they can stimulate the infiltration of T-reg cells into the TME. Together, CSC-secreted TGF-β and specific chemokines play key roles in T-reg cell recruitment and expansion in the TME.
T Cells Regulate CSC Stemness
Emerging evidence demonstrates that different subsets of T cells can regulate CSC stemness (Figure 4; Table 4). In GBM, T-cell-conditioned medium inhibits GSC self-renewal via secretion TNF-α and interferon (IFN)-γ (Mirzaei et al., 2018). In NSCLC, CD8+ T cells are the main sources of IFN-γ, where low levels of IFN-γ promote CSC stemness via activation of the PI3K/AKT/ NOTCH1 pathway and high levels of IFN-γ induce cancer cell apoptosis via activation of the Janus kinase 1 (JAK1)/STAT1/caspase pathway (Song et al., 2019). In pancreatic cancer, infiltrating Th2 cells produce cytokines IL-4 and IL-13 to activate the JAK1/STAT6 pathway in cancer cells, which in turn increases MYC-driven glycolysis (Dey et al., 2020), an anabolic process that supports CSC stemness (Chen et al., 2020b; Sancho et al., 2015). In addition to secretion of soluble factors, T cells can regulate CSC stemness via a direct cell-to-cell contact mechanism in breast cancer where cognate non-lytic interactions between CD8+ T cells and cancer cells can promote cancer cell stemness (Stein et al., 2019).
In addition to effector T cells, T-reg cells and Th17 cells can also regulate stemness. For example, in AML, T-reg cells secrete IL-10 to promote the stemness of leukemic stem cells via activation of the PI3K/AKT/OCT4/NANOG pathway (Xu et al., 2017b). The stemness-promoting effect of T-reg cells is also observed in breast cancer, where unknown T-reg cell soluble factors upregulate stemness-related pathways: SOX2; NANOG; and OCT4 (Xu et al., 2017a). In HCC, T-reg cells secrete TGF-β to support CSC stemness by promoting EMT (Shi et al., 2018a; Xu et al., 2009), whereas in CRC, T-reg-cell-derived TGF-β drives cancer cell dedifferentiation (Nakano et al., 2019), suggesting context-specific actions of TGF-β in CSCs. These observations are consistent with the known highly contextual functions of TGF-β in cancer (Massagué, 2008). Although Th17 is a subset of T helper cells that mediate anti-tumor immune responses (Guéry and Hugues, 2015), IL-17 from Th17 cells or CD4+ T cells also promote CSC stemness through activation of NF-κB and p38 MAPK pathways in ovarian and pancreatic cancers (Xiang et al., 2015; Zhang et al., 2018) and STAT3 pathway in gastric cancer (Jiang et al., 2017). In addition, IL-17 can be upregulated in T-reg cells under hypoxic conditions, which in turn fosters CSC stemness in CRC (Yang et al., 2011). Thus, different T cell subsets contribute to maintenance of CSCs via a variety of mechanisms involving soluble factors and cell-to-cell contact.
Therapeutic Potential of Intercepting CSC-Immune Cell Crosstalk
Targeting CSC-TAM Crosstalk
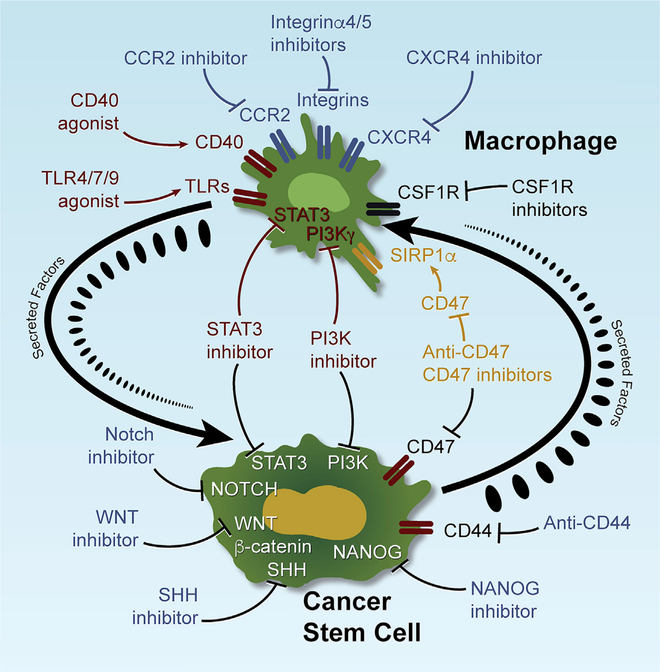
Major preclinical and clinical efforts have sought to target the distinct biologic characteristics and crucial signaling pathways of CSCs and TAMs, as reviewed previously (Agliano et al., 2017; Pathria et al., 2019; Zhao et al., 2018). As summarized in Figure 5, clinical trials that can target CSC biology include inhibitors of the SHH, NOTCH, WNT/β-catenin, STAT3, and NANOG pathways (Agliano et al., 2017; Zhao et al., 2018) and anti-CD44 antibodies (Menke-van der Houven van Oordt et al., 2016). To date, the SHH inhibitor, vismodegib, has been approved for metastatic or locally advanced basal cell carcinoma (Sekulic et al., 2012), and clinical trials are underway for agents targeting macrophage recruitment (CCR2, CXCR4, integrin subunit alpha 4 [ITGA4] and ITGA5 inhibitors), polarization (toll-like receptor 4 [TLR4], TLR7, TLR9 and CD40 activators and PI3Kγ inhibitor), and survival (CSF1R inhibitors; Grégoire et al., 2020; Pathria et al., 2019). However, despite the appeal of CSC-targeting agents, dramatic responses have not been observed, perhaps owing to a lack of truly specific CSC targets (Agliano et al., 2017; Turdo et al., 2019) as well as the high plasticity of CSC, which enables loss and reacquisition of stemness under varying TME conditions (Agliano et al., 2017; Müller et al., 2020; Plaks et al., 2015). Similarly, TAM-targeted therapies, such as CSF1R inhibition, have shown meager anti-tumor responses in GBM due in part to resistance conferred by activated PI3K signaling in glioma cells (Quail et al., 2016).
Figure 5. CSC and TAM Targeting Strategies in Cancer Therapy.
Preclinical studies with mice have identified critical pathways that regulate CSC stemness and TAM biology (including recruitment, polarization, survival, and phagocytosis) during tumor progression. Targeting these key pathways can inhibit the properties of CSCs and TAMs and impair tumor progression. In addition to the pathways targeting either CSCs or TAMs, several pathways, including STAT3, PI3K, and CD47-SIRP1α, are involved in regulating the properties of both CSCs and TAMs, thus providing more-effective therapeutic strategies. Given the existence of CSC-TAM crosstalk during tumor progression, targeting CSC-TAM co-dependency is another promising cancer therapy strategy.
It is tempting to speculate that targeting more deliberately the entwined co-dependencies of CSCs and TAMs and their plasticity in specific contexts could yield more robust responses. For example, IL-6/STAT3 and PI3K are essential for the regulation of both CSC stemness and macrophage pro-tumor polarization in many tumor types; inhibition of these pathways has shown anti-tumor activity (Agliano et al., 2017; Kobatake et al., 2020; Pathria et al., 2019; Weng et al., 2019). Moreover, selecting patients with high CSC and TAM signatures could enhance responses to agents targeting these pathways (Agliano et al., 2017; Pathria et al., 2019). Such trials could benefit further from pharmacodynamic assessment of whether these agents can modulate these pathways and their associated tumor biology (Figure 5). Finally, given the plasticity of this system, these pharmacodynamic studies should be complemented by integrated omic analyses of the adaptive responses to these therapeutic interventions, which may further inform combination trials of synergistic agents.
