Summary
Background
The gut microbiome plays an important role in human health and disease. Gnotobiotic animal and in vitro cell-based models provide some informative insights into mechanistic crosstalk. However, there is no existing system for a long-term co-culture of a human colonic mucosal barrier with super oxygen-sensitive commensal microbes, hindering the study of human-microbe interactions in a controlled manner.
Methods
Here, we investigated the effects of an abundant super oxygen-sensitive commensal anaerobe, Faecalibacterium prausnitzii, on a primary human mucosal barrier using a Gut-MIcrobiome (GuMI) physiome platform that we designed and fabricated.
Findings
Long-term continuous co-culture of F. prausnitzii for two days with colon epithelia, enabled by continuous flow of completely anoxic apical media and aerobic basal media, resulted in a strictly anaerobic apical environment fostering growth of and butyrate production by F. prausnitzii, while maintaining a stable colon epithelial barrier. We identified elevated differentiation and hypoxia-responsive genes and pathways in the platform compared with conventional aerobic static culture of the colon epithelia, attributable to a combination of anaerobic environment and continuous medium replenishment. Furthermore, we demonstrated anti-inflammatory effects of F. prausnitzii through HDAC and the TLR-NFKB axis. Finally, we identified that butyrate largely contributes to the anti-inflammatory effects by downregulating TLR3 and TLR4.
Conclusions
Our results are consistent with some clinical observations regarding F. prausnitzii, thus motivating further studies employing this platform with more complex engineered colon tissues for understanding the interaction between the human colonic mucosal barrier and microbiota, pathogens, or engineered bacteria.
eTOC blurb
Many human gut bacteria of clinical relevance are extremely oxygen sensitive, hampering the investigation of crosstalk with host cells. Zhang et al. developed a gut-microbe physiomimetic platform for long-term continuous co-culture of super oxygen sensitive bacterial species with primary human colon epithelium in the context of inflammation.
Graphical Abstract
Introduction
The gut microbiome has emerged as a key factor regulating and responding to human health and disease. Altered gut microbiome composition has been linked to numerous diseases including autoimmunity, inflammatory bowel diseases (IBD), neurodevelopmental disease, metabolic disorders, cancer, and more recently behavior learning.1–8 For example, reduction of a major member of the phylum Firmicutes, Faecalibacterium prausnitzii, is strongly associated with a higher risk of ileal Crohn’s disease.9 In a chemically-induced colitis mouse model, F. prausnitzii and its supernatant markedly alleviated the severity of the colitis.9 In a mouse model of autism, neurodevelopmental symptoms have been demonstrated to coincide with microbiota alterations and a “leaky gut” in which the integrity of the epithelial barrier is compromised. Bacteroides fragilis was demonstrated to ameliorate gut permeability as well as communicative and sensorimotor behavioral defects in this animal model.10
While studies in mice provide some informative insights into the mechanisms by which F. prausnitzii, B. fragilis and other organisms exert their beneficial effects, they also have limitations: inconsistent translation to human physiology; limited ability to control microbe-gut interactions; and relatively low throughput. Moreover, many bacterial species implicated in human health are highly specific to the human host and do not appear in common mice fecal microbiota; nor do humans host many of the microbes found naturally in mice.11 Furthermore, microbes colonize the gut in a microenvironment-specific manner related to nutrient load, mucus properties, and oxygen tension. For example, F. prausnitzii is a super oxygen-sensitive anaerobe12 (i.e., it is on the extreme end of oxygen intolerance among “obligate anaerobes”) that cannot colonize the microaerobic environment 13 of the upper gastrointestinal (GI) tract, but robustly colonizes the anaerobic environment of the human large intestine.14 However, these microenvironments are difficult to control and study in animal models. These concerns, together with an increasing emphasis on replacement, reduction, and refinement of animal use15 motivate the development of in vitro microphysiological systems (MPSs) representing the unique environment of the human colon to enable studies of human-microbe interactions in a controlled manner.
Among the organisms significantly implicated in human health and disease, F. prausnitzii is particularly interesting for in vitro mechanistic studies, as it is so fastidious that it fails to colonize gnotobiotic mice unless the mice are first colonized with a commensal such as B. thetaiotaomicron, a more oxygen-tolerant organism than F. prausnitzii;12,16 further, sustained colonization typically requires repeated inoculations.17 In humans, the relative deficiency of F. prausnitzii in both UC and CD patients concurrent with a robust increase in TLR4 observed in the intestinal epithelial cells of these patient populations18 suggests a possible role for F. prausnitzii in modulation of TLRs in the human intestine. TLR receptors are critical for intestinal recognition of bacteria, but activation of TLRs is also associated with an increase of NFKB signaling and inflammation. Imbalanced relationships within this triad may promote aberrant TLR signaling, contributing to acute and chronic intestinal inflammatory processes in IBD, colitis and associated cancer.19,20 The responsiveness of intestinal cells to LPS is positively correlated with TLR4 expression.21 Thus reduction of factors that keep TLR expression in check – potentially, F. prausnitzii –may contribute to chronic intestinal inflammatory diseases.
In vitro studies to test hypotheses about F. prausnitzii-mediated regulation of colon epithelial function or immune functions have been limited to <12-hr static culture by technical difficulties in maintaining an anaerobic apical environment supporting viable microbes in the context of an aerobic basal environment to maintain epithelial cell integrity.22–24 As appreciated by these previous investigators, the influences of F. prausnitzii on cellular phenotype may arise from relatively long (>24 hr) time scale phenomena, such as accumulation of butyrate and other microbial metabolites,25 in addition to other transcriptionally-controlled behaviors. Recent technological innovations to culture obligate anaerobes in modified Transwell formats are restricted to static cultures – where microbial nutrition become limiting – and have so far been used to culture anaerobes that are not as strictly oxygen sensitive as F. prausnitzii.26,27
In order to study the long-term effects of live F. prausnitzii on human primary colon epithelial barriers, we engineered a microfluidic platform to allow long-term (days) co-culture of super oxygen-sensitive microbes with a primary human colon epithelial barrier maintained on a standard cell culture membrane insert (Transwell). Recognizing that maintenance of a robust microbial population for long culture periods requires frequent refreshment of the microbial culture medium, a crucial platform feature is programmable apical fluid flow, which can provide a continuous flow of completely anoxic inlet feed media at a desired rate and maintain anaerobic conditions in the culture region. Further, to enable quantitative analysis of steroids and lipophilic drugs, the platform is constructed from materials that inhibit adsorption of lipophilic compounds. Several continuous microfluidic devices that meet some of these criteria have been reported in the literature,28–30 and though some have supported culture of some species of obligate anaerobes,30,31 none have reported culture of F. prausnitzii and it is unclear whether these devices support microbes that fall into the most super oxygen-sensitive range. Further, most microfluidic devices are fabricated from polydimethylsiloxane (PDMS), which is highly adsorptive of lipophilic compounds and oxygen permeable.29,30,32 Approaches to mitigate the high oxygen permeability of PDMS include placement of devices in a custom anaerobic chamber 29 and increasing the thickness of the PDMS. 30
Here, we describe the design and implementation Gut-MIcrobiome (GuMI) mesofluidic culture platform for analysis of interactions between F. prausnitzii and a primary human colon mucosal barrier over a 4-day period. We first examined the effects of platform culture on the phenotype of the colon mucosal barrier compared to traditional static culture in the absence of bacteria, assessing morphological criteria, barrier function, and distribution of cell types via immunostaining, along with RNA sequencing analysis for transcriptional regulation. We then studied the behavior of F. prausnitzii in multi-day co-culture with the human primary colon mucosal barrier, measuring microbial localization, growth kinetics, and short chain fatty acid (SCFA) production rates. In addition, using RNA sequencing, pathway analyses, and quantitative PCR, we revealed specific responses of colon epithelium to luminal hypoxia and the continuous growth of and butyrate production by F. prausnitzii. In addition, extended co-culture with F. prausnitzii was associated with downregulation of toll-like receptor 3 (TLR3) and 4 (TLR4) in the epithelia, consistent with expressional changes strongly associated with IBD in humans.18 Downstream signaling pathways NFKB1 and NFKB-activating pathway were also downregulated concomitant with upregulation of the NFKB-inhibitory pathway. We further parsed these F. prausnitzii-mediated changes to identify those attributable primarily to its production of butyrate.
Results and Discussion
GuMI physiome platform design and fabrication
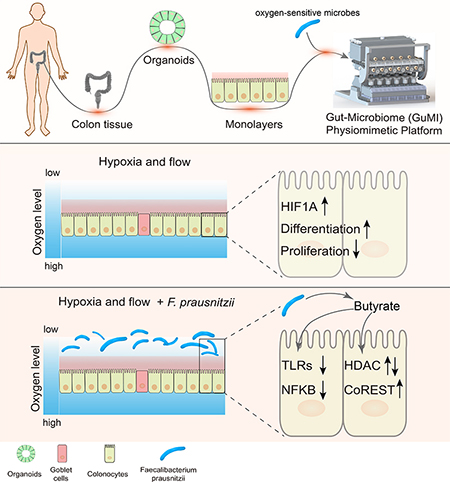
Transwell® and related commercially-available membrane culture inserts remain widely used for building in vitro models to study epithelial barriers, including gut, lung, cervix and endometrium, as they foster robust monolayer formation, are highly reproducible, and easy to use.33–36 We therefore designed the GuMI device to modulate the apical and basolateral microenvironments of these standardized membrane culture inserts individually, by controlling the flow rates and oxygen concentrations separately on the apical and basolateral sides to establish a steep oxygen gradient across the epithelial barrier resembling that in the colon mucosa.37
The GuMI physiome platform, designed to maintain six cultures with controlled microenvironments, comprises separate apical and basolateral modules (Figure 1). Each module is assembled in three layers: a fluidic plate, a flexible membrane, and a pneumatic plate. Since the fluidic plate is machined from a monolithic block of polysulfone, in contrast to oxygen-permeable PDMS commonly used in microfluidic devices, GuMI is designed to operate in a standard cell culture incubator. The polysulfone plate is both oxygen impermeable and sterilizable through autoclaving. Microfluidic channels and pumps are machined at the bottom of the fluidic plate, where each pump comprises three micro-chambers in line with a fluidic channel. Each chamber can be set to open or close by applying either vacuum or pressure to the pneumatic line to flex the membrane located in between the fluidic and pneumatic plates. By operating the chambers in a specific sequence and frequency, the fluidic direction and flow rate can be precisely controlled as described previously.38,39 Driving flow with pneumatic control provides a great advantage in parallelism and scalability, and it also allows GuMI to process multiple samples without additional pneumatic lines.
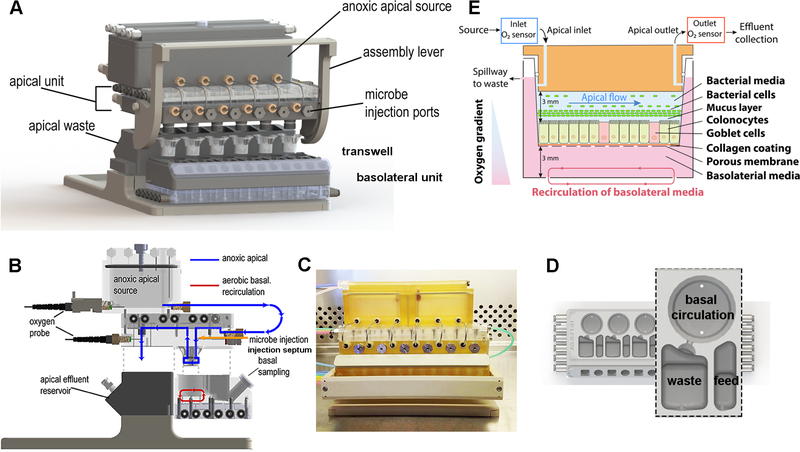
Figure 1.
Schematic illustration of GuMI physiome platform. (A) The GuMI physiome platform recapitulates a steep oxygen gradient across intestinal epithelium cultured in standardized Transwells to support the co-culture of the intestinal epithelium and obligate anaerobes. The platform is composed of an apical and basolateral unit with individual flow control and is capable of supporting six samples at once. Individual microbe injection ports capped with an oxygen impermeable septum allows obligate anaerobes of interest to be introduced into each sample independently. (B) Side illustration of the GuMI physiome platform. Anoxic apical media purged with inert gas is delivered into each Transwell by pneumatic pumps located in the apical units, where the effluent is collected at the apical effluent reservoir. Meanwhile, continuous recirculation of basolateral media by pneumatic pumps inside the basolateral unit promotes oxygenation and supply the oxygen needed for epithelial cells. Oxygen probes at the apical medial reservoir and near the apical effluent exit measure oxygen concentration at apical inlet and outlet respectively. (C) Photo of GuMI physiome platform assembled in a sterile cell culture hood. (D) Schematic illustration of the basolateral unit of the GuMI platform. Each basolateral unit contains six replicates of a basolateral module. A Transwell is located inside the circular well at the top of the module, with a set of pneumatic pumps underneath providing oxygen through circulation. Another set of pneumatic pumps located underneath the feeding compartment supply fresh media to the basolateral side of the sample, where the spent media gets displaced to the waste compartment via the spillway. (E) Schematic illustration of a Transwell located inside the GuMI physiome platform. Differentiated colonic epithelial cells seeded inside a Transwell are cultured inside the GuMI platform to study host-microbe interaction in vitro.