An appealing strategy to disrupt CSC-TAM crosstalk may include blockade of the CD47-signal regulatory protein alpha (SIRPα) pathway. CD47 is a transmembrane protein expressed on CSCs and cancer cells that functions as a “don’t eat me” signal (Cioffi et al., 2015); interaction of CD47 with SIRPα on macrophages results in inhibition of phagocytosis by TAMs (Matozaki et al., 2009). Anti-CD47 therapy increases CSC phagocytosis in vitro and decreases tumor burden in vivo (Chan et al., 2009; Cioffi et al., 2015; Majeti et al., 2009), which are further augmented when this therapy is combined with chemotherapy (Cioffi et al., 2015). Several clinical trials testing monoclonal anti-CD47 antibodies (Hu5F9-G4, SFR231, CC-90002, and IBI188) and small-molecule inhibitors (TTI-621 and ALX148) are underway (Figure 5; Grégoire et al., 2020; Pathria et al., 2019). Another opportunity to disrupt CSC-TAM crosstalk centers on targeting soluble factors that reciprocally support each cell type. For example, inhibition of the CSC-specific POSTN and its related pathway (Zhou et al., 2015) or TAM-derived CCL5 (Huang et al., 2020) have been shown to interrupt CSC-TAM crosstalk, suppress tumor growth, and extend survival in mouse models of GBM and prostate cancer.
On a more conventional note, standard of care chemotherapies have shown limited promise for advanced metastatic disease due to severe toxicity and rapid development of resistance. As CSCs and TAMs play critical roles in the development of drug resistance (Agliano et al., 2017; De Palma and Lewis, 2013), disruption of CSC-TAM crosstalk could improve its response to chemotherapy. Along these lines, CSCs in chemoresistant tumors secrete cytokines to create a pro-tumorigenic microenvironment by skewing macrophages toward a pro-tumor phenotype (Yamashina et al., 2014). Indeed, depletion of TAMs reduces CSC stemness, inhibits metastasis, and improves chemotherapeutic responsiveness in pancreatic cancer (Mitchem et al., 2013). Mechanistically, TAMs promote CSC stemness and chemoresistance via release of MFG-E8, which can trigger activation of STAT3 and SHH pathways in CSCs of CRC (Jinushi et al., 2011). Thus, mounting evidence points to the potential of targeting the CSC-TAM circuits for novel cancer treatments as well as for enhancement of chemotherapy effectiveness.
Targeting CSC-MDSC Crosstalk
The importance of MDSCs in promoting tumor growth, metastasis, angiogenesis, CSC stemness, and immune suppression has motivated the testing agents that inhibit MDSCs (Fleming et al., 2018). MDSC inhibition strategies target recruitment (e.g., inhibition of CCR5 and CXCR2), promote depletion (e.g., tyrosine-kinase inhibitors and chemotherapeutic agents), and block immunosuppressive activity (e.g., inhibition of STAT3, phosphodiesterase-5, and class I histone deacetylases; Fleming et al., 2018). However, the development of MDSC-targeted therapies is hampered by heterogeneity of MDSCs and lack of cellular markers (Lu et al., 2019). That is, MDSCs are heterogeneous immature myeloid cells composed of PMN-MDSCs and M-MDSCs, which possess distinct biological functions. Current MDSC-targeted agents may target both MDSC subgroups and other cell types in the TME. In addition, the lack of specific markers for human MDSCs and identification of the equivalent murine MDSCs has impeded translational research. Finally, MDSC density and activation states can change dynamically in the TME.
Notwithstanding these challenges, several strategies targeting CSC-MDSC crosstalk are worth considering. One strategy would be to target STAT3, which is dually essential for CSC maintenance and MDSC infiltration/activation, and inhibition of STAT3 has demonstrated potent anti-tumor activity associated with impaired CSC stemness and MDSC infiltration/activation in cancer mouse models (Fleming et al., 2018; Peng et al., 2016). A second strategy would be to target soluble factors fostering CSC-MDSC crosstalk. For example, inhibition of CSC-derived MIF or MDSC-derived IL-6 extends survival in mouse models of GBM (Otvos et al., 2016) and breast cancer (Peng et al., 2016). A third strategy would involve neutralization of exosome-stimulated CSC-MDSC symbiosis. Specifically, knockdown of S100A9 in MDSC exosomes impairs STAT3 activation and inhibits tumor growth in mouse models of CRC (Wang et al., 2019b). Together, these various mechanistic insights point to disruption of the IL-6/STAT3 pathway as a strategy to interfere with CSC-MDSC crosstalk and inhibit tumor growth.
Targeting CSC-T Cell Crosstalk
The symbiotic interactions between CSCs and T cells may also offer several precision therapeutic strategies. First, in breast cancer, blockade of CSC-derived T-reg cell supporting factors, such as CCL1, has been shown to significantly inhibit tumor growth and T-reg cell infiltration (Xu et al., 2017a). A similar anti-tumor effect has been observed in a mouse model of pancreatic cancer by inhibition of T-cell-derived stemness supporting factors, such as Th17 cell-derived IL-17, which dramatically impaired tumor growth and CSC stemness (Xiang et al., 2015). The second approach is to harness the potential of T-cell-based immunotherapies, especially immune checkpoint inhibition (ICI). The anti-tumor effectiveness of ICIs relates to the expression of immune checkpoint molecules, such as PD-L1, in the TME (Ravindran et al., 2019). Following activation of the STAT3 and NOTCH3/mTOR pathways (Lee et al., 2016; Mansour et al., 2020), CSCs express higher PD-L1 level compared to non-CSCs in many cancer types, including GBM, melanoma, SCCHN, CRC, breast cancer, gastric cancer, and ovarian cancer, in which PD-L1 can further promote CSC stemness, thus inducing a positive feedback loop (Gao et al., 2019; Gupta et al., 2016; Ravindran et al., 2019; Wei et al., 2019a). In addition, PD-L1 level can be amplified following symbiotic CSC-immune cell interactions. For example, MDSCs can promote stemness and upregulate PD-L1 in CSCs via activation of the PI3K/AKT/mTOR pathway (Komura et al., 2020). Consequently, CSCs secrete exosomes to upregulate PD-L1 in macrophages via activation of the STAT3 pathway (Gabrusiewicz et al., 2018). Together, these findings point to the potential utility of ICI agents, a concept supported by the anti-PD1 therapy enhancement of the anti-tumor activity of a CSC vaccine in a mouse model of bladder cancer (Shi et al., 2018b). The third approach would be the development of combination therapies targeting CSC-immune cell crosstalk and immune checkpoints. ICIs produce remarkable responses in some cancer patients; however, the majority of patients do not have responses. Mechanistic studies have shown that the effectiveness of ICIs is highly dependent on the TME (Murciano-Goroff et al., 2020). TAMs, MDSCs, and T-reg cells are the most prominent immune cells in the TME, where they form symbiotic interactions with CSCs, interact with each other, and suppress T cell function (Figure 4). Mechanistically, TAMs and MDSCs can suppress T-cell-mediated anti-tumor immune responses by high expression of immune checkpoint molecules (e.g., PD-L1, PD-L2, CD80, and CD86), production of immunosuppressive cytokines (e.g., IL-10 and TGF-β), and recruitment of immunosuppressive T-reg cells into the TME (Engblom et al., 2016; Kumar et al., 2016; Mantovani et al., 2017). These studies highlight the promise of TAMor MDSC-targeted therapies for improved ICI effectiveness. Indeed, a growing body of evidence demonstrates that macrophage-targeted therapies, such as activation of macrophage phagocytosis (Lian et al., 2019; Liu et al., 2018) or reprogramming of TAMs from pro- to anti-tumor phenotype (Baer et al., 2016; Guerriero et al., 2017; Kaneda et al., 2016; Zhu et al., 2014), synergize with ICIs in multiple cancer mouse models. Similarly, MDSC-targeted therapies, by inhibiting MDSC infiltration (Flores-Toro et al., 2020; Highfill et al., 2014; Liao et al., 2019; Zhao et al., 2020) or blocking MDSC activation (Davis et al., 2017; Liu et al., 2020; Lu et al., 2017), show robust synergy with ICIs in mouse models. These preclinical studies have prompted combination therapeutic trials for many cancer types (Hou et al., 2020; Pathria et al., 2019).
Concluding Remarks and Future Perspectives
The genetic paradigm has dominated our approach to cancer therapy, generating many agents targeting driver oncogenic events in cancer cells. In recent years, the success of targeting immunity and angiogenesis has heightened interest in targets operating within the TME ecosystem. This review specifically has cataloged the molecular circuitry underlying reciprocal interactions between CSCs and immune cells, including TAMs, MDSCs, and T cells, in tumor maintenance. This bidirectional crosstalk is manifested on several levels, including CSC-directed immune cell recruitment and activation and the role of these immune cells in promoting cancer cell stemness and establishing a supportive CSC niche. The molecular characterization of CSC-immune cell symbiosis has uncovered potential therapeutic strategies, including dual targeting of vital pathways activated in both CSCs and immune cells (e.g., STAT3 and PI3K), disrupting the molecules responsible for physical CSC-immune cell interactions (e.g., CD47-SIRPα), and neutralizing soluble factors that reciprocally support both CSCs and immune cells (e.g., IL-6). Collectively, elucidation of these symbiotic CSC-immune cell interactions has also revealed the centrality of these novel molecular mechanisms in driving tumorigenesis, metastasis, and chemotherapy resistance. Thus, targeting of this molecular circuit has the potential to disrupt CSC-immune cell co-dependencies and enhance the effectiveness of conventional therapies.