In order to keep the apical environment anoxic, the apical medium was first purged with a gas mixture of 5% CO2 and 95% N2 to remove any dissolved oxygen in the apical medium. Then the medium was drawn into the apical module by the pneumatic pump via stainless steel tubing (Figure 1B). To ensure the apical medium stays anoxic in the module, pump chambers were actuated with nitrogen and vacuum, and an additional stream of nitrogen was applied to the space between the membrane and the pneumatic plate to displace any remaining oxygen in the device. An assembly lever located on the apical module is designed to mechanically hold the apical and basolateral modules together (Figure 1C). Upon activation of the lever, the compression force helps form a tight seal between the O-ring and the sidewall inside a Transwell, allowing apical effluent to be collected at the apical effluent reservoir (Figure 1D).
In vivo, oxygen and nutrients supporting the metabolism of cells are delivered to the colon epithelial cells from the submucosa (basolateral side).37 Therefore, a recirculation pump is included in the bottom of the basolateral module to enhance oxygen exchange between the basolateral medium and air inside the incubator. The basolateral module also contains a feed compartment with an additional set of pneumatic pumps to supply fresh medium to the basolateral compartment in a programmable fashion, where the spent medium is spilled and collected at the waste compartment through the spillway design (Figure 1D).
The continuous perfusion of anoxic apical medium and recirculation of aerobic basolateral medium maintains a physiologic hypoxia with a steep oxygen gradient across the epithelial layer as shown in Figure 1E. Ruthenium-based optical oxygen sensors are incorporated near the apical inlet and outlet for real-time monitoring of oxygen concentration. Once an anoxic condition is reached and maintained, obligate anaerobes grown in the log phase are injected into the lumen via the septum located at the front of GuMI (Figure 1B and 1E).
Prediction and validation of oxygen tensions
After conducting pilot experiments using Caco-2 monolayers40 to establish that the GuMI device operated as designed, i.e., maintained a viable mucosal barrier with a steep oxygen gradient between the aerobic basolateral compartment and the anoxic apical compartment (data not shown), we focused on using primary human colon epithelial cells. Epithelial cells obtained from a healthy region of a colon biopsy were expanded as intestinal organoids and seeded as monolayers in Transwells prior to each experiment following the protocol described earlier.41 Briefly, primary epithelial cells seeded in a Transwell at 268,000 cells per cm2 (300,000 per well) were allowed to reach confluency in seeding medium for three days, then the cells were switched to differentiation medium for four days before being transferred to GuMI for experiments (Figure 2A). Inside the GuMI platform, the apical compartment was perfused with anoxic apical medium during operation with mucosal barrier-microbe cultures. The oxygen levels at points in the feed and effluent fluidic lines for the apical compartment were constantly measured at one-minute intervals using ruthenium-based probes (Figure 2B). During routine culture, the outlet probe was used to measure approach to and deviation from steady state, as its position in the fluidic line was sufficiently close to effluent exit drip tube for back diffusion of oxygen from the atmosphere. Computational simulation suggested that the apical compartment would reach equilibrium at a nearly complete anoxic state after ~2.5 h (Figure S1A and S1B) using parameters in Table S1. A companion simulation of oxygen distribution at steady state indicates that 86.6% of total volume of the apical compartment is <1 kPa, Figure S1C and S1D), a prediction supported by the ability of the apical compartment to support robust growth of strict anaerobes throughout the device, as described in subsequent sections. Experimentally, we observed that for an oxygen level of 0 kPa in the inlet reservoir, the outlet concentration measured at the point of the drip tube typically approached steady state after about 24 hours of perfusion at 10 μl min−1 across three different samples as shown by the closely overlapping oxygen measurements (Figure 2B). Further, the more extended time to approach homeostasis than predicted is likely due to the almost unavoidable appearance of small bubbles in the apical region when placing the Transwell (see the quantitative estimation of bubble purge times in Methods), leading to some variability in the time scale to approach steady state. Functionally, we observed in pilot experiments that F. prausnitzii grew robustly when injected at 16 hr or 24 hr, suggesting the bulk of the compartment was nearly anoxic by 16 hr in typical cases (data not shown); a 24-hr injection time for bacteria was routinely used thereafter (see subsequent section).
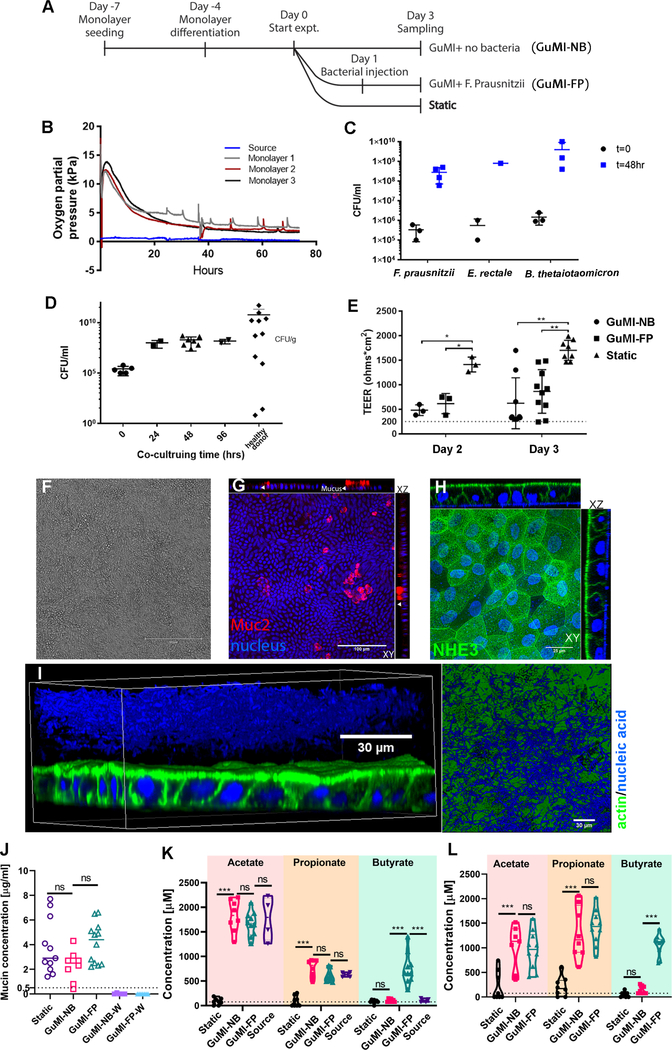
Figure 2.
Characterizations of colon epithelial monolayers and their co-culture with obligate anaerobes in GuMI physiome platform. (A) Timeline overview of experimental design. For each experiment, monolayers are treated in three conditions: anoxic apical flow (GuMI-NB), anoxic apical flow with F. prausnitzii (GuMI-FP) and normoxic static culture in standard cell-culture incubator (Static). (B) Representative oxygen measurements of monolayers cultured in GuMI physiome platform. Oxygen concentration is measured at source and near the apical effluent exit for each monolayer to monitor oxygen concentration on the apical side. (C) Initial and final concentration (48 hours) of three species of obligate anaerobes (F. prausnitzii, Eubacterium rectale, B. thetaiotaomicron) co-cultured with colon epithelium inside GuMI physiome platform. Data are presented as mean ± standard deviation (SD). CFU: colony-forming unit. (D) The concentration of F. prausnitzii cells co-cultured with colon epithelium inside GuMI at different time points compared to values reported in vivo. Data are presented as mean ± standard deviation (SD). CFU: colony-forming unit. (E) Transepithelial electrical resistance (TEER) measurements of monolayers on day 2 and 3. GuMI-NB and GuMI-FP have comparable TEER values and are both significantly higher than 300 ohms · cm2 (dotted line), suggesting epithelial barriers are intact. Monolayers cultured in GuMI-NB and GuMI-FP have significantly (p<0.05) lower TEER values compared to Static on both day 2 and 3. Data are presented as mean ± SD. (F-H) Phase contrast and confocal immunofluorescent staining of monolayers cultured in GuMI-NB for three days. The monolayer is intact and functional and stained positively for different epithelial cell types (G-H) with proper apical polarization of NHE3 (H). (I) Bacterial cells (F. prausnitzii, blue) located on top of the colon epithelia (green/blue) from GuMI-FP in 3D reconstruction and 2D top-down view. Green: actin filament staining, blue: 4’,6-diamidino-2-phenylindole (DAPI) nuclei/nucleic acid staining. A ~15 μm gap in DAPI staining is apparent between the colon epithelia and the zone positive for bacteria. This gap zone likely corresponds to the firm inner mucus layer observed in vivo in a mouse distal colon.55 (J) Concentration of mucin (μg/ml) in the apical media collected directly above the monolayer (GuMI-NB, GuMI FP, and Static) or at the effluent waste (GuMI-NB-W and GuMI-FP-W). Concentration of acetate, propionate, and butyrate in apical side (K) and basolateral side (L). Source: the apical media from the anoxic apical source unit in Figure 1A. Data are presented as mean ± SD. Dotted line in (J-K) indicates the limit of quantification. Two-way ANOVA with Dunnett’s multiple comparisons test was used in (k-l). One-way ANOVA with Dunn’s multiple comparisons test was used for Static, GuMI-NB, and GuMI-FP in (E) and (J). ns: not significant; *: p<0.05; ** p <0.01; ***: p<0.001. See also Figure S1, Figure S2, Table S1, and Table S2.
At this flow rate of 10 μl min−1 the maximum time-average shear stress that the monolayer experienced is up to 11 μPa, with the highest shear stress at the inlet and outlet ports (Figure S1E and S1F). This agrees with the flow distribution pattern across the monolayer (Figure S1G and S1H) and the estimated shear stress of 0.7–6.0 μPa in human colon based on the flow rate (1.1–20 ml/min)42 and colonic diameter of 3.5–7.6 cm,43 although transient mechanical stresses resulting from peristaltic pumping in the GI tract likely exceeds these estimates.44 It is unlikely that the pulsatility of the pump captures the kind of transient mechanics from peristalsis in vivo, as video images of dye penetration into the apical chamber over multiple pump cycles (not shown) did not exhibit significant pulsatile behavior, suggesting the system capacitance damped the pump action, resulting in approximately constant shear. The relatively low average shear stress for a relatively high volumetric flow rate is achieved by a meso-scale height of the upper chamber, 3 mm, compared to the microscale of typical microfluidic culture devices ( ~ 0.15 mm high).32 Oxygen concentrations measured in the constantly-mixed basal compartment during microbial-mucosal barrier culture during pilot experiments to establish parameters were typically ~16 kPa (n = 6 Caco2-HT29 cultures and 6 primary cultures).
Obligate anaerobe co-culture with colon epithelia in GuMI
The most rigorous test of oxygen levels at the apical surface of the mucosal barrier is maintenance and/or growth of super oxygen-sensitive anaerobes in the apical microenvironment. Therefore, to validate the anoxic apical environment that GuMI is designed to accomplish, three species of obligate anaerobes with varying degrees of oxygen sensitivity (F. prausnitzii, E. rectale and B. thetaiotaomicron, on the order of most oxygen-sensitive to least oxygen-sensitive), each with significant relevance to human health, were selected to be cultured with colon monolayers independently inside GuMI (Figure 2C). The experiments were designed to test the robustness of the platform in supporting oxygen-sensitive anaerobes, thereby establishing the system as an in vitro mimic of gnotobiotic conditions useful for mechanistic studies. F. prausnitzii, in particular, is characterized as an extremely oxygen-sensitive species, and is strongly implicated in inflammatory bowel diseases;45–47 however, mechanistic insights of its effects on human colon remain lacking partly due to its extreme sensitivity to oxygen.12,48
F. prausnitzii, E. rectale and B. thetaiotaomicron at 1–10 × 105 colony forming unit per ml (CFU ml−1) were each injected into the apical compartments of separate GuMI Transwell cultures following 24 hours of anoxic perfusion. The microbes were then co-cultured on the apical side of the colon epithelial cells for 2–4 days using standard YCFA bacterial culture medium diluted to 10% (v/v) with phosphate buffered saline flowing continuously at 10 μl/min. The final density of the selected anaerobes measured at the end of the experiment demonstrates that GuMI is capable of supporting the growth of even the most oxygen-sensitive species in isolation, without the support of any facultative anaerobes. Final densities of F. prausnitzii, E. rectale and B. thetaiotaomicron all reached ~109 CFU ml−1 after 48 hours of co-culturing (Figure 2C). This is in stark contrast with control cultures of F. prausnitzii maintained in co-culture with an epithelial monolayer that is deliberately perforated via a mechanical scraping away of a small section of epithelia to make it leaky to oxygen (“leaky epithelia”) in GuMI, or cultures inoculated into the apical side of static Transwell co-culture and maintained in the standard incubator environment. In both of these control cases, the density of microbes was undetectable due to the presence of oxygen (data not shown). A similar trend was observed for E. rectale, but B. thetaiotaomicron tolerated standard incubator co-culture with colon epithelia, overgrowing and killing the epithelial monolayer within 24 h.
We then focused on the most oxygen-sensitive microbe, F. prausnitzii, and determined the growth properties at finer temporal resolution. The bacterial population reached 109 CFU ml−1 within 24 hours of inoculation into the apical compartment and remained stable at this concentration thereafter (Figure 2D). This density is consistent with the density observed in healthy human49 and mice colon or feces associated with F. prausnitzii.14 The washout of viable bacterial cells was not monitored as they cannot survive in the effluent reservoir connecting to the incubator atmosphere; however, we presume that the stable concentration reflects a dynamic equilibrium with continued growth accompanied by washout.
Effects of F. prausnitzii on the phenotype of colon epithelial monolayers
After showing the GuMI device supported the growth of F. prausnitzii on the apical side of colon epithelial cells cultured in Transwells, we examined the effects of F. prausnitzii on the functions of the colon epithelium. One of the most important functions of colon epithelium is providing a physical and biological barrier against pathogenic species.50 The tight junctions formed between densely packed epithelial cells contribute to the physical barrier, whereas the mucus and anti-bacterial peptides secreted by the cells create a biological barrier.
Measurement of transepithelial electrical resistance (TEER) is a widely adopted metric for evaluating the physical barrier of an epithelium. TEER provides a relatively non-invasive measurement of average barrier integrity by reflecting the ionic conductance of the paracellular pathway.51 TEER values are difficult to compare directly to in vivo behavior, especially since permeability is not uniform in vivo and local state of differentiation in a monolayer may affect average TEER values. TEER values also varied among different in vitro models. It has been shown that the bioprinted bilayer co-culture of primary human intestinal and myofibroblasts has similar chemical permeability but much lower TEER values comparing to Caco-2 monolayer.52 TEER values above 300–400 Ω · cm2 are generally considered to reflect an intact mucosal barrier.51,53 To evaluate the effects of F. prausnitzii on the physical barrier of colon epithelium, we measured the TEER of epithelia cultured either in GuMI with no bacteria (GuMI-NB), in GuMI with F. prausnitzii (GuMI-FP), or in a standard incubator under static conditions (Static) (Figure 2E). We observed that as early as day 2, the static group maintained significantly higher TEER values (1412 ± 150 Ω · cm2) compared to GuMI-NB (482 ± 108 Ω · cm2, P=0.01) and GuMI-FP (616 ± 205 Ω · cm2, P=0.04), and a similar trend obtained at day 3. Importantly, despite the differences, the epithelial barriers remained intact in each condition with average TEER values greater than 480 Ω · cm2, i.e., significantly above the standard threshold reflective of intact monolayers.
The integrity of the physical barrier was further validated by inspecting the monolayers under phase-contrast microscopy and immunofluorescent confocal microscopy (Figure 2F–I). Colon epithelial monolayers maintained inside GuMI with constant apical flow, in both the absence and presence of F. prausnitzii, were intact and qualitatively similar in appearance to those maintained in Static conditions (Figure S2A–C), containing differentiated cells as evidenced by staining for MUC2 (goblet cells), and NHE3 (colonocytes) (Figure 2F–H, Figure S2D–E). The epithelium also had proper polarization of sodium-hydrogen antiporter (NHE3) on the apical surface (Figure 2H. XZ projection), critical for transepithelial Na+ absorption, intracellular pH, and nutrient absorption.54 These phenotypic markers revealed by immunofluorescence staining were not quantified, hence there may be differences in the number of cells expressing them or the levels of expression in the two conditions that are not obvious in this visual comparison.
In the GuMI-FP condition, a large cloud of F. prausnitzii cells (blue) about ~40 μm thick was observed on top of colon epithelial cells with a ~15 μm gap area between the microbes and epithelial surface (Figure 2I). The gap area is likely the tightly-crosslinked inner mucus gel that prevents the bacterial cells from being direct contact with the colon cell membrane.55 These results, together with the muc2 staining (Figure 2G), suggest the bacterial cells in the GuMI device reside relatively close to the colon epithelia in an outer diffuse mucus layer, in an arrangement similar to mucus-associated microbes in vivo. The relatively low shear in the GuMI flow arrangement may facilitate the development of this robust mucus layer.
We next confirmed the biological barrier of the epithelium by measuring the concentration of apical mucin in GuMI-NB, GuMI-FP, and Static conditions. Although samples cultured in GuMI were constantly being perfused with anoxic apical medium, the concentration of mucin measured inside Transwells harvested at the end of the 3-day experiment was comparable between GuMI-NB (2.3±1.2 μg/ml), GuMI-FP (4.1 ± 1.6 μg/ml), and Static (3.8 ± 2.2 μg/ml) (Figure 2J). No mucin was detected in the effluent collected at the apical effluent reservoirs of GuMI-NB and GuMI-FP samples (Figure 2J), although it may have been present below the detection limit of 0.5 μg/ml. Taken together, our data suggest that GuMI addresses the metabolic needs of both obligate anaerobes and colon epithelial cells and allows epithelium to maintain its physical and biological barrier properties.
The GuMI physiome platform supports fermentation by the super oxygen-sensitive bacterium F. prausnitzii
After verifying that GuMI can maintain both the colon epithelial barrier and obligate anaerobes, we next asked if bacterial fermentation occurs. F. prausnitzii is one of the most abundant species in the human fecal microbiota56 and the major producer of butyrate.57 It produces butyrate abundantly (>10 mM) in vitro.12,58 To determine if F. prausnitzii cells actively produce butyrate when co-cultured with human colon epithelia, we compared the concentration of butyrate in the apical medium and the GuMI effluent in the absence (GuMI-NB) and presence of F. prausnitzii (GuMI-FP). As expected, the concentration of butyrate in the apical medium in GuMI-NB (0.08±0.04 mM) or the inlet (source) medium (0.10±0.04 mM) was close to or below the detection limit 0.08 mM (Figure 2K). In contrast, butyrate in the bulk apical medium collected from the GuMI-FP condition increased significantly (p < 0.0001), to 0.75±0.31 mM (Figure 2K). We attribute this increase to the fermentation of apical medium substrates by F. prausnitzii. Conversely, no statistically relevant change was observed for the other two SCFA, acetate and propionate, between GuMI-NB and GuMI-FP (Figure 2K). A similar trend – i.e., a dramatic increase in butyrate for the GuMI-FP condition, with no or minor discernible changes in other SCFA – was observed in the GuMI-NB and GuMI-FP effluents (Figure S2F). These data suggest that F. prausnitzii is functionally active without compromising colon epithelial barrier functions (Figure 2E–J), as the compromise of barrier function would cause an influx of oxygen and death of the bacteria.
In vivo, SCFAs are transported apically from the luminal surface into colonocytes by the monocarboxylate transporter 1 (MCT1, also known as SLC16A1), where they are partially metabolized before excretion on the basolateral surface by SLC16A1 and possibly other transporters.59 We observed high levels of butyrate in the basolateral medium of the monolayers only from GuMI-FP, indicating that butyrate was actively taken up from the apical side and transported to the basal compartment in the presence of F. prausnitzii (Figure 2L). Under Static conditions, SCFAs were at low levels in either the basolateral or apical medium (Figure 2K and 2L), suggesting that SCFAs are consumed by colon epithelium during transport as we have previously reported for microbe-free Transwell cultures of primary human colon supplemented with apical SCFAs.60 The relatively low concentrations in Static likely reflect a general nutrient depletion state relative to GuMI-NB and GuMI-FP, as the total volume of apical medium to which the epithelial barrier in GuMI-NB or GuMI-FP is exposed during the culture period is 28.8 mL (10 μl/min × 48 h × 60 min/h) compared to 0.5 mL over the same time period in Static culture. In the GuMI-FP condition, in contrast, the measured final concentrations of propionate and butyrate in the apical compartment were at or above the source concentration, and concentrations in the basolateral medium were unexpectedly slightly higher than those in the apical media for GuMI-FP, in contrast to the descending SCFA gradient observed from colon to peripheral blood in vivo.61 This observed phenomenon likely reflects the highly inhomogeneous distribution of microbes in the apical compartment, with microbes concentrated in the mucus region associated with the monolayer (Figure 2I), causing SCFA fermentation products to reach locally high concentrations compared to that in the apical medium flowing above the mucus layer and driving active transport across a physiologically-normal descending gradient. The characteristic diffusion time for butyrate from the apical surface to the upper barrier in the GuMI device, a distance of 3 mm, is ~ 2–5 hr (see Supplementary Method S1) compared to the average time of 35 min required to replace medium in the apical compartment (volume ~350 μl) at the standard flow rate of 10 μl/min. Thus, the collection of the bulk apical medium for analysis likely dilutes the local peri-epithelial concentrations significantly.
Differences in the total volume of medium that cells experience in Static compared to GuMI culture made it difficult to directly compare the overall contribution of F. prausnitzii to the concentration of SCFAs over the course of the experiment using single time-point concentrations, we therefore calculated the net consumption or production (μmole) of individual SCFA over the entire course of the experiment, summarized in Table 1. Net consumption and transport of both acetate and propionate were observed across all conditions (i.e., Static, GuMI-NB, GuMI-FP). Net production of butyrate, however, was observed only in the presence of F. prausnitzii (Table 1). These results indicate a significant amount of butyrate was produced in apical media, while similar amounts of acetate and propionate were transported from the apical to the basolateral medium in the presence of F. prausnitzii. The total amounts of butyrate present in the (1) apical, (2) basolateral media, and (3) apical effluent at the end of the experiment in GuMI-FP were 0.26, 1.6, and 22.7 μmole, respectively, representing 5–8 times higher amounts compared to these three compartments in the GuMI-NB.
Table 1.
Net consumption or production (μmole) of SCFA acetate, propionate, and butyrate in the apical medium, basolateral medium, and apical effluent under Static, GuMI-NB, and GuMI-FP conditions.
| SCFA | Apical medium | Basolateral medium | Apical effluent | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Static | GuMI-NB | GuMI-FP | Static | GuMI-NB | GuMI-FP | Static | GuMI-NB | GuMI-FP | |
| Acetate | −0.8±0.02a | −0.2±0.1a | −0.2±0.08a | 0.3±0.4a | 1.5±0.6b | 1.4±0.6b | NA | −24±45a | −21±38a |
| Propionate | −0.3±0.03a | −0.05±0.06a | −0.08±0.07a | 0.3±0.3a | 2.0±0.7b | 2.1±0.5b | NA | −13±15a | −4.6±16a |
| Butyrate | −0.02±0.01a | −0.001±0.03a | 0.2±0.1b | 0.07±0.08a | 0.2±0.1b | 1.6±0.3c | NA | 0.1±3a | 19±10b |
| Total | −1.1±0.05a | −0.2±0.1b | −0.08±0.1c | 0.7±0.6a | 3.5±0.8b | 5±0.6c | NA | −37±48a | −6±60a |
Negative values represent consumption, and positive values represent production. The standard deviation represents the variation of the original amount (Table S2), not the net changes, of individual SCFA in 6–8 replicate samples from 3–4 independent experiments. Two-way ANOVA with Dunnett’s multiple comparisons test was used to compare the different conditions within the apical medium and basolateral medium, respectively. Multiple comparison Dunn’s test was used to compare the apical effluent of GuMI-NB and GuMI-FP, respectively. Different letters indicate a significant difference in GuMI-NB vs Static, or GuMI FP vs GuMI-NB.
NA: not applicable since there is no flow in the Static culture, thus no effluent exists to be collected. SCFA: short chain fatty acid. Data are presented as mean ± SD. See also Table S2.
In the human large intestine, the molar ratio of acetate, propionate, and butyrate is around 3:1:1.62 A mixture of these SCFA, especially propionate and butyrate, may have additive effects on their biological activities in human colon cells.63 While the total cumulative concentrations of SCFA in the apical GuMI-FP compartment (4 mM) were lower than in vivo (90 mM),64 F. prausnitzii adjusted the molar ratio of acetate, propionate, and butyrate from its original value in the source, 2.4:1:0.15 (Figure 2K, Table S2), to the 3:1:1 ratio observed in vivo in human large intestine,61 suggesting that GuMI-FP can faithfully capture some features of SCFA exposure in human colon. Finally, we note that the production of butyrate is not likely limited by the diffusion rate of glucose into the microbial layer associated with the mucus (see Supplementary Method S2).
Addition of bacteria in GuMI culture induces profound changes in global epithelial gene expression compared to only moderate changes induced by GuMI vs. Static
Physiological cues such as oxygen gradients, luminal flow, commensal bacteria, and SCFA created in the GuMI physiome platform contribute to the phenotype of the epithelial barrier. To further understand the molecular effects of these microenvironmental cues on human colon epithelia, we performed mRNA sequencing on the cells harvested from monolayers under three conditions: Static, GuMI-NB, and GuMI-FP.