Although our knowledge of CSC-immune cell crosstalk is maturing, multiple questions will need to be answered in order to effectively and systematically convert mechanistic insights into new therapeutic interventions. First, many studies investigating CSC-immune cell crosstalk have relied on cell line co-culture models or isolated cells from tumor tissues, which highlights the need for complementary studies using in vivo models, genetic validation, and dynamic analyses of the TME using lineage tracing and live micro positron emission tomography (microPET)/computed tomography (CT) imaging technologies. Such in vivo models could be complemented by organoid cultures, which appear to more faithfully recapitulate the features of their source tissues (Baker, 2018). That is, cancer cell organoids and immune cell co-cultures could serve as more robust and higher throughput model systems to study the dynamic and reciprocal interactions between CSCs and immune cells and to test therapeutic agents targeting CSC-immune cell crosstalk. Second, the remarkable plasticity and heterogeneity of both CSCs (transitioning between stem versus non-stem states) and immune cells (including the transitions within and across cell types, such as the transition across the phenotypic spectrum in TAMs, differentiation of MDSC to PMN-MDSC and M-MDSC subgroups, and differentiation of M-MDSCs to TAMs) highlight the challenges in identifying the context-specific nature of distinct critical CSC-immune cell circuits at different tumor stages and in different cancer types, as well as changes in CSC-immune cell interactions resulting from therapeutic interventions. Thus, harnessing the full potential of targeting CSC-immune cell crosstalk will require extensive use of scRNAseq to identify new subpopulations and define the physiological states of CSCs and immune cells, as well as their crosstalk in specific tumor contexts and under exposure to certain therapies. Such single-cell auditing must be complemented by functional and genetic analyses using in vivo model systems to identify and validate targets and mechanisms governing CSC-immune cell co-dependencies. Given the number of factors involved, bispecific antibodies dually targeting key factors acting in concert in the CSC-immune cell circuit should be considered. Third, across many tumor types, the IL-6/IL-6R/STAT3 pathway appears to be the most prominent and important driver of CSC-immune cell crosstalk, as evidenced by the finding that pharmacological inhibition of the IL-6R/STAT3 pathway impairs tumor progression and reduces CSC stemness, TAMs, and MDSCs in bladder cancer, breast cancer, and HCC mouse models (Kobatake et al., 2020; Peng et al., 2016; Wan et al., 2014). However, a more-detailed investigation of the actions of these drugs is needed, as they also target other stromal cells in the TME. That is, the anti-tumor actions may not relate to CSC-immune cell crosstalk and/or may target stromal cells with opposing actions to CSCs, TAMs, and MDSCs. In this regard, genetically engineered mouse models would be useful to more precisely dissect the myriad roles of the IL-6/IL-6R/STAT3 pathway specifically in CSCs, TAMs, MDSCs, and/or T cells versus other cells within the TME ecosystem. Finally, in addition to TAMs, MDSCs, and T cells, unbiased analyses on TGGA datasets demonstrated that high cancer cell stemness is associated with reduced NK cells (Miranda et al., 2019), suggesting a potential CSC-NK cell crosstalk. CSCs are generally susceptible to killing by activated NK cells; however, a growing body of evidence shows that CSCs may be resistant to NK cells in some cancer types, such as GBM, AML, and breast cancer (Sultan et al., 2017). Emerging evidence demonstrates that the anti-CSC activity of NK cells is largely dependent on the TME and that NK cell activation can be suppressed by TAMs, MDSCs, and T-reg cells (Bruno et al., 2019; Ghiringhelli et al., 2006; Krneta et al., 2017). In addition to NK cells, very limited evidence demonstrates that cancer cell stemness is related to the presence of dendritic cells (DCs), B cells (Hsu et al., 2018; Miranda et al., 2019), and neutrophils (Hira et al., 2015; Hwang et al., 2019). However, the nature of the crosstalk between CSCs and these four types of immune cells (NK cells, B cells, DCs, and neutrophils) is largely unknown. Therefore, further studies characterizing such crosstalk, as well as the relationship of these four cell types with other immune cells, including TAMs, MDSCs, and T cells, will pave the way for developing novel and more effective immunotherapies.
In summary, we have presented mounting evidence implicating CSC-immune cell interactions as drivers of tumor development involving many hallmarks of cancer and as modulators of the response to therapeutic interventions. Harnessing the therapeutic potential of these interactions will require rigorous validation of the targets and mechanisms underlying this symbiotic relationship as well as a deeper understanding of the specific biological contexts in which they play essential rate-limiting roles in tumor maintenance. Successful achievement of this goal would greatly benefit cancer patients.
ACKNOWLEDGMENTS
We thank Drs. Denise Spring, Raghu Kalluri, Linghua Wang, Shabnam Shalapour, Jian Hu, Kyle LaBella, and Deepavali Chakravarti, as well as Scientific Publications, Research Medical Library (Donald R. Norwood), and Creative Communications (David M. Aten) for their help and advice. This work was supported in part by NIH R00 CA240896 (to P.C.), NIH P50CA221747 (to P.C.), the Cancer Research Institute Irvington Postdoctoral Fellowship (to P.C.), the Harter Prize (to P.C.), the Caroline Ross Endowed Fellowship (to P.C.), The Harold C. and Mary L. Daily Endowment Fellowship (to P.C.), the Burkhart III Distinguished University Chair in Cancer Research Endowment (to R.A.D.), NIH P01 CA117969 (to R.A.D.), NIH R01 CA225955 (to R.A.D.), and NIH R01 CA231360 (to R.A.D.).
Footnotes
DECLARATION OF INTERESTS
R.A.D. is a co-founder, advisor, and director of Tvardi Therapeutics focused on the development of STAT3 inhibitors. R.A.D. is also co-founder and advisor of Asylia Therapeutics, Nirogy Therapeutics, and Stellanova Therapeutics. The other authors declare no competing interests.