We first compared the magnitudes of global changes in gene expression caused by the switch from Static to the GuMI physiome platform, and then from GuMI with no bacteria present to GuMI with F. prausnitzii; i.e, we carried out differential gene expression analysis for the comparison of two groups: GuMI-NB vs. Static, and GuMI-FP vs. GuMI-NB (Figure S3). More than 24000 genes were included for both comparisons, with the exclusion of zero-expression genes [Transcripts Per kilobase Million (TPM) = 0]. Compared to the Static, the expression levels of 1627 genes were significantly changed (adj. p<0.05, |log2Fold Change| >0.5. Figure S3A and Table S3A) in GuMI-NB, accounting for 6.7% of the expressed genes. Among the genes changed in the GuMI-NB condition, 796 genes were increased, while 831 genes were decreased (Figure S3C). Unexpectedly, the addition of bacteria to GuMI caused a much more profound change in global epithelial gene expression: 4834 genes were significantly higher in cells in GuMI-FP relative to GuMI-NB (Figure S3B and Table S3B), accounting for 20% of the expressed genes. Of these 4834 genes, more than 4000 were changed solely by the presence of F. prausnitzii (Figure S3C), as they were unchanged between GuMI-NB and Static. Only a small fraction (<16%) of the F. prausnitzii-influenced genes overlapped with GuMI-influenced genes (Figure S3C), indicating that the effects of culture alone on colon epithelia in the GuMI platform are very different from that of co-culture with F. prausnitzii at the transcriptional level. The altered genes are broadly distributed to all chromosomes, suggesting an extensive and non-chromosome-specific influence on gene expression (Figure S3D and S3E). Comparative changes in gene expression in germ-free animal models in vivo compared to those colonized with F. prausnitzii are not available, as efforts to colonize germ-free animals with F. prausnitzii as monocultures have been unsuccessful.17 Hence, GuMI culture offers a unique opportunity to explore the effects of strict anaerobes on transcriptional responses.
GuMI culture accelerates cell differentiation pathways and represses proliferation pathways with no evidence of apoptosis
Next, we probed the transcriptional changes in more detail to uncover specific categories of molecular pathways in the colon epithelia altered by these different conditions, starting with analysis of differentiation and proliferation changes induced by physiologically relevant stimuli, i.e., apical hypoxia and continuous flow of the fresh apical medium, in the absence of bacteria. For example, fluid flow is associated with enhanced differentiation of some stem cells.65 At the same time, SCFAs, particularly butyrate, also suppress cell proliferation in colon epithelial cells and the greater exposure to butyrate in GuMI culture via continuous apical medium replenishment of butyrate-containing medium may also alter differentiation and proliferation.66
Focusing on pathways involved in proliferation and differentiation, differential gene expression analysis of GuMI-NB vs. Static indicates a significant upregulation of representative cell differentiation marker genes, e.g., SLC26A3, KLF4, CDX1, DLL1, CEACAM7, and CEACAM6 (Figure 3A). Consistently, most of these genes, SLC26A3, CDX1, DLL1, and CEACAM7 were also found to be increased in more differentiated colon monolayers derived from other human donors.33 Meanwhile, proliferative or stem cell marker genes such as MKI67, LGR5, ASCL2, and MYBL2 were significantly decreased (Figure 3A). RT-qPCR confirmed a subset of these proliferative markers, MKI67 and LGR5, were decreased, while differentiation markers SLC26A3, CDX1, and CEACAM6 were increased (Figure 3B). These results suggest that cells in the GuMI-NB culture format are more differentiated than cells cultured in Static condition. To further support this idea, gene set enrichment analysis (GSEA) was performed. GSEA is designed to detect coordinate changes in the expression of genes that are related in terms of biological function, chromosomal location, or regulation.67,68 This approach has been widely used since its introduction in 2003.68,69 Herein, GSEA takes into consideration the genes that are known to be related to cell differentiation and therefore gives a comprehensive evaluation of the cell differentiation status. GSEA revealed that the cell differentiation gene set is over-represented in GuMI-NB cells, with more than 150 genes being enriched in GuMI-NB (Figure 3C and Table S3C).
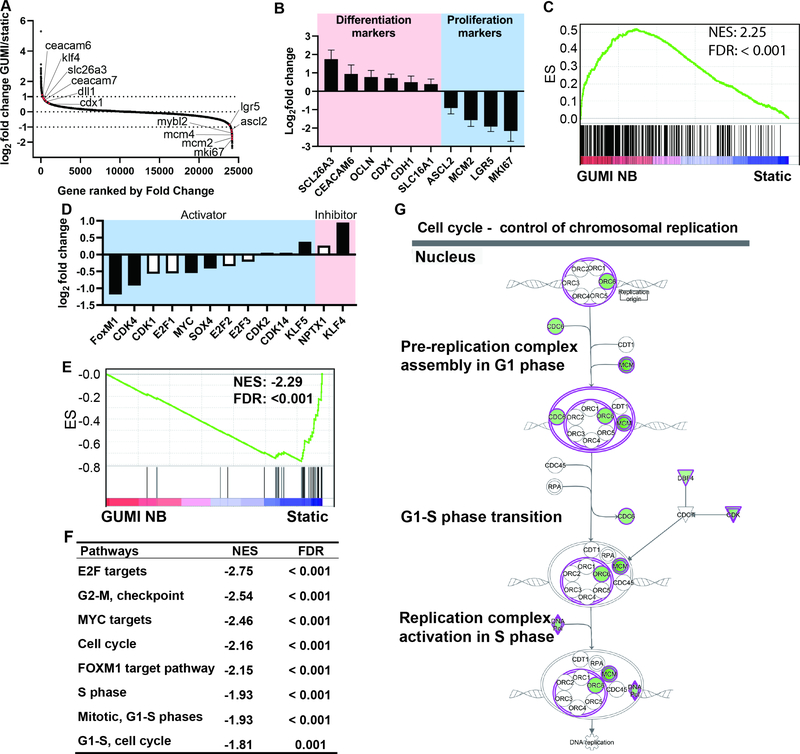
Figure 3.
GuMI promotes cell differentiation and represses cell proliferation. (A) Increased expression of representative differentiation marker genes (left upper edge) and decreased expression of representative stem cell or cell proliferation marker genes (bottom right edge) in GuMI vs Static cells. All marker genes are highlighted in red dots; all the gene symbols were in small letters due to space limit. (B) RT-qPCR confirmation of cell differentiation and cell proliferation marker genes, bars represent mean values from 6–8 replicate samples from 3–4 independent experiments. (C) Gene set enrichment analysis (GSEA) for the cell differentiation gene set revealed an over-representation in GuMI over Static monolayers. NES: normalized enrichment score. FDR: false discovery rate. (D) Fold change in the expression of cell proliferation regulatory genes in GuMI over Static monolayers. Black-filled bar indicates a significant increase or decrease; white bar indicates a non-significant increase or decrease. (E) GSEA for DNA synthesis (cell proliferation) is repressed in GuMI cells. NES: normalized enrichment score. FDR: false discovery rate. (F) Pathways related to the control of cell cycle and targeted by transcriptional factors MYC, FOXM1, and E2F were under-represented in GuMI cells, analyzed by GSEA. (G) Ingenuity Pathway Analysis (QIAGEN Bioinformatics) revealed the repression of DNA replication processes. Green background color indicates a significant downregulation, and white refers to no significant change. See also Figure S3 and Table S3.
In the normal healthy colon, the more differentiated cells in the lumen are non-proliferative relative to the stem and progenitor compartment in the crypt, suggesting that GuMI-NB culture conditions may suppress proliferation. Cell proliferation is regulated by multiple transcriptional factors and kinases.70 Some transcriptional factors such as MYC, FOXM1,71 MYBL2, SOX4, SOX9,72 CDK4,73 and KLF5 act as activators of cell proliferation, while others such as KLF4 and LGALS174 are inhibitors of cell proliferation. Indeed, the proliferation marker genes were significantly reduced in GuMI-NB culture (Figure 3B), suggesting that the promotion of cell differentiation was accompanied by the inhibition of cell growth and proliferation. We observed a significant reduction of activators such as FOXM1 and CDX1, and an increase of cell proliferation inhibitors such as KLF4 (Figure 3D). For example, FOXM1 stimulates proliferation by promoting S-phase entry as well as M-phase entry and is involved in the proper execution of mitosis.75 These processes involve DNA synthesis, which is a critical preparation process for cell proliferation. Therefore, we performed GSEA for the genes in DNA synthesis machinery. The results clearly indicated that the DNA synthesis pathway is over-represented in Static cells and dramatically repressed in GuMI-NB cells (Figure 3E). The core genes for the DNA synthesis pathway were significantly decreased in GuMI-NB cells. These genes are responsible for the pre-initiation and initiation of DNA synthesis, elongation and maturation of newly synthesized DNA (Figure S3F). In addition, other pathways related to cell cycle regulation and MYC/FOXM1/E2F-target pathways were found to be significantly repressed in GuMI-NB cells (Figure 3F). Ingenuity Pathway Analysis (IPA) confirmed that the processes for controlling chromosomal replication in the nucleus were decreased in GuMI-NB (Figure 3G). This is potentially due to the downregulation of proliferation-associated transcription factors. Indeed, using upstream regulator analysis in IPA, an algorithm that identifies molecules upstream of a gene set or pathway, we identified three proliferation-activating transcription factors, MYBL2, FOXM1, and MYC (Table S3D) and proliferation-inhibitory factors LGALS1 and KLF4 (Table S3D). In addition, the WNT-activating gene SOX476 is decreased in GuMI cells. SOX family actively interacts with the WNT/β-catenin pathway, which is key to maintaining the colonic stem cells.77
We did not see enhancement of apoptotic genes (e.g., apoptosis-inducing factor AIFM1, AIFM2, AIFM3, DIABLO [also known as SMAC], apoptosis regulator BCL2) nor did we observe evidence of death or apoptosis in the monolayers via microscopy. Together, these results suggest that the physiological stimuli maintained by the GuMI physiome platform promotes cell differentiation and represses cell proliferation, likely owing to the changes in multiple transcription and growth factors. The alterations in gene expression due to introduction of F. prausnitzii are considered separately below, after we consider additional pathways altered by the change from Static to GuMI culture.
GuMI recapitulates cell responses to hypoxia
Colon epithelial cells are exposed to a uniquely steep oxygen gradient. This oxygen gradient helps maintain a healthy colon microbiota comprising many oxygen-sensitive commensals,78 preventing aerobic pathogen expansion,79 and maintaining host homeostasis.78 Hypoxia-induced factor-1 alpha (HIF1A) is a master transcriptional regulator of cellular response to hypoxia.80,81 Thus, we hypothesized that apical hypoxia in GuMI culture would induce hypoxia-related responses in colon epithelia and promote epithelial barrier functions, and that we could find evidence of this by probing the transcriptional profiles further. First, we found under hypoxic conditions HIF1A gene expression was significantly increased (p = 0.0001) in GuMI-NB compared with Static cells cultured in normoxia, with a fold change of 1.52 (Figure 4A and 4b). RT-qPCR validation confirmed the fold change is about 1.80 (Figure 4C). The stability of HIF1A protein is regulated by an ubiquitin-proteasome-based degradation, which mandatorily requires oxygen and HIF1A hydroxylases (EGLN1–3 and HIF1AN).82 The transcription of HIF hydroxylases EGLN1–3 and HIF1AN was not significantly changed in GuMI-NB (p > 0.4 for all genes in Supplementary Figure S4A), together with the hypoxic conditions maintained in GuMI-NB (Figure 1B), suggested that HIF1A is likely stabilized.
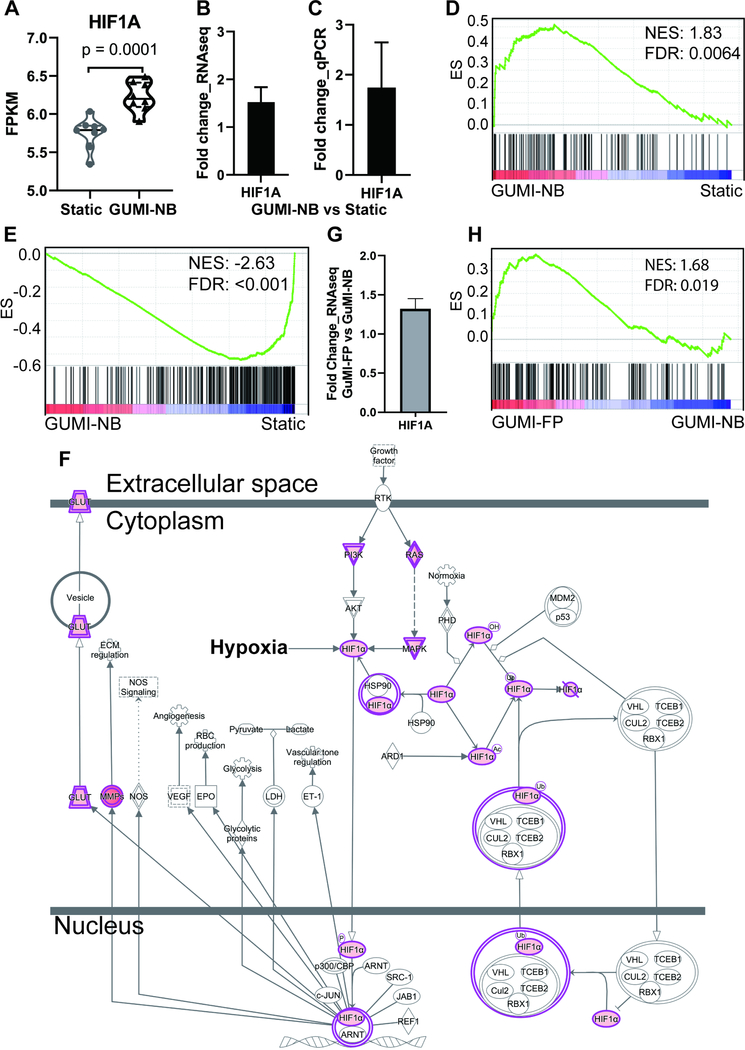
Figure 4.
The physiological oxygen gradient increases hypoxia sensing pathway. (A) absolute values of expression of hypoxia-inducible factor 1 alpha (HIF1A), a hypoxia response gene from RNA-seq expressed as FPKM: fragments per kilobase of exon model per million reads mapped. (B-C) Fold change of HIF1A mRNA in GuMI-NB vs Static based on (B) RNA-seq and (C) RT-qPCR. Bars in (B-C) represent the mean±SD of 6–8 replicate samples from 3–4 independent experiments. Black-filled bars indicate a significant change (log2FC>0.5, adj. p<0.05). (D) GSEA revealing that genes in response to HIF1A are overrepresented in GuMI cells. (E) GSEA revealing that genes downregulated in response to HIF1A are underrepresented in GuMI cells. (F) IPA revealing the curated pathways responding to HIF1A are increased in response to increased expression of HIF1A. (G) Increased expression of HIF1A in GuMI-FP vs GuMI-NB. Grey bar indicates a significant change (adj p<0.05) but log2FC<0.5. (H) GSEA revealing that genes in response to HIF1A are overrepresented in cells exposed to F. prausnitzii (GuMI-FP vs GuMI-NB). See also Figure S4.
HIF1A is a heterodimeric DNA-binding complex that regulates an extensive transcriptional response to hypoxia.83 The activation of HIF1A induced by hypoxia has a protective role in mucosal barrier function in vivo and on cells in vitro.84 We thus asked if the target genes or pathways of HIF1A are changed due to elevated expression of HIF1A. To test this, we performed GSEA using the compiled gene sets that are known to respond to HIF1A or HIF1A inducers in different types of cells.85,86 Genes known to be increased in response to HIF1A are indeed over-represented in GuMI-NB, while genes known to be decreased by HIF1A are underrepresented in GuMI-NB (Figure 4D and 4E). This response agrees with Ingenuity Pathway Analysis, which revealed that pathways responding to HIF1A are significantly upregulated (Figure 4F). In the presence of F. prausnitzii (GuMI-FP), the transcription of HIF1A was slightly but significantly (log2FC=0.4, adj. p = 0.0077) increased in GuMI-FP over GuMI-NB (Figure 4G). In agreement, gene set responding to HIF1A was slightly enriched (NES =1.68, FDR = 0.019, Figure 4H). Together, these results suggest that hypoxia conditions maintained by GuMI are able to recapitulate colonic cellular responses to a physiologically-steep oxygen gradient. The increase of HIF1A mRNA, and suppression of cell cycle related pathways was also observed in Caco-2 monolayers cultured anaerobically with F. prausnitzii for 12 h.23
Having established the differentiational regulation of proliferation, differentiation, and hypoxia pathways by the switch to GuMI culture from Static, we next examined pathways regulated by introduction of bacteria into GuMI culture. The number of global gene expression changes was far greater in the GuMI-FP vs. GuMI-NB compared to the GuMI-NB vs. static, suggesting potential alterations in several classes of pathways.
F. prausnitzii exerts anti-inflammatory effects on the epithelial monolayer and represses TLR3 and TLR4 expression
The GuMI device maintains a microenvironment that supports long-term (2–4 days) co-culture of a primary human colon monolayer with a continuously-growing commensal bacterial population including the strictest of anaerobes F. prausnitzii (Figure 2), one the most abundant species in the human colon microbiome. In humans, F. prausnitzii is the major producer of butyrate (Figure 2K) which, together with other metabolites,14 mediates an array of effects on host cells including modulation of immune cell behavior and inhibition of proliferation and inflammation.78,87–89 Secreted products of F. prausnitzii have been shown to modulate the NFKB signaling pathway in both cell culture and mouse colon injury models.9,48 F. prausnitzii has been identified as an anti-inflammatory bacterium, whose abundance was significantly decreased in IBD patients.9,90
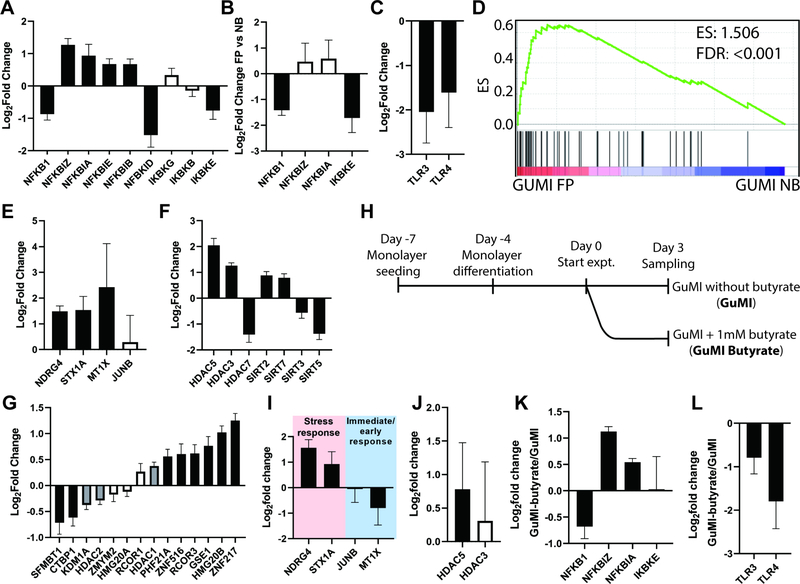
We therefore probed the transcriptional profiles of GuMI-FP vs. GuMI-NB in detail, to illuminate whether GuMI-FP was able to recapitulate reported anti-inflammatory effects of F. prausnitzii. We first focused on the transcription of NFKB1, its upstream regulation pathway and its downstream target genes in GuMI-FP over GuMI-NB. No comparison of GuMI-FP over Static was carried out as any difference cannot be solely attributed to F. prausnitzii. First, NFKB1, the key subunit of the NFKB complex, was significantly decreased by two-fold in GuMI-FP over GuMI-NB cells (Figure 5A). The NFKB complex is regulated by multiple mechanisms, including NFKB inhibitors, NFKB activators, and TLR-NFKB signaling. We therefore next examined the gene expression of NFKB inhibitor (NFKBI) genes, which encode NFKBI proteins to form a complex with RELA and NFKB1, preventing the activation of NFKB. To activate NFKB, IkappaB kinase (IKBK) phosphorylates NFKI protein, which then is ubiquitinated and degraded by proteasome.91 Herein we show that four out of five NFKBI genes, i.e. NFKBIZ, NFKBIA, NFKBIE, and NFKBIB, were increased (Figure 5A and 5B). On the other hand, IKBKE, one of the three NFKB activating genes, was decreased, while the other two were not significantly changed (Figure 5A and 5B).
Figure 5.
Effects of F. prausnitzii on colon epithelia in GuMI. (A) RNA-seq analysis reveals changes in gene expression of NFKB1, its inhibitors (NFKBI) and activators (IKBK) induced by F. prausnitzii. (B) Validation by RT-qPCR of changes in NFKB1, NFKBIZ, NFKBIA, and IKBKE induced by F. prausnitzii. (C) Validation by RT-qPCR of changes in TLR3, TLR4 induced by F. prausnitzii. (D) GSEA revealing that genes that respond to butyrate are overrepresented in GuMI-FP cells over GuMI-NB cells. Differentiation and proliferation gene sets are not overrepresented. (E) Validation by RT-qPCR of a subset of butyrate responsive genes. (F) RNA-seq analysis reveals HDAC genes that are significantly (log2FC ≥ 0.5, adj. p <0.05) changed by F. prausnitzii. (G) RNA-seq analysis reveals CoREST complex that are changed by F. prausnitzii. Data presented in (A-G) are comparison of GuMI-FP vs GuMI-NB. Black-filled bar in (A-C, E-G) indicates a significant increase or decrease; grey bar indicates a statistically significant (adj. p< 0.05) but log2FC <0.5, white bar indicates a non-significant increase or decrease. Bars in (A-C, E-G) represent the Mean±SD of 4–8 replicate samples from 3–4 independent experiments. (H) Timeline of the experiment to determine the effects of butyrate on colon epithelial cells in GuMI. (I-L) RT-qPCR analysis of (I) a subset of butyrate-responding genes NDRG4, STX1A, JUNB, and MT1X; (J) HDAC3 and HDAC5; (K) NFKB1, NFKBIZ, NFKBIA, and IKBKE; (L) TLC3 and TLR4 in GuMI-butyrate over GuMI. Bars in (I-L) represent the Mean±SD of 3 replicate samples from 2 independent experiments. Black-filled bar in (H-K) indicates a significant increase or decrease; white bar indicates a non-significant increase or decrease. See also Table S4.
We next looked at the toll-like receptors (TLR), including TLR2, TLR3, and TLR4, which are directly involved in the molecular recognition of bacteria and regulation of the NFKB pathway92 and which show dysregulated expression in UC and CD patients.18 We find that F. prausnitzii down-regulates TLR3 and TLR4 expression and the NFKB pathway in primary colon epithelia (Figure 5C). This finding of reduction in TLR4 expression is consistent with F. prausnitzii as more abundant in healthy individuals than patients with UC or Crohn’s;9,90 however, TLR3 is reduced in active CD compared to healthy, and not changed in UC patients.18 Using Caco-2 cells, it was found that F. prausnitzii activates TLR3 in Caco-2 cells during their short-term and aerobic co-culture.93 Further studies are needed to illuminate the discrepancies in TLR3.
Two pathways can respond to changes in TLR3 and TLR4. One is dependent on MYD88, TIRAP, and IRAK, and the other one is dependent on TICAM and MAP3K14.94,95 These two responding pathways upregulate the same downstream factors, i.e. MAP3K7 and TAB, which then regulate the phosphorylation of NFKB. Interestingly, with the decrease of TLR3 and TLR4 (Figure 5C), the responding pathway MYD88-TIRAP-IRAK was downregulated, whereas the other responding pathway TICAM1-MAP3K14 was up-regulated. As a result, MAP3K7-TAB was not significantly changed (Figure S4B). Together, these results suggest that F. prausnitzii inhibits NFKB, potentially through upregulation of NFKB inhibitors, downregulation of NFKB activator and TLR3/4, but with no change in expression of MAP3K7-TAB.
In our experimental setting, the genes known to be induced by butyrate96–98 are overrepresented in GuMI-FP over GuMI-NB cells (Figure 5D), with a subset of genes, i.e., NDRG4, STX1A, and MT1X, in the gene set confirmed by RT-qPCR (Figure 5E). Consistently, IPA identified butyric acid as an upstream regulator which activates several pathways, including histone deacetylases (HDAC) (Table S4). The transcription of genes is largely regulated by epigenetic modification mediated by histone acetylases (HAT) and HDAC. Therefore, we examined the changes of HAT and HDAC genes influenced by F. prausnitzii. Interestingly, no HAT was changed in the presence of F. prausnitzii, whereas seven out of eighteen HDAC (HDAC1–11, SIRT1–7) were significantly changed (adj. p <0.05, log2FC ≥ 0.5, Figure 5F). HDAC3, HDAC5, SIRT2 and SIRT7 were increased, while HDAC7, SIRT3 and SIRT5 were decreased (Figure 5F). Moreover, the HDAC-family CoREST complex, which is commonly associated with gene expression silencing, was significantly changed in the presence of F. prausnitzii (Figure 5G). These results indicate global and differential changes across different HDAC by F. prausnitzii.
Despite the relatively greater exposure to butyrate produced by F. prausnitzii, cell differentiation was not further enhanced in GuMI-FP over GuMI-NB, as most of the differentiation marker genes did not change significantly (adj. p >0.05) in GuMI FP over GuMI-NB. These genes include SLC26A3, KLF4, CDX1, DLL1, CEACAM7, and CEACAM6. Consistently, proliferative markers were also relatively unchanged between GuMI-NB and GuMI-FP, i.e. MKI67, LGR5, and ASCL2, with one exception, MYBL2, which is significantly increased in GuMI-FP. These results indicate that the monolayer in the platform was at a very high differentiation state.