REFERENCES
- Agliano A, Calvo A, and Box C (2017). The challenge of targeting cancer stem cells to halt metastasis. Semin. Cancer Biol 44, 25–42. [DOI] [PubMed] [Google Scholar]
- Ai L, Mu S, Sun C, Fan F, Yan H, Qin Y, Cui G, Wang Y, Guo T, Mei H, et al. (2019). Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol. Cancer 18, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Z, Knobbe-Thomsen CB, Wan X, Fan QW, Reifenberger G, and Weiss WA (2018). EGFR cooperates with EGFRvIII to recruit macrophages in glioblastoma. Cancer Res. 78, 6785–6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayob AZ, and Ramasamy TS (2018). Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci 25, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer C, Squadrito ML, Laoui D, Thompson D, Hansen SK, Kiialainen A, Hoves S, Ries CH, Ooi CH, and De Palma M (2016). Suppression of microRNA activity amplifies IFN-γ-induced macrophage activation and promotes anti-tumour immunity. Nat. Cell Biol 18, 790–802. [DOI] [PubMed] [Google Scholar]
- Baker K (2018). Organoids provide an important window on inflammation in cancer. Cancers (Basel) 10, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, and Weinberg RA (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet 40, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle A, and Mackenzie IC (2012). Cancer stem cells and EMT in carcinoma. Cancer Metastasis Rev. Published online February 3, 2012. 10.1007/s10555-012-9345-0. [DOI] [PubMed] [Google Scholar]
- Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, et al. (2014). SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511, 246–250. [DOI] [PubMed] [Google Scholar]
- Bruno A, Mortara L, Baci D, Noonan DM, and Albini A (2019). Myeloid derived suppressor cells interactions with natural killer cells and pro-angiogenic activities: roles in tumor progression. Front. Immunol 10, 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S, et al. (2017). CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KS, Espinosa I, Chao M, Wong D, Ailles L, Diehn M, Gill H, Presti J Jr., Chang HY, van de Rijn M, et al. (2009). Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc. Natl. Acad. Sci. USA 106, 14016–14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TS, Shaked Y, and Tsai KK (2019). Targeting the interplay between cancer fibroblasts, mesenchymal stem cells, and cancer stem cells in desmoplastic cancers. Front. Oncol 9, 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zuo H, Xiong H, Kolar MJ, Chu Q, Saghatelian A, Siegwart DJ, and Wan Y (2017). Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis. Proc. Natl. Acad. Sci. USA 114, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zhao D, Li J, Liang X, Li J, Chang A, Henry VK, Lan Z, Spring DJ, Rao G, et al. (2019a). Symbiotic macrophage-glioma cell interactions reveal synthetic lethality in PTEN-null glioma. Cancer Cell 35, 868–884.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wen H, Zhou C, Su Q, Lin Y, Xie Y, Huang Y, Qiu Q, Lin J, Huang X, et al. (2019b). TNF-α derived from M2 tumor-associated macrophages promotes epithelial-mesenchymal transition and cancer stemness through the Wnt/β-catenin pathway in SMMC-7721 hepatocellular carcinoma cells. Exp. Cell Res 378, 41–50. [DOI] [PubMed] [Google Scholar]
- Chen L, Yang QC, Li YC, Yang LL, Liu JF, Li H, Xiao Y, Bu LL, Zhang WF, and Sun ZJ (2020a). Targeting CMTM6 suppresses stem cell-like properties and enhances antitumor immunity in head and neck squamous cell carcinoma. Cancer Immunol. Res 8, 179–191. [DOI] [PubMed] [Google Scholar]
- Chen P, Hsu WH, Chang A, Tan Z, Lan Z, Zhou A, Spring DJ, Lang FF, Wang YA, and DePinho RA (2020b). Circadian regulator CLOCK recruits immune-suppressive microglia into the GBM tumor microenvironment. Cancer Discov. 10, 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Huang Z, Zhou W, Wu Q, Donnola S, Liu JK, Fang X, Sloan AE, Mao Y, Lathia JD, et al. (2013). Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell 153, 139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia K, Mazzolini J, Mione M, and Sieger D (2018). Tumor initiating cells induce Cxcr4-mediated infiltration of pro-tumoral macrophages into the brain. eLife 7, e31918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikamatsu K, Takahashi G, Sakakura K, Ferrone S, and Masuyama K (2011). Immunoregulatory properties of CD44+ cancer stem-like cells in squamous cell carcinoma of the head and neck. Head Neck 33, 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi M, Trabulo S, Hidalgo M, Costello E, Greenhalf W, Erkan M, Kleeff J, Sainz B Jr., and Heeschen C (2015). Inhibition of CD47 effectively targets pancreatic cancer stem cells via dual mechanisms. Clin. Cancer Res 21, 2325–2337. [DOI] [PubMed] [Google Scholar]
- Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, et al. (2014). Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui TX, Kryczek I, Zhao L, Zhao E, Kuick R, Roh MH, Vatan L, Szeliga W, Mao Y, Thomas DG, et al. (2013). Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity 39, 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, Moore EC, Clavijo PE, Friedman J, Cash H, Chen Z, Silvin C, Van Waes C, and Allen C (2017). Anti-PD-L1 efficacy can be enhanced by inhibition of myeloid-derived suppressor cells with a selective inhibitor of PI3Kδ/γ. Cancer Res. 77, 2607–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma M, and Lewis CE (2013). Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell 23, 277–286. [DOI] [PubMed] [Google Scholar]
- de Sousa E Melo F, and Vermeulen L (2016). Wnt signaling in cancer stem cell biology. Cancers (Basel) 8, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H, et al. (2017). A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680. [DOI] [PubMed] [Google Scholar]
- Deng X, Zhang P, Liang T, Deng S, Chen X, and Zhu L (2015). Ovarian cancer stem cells induce the M2 polarization of macrophages through the PPARγ and NF-κB pathways. Int. J. Mol. Med 36, 449–454. [DOI] [PubMed] [Google Scholar]
- Dey P, Li J, Zhang J, Chaurasiya S, Strom A, Wang H, Liao WT, Cavallaro F, Denz P, Bernard V, et al. (2020). Oncogenic KRAS-driven metabolic reprogramming in pancreatic cancer cells utilizes cytokines from the tumor microenvironment. Cancer Discov. 10, 608–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenis R, Cesselli D, Toffoletto B, Bourkoula E, Caponnetto F, Manini I, Beltrami AP, Ius T, Skrap M, Di Loreto C, and Gri G (2017). Systemic T cells immunosuppression of glioma stem cell-derived exosomes is mediated by monocytic myeloid-derived suppressor cells. PLoS ONE 12, e0169932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongre A, and Weinberg RA (2019). New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol 20, 69–84. [DOI] [PubMed] [Google Scholar]
- Engblom C, Pfirschke C, and Pittet MJ (2016). The role of myeloid cells in cancer therapies. Nat. Rev. Cancer 16, 447–462. [DOI] [PubMed] [Google Scholar]
- Fan QM, Jing YY, Yu GF, Kou XR, Ye F, Gao L, Li R, Zhao QD, Yang Y, Lu ZH, and Wei LX (2014). Tumor-associated macrophages promote cancer stem cell-like properties via transforming growth factor-beta1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 352, 160–168. [DOI] [PubMed] [Google Scholar]
- Fang M, Li Y, Huang K, Qi S, Zhang J, Zgodzinski W, Majewski M, Wallner G, Gozdz S, Macek P, et al. (2017). IL33 promotes colon cancer cell stemness via JNK activation and macrophage recruitment. Cancer Res. 77, 2735–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming V, Hu X, Weber R, Nagibin V, Groth C, Altevogt P, Utikal J, and Umansky V (2018). Targeting myeloid-derived suppressor cells to bypass tumor-induced immunosuppression. Front. Immunol 9, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Toro JA, Luo D, Gopinath A, Sarkisian MR, Campbell JJ, Charo IF, Singh R, Schall TJ, Datta M, Jain RK, et al. (2020). CCR2 inhibition reduces tumor myeloid cells and unmasks a checkpoint inhibitor effect to slow progression of resistant murine gliomas. Proc. Natl. Acad. Sci. USA 117, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI (2017). Myeloid-derived suppressor cells. Cancer Immunol. Res 5, 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI, and Nagaraj S (2009). Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol 9, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrusiewicz K, Li X, Wei J, Hashimoto Y, Marisetty AL, Ott M, Wang F, Hawke D, Yu J, Healy LM, et al. (2018). Glioblastoma stem cell-derived exosomes induce M2 macrophages and PD-L1 expression on human monocytes. OncoImmunology 7, e1412909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galoczova M, Coates P, and Vojtesek B (2018). STAT3, stem cells, cancer stem cells and p63. Cell. Mol. Biol. Lett 23, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Guo Q, Li X, Yang X, Ni H, Wang T, Zhao Q, Liu H, Xing Y, Xi T, and Zheng L (2019). MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine 41, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiringhelli F, Ménard C, Martin F, and Zitvogel L (2006). The role of regulatory T cells in the control of natural killer cells: relevance during tumor progression. Immunol. Rev 214, 229–238. [DOI] [PubMed] [Google Scholar]
- Grégoire H, Roncali L, Rousseau A, Chérel M, Delneste Y, Jeannin P, Hindré F, and Garcion E (2020). Targeting tumor associated macrophages to overcome conventional treatment resistance in glioblastoma. Front. Pharmacol 11, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero JL, Sotayo A, Ponichtera HE, Castrillon JA, Pourzia AL, Schad S, Johnson SF, Carrasco RD, Lazo S, Bronson RT, et al. (2017). Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature 543, 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guéry L, and Hugues S (2015). Th17 cell plasticity and functions in cancer immunity. BioMed Res. Int 2015, 314620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhao Y, Yan H, Yang Y, Shen S, Dai X, Ji X, Ji F, Gong XG, Li L, et al. (2017). Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 31, 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Cheng X, Chen H, Chen C, Xie S, Zhao M, Liu D, Deng Q, Liu Y, Wang X, et al. (2019a). Induction of breast cancer stem cells by M1 macrophages through Lin-28B-let-7-HMGA2 axis. Cancer Lett. 452, 213–225. [DOI] [PubMed] [Google Scholar]
- Guo X, Pan Y, and Gutmann DH (2019b). Genetic and genomic alterations differentially dictate low-grade glioma growth through cancer stem cell-specific chemokine recruitment of T cells and microglia. Neuro-oncol. 21, 1250–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta HB, Clark CA, Yuan B, Sareddy G, Pandeswara S, Padron AS, Hurez V, Conejo-Garcia J, Vadlamudi R, Li R, and Curiel TJ (2016). Tumor cell-intrinsic PD-L1 promotes tumor-initiating cell generation and functions in melanoma and ovarian cancer. Signal Transduct. Target. Ther 1, 16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Qu Y, Jin Y, Yu Y, Deng N, Wawrowsky K, Zhang X, Li N, Bose S, Wang Q, et al. (2015). FOXC1 activates Smoothened-independent hedgehog signaling in basal-like breast cancer. Cell Rep. 13, 1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatina J (2012). The dynamics of cancer stem cells. Neoplasma 59, 700–707. [DOI] [PubMed] [Google Scholar]
- Hide T, Komohara Y, Miyasato Y, Nakamura H, Makino K, Takeya M, Kuratsu JI, Mukasa A, and Yano S (2018). Oligodendrocyte progenitor cells and macrophages/microglia produce glioma stem cell niches at the tumor border. EBioMedicine 30, 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, Kaplan RN, and Mackall CL (2014). Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med 6, 237ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira VVV, Ploegmakers KJ, Grevers F, Verbovšek U, Silvestre-Roig C, Aronica E, Tigchelaar W, Turnšek TL, Molenaar RJ, and Van Noorden CJF (2015). CD133+ and Nestin+ glioma stem-like cells reside around CD31+ arterioles in niches that express SDF-1α, CXCR4, osteopontin and cathepsin K. J. Histochem. Cytochem 63, 481–493. [DOI] [PubMed] [Google Scholar]
- Hirata N, Yamada S, Shoda T, Kurihara M, Sekino Y, and Kanda Y (2014). Sphingosine-1-phosphate promotes expansion of cancer stem cells via S1PR3 by a ligand-independent Notch activation. Nat. Commun 5, 4806. [DOI] [PubMed] [Google Scholar]
- Hou YC, Chao YJ, Hsieh MH, Tung HL, Wang HC, and Shan YS (2019). Low CD8+ T cell infiltration and high PD-L1 expression are associated with level of CD44+/CD133+ cancer stem cells and predict an unfavorable prognosis in pancreatic cancer. Cancers (Basel) 11, 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou A, Hou K, Huang Q, Lei Y, and Chen W (2020). Targeting myeloid-derived suppressor cell, a promising strategy to overcome resistance to immune checkpoint inhibitors. Front. Immunol 11, 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YL, Chen YJ, Chang WA, Jian SF, Fan HL, Wang JY, and Kuo PL (2018). Interaction between tumor-associated dendritic cells and colon cancer cells contributes to tumor progression via CXCL1. Int. J. Mol. Sci 19, 2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wang Q, Wang YA, Hua S, Sauvé CG, Ong D, Lan ZD, Chang Q, Ho YW, Monasterio MM, et al. (2016). Epigenetic activation of WNT5A drives glioblastoma stem cell differentiation and invasive growth. Cell 167, 1281–1295.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Wang S, Wang N, Zheng Y, Zhou J, Yang B, Wang X, Zhang J, Guo L, Wang S, et al. (2020). CCL5 derived from tumor-associated macrophages promotes prostate cancer stem cells and metastasis via activating β-catenin/STAT3 signaling. Cell Death Dis. 11, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WL, Lan HY, Cheng WC, Huang SC, and Yang MH (2019). Tumor stem-like cell-derived exosomal RNAs prime neutrophils for facilitating tumorigenesis of colon cancer. J. Hematol. Oncol 12, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachetti E, Caputo S, Mazzoleni S, Brambillasca CS, Parigi SM, Grioni M, Piras IS, Restuccia U, Calcinotto A, Freschi M, et al. (2015). Tenascin-C protects cancer stem-like cells from immune surveillance by arresting T-cell activation. Cancer Res. 75, 2095–2108. [DOI] [PubMed] [Google Scholar]
- Jiang YX, Yang SW, Li PA, Luo X, Li ZY, Hao YX, and Yu PW (2017). The promotion of the transformation of quiescent gastric cancer stem cells by IL-17 and the underlying mechanisms. Oncogene 36, 1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinushi M, Chiba S, Yoshiyama H, Masutomi K, Kinoshita I, Dosaka-Akita H, Yagita H, Takaoka A, and Tahara H (2011). Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc. Natl. Acad. Sci. USA 108, 12425–12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S, Woo G, Nguyen AV, Figueiredo CC, Foubert P, et al. (2016). PI3Kγ is a molecular switch that controls immune suppression. Nature 539, 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobatake K, Ikeda K-I, Nakata Y, Yamasaki N, Ueda T, Kanai A, Sentani K, Sera Y, Hayashi T, Koizumi M, et al. (2020). Kdm6a deficiency activates inflammatory pathways, promotes M2 macrophage polarization, and causes bladder cancer in cooperation with p53 dysfunction. Clin. Cancer Res 26, 2065–2079. [DOI] [PubMed] [Google Scholar]
- Kodama H, Murata S, Ishida M, Yamamoto H, Yamaguchi T, Kaida S, Miyake T, Takebayashi K, Kushima R, and Tani M (2017). Prognostic impact of CD44-positive cancer stem-like cells at the invasive front of gastric cancer. Br. J. Cancer 116, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura N, Mabuchi S, Shimura K, Yokoi E, Kozasa K, Kuroda H, Takahashi R, Sasano T, Kawano M, Matsumoto Y, et al. (2020). The role of myeloid-derived suppressor cells in increasing cancer stem-like cells and promoting PD-L1 expression in epithelial ovarian cancer. Cancer Immunol. Immunother 69, 2477–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krneta T, Gillgrass A, Poznanski S, Chew M, Lee AJ, Kolb M, and Ashkar AA (2017). M2-polarized and tumor-associated macrophages alter NK cell phenotype and function in a contact-dependent manner. J. Leukoc. Biol 101, 285–295. [DOI] [PubMed] [Google Scholar]
- Kumar V, Patel S, Tcyganov E, and Gabrilovich DI (2016). The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 37, 208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Mabuchi S, Yokoi E, Komura N, Kozasa K, Matsumoto Y, Kawano M, Takahashi R, Sasano T, Shimura K, et al. (2018). Prostaglandin E2 produced by myeloid-derived suppressive cells induces cancer stem cells in uterine cervical cancer. Oncotarget 9, 36317–36330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, and Dick JE (1994). A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648. [DOI] [PubMed] [Google Scholar]
- Lee Y, Shin JH, Longmire M, Wang H, Kohrt HE, Chang HY, and Sunwoo JB (2016). CD44+ cells in head and neck squamous cell carcinoma suppress T-cell-mediated immunity by selective constitutive and inducible expression of PD-L1. Clin. Cancer Res 22, 3571–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, Reinhardt F, Herschman HR, and Weinberg RA (2012). Cancerstimulated mesenchymal stem cells create a carcinoma stem cell niche via prostaglandin E2 signaling. Cancer Discov. 2, 840–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XF, Chen C, Xiang DM, Qu L, Sun W, Lu XY, Zhou TF, Chen SZ, Ning BF, Cheng Z, et al. (2017). Chronic inflammation-elicited liver progenitor cell conversion to liver cancer stem cell with clinical significance. Hepatology 66, 1934–1951. [DOI] [PubMed] [Google Scholar]