Butyrate contributes to the anti-inflammatory effect induced by F. prausnitzii
The analysis of transcriptional changes in GuMI-FP compared to GuMI-NB implicates butyrate as a major contributor. To isolate the effects butyrate from all other changes brought about by co-culture with F. prausnitzii, we carried out a new experiment in which we exposed cells in GuMI-NB to 1 mM butyrate in the anaerobic apical medium (Figure 5H). The concentration used was to mimic the butyrate production of F. prausnitzii in GuMI-FP, with default presence of acetate and propionate (Figure 2K). We first determined the expression of multiple stress response genes (NDGR4, STX1A, JUNB, and MT1X), which appears in enriched butyrate-responding pathway (Figure 5I), and found NDRG4 and MT1X were increased by up to three-fold, while the immediate-early response gene JUNB was not changed and MT1X was decreased (Figure 5I). This data confirmed that butyrate contributes to the effects produced by F. prausnitzii, but the pattern is not exactly the same as for F. prausnitzii. Similar discrepancies were observed for HDAC, with a significant increase of HDAC5 and no change of HDAC3 in GuMI-Butyrate comparing to GuMI (Figure 5J). Interestingly, we found NFKB pathway was modulated by butyrate in a similar pattern as by F. prausnitzii (Figure 5J and 5A). Finally, TLR3 and TLR4 were decreased by butyrate (Figure 5L) in a similar manner to the exposure to F. prausnitzii, suggesting that butyrate largely contributes to the TLR and NFKB regulation by F. prausnitzii. These results together confirmed that butyrate contributes to the gene-regulatory effects, especially TLR downregulation, induced by F. prausnitzii.
Butyrate (1 mM) decreases TLR4 expression in the colon cancer cell line HCT116 at 48–72 hours post-exposure.99 This inhibitory effect of butyrate on TLR4 expression was also observed in mouse adipose tissue in vivo.100 Sodium butyrate significantly decreased the TLR4 and NFKB signaling pathways in lipopolysaccharide-induced acute lung injury in mice.101 In addition, butyrate inhibits the NF-kB pathway in the HT-29 colon cancer cell line.102 However, this effect might be tissue- or cell type-specific. In SW480 cells and mouse colon cancer CT26 cells, butyrate upregulates TLR4 expression.103
The similarity in the transcriptional changes that occurred in response to butyrate and F. prausnitzii indicates that butyrate is an important contributor to the effects of F. prausnitzii. However, there are also discrepancies observed between butyrate and F. prausnitzii, suggesting that there are other effectors (metabolites, secreted proteins, or bacterial cell components) that contribute to the effects of F. prausnitzii. Indeed, F. prausnitzii can secrete the anti-inflammatory protein MAM,48 which downregulates the NFKB pathway when it is transfected into a colon cancer cell line.48 In another study using a gnotobiotic mouse colitis model, many metabolites in the gastrointestinal tract were associated with colitis-protective effects by F prausnitzii.14 F prausnitzii-produced salicylic acid, in addition to butyrate, was shown to contribute to the attenuation of inflammation.14
Butyrate is also a known modulator of HDAC genes in colon cancer cell lines.88,98,104 It has been shown that butyrate regulates the SIRT family of deacetylases in human neuronal cells, with increased expression of SIRT1, SIRT5, and SIRT6 and downregulation of SIRT2, SIRT4 and SIRT7.105 The regulation pattern of SIRT, the class III HDAC, is consistent with that of butyrate.105 However, the activation of HDAC3 and HDAC5 by F. prausnitzii and/or butyrate does not agree with previous observations, where both HDAC3 and HDAC5 were inhibited by butyrate.106 While the detailed mechanisms require further research, it is worth noting that most of these studies were carried out with cancer cell lines.106 Indeed, even with different colon cancer cell lines, the response to the same amount of butyrate is different, with some cell lines being very sensitive to butyrate, and others resistant.97 The regulation of HDACs by butyrate in other cell types is similarly heterogeneous: 1 mM of butyrate was shown to induce the expression of HDAC1 and HDAC3 in human PBMCs after 6 and 48 h, but repress the expression of HDAC2 after 48 h;107 oral butyrate for 8 weeks decreased HDAC2 expression in cardiac cells from Wistar rats;108 and sub-mM butyrate inhibits the NFKB signaling pathway and histone deacetylation in intestinal epithelial cells and macrophages.109
Conclusions
This report is, to the best of our knowledge, the first to describe long-term co-culture of the super oxygen-sensitive anaerobe F. prausnitzii with a primary human colon mucosal barrier using continuous flow in the apical compartment to foster robust growth and metabolic activity of the microbial population. To accomplish this, we designed and fabricated a GuMI physiome platform that maintains a mucosal barrier-anaerobe interaction in a manner that allows for continuous media exchange in both compartments, along with the introduction of microbes, including pathogens, following stabilization of the epithelial barrier. This platform houses six independent culture chambers with associated independent media flow circuits. Using primary human colon epithelial monolayers co-cultured continuously with super oxygen-sensitive bacterium F. prausnitzii for two days, we demonstrated the epithelium experiences a steep oxygen gradient with an apical environment of sufficiently anaerobic nature to foster the growth and active fermentation of F. prausnitzii. Using transcriptomics, GSEA, and RT-qPCR, we identified elevated differentiation and hypoxia-responding genes and pathways in the platform over conventional static culture. We further used this platform to elucidate the responses of primary colon epithelia to commensal F. prausnitzii and demonstrated an anti-inflammatory effect of F. prausnitzii through the HDAC, TLR-NFKB axis. Finally, we identified butyrate largely contributes to these anti-inflammatory effects by downregulating TLR3 and TLR4.
Limitations of Study
While the conclusions of this study relating to inflammatory pathways are limited by the use of a single donor, donor choice is unlikely to influence a major outcome of this work: demonstration of long-term co-culture of F. prausnitzii with a human primary colon monolayer. We anticipate further studies with additional donors will illustrate generality of the inflammation findings, and anticipate that donor choice may influence more complex microbial communities involving F. prausnitzii, as seen clinically. Although monolayers were used for this initial demonstration, the device can also accommodate 3D crypt-containing tissue-engineered mucosal barrier structures such as those previously described.110 Overall, this platform faithfully recapitulates several important features of the physiological microenvironment in the colon in vivo and has the potential to enable a better understanding of human colon mucosal barrier-bacteria interactions. Given the reported difficulties on translating microbiome/bacteria-based therapy from animal models to human recently,111,112 GuMI could help better understanding the inconsistency on the translation.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Linda G. Griffith (griff@mit.edu)
Materials Availability:
All unique reagents and materials generated in this study are available from the Lead Contact upon request.
Data and Code Availability:
Raw RNAseq data of the colon epithelial donor could not be included due to sharing restrictions but can be shared upon request to the Lead Contact (Linda G. Griffith, griff@mit.edu).
Experimental Model and Subject Details
Colon organoid culture and monolayer establishment
Colon organoids used in this study were established by the Yilmaz lab at the Koch Institute/MIT (HC2978 – 30 yr male patient for diverticulosis and diverticulitis, the normal appearing region of rectosigmoid sample used). Endoscopic tissue biopsies were collected from the ascending colon of de-identified individuals at either Massachusetts General Hospital upon the donors informed consent. Methods were carried out in accordance with the Koch Institute Institutional Review Board Committee as well as the Massachusetts Institute of Technology Committee on the use of humans as experimental subjects. The colon monolayer model was derived from primary human colon organoids. The medium used for maintaining organoids and monolayers include a base medium, organoid growth medium, seeding medium, and differentiation medium. The recipe for each medium is listed in Table S5A.
The establishment and maintenance of the organoids were done according to the protocols previously described.41,60 In brief, organoids grown in Matrigel (growth factor reduced, phenol red free; Corning, 356231) droplets were passaged every seven days at a 1:3 split ratio. A medium change was performed on day four after passaging. To prepare the monolayer, organoids were collected at day 7 and pelleted by centrifugation (1000 g × 5min, 4°C). After that, organoid pellet was disrupted using Cell Recovery Solution (Corning, 354253; 1mL per 100 μL Matrigel). The resulting organoid suspension was then incubated on ice for 45–60 min, pelleted, and resuspended with 1 mL pre-warmed PBS without calcium and magnesium (PBS−/−, Gibco, 10010–023) containing 2.5mg/mL Trypsin (Sigma, T4549) and 0.45 mM EDTA (Ambion, AM9260G). The resuspended organoids were warmed up (37 °C water bath for 5 min) and then manually dissociated into single cells using a 1000-μL pipette with a bent tip. Trypsin was neutralized with 10% FBS in base medium. The cell suspension was then pelleted at 300 g × 5 min, 4°C. Finally, then cell pellet was resuspended in the seeding medium. Cell density and viability were determined using an automated cell counter (Invitrogen) and TrypanBlue. Before seeding, Transwells were coated with rat tail collagen I (Gibco, A10483–01, 50 μg mL−1 in PBS) for 1–2 hours in the incubator, then were washed with PBS right before adding the cells. For seeding, cells were diluted to a density of 600,000 cells per mL in seeding medium and seeded (500 μL) into the apical side of each 12-well collagen-coated Transwell (surface area: 1.12 cm2) and 1.5 mL cell-free seeding medium was added to the basolateral side. On day three after seeding, the monolayers were differentiated by switching to the antibiotic-free base medium on the apical side and differentiation medium on the basolateral side. After switching to differentiation medium, the monolayers were further cultured for four days (total seven days), with medium change on day five. On day seven after seeding (day four after differentiation), the monolayers were used for experiments.
Bacteria culture and maintenance
E. rectale ATCC33656, B. thetaiotaomicron VPI-5482 were purchased from ATCC. Faecalibacterium prausnitzii DSM17677 was obtained from the Harvard Digestive Disease Center. The identity of all strains was confirmed using Sanger sequencing (see below). Bacteria from glycerol stock were plated in yeast casitone fatty acid (YCFA) agar (Anaerobe Systems, AS-675), 24–48 h after being cultured at 37 °C in the incubator inside the anaerobic chamber (Coy Laboratory), a colony was picked and cultured in Hungate tubes containing liquid YCFA medium (Anaerobe Systems, AS-680). Standard YFCA medium contains 33 mM acetate, 9 mM propionate and no butyrate.12 O2 in the anaerobic chamber was constantly removed by the Palladium Catalyst (Coy Laboratory, #6501050), which was renewed biweekly by incubating in the 90 °C oven for two days.
METHOD DETAILS
Colon epithelial monolayer culture in the GuMI device
All components of the GuMI device (Figure 1) were sterilized by autoclave (121 °C, 45 min), except the pneumatic plates, oxygen probes, and probe controlling boxes, which were sterilized with ethylene oxide. Then the device was assembled under sterile conditions. Antibiotic-free base medium (1.5 mL; see Table S5A for composition) was pipetted into the basal compartment. GuMI apical medium (110 mL), comprising filter-sterilized diluted YCFA medium (10% YFCA in PBS+/+) was added to the apical source reservoir on top of the GuMI device (total capacity 150 mL). The medium in the apical source reservoir was deoxygenized with 5% CO2, 95% N2 for 45–60 min before being introduced into the apical inlet through stainless steel tubing (Figure 1). After that, the apical inlet of the Transwell was temporally blocked with a 200-μl pipette tip to force the deoxygenized apical medium to flow out of the injection port, which was then sealed with an injection septum and a customized stainless-steel hollow screw. The pipette tips were then removed. The colon epithelial monolayers were transferred to each of the 6 basolateral reservoirs designed to accommodate standardized Transwells and the apical medium of the monolayers was replaced with the 10% diluted YCFA in PBS+/+. Then the entire basal plate was integrated with the apical plate using the lever (Figure 1). The system was primed 24 h in a cell culture incubator while the medium in the apical source reservoir was constantly purged with 5% CO2, 95% N2. The recirculation flow rate in the basal compartment was 5 μl/min and the apical flow rate was 10 μl/min. The effluent was cleared every 24 h with a 10-ml syringe (302995, BD Biosciences) throughout the experiments.