- Li X, Bu W, Meng L, Liu X, Wang S, Jiang L, Ren M, Fan Y, and Sun H (2019). CXCL12/CXCR4 pathway orchestrates CSC-like properties by CAF recruited tumor associated macrophage in OSCC. Exp. Cell Res 378, 131–138. [DOI] [PubMed] [Google Scholar]
- Li X, Wang J, Wu W, Gao H, Liu N, Zhan G, Li L, Han L, and Guo X (2020). Myeloid-derived suppressor cells promote epithelial ovarian cancer cell stemness by inducing the CSF2/p-STAT3 signalling pathway. FEBS J. 287, 5218–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian S, Xie R, Ye Y, Lu Y, Cheng Y, Xie X, Li S, and Jia L (2019). Dual blockage of both PD-L1 and CD47 enhances immunotherapy against circulating tumor cells. Sci. Rep 9, 4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, Li J, Wang G, Lan Z, Li J, et al. (2019). KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell 35, 559–572.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Liu L, Ren Z, Yang K, Xu H, Luan Y, Fu K, Guo J, Peng H, Zhu M, and Fu YX (2018). Dual targeting of innate and adaptive checkpoints on tumor cells limits immune evasion. Cell Rep. 24, 2101–2111. [DOI] [PubMed] [Google Scholar]
- Liu D, Lu Q, Wang X, Wang J, Lu N, Jiang Z, Hao X, Li J, Liu J, Cao P, et al. (2019). LSECtin on tumor-associated macrophages enhances breast cancer stemness via interaction with its receptor BTN3A3. Cell Res. 29, 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhou J, Liu X, Feng Y, Yang W, Wu F, Cheung OK, Sun H, Zeng X, Tang W, et al. (2020). Targeting monocyte-intrinsic enhancer reprogramming improves immunotherapy efficacy in hepatocellular carcinoma. Gut 69, 365–379. [DOI] [PubMed] [Google Scholar]
- Lu H, Clauser KR, Tam WL, Fröse J, Ye X, Eaton EN, Reinhardt F, Donnenberg VS, Bhargava R, Carr SA, and Weinberg RA (2014). A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat. Cell Biol 16, 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Horner JW, Paul E, Shang X, Troncoso P, Deng P, Jiang S, Chang Q, Spring DJ, Sharma P, et al. (2017). Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature 543, 728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CH, Yeh DW, Lai CY, Liu YL, Huang LR, Lee AY, Jin SC, and Chuang TH (2018). USP17 mediates macrophage-promoted inflammation and stemness in lung cancer cells by regulating TRAF2/TRAF3 complex formation. Oncogene 37, 6327–6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LC, Chang CJ, and Hsu CH (2019). Targeting myeloid-derived suppressor cells in the treatment of hepatocellular carcinoma: current state and future perspectives. J. Hepatocell. Carcinoma 6, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD Jr., van Rooijen N, and Weissman IL (2009). CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 138, 286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanchi I, Santamaria-Martínez A, Susanto E, Peng H, Lehr HA, Delaloye JF, and Huelsken J (2011). Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 481, 85–89. [DOI] [PubMed] [Google Scholar]
- Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J, Omberg L, Gevaert O, et al. ; Cancer Genome Atlas Research Network (2018). Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173, 338–354.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyshev I, and Malyshev Y (2015). Current concept and update of the macrophage plasticity concept: intracellular mechanisms of reprogramming and M3 macrophage “switch” phenotype. BioMed Res. Int 2015, 341308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour FA, Al-Mazrou A, Al-Mohanna F, Al-Alwan M, and Ghebeh H (2020). PD-L1 is overexpressed on breast cancer stem cells through notch3/mTOR axis. OncoImmunology 9, 1729299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Marchesi F, Malesci A, Laghi L, and Allavena P (2017). Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol 14, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J (2008). TGFbeta in cancer. Cell 134, 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Martin-Jaular L, Lavieu G, and Théry C (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol 21, 9–17. [DOI] [PubMed] [Google Scholar]
- Matozaki T, Murata Y, Okazawa H, and Ohnishi H (2009). Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway. Trends Cell Biol. 19, 72–80. [DOI] [PubMed] [Google Scholar]
- McCann AP, Smyth P, Cogo F, McDaid WJ, Jiang L, Lin J, Evergren E, Burden RE, Van Schaeybroeck S, Scott CJ, and Burrows JF (2018). USP17 is required for trafficking and oncogenic signaling of mutant EGFR in NSCLC cells. Cell Commun. Signal 16, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke-van der Houven van Oordt CW, Gomez-Roca C, van Herpen C, Coveler AL, Mahalingam D, Verheul HMW, van der Graaf WTA, Christen R, Rüttinger D, Weigand S, et al. (2016). First-in-human phase I clinical trial of RG7356, an anti-CD44 humanized antibody, in patients with advanced, CD44-expressing solid tumors. Oncotarget 7, 80046–80058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland I, et al. (2017). Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549, 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda A, Hamilton PT, Zhang AW, Pattnaik S, Becht E, Mezheyeuski A, Bruun J, Micke P, de Reynies A, and Nelson BH (2019). Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc. Natl. Acad. Sci. USA 116, 9020–9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei R, Sarkar S, Dzikowski L, Rawji KS, Khan L, Faissner A, Bose P, and Yong VW (2018). Brain tumor-initiating cells export tenascin-C associated with exosomes to suppress T cell activity. OncoImmunology 7, e1478647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchem JB, Brennan DJ, Knolhoff BL, Belt BA, Zhu Y, Sanford DE, Belaygorod L, Carpenter D, Collins L, Piwnica-Worms D, et al. (2013). Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 73, 1128–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RG, Mortensson E, and Williams AC (2018). Targeting LGR5 in colorectal cancer: therapeutic gold or too plastic? Br. J. Cancer 118, 1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller L, Tunger A, Plesca I, Wehner R, Temme A, Westphal D, Meier F, Bachmann M, and Schmitz M (2020). Bidirectional crosstalk between cancer stem cells and immune cell subsets. Front. Immunol 11, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murciano-Goroff YR, Warner AB, and Wolchok JD (2020). The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res. 30, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair N, Calle AS, Zahra MH, Prieto-Vila M, Oo AKK, Hurley L, Vaidyanath A, Seno A, Masuda J, Iwasaki Y, et al. (2017). A cancer stem cell model as the point of origin of cancer-associated fibroblasts in tumor microenvironment. Sci. Rep 7, 6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Kikushige Y, Miyawaki K, Kunisaki Y, Mizuno S, Takenaka K, Tamura S, Okumura Y, Ito M, Ariyama H, et al. (2019). Dedifferentiation process driven by TGF-beta signaling enhances stem cell properties in human colorectal cancer. Oncogene 38, 780–793. [DOI] [PubMed] [Google Scholar]
- Nomura A, Gupta VK, Dauer P, Sharma NS, Dudeja V, Merchant N, Saluja AK, and Banerjee S (2018). NFκB-mediated invasiveness in CD133+ pancreatic TICs is regulated by autocrine and paracrine activation of IL1 signaling. Mol. Cancer Res 16, 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos B, Silver DJ, Mulkearns-Hubert EE, Alvarado AG, Turaga SM, Sorensen MD, Rayman P, Flavahan WA, Hale JS, Stoltz K, et al. (2016). Cancer stem cell-secreted macrophage migration inhibitory factor stimulates myeloid derived suppressor cell function and facilitates glioblastoma immune evasion. Stem Cells 34, 2026–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounova M, Lee E, Piranlioglu R, El Andaloussi A, Kolhe R, Demirci MF, Marasco D, Asm I, Chadli A, Hassan KA, et al. (2017). Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat. Commun 8, 14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczulla AM, Rothfelder K, Raffel S, Konantz M, Steinbacher J, Wang H, Tandler C, Mbarga M, Schaefer T, Falcone M, et al. (2019). Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature 572, 254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panni RZ, Sanford DE, Belt BA, Mitchem JB, Worley LA, Goetz BD, Mukherjee P, Wang-Gillam A, Link DC, Denardo DG, et al. (2014). Tumor-induced STAT3 activation in monocytic myeloid-derived suppressor cells enhances stemness and mesenchymal properties in human pancreatic cancer. Cancer Immunol. Immunother 63, 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathria P, Louis TL, and Varner JA (2019). Targeting tumor-associated macrophages in cancer. Trends Immunol. 40, 310–327. [DOI] [PubMed] [Google Scholar]