Bacteria co-culture with colon epithelial monolayers
Colon epithelial monolayers were cultured in the GuMI device for 24 h before the addition of bacteria. The overnight grown bacterial cultures were diluted 1000 times with pre-reduced YCFA medium. After that, 0.8–1 ml of the diluted bacterial cells were slowly injected into the apical channel through the injection port (Figure 1) using a 1-ml syringe (309659, BD Biosciences) with a needle (305127, BD Biosciences). Before bacteria injection, the flow was paused to ensure the bacterial culture going through in one direction from the inlet and outlet of the apical channel. After one-hour settling of the bacterial cells, the flow was resumed in both apical and basolateral sides. At the end of the experiment, the whole device was transferred to a biosafety cabinet and the basal plate was carefully disassembled using the lever. The sealed Transwells were individually take off from the apical plate and placed onto a new 12-well plate. Immediately after that, the apical medium was collected using a 1-ml syringe with a short needle (305122, BD Biosciences), and then immediately injected into a 20-ml pre-reduced and autoclaved HDSP vial (C4020–201, Thermo Scientific) sealed with 20-mm Crimp Cap (9502501–1S, MicroSolv). Then all the vials were transferred into an anaerobic chamber, where 10 μl of the apical medium was used for CFU counting on agar plates. The rest of the medium was transferred into a 1.5-ml polypropylene tube, where bacterial cells were pelleted in a microcentrifuge (14000 g × 5 min). The supernatant was transferred into a new tube. All samples were stored at −80 °C until further analysis.
The Transwells were washed with PBS+/+ (14040182, Thermo Scientific) twice in both apical and basolateral sides to completely remove cell-culture medium prior to bright field images and TEER measurement. After aspirating PBS, 350 μl of 1% 2-mercaptoethanol solution was added into the apical side, followed by incubation for 10 min at room temperature. One volume of 70% ethanol was then added and mixed homogeneously with pipetting. The mixture was collected and stored at −80 °C until further analysis.
Immunofluorescence staining
Monolayers seeded in Transwells were washed with PBS+/+ and fixed in 4% formaldehyde for 10 minutes. The samples were then washed three times with PBS+/+ and permeabilized with 0.2% Triton-X for 10 minutes. After permeabilization, the wells were washed twice in PBS and blocked with Blockaid (Thermo Scientific B10710) for one hour. Primary antibodies diluted in Blockaid were incubated with samples overnight at 4°C. The following antibodies were used in the experiments at 1:200 dilution: anti-Muc2 (Abcam ab90007), anti-NHE3 (Novus, NBP1–82574). The samples were then washed three times in DPBS and incubated with secondary antibodies –Alexa Fluor 568 (1:200), phalloidin-Alexa Fluor 488 (1:20, ab176753) and DAPI (1:1000) diluted in Blockaid for one hour at room temperature. After washing the samples with PBS+/+ for three times, the monolayers were excised and mounted on coverslip using ProLong Gold antifade reagent (Thermo Fisher). Mounted samples were imaged with Zeiss LSM800 confocal microscope.49
The staining procedures were modified to preserve the bacterial cells attached to the colon epithelia (specifically for Figure 2h). Briefly, monolayers taken off from the platform were immediately fixed with 4% formaldehyde for 10 minutes following a very gentle sampling of the apical medium. The samples were then permeabilized with 0.2% Triton-X for 10 minutes. After permeabilization, the wells were washed once with PBS+/+ and immediately stained overnight with Phalloidin-iFluor 488 Reagent (ab176753–300TEST) and DAPI (1:1000) in Blockaid at 4 °C. After washing the samples with PBS+/+ for two times, the monolayers were excised, mounted, and imaged as described above.
PCR and Sanger sequencing for bacteria
Bacteria identity was confirmed by Sanger sequencing by adapting the established protocol.113 Briefly, bacterial cells collected from the apical side of GuMI were collected and pelleted by centrifugation (12000 g × 5 min). The DNA was extracted using GeneElute bacterial DNA kit (NA2110, Sigma-Aldrich) by following the manufacturer protocol. Afterward, PCR was performed in triplicate to amplify 16s rDNA using DreamTaq Green PCR Master Mix (K1081, Thermo Fisher Scientific Inc.) with primers F8 (5’-AGTTTGATCCTGGCTCAG-3’) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3’) by following the procedures described elsewhere.58 PCR products were purified using DNA purification solid-phase reversible immobilization magnetic beads (G95, Applied Biological Materials Inc.) and the purified products were sent out for Sanger sequencing (Genewiz Inc.).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed to quantify gene expression. Briefly, the mRNA was converted to cDNA using the High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, 4387406). TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific, 4444557) and TaqMan probe (Table S5B) were mixed in MicroAmp EnduraPlate Optical 96-well fast clear reaction plate with barcode (Thermo Fisher Scientific, 4483485) according to manufacture protocol. TaqMan probes used in this study are available in Table S5B.
Transepithelial electrical resistance measurement
EndOhm-12 chamber with an EVOM2 meter (World Precision Instruments) was used to measure the transepithelial electrical resistance (TEER) values.
Mucin measurement
Mucin measurement is performed according to the method previously reported114 and adapted to the 96-well plate. Briefly, Dye stock solution contains 1% (w/v) alcian blue (A5268, Sigma-Aldrich) and 0.098% (v/v) H2SO4. Before diluting to dye working solution, dye stock solution was syringe filtered (0.22 μm PVDF, Cat No 09–720-3, Fisher Scientific) to remove the particles. Dye working solution containing 0.25% Triton X-100, 0.098% H2SO4, and 5% dye stock solution was syringe filtered. Ten microliters of reaction solution (0.147% v/v H2SO4, 0.375% Triton X-100, and 4 M guanidine HCl) was added to each 10 μl of standard series (0, 4.84, 9.68, 19.37, 38.75, 67.5, 125, and 250 μg/mL) in bacterial medium (10% YCFA in PBS+/+) along with 100 μl of dye working solution. After centrifugation for 20 min at 2400 g, the supernatant was removed by reverting the 96-well plate to the bed of tissue papers. Then, an 8M guanidine HCl solution was added to dissolve the pellet. The absorbance at 600 nm was recorded and mucin concentration was determined from a calibration curve. Technical duplicates were included for all the samples and standards.
Liquid chromatography-mass spectrometry (LC-MS) analysis
Short chain fatty acids, butyrate, propionate, and acetate, were quantified by LC-MS using the derivatization method reported elsewhere.115 Briefly, SCFA in the samples were derivatized by mixing with 20 μl sample with 20 μl 3-nitrophenylhydrazine, and 20 μl 120 mM N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide HCl. The reaction mixture was incubated at 37 °C for 30 min before being diluted 20 times with 9:1-water:acetonitrile mixture. The diluted samples were then filtered with 0.22 μm PTFE membrane. The filtrate was collected and immediately injected for LC-MS analysis or store at −20 °C until analysis within a week. An Agilent 1200 HPLC system (Agilent) coupled to a Q-TOF mass spectrometry (Agilent) and an electrospray ionization (ESI) source was used. Chromatographic separation was carried out using a Phenonemix NH2 column (4.6 ×150 mm, 4.6 μm). Analytes were eluted with mobile phase A (water with acetic acid) and B (acetonitrile with acetic acid) at 0.35 ml/min with the following gradient program: 0–2 min 35% B; 15.5–17 min 98% B; 17.5–25 35% B. The elute was ionized in negative mode with following parameters: gas temperature 300 °C, drying gas 8 l/min, nebulizer 30 psi, sheath as temperature 350 °C, sheath gas flow 12 l/min, MS TOF fragmentor at 70 V, skimmer 65 V. Targeted m/z for acetate, propionate, and butyrate, and isotope labeled butyrate were 194.0571, 208.0728, 222.0884, and 229.1324, respectively. The detection limit for all analytes was 0.08 mM.
RNA extraction and quality control
Prior to extraction, the cell lysate in 1% 2-mercaptoethanol solution was mixed with one volume 350 μl of 70% ethanol and pipetted to a homogeneous mixture. Then total RNA was extracted using PureLink RNA mini kit (ThermoFisher, 12183020) by following the manufacture protocol, except treating samples with PureLink DNase (ThermoFisher, 12185010) during one of the wash steps to remove DNA. The total RNA was analyzed by Bioanalyzer in BioMciroCenter at MIT. All the RNA samples passed the QC with RNA quality number (RQN) 9.8–10.0.
Library preparation and Illumina sequencing
The library preparation, sequencing, and analysis were carried out in BioMicro Center at MIT. Briefly, 50ng of RNA was confirmed for quality using the Agilent Fragment Analyzer and 50ng of material was polyA selected using NEBNext® Poly(A) mRNA Magnetic Isolation Module (E7490) modified to include two rounds of polyA binding and 10-minute incubations. cDNA was generated using the NEB Ultra II directional kit (E7760) following the manufacturer’s protocol using 15 cycles of PCR and a 0.9X SPRI clean. The resulting libraries were quality assessed using the Agilent Fragment Analyzer and quantified by qPCR prior to being sequenced on the Illumina HiSeq2000. The 40nt single-end reads with an average depth of 5 million reads per sample were sequenced for all conditions.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA-seq data analysis
Differential gene expression.
To guarantee data quality for downstream analyses, QC was performed by MIT BioMicro Center in-house QC software (script upon request) to monitor sequencing error rate, GC bias, sequence count, mapping rate, contamination, CDS percentage, UTR percentage, intro percentage, intergenic percentage, exon to intron ratio, 5’ to 3’ ratio, sense to antisense ratio, rRNA percentage, sequence complexity, and number of genes detected. The single-end sequences were mapped to GRCh38 reference sequence by STAR 2.5.3a116 using the following parameters: --outFilterType BySJout: keep only those reads that contain junctions that passed filtering into SJ.out.tab; --outFilterMultimapNmax 20: alignment will be output only if it has fewer mismatches than 20; --alignSJoverhangMin 8: minimum overhang for unannotated junctions 8bp; --alignSJDBoverhangMin 1: minimum overhang for annotated junctions 1bp; --outFilterMismatchNmax 999: maximum number of mismatches 999. Large number switches off this filter; --alignIntronMin 10: minimum intron size: genomic gap is considered intron if its length>=10, otherwise it is considered Deletion; --alignIntronMax 1000000: maximum intron size 1000000 bp; --alignMatesGapMax 1000000: maximum gap between two mate 1000000bp; --outSAMtype BAM SortedByCoordinate: output sorted by coordinate Aligned.sortedByCoord.out.bamfile, similar to samtools sort command; --quantMode TranscriptomeSAM: outputs alignments translated into transcript coordinates in the Aligned.toTranscriptome.out.bam file (in addition to alignments in genomic coordinates in Aligned.*.sam/bamfiles).
The sorted bam files were further indexed using samtools 1.3. Gene expression was estimated by calculating the gene level raw read counts, FPKM, and TPM using rsem 1.3.0 rsem-calculate-expression. Due to the strand specificity of the library, only reverse strand was counted using –forward-prob 0 option. In addition, the command –calc-pme option was applied to run collapsed Gibbs sampler for posterior mean estimation. The raw counts, log2(FPKM+1), and log2(TPM+1) from all samples were merged into 3 corresponding tables using MIT BioMicro Center in-house tools (script available upon request). Differential expression was performed using DESeq2 1.10.1 based on gene level raw counts. Pair-wise comparisons were performed across two conditions: NB versus S, and FP versus NB. To reduce the burder for multi-testing correction, genes with no expression in any samples during comparison were filtered away. Up-regulated and the down-regulated genes were further selected by MIT BioMicro Center in-house tools. Up-regulation is defined as: baseMean>10, log2FoldChange>0.5, and adjusted p <0.05. Down-regulation is defined as: baseMean>10, log2FoldChange<−0.5, and adjusted p <0.05.
Gene set enrichment analysis.
Gene set enrichment analysis were performed by GSEA 4.0.3.67 log2(TPM+1) of the expressed genes in each comparison were project to compiled gene sets from literature or databases that have curated gene sets (Hallmarks and MSigDB). The significantly enriched gene set is defined as nominal p < 0.05, and FDR q-value <0.05.
Ingenuity Pathway Analysis (IPA).
The identified differentially expressed genes was uploaded in IPA to identify the networks and upstream regulators that are most significant in GuMI-NB vs Static or GuMI-FP vs GuMI-NB by Causal Networks Analysis and Upstream Regulators Analysis.117
Computational Simulation
A finite element method was performed using COMSOL Multiphysics 5.4 (COMSOL Inc.). Two modules (laminar flow fluid dynamics and transport of diluted species) were coupled to compute the flow distribution and the profile of oxygen concentration inside the apical and basal compartment of the MPS. The 3D CAD files of the MPS were drawn using Solidworks (Dassaut Systèmes®) and exported into COMSOL. Only half of the 3D MPS was simulated given the plane of symmetry along the feeding axis. First, the steady state solution for the computational fluid dynamics (CFD) was calculated using the Navier-Stokes equation assuming incompressible fluid. As boundary condition for the CFD, the interfaces between the medium and walls of the MPS (solid walls as well as on the Transwell membrane on both sides) were set as no slip conditions. The interface between the medium and air was set as a stationary free interface with intrinsic surface tension. The linear flow rates (μL min−1) at both inlets (basal in recirculation and apical for feeding) were set according to experimental measurements, and were assumed to be fully developed. The two outlets (basal for recirculation and apical for effluent) were set as outflows for the simulation (degree of freedom for pressure). To simulate the transport of oxygen molecules, a diffusion/convection model was applied using Fick’s 2nd law. The oxygen consumption rate (OCR) was calculated based on published OCR from intestinal epithelial cells. The total number of epithelial cells on the Transwell was estimated and the thickness of the tissue was measured using confocal microscopy. The flux of oxygen through the membrane of the Transwell was allowed through passive diffusion using the manufacturers porosity. The inlet on the apical side was set at a fixed oxygen concentration (O2 deprived medium). On the basal compartment, the inlet and outlet coming in and out of the pump are constrained to the same oxygen concentration using a continuity condition. The standard mesh was applied, and all simulations were performed with the assumption that the system is at 37 °C, in an air composed of 5% CO2 and with 100% humidity.