- Peng D, Tanikawa T, Li W, Zhao L, Vatan L, Szeliga W, Wan S, Wei S, Wang Y, Liu Y, et al. (2016). Myeloid-derived suppressor cells endow stem-like qualities to breast cancer cells through IL6/STAT3 and NO/NOTCH crosstalk signaling. Cancer Res. 76, 3156–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaks V, Kong N, and Werb Z (2015). The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 16, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian BZ, and Pollard JW (2010). Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail DF, Bowman RL, Akkari L, Quick ML, Schuhmacher AJ, Huse JT, Holland EC, Sutton JC, and Joyce JA (2016). The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science 352, aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, and Morrison SJ (2008). Efficient tumour formation by single human melanoma cells. Nature 456, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggi C, Correnti M, Sica A, Andersen JB, Cardinale V, Alvaro D, Chiorino G, Forti E, Glaser S, Alpini G, et al. (2017). Cholangiocarcinoma stem-like subset shapes tumor-initiating niche by educating associated macrophages. J. Hepatol 66, 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan S, Mehta P, Xie Y, Lei YL, and Mehta G (2019). Ovarian cancer stem cells and macrophages reciprocally interact through the WNT pathway to promote pro-tumoral and malignant phenotypes in 3D engineered microenvironments. J. Immunother. Cancer 7, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran S, Rasool S, and Maccalli C (2019). The cross talk between cancer stem cells/cancer initiating cells and tumor microenvironment: the missing piece of the puzzle for the efficient targeting of these cells with immunotherapy. Cancer Microenviron. 12, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, Maira G, Parati EA, Stassi G, Larocca LM, and De Maria R (2010). Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature 468, 824–828. [DOI] [PubMed] [Google Scholar]
- Rutkowska A, Stoczyńska-Fidelus E, Janik K, Włodarczyk A, and Rieske P (2019). EGFRvIII: an oncogene with ambiguous role. J. Oncol 2019, 1092587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B Jr., Alcala S, Garcia E, Sanchez-Ripoll Y, Azevedo MM, Cioffi M, Tatari M, Miranda-Lorenzo I, Hidalgo M, Gomez-Lopez G, et al. (2015). Microenvironmental hCAP-18/LL-37 promotes pancreatic ductal adenocarcinoma by activating its cancer stem cell compartment. Gut 64, 1921–1935. [DOI] [PubMed] [Google Scholar]
- Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, Barneda D, Sellers K, Campos-Olivas R, Graña O, et al. (2015). MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 22, 590–605. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Döring A, Zemp FJ, Silva C, Lun X, Wang X, Kelly J, Hader W, Hamilton M, Mercier P, et al. (2014). Therapeutic activation of macrophages and microglia to suppress brain tumor-initiating cells. Nat. Neurosci 17, 46–55. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Yang R, Mirzaei R, Rawji K, Poon C, Mishra MK, Zemp FJ, Bose P, Kelly J, Dunn JF, and Yong VW (2020). Control of brain tumor growth by reactivating myeloid cells with niacin. Sci. Transl. Med 12, eaay9924. [DOI] [PubMed] [Google Scholar]
- Schatton T, Schütte U, Frank NY, Zhan Q, Hoerning A, Robles SC, Zhou J, Hodi FS, Spagnoli GC, Murphy GF, and Frank MH (2010). Modulation of T-cell activation by malignant melanoma initiating cells. Cancer Res. 70, 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, Solomon JA, Yoo S, Arron ST, Friedlander PA, et al. (2012). Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N. Engl. J. Med 366, 2171–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Chen Y, Chen Y, Yang Y, Bing W, and Qi J (2018a). CD4+ CD25+ regulatory T cells promote hepatocellular carcinoma invasion via TGF-β1-induced epithelial-mesenchymal transition. OncoTargets Ther. 12, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Zhang X, Li J, Mo L, Zhao H, Zhu Y, Hu Z, Gao J, and Tan W (2018b). PD-1 blockade enhances the antitumor efficacy of GM-CSF surface-modified bladder cancer stem cells vaccine. Int. J. Cancer 142, 2106–2117. [DOI] [PubMed] [Google Scholar]
- Shidal C, Singh NP, Nagarkatti P, and Nagarkatti M (2019). MicroRNA-92 expression in CD133+ melanoma stem cells regulates immunosuppression in the tumor microenvironment via integrin-dependent activation of TGFβ. Cancer Res. 79, 3622–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozawa Y, Nie B, Pienta KJ, Morgan TM, and Taichman RS (2013). Cancer stem cells and their role in metastasis. Pharmacol. Ther 138, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Ping Y, Zhang K, Yang L, Li F, Zhang C, Cheng S, Yue D, Maimela NR, Qu J, et al. (2019). Low-dose IFNγ induces tumor cell stemness in tumor microenvironment of non-small cell lung cancer. Cancer Res. 79, 3737–3748. [DOI] [PubMed] [Google Scholar]
- Stein RG, Ebert S, Schlahsa L, Scholz CJ, Braun M, Hauck P, Horn E, Monoranu CM, Thiemann VJ, Wustrow MP, et al. (2019). Cognate nonlytic interactions between CD8+ T cells and breast cancer cells induce cancer stem cell-like properties. Cancer Res. 79, 1507–1519. [DOI] [PubMed] [Google Scholar]
- Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, Deng X, Li H, Huang Y, Gao L, et al. (2020). Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell 38, 79–96.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Coyle KM, Vidovic D, Thomas ML, Gujar S, and Marcato P (2017). Hide-and-seek: the interplay between cancer stem cells and the immune system. Carcinogenesis 38, 107–118. [DOI] [PubMed] [Google Scholar]
- Tabu K, Liu W, Kosaku A, Terashima K, Murota Y, Aimaitijiang A, Nobuhisa I, Hide T, and Taga T (2020). Glioma stem cell (GSC)-derived autoschizis-like products confer GSC niche properties involving M1-like tumor-associated macrophages. Stem Cells 38, 921–935. [DOI] [PubMed] [Google Scholar]
- Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, and Ivy SP (2015). Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol 12, 445–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Chu C, Zhou W, Huang Z, Zhai K, Fang X, Huang Q, Zhang A, Wang X, Yu X, et al. (2020). Dual Role of WISP1 in maintaining glioma stem cells and tumor-supportive macrophages in glioblastoma. Nat. Commun 11, 3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi Y, Shitara K, and Nishikawa H (2019). Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat. Rev. Clin. Oncol 16, 356–371. [DOI] [PubMed] [Google Scholar]
- Turdo A, Veschi V, Gaggianesi M, Chinnici A, Bianca P, Todaro M, and Stassi G (2019). Meeting the challenge of targeting cancer stem cells. Front. Cell Dev. Biol 7, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeta-Magara A, Gadi A, Volta V, Walters B, Arju R, Giashuddin S, Zhong H, and Schneider RJ (2019). Inflammatory breast cancer promotes development of M2 tumor-associated macrophages and cancer mesenchymal cells through a complex chemokine network. Cancer Res. 79, 3360–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia S, Verga JA, Stephansky J, Pastika TJ, Lombardi Story J, et al. (2019). Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S, Zhao E, Kryczek I, Vatan L, Sadovskaya A, Ludema G, Simeone DM, Zou W, and Welling TH (2014). Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology 147, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, Fligelman B, Leversha M, Brennan C, and Tabar V (2010a). Glioblastoma stem-like cells give rise to tumour endothelium. Nature 468, 829–833. [DOI] [PubMed] [Google Scholar]
- Wang YK, Zhu YL, Qiu FM, Zhang T, Chen ZG, Zheng S, and Huang J (2010b). Activation of Akt and MAPK pathways enhances the tumorigenicity of CD133+ primary colon cancer cells. Carcinogenesis 31, 1376–1380. [DOI] [PubMed] [Google Scholar]
- Wang D, Fu L, Sun H, Guo L, and DuBois RN (2015). Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology 149, 1884–1895.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang M, Lin L, Ren H, Lin C, Lin S, Shen G, Ji B, and Meng C (2016). HepG2 cells acquire stem cell-like characteristics after immune cell stimulation. Cell Oncol. (Dordr.) 39, 35–45. [DOI] [PubMed] [Google Scholar]
- Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, et al. (2017). Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jin J, Wan F, Zhao L, Chu H, Chen C, Liao G, Liu J, Yu Y, Teng H, et al. (2019a). AMPK promotes SPOP-mediated NANOG degradation to regulate prostate cancer cell stemness. Dev. Cell 48, 345–360.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yin K, Tian J, Xia X, Ma J, Tang X, Xu H, and Wang S (2019b). Granulocytic myeloid-derived suppressor cells promote the stemness of colorectal cancer cells through exosomal S100A9. Adv. Sci. (Weinh.) 6, 1901278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Barr J, Kong LY, Wang Y, Wu A, Sharma AK, Gumin J, Henry V, Colman H, Priebe W, et al. (2010). Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol. Cancer Ther 9, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Jiang Y, Zou F, Liu Y, Wang S, Xu N, Xu W, Cui C, Xing Y, Liu Y, et al. (2013). Activation of PI3K/Akt pathway by CD133-p85 interaction promotes tumorigenic capacity of glioma stem cells. Proc. Natl. Acad. Sci. USA 110, 6829–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Zhang T, Deng SC, Wei JC, Yang P, Wang Q, Chen ZP, Li WL, Chen HC, Hu H, and Cao J (2019a). PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. 450, 1–13. [DOI] [PubMed] [Google Scholar]
- Wei X, Yang S, Pu X, He S, Yang Z, Sheng X, Meng X, Chen X, Jin L, Chen W, and Zhang Y (2019b). Tumor-associated macrophages increase the proportion of cancer stem cells in lymphoma by secreting pleiotrophin. Am. J. Transl. Res 11, 6393–6402. [PMC free article] [PubMed] [Google Scholar]
- Wei J, Chen P, Gupta P, Ott M, Zamler D, Kassab C, Bhat KP, Curran MA, de Groot JF, and Heimberger AB (2020). Immune biology of glioma-associated macrophages and microglia: functional and therapeutic implications. Neuro-oncol. 22, 180–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng YS, Tseng HY, Chen YA, Shen PC, Al Haq AT, Chen LM, Tung YC, and Hsu HL (2019). MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol. Cancer 18, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphalen CB, Asfaha S, Hayakawa Y, Takemoto Y, Lukin DJ, Nuber AH, Brandtner A, Setlik W, Remotti H, Muley A, et al. (2014). Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J. Clin. Invest 124, 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Wei J, Kong LY, Wang Y, Priebe W, Qiao W, Sawaya R, and Heimberger AB (2010). Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro-oncol. 12, 1113–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Qu D, Weygant N, Peng J, and Houchen CW (2020). Cancer stem cell marker DCLK1 correlates with tumorigenic immune infiltrates in the colon and gastric adenocarcinoma microenvironments. Cancers (Basel) 12, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Long H, He L, Han X, Lin K, Liang Z, Zhuo W, Xie R, and Zhu B (2015). Interleukin-17 produced by tumor microenvironment promotes self-renewal of CD133+ cancer stem-like cells in ovarian cancer. Oncogene 34, 165–176. [DOI] [PubMed] [Google Scholar]
- Xu J, Lamouille S, and Derynck R (2009). TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 19, 156–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Dong X, Qi P, Ye Y, Shen W, Leng L, Wang L, Li X, Luo X, Chen Y, et al. (2017a). Sox2 Communicates with Tregs through CCL1 to promote the stemness property of breast cancer cells. Stem Cells 35, 2351–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wan Y, Liu J, Wang Y, Li S, Xing H, Tang K, Tian Z, Rao Q, Wang M, and Wang J (2017b). Regulatory T cells promote the stemness of acute myeloid leukemia cells through IL10 cytokine related signaling pathway. Blood 130, 2727. [DOI] [PubMed] [Google Scholar]
- Xu Y, Xu Z, Li Q, Guo L, Wang Y, Zhou J, Wang G, and Liu Y (2019). Mutated p53 promotes the symmetric self-renewal of cisplatin-resistant lung cancer stem-like cells and inhibits the recruitment of macrophages. J. Immunol. Res 2019, 7478538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashina T, Baghdadi M, Yoneda A, Kinoshita I, Suzu S, Dosaka-Akita H, and Jinushi M (2014). Cancer stem-like cells derived from chemoresistant tumors have a unique capacity to prime tumorigenic myeloid cells. Cancer Res. 74, 2698–2709. [DOI] [PubMed] [Google Scholar]
- Yang S, Wang B, Guan C, Wu B, Cai C, Wang M, Zhang B, Liu T, and Yang P (2011). Foxp3+IL-17+ T cells promote development of cancer-initiating cells in colorectal cancer. J. Leukoc. Biol 89, 85–91. [DOI] [PubMed] [Google Scholar]
- Yang J, Liao D, Chen C, Liu Y, Chuang TH, Xiang R, Markowitz D, Reisfeld RA, and Luo Y (2013). Tumor-associated macrophages regulate murine breast cancer stem cells through a novel paracrine EGFR/Stat3/Sox-2 signaling pathway. Stem Cells 31, 248–258. [DOI] [PubMed] [Google Scholar]
- Yang Z, Xie H, He D, and Li L (2016). Infiltrating macrophages increase RCC epithelial mesenchymal transition (EMT) and stem cell-like populations via AKT and mTOR signaling. Oncotarget 7, 44478–44491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Ye H, Qi Z, Mo L, Yue Q, Baral A, Hoon DSB, Vera JC, Heiss JD, Chen CC, et al. (2016). B7-H4(B7x)-mediated cross-talk between glioma-initiating cells and macrophages via the IL6/JAK/STAT3 pathway lead to poor prognosis in glioma patients. Clin. Cancer Res. 22, 2778–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi E, Mabuchi S, Komura N, Shimura K, Kuroda H, Kozasa K, Takahashi R, Sasano T, Kawano M, Matsumoto Y, et al. (2019). The role of myeloid-derived suppressor cells in endometrial cancer displaying systemic inflammatory response: clinical and preclinical investigations. OncoImmunology 8, e1662708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Li Y, Li M, Lei M, Wu M, Qu Y, Yuan Y, Chen T, and Jiang H (2018). Ovarian cancer stem cells promote tumour immune privilege and invasion via CCL5 and regulatory T cells. Clin. Exp. Immunol 191, 60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Liu Z, Sun S, Xie J, Cao L, Lv P, Nie S, Zhang B, Xie B, Peng S, and Jiang B (2018). Tumor-associated macrophages recruited by periostin in intrahepatic cholangiocarcinoma stem cells. Oncol. Lett 15, 8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zoltan M, Riquelme E, Xu H, Sahin I, Castro-Pando S, Montiel MF, Chang K, Jiang Z, Ling J, et al. (2018). Immune cell production of interleukin 17 induces stem cell features of pancreatic intraepithelial neoplasia cells. Gastroenterology 155, 210–223.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Ye H, Ren X, Zheng S, Zhou Q, Chen C, Lin Q, Li G, Wei L, Fu Z, et al. (2019a). Macrophage-expressed CD51 promotes cancer stem cell properties via the TGF-b1/smad2/3 axis in pancreatic cancer. Cancer Lett. 459, 204–215. [DOI] [PubMed] [Google Scholar]
- Zhang H, Brown RL, Wei Y, Zhao P, Liu S, Liu X, Deng Y, Hu X, Zhang J, Gao XD, et al. (2019b). CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev. 33, 166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen M, Zhu Y, Dai X, Dang F, Ren J, Ren S, Shulga YV, Beca F, Gan W, et al. (2019c). SPOP promotes Nanog destruction to suppress stem cell traits and prostate cancer progression. Dev. Cell 48, 329–344.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen L, Dang WQ, Cao MF, Xiao JF, Lv SQ, Jiang WJ, Yao XH, Lu HM, Miao JY, et al. (2020). CCL8 secreted by tumor-associated macrophages promotes invasion and stemness of glioblastoma cells via ERK1/2 signaling. Lab. Invest 100, 619–629. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dong Q, Li J, Zhang K, Qin J, Zhao J, Sun Q, Wang Z, Wartmann T, Jauch KW, et al. (2018). Targeting cancer stem cells and their niche: perspectives for future therapeutic targets and strategies. Semin. Cancer Biol 53, 139–155. [DOI] [PubMed] [Google Scholar]
- Zhao D, Cai L, Lu X, Liang X, Li J, Chen P, Ittmann M, Shang X, Jiang S, Li H, et al. (2020). Chromatin regulator CHD1 remodels the immunosuppressive tumor microenvironment in PTEN-deficient prostate cancer. Cancer Discov. 10, 1374–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, and Dirks PB (2009). Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat. Rev. Drug Discov 8, 806–823. [DOI] [PubMed] [Google Scholar]
- Zhou W, Ke SQ, Huang Z, Flavahan W, Fang X, Paul J, Wu L, Sloan AE, McLendon RE, Li X, et al. (2015). Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat. Cell Biol 17, 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, Wang-Gillam A, Goedegebuure SP, Linehan DC, and DeNardo DG (2014). CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 74, 5057–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]