A first simulation was performed as time-dependent to characterize how fast the system reaches equilibrium (steady state oxygen delivery). A second simulation was performed at steady state to fully characterize oxygen and shear stress distribution in the MPS. Parameters used in the simulations are provided in Table S1.
An estimate of the purge time of tiny bubbles entrained during placement of the Transwell was carried out as follows: Assuming a 50-μl air bubble underneath the upper insert into the apical compartment that forms the upper wall of the apical flow channel, with 0.4 cm2 of contact area with liquid medium. The half-life of diffusion from the entrained air bubble to the liquid medium is about 5.6 h. At flow rate 10 μl min−1, the speed of purging is 0.183 × 10−8 mol min−1. As the concentration of oxygen in the bubble is equal to m/v = 0.2 × 1.01 × 105/8.314/310 = 7.84 mol m-3. The time required for purging the medium is C × V/ Speed = 7.84 mol m−3 × 50 × 10−9 m3/ (0.183 × 10−8 mol/min) = 3.5 h. Diffusion is longer than purging. Therefore, diffusion from the air bubble to apical medium is the decisive factor for reaching equilibrium. Generally, it takes about five half-lives to replace 99% of oxygen in the air bubble, (about 18.5 h). Together with 2.5 h to purge liquid medium, the total time is very close to the experimentally observed time for the oxygen reading to reach equilibrium.
Supplementary Material
Table S1. Parameters used for the simulation of flow pattern and oxygen distribution. Related to Figure 2 and Figure S1.
Table S2. Absolute amount of SCFA under Static, GuMI-NB, and GuMI-FP conditions. Related to Figure 2 and Table 1.
Table S4. IPA revealing upstream regulators in GuMI-FP vs GuMI-NB cells. Related to Figure 5.
Table S5. (A) Composition of all media used for colon organoids and monolayers. Related to STAR Methods Experimental Model and Subject Details, and Key Resource Table. (B) TaqMan probes used for RT-qPCR. Related to STAR Methods Method Details, and Key Resource Table.
Table S3. (A) The list of genes that are significantly changed in GuMI-NB compared with Static cells. Related to Figure 3 and Figure S3. (B) The list of genes that are significantly changed in GuMI-FP compared with GuMI-NB. Related to Figure 3 and Figure S3. (C) The list of core genes in the cell differentiation gene set enrichment analysis. Related to Figure 3. (D) IPA revealing upstream regulators in GuMI-NB compared with Static cells. Related to Figure 3.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Muc2 | Abcam | ab90007, RRID:AB_10713220 |
| anti-NHE3 | Novus | NBP1–82574, RRID:AB_11038394 |
| Bacterial and Virus Strains | ||
| Bacteroide thetaiotaomicron | ATCC | VPI-5482 |
| Eubacterium rectale | ATCC | ATCC33656 |
| Faecalibacterium prausnitzii | Harvard Digestive Disease Center | DSM17677 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Components for media, see Table S5A | N/A | N/A |
| BlockAid blocking solution | Thermo Fisher | B10710 |
| DAPI | Thermo Scientific | 62248 |
| Alcian blue | Sigma-Aldrich | A5268 |
| Yeast casitone fatty acid (YCFA) agar | Anaerobe Systems | AS-675 |
| Liquid YCFA medium | Anaerobe Systems | AS-680 |
| 3-Nitrophenylhydrazine | Fisher Scientific | N02325G |
| N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide HCl | Fisher Scientific | 50–848-678 |
| Acetonitrile | Sigma-Aldrich | 34998 |
| Butyric acid | Sigma-Aldrich | B103500–100ML |
| Butyric acid-d7 | CDN isotope | D-0171 |
| Acetic acid | Sigma-Aldrich | 338826 |
| Propionic acid | Sigma-Aldrich | P1386 |
| Phalloidin-iFluor 488 Reagent | Abcam | ab176753 |
| DMEM/F12 | Gibco | 12634–010 |
| Glutamax–I | Gibco | 35050–061 |
| HEPES | Gibco | 15630–080 |
| Pen-Strep | Gibco | 15140–148 |
| WRN conditioned medium | Boston Children’s Hospital (This study) | N/A |
| R-spondin 1 conditioned medium | Boston Children’s Hospital (This study) | N/A |
| B-27 Supplement 50X | Gibco | 17504–001 |
| N-2 Supplement 100X | Gibco | 17502–001 |
| Nicotinamide | Sigma-Aldrich | N0636 |
| N-acetyl cysteine | Sigma-Aldrich | A9165 |
| Y-27632 dihydrochloride | Biogems | 1293823 |
| SB202190 | Biogems | 1523072 |
| A 83–01 | Biogems | 9094360 |
| Murine EGF | PeproTech | AF-315–09 |
| Human [Leu15]-Gastrin I | Sigma-Aldrich | G9145 |
| Prostaglandin E2 | Biogems | 3632464 |
| Thiazovivin | Biogems | 1293823 |
| Human noggin | PeproTech | 120–10C |
| Critical Commercial Assays | ||
| High-Capacity RNA-to-cDNA Kit | 4387406 | |
| TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | 4444557 |
| NEBNext® Poly(A) mRNA Magnetic Isolation Module | New England Biolabs | E7490 |
| NEB Ultra II directional kit | New England Biolabs | E7760 |
| PureLink RNA mini kit | Thermo Fisher Scientific | 12183020 |
| PureLink DNase | Thermo Fisher Scientific | 12185010 |
| Customized MILLIPLEX MAP assays, 47-plex human cytokine/TH17 panel | EMD Millipore | N/A |
| DreamTaq Green PCR Master Mix | Thermo Fisher Scientific | K1081 |
| DNA purification solid-phase reversible immobilization magnetic beads | Applied Biological Materials Inc. | G95 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Human primary colon organoids | This study | N/A |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Taqman Probes, see Table S5B | Thermo Fisher Scientific Inc. | N/A |
| F8 (5’-AGTTTGATCCTGGCTCAG-3’) | Zhang et al. 2019 | N/A |
| 1492R (5’-TACGGYTACCTTGTTACGACTT-3’) | Zhang et al. 2019 | N/A |
| Recombinant DNA | ||
| Software and Algorithms | ||
| COMSOL Multiphysics 5.4 | COMSOL Inc. | RRID:SCR_014767 |
| FLEXMAP 3D software | Luminex Corporation | Version 4.2 |
| Graphpad prism 8.4 | GraphPad Software | Version 8.3.0. RRID:SCR_002798 |
| GSEA 4.0.0 | Broad Institute | RRID: SCR_003199 |
| ImageJ | Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA | 1.52p. RRID:SCR_003070 |
| Ingenuity Pathway Analysis | QIAGEN | RRID: SCR_008653 |
| STAR 2.5.3a | Dobin et al., 2013 | RRID: SCR_015899 |
| StepOne Software | Life Technologies | V2.3. RRID:SCR_014281 |
| Zen 2.3 SP1 FP3 | Zeiss | 14.0.20.201 |
| Other | ||
| HDSP vial | Thermo Scientific | C4020–201 |
| 20-mm Crimp Cap | MicroSolv | 95025–01-1S |
| 10-ml syringe | BD Biosciences | 302995 |
| 1-ml syringe | BD Biosciences | 309659 |
| Needle | BD Biosciences | 305127 |
| Short needle | BD Biosciences | 305122 |
| Palladium Catalyst | Coy Laboratory | 6501050 |
| Ultrapure water | MilliporeSigma | MilliQ purification system |
Highlights.
Development of a model of the primary human colon mucosal barrier
Super oxygen-sensitive F. prausnitzii and colon epithelium co-cultured for up to 4d
F. prausnitzii exerts anti-inflammatory effects through HDAC and the TLR-NFKB axis
F. prausnitzii reduces TLR3/TLR4 expression in colon epithelium via butyrate
Context and Significance.
In humans, various gut microbes are associated with disease protection and others with disease symptoms. Exactly how microbes exert these effects is difficult to study, as many gut microbes are so sensitive to oxygen that they cannot survive in co-culture with human gut cells. Authors from the Massachusetts Institute of Technology developed a fluidic platform that creates a steep oxygen gradient across colon epithelial cells, allowing both human epithelia and bacteria to survive together in long-term co-culture. They demonstrate that F. prausnitzii, a beneficial bacterium in human inflammatory bowel disease, exerts anti-inflammatory effects, as seen clinically, and confirm that butyrate contributes largely to this effect. This platform could be useful for understanding the interaction between the human colonic mucosal barrier and microbiota, pathogens, or bacteriotherapeutics.
Acknowledgment
This study was supported by the NIH R01EB021908 and the Boehringer Ingelheim SHINE Program. This work was supported in part by the National Institute of Environmental Health Sciences of the NIH under award P30-ES002109. We are grateful for S. Levine and D. Ma at MIT BioMicro Center for their help with RNA-seq data analysis. We thank B. Joughin for helpful discussion, and S. Manalis and the MIT 20.330 students for inspiration to analyze diffusion and reaction in the mucus layer.
Funding
US National Institute of Biomedical Imaging and Bioengineering. Boehringer Ingelheim SHINE Program. US National Institute of Environmental Health Sciences.
Footnotes
Declaration of Interests: A provisional patent was filed.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang Y & Kasper LH The role of microbiome in central nervous system disorders. Brain, Behavior, and Immunity 38, 1–12, doi: 10.1016/j.bbi.2013.12.015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clemente JC, Ursell LK, Parfrey LW & Knight R The impact of the gut microbiota on human health: An integrative view. Cell 148, 1258–1270, doi: 10.1016/j.cell.2012.01.035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemente JC, Manasson J & Scher JU The role of the gut microbiome in systemic inflammatory disease. BMJ 360, j5145, doi: 10.1136/bmj.j5145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flint HJ, Scott KP, Louis P & Duncan SH The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9, 577–589, doi: 10.1038/nrgastro.2012.156 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Janssen AW & Kersten S The role of the gut microbiota in metabolic health. FASEB J 29, 3111–3123, doi: 10.1096/fj.14-269514 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Keku TO, Dulal S, Deveaux A, Jovov B & Han X The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver Physiol 308, G351–363, doi: 10.1152/ajpgi.00360.2012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLean MH, Dieguez D Jr., Miller LM & Young HA Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut 64, 332–341, doi: 10.1136/gutjnl-2014-308514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu C et al. The microbiota regulate neuronal function and fear extinction learning. Nature 574, 543–548, doi: 10.1038/s41586-019-1644-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokol H et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America 105, 16731–16736, doi: 10.1073/pnas.0804812105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsiao EY et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463, doi: 10.1016/j.cell.2013.11.024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen TL, Vieira-Silva S, Liston A & Raes J How informative is the mouse for human gut microbiota research? Dis Model Mech 8, 1–16, doi: 10.1242/dmm.017400 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan SH, Hold Georgina L, Harmsen Hermie J M, Stewart Colin S & Flint HJ Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. International Journal of Systematic and Evolutionary Microbiology 52, 2141–2146, doi: 10.1099/ijs.0.02241-0 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Zheng L, Kelly CJ & Colgan SP Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A Review in the Theme: Cellular Responses to Hypoxia. Am J Physiol Cell Physiol 309, C350–360, doi: 10.1152/ajpcell.00191.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miquel S et al. Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. MBio 6, doi: 10.1128/mBio.00300-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flecknell P Replacement, reduction and refinement. ALTEX 19, 73–78 (2002). [PubMed] [Google Scholar]
- 16.Mishra S & Imlay JA An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Molecular Microbiology 90, 1356–1371, doi: 10.1111/mmi.12438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laura Wrzosek SM, Noordine Marie-Louise, Bouet Stephan, Chevalier-Curt Marie Joncquel, Robert Véronique, Philippe Catherine, Bridonneau Chantal, Cherbuy Claire, Robbe-Masselot Catherine, Langella Philippe and Thomas Muriel. Bacteroides thetaiotaomicron and Faecalibacterium prausnitziiinfluence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol, 61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cario E & Podolsky DK Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun 68, 7010–7017, doi: 10.1128/iai.68.12.7010-7017.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cario E Toll-like receptors in inflammatory bowel diseases: a decade later. Inflamm Bowel Dis 16, 1583–1597, doi: 10.1002/ibd.21282 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abreu MT Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nature Reviews Immunology 10, 131–143, doi: 10.1038/nri2707 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Bocker U et al. Responsiveness of intestinal epithelial cell lines to lipopolysaccharide is correlated with Toll-like receptor 4 but not Toll-like receptor 2 or CD14 expression. Int J Colorectal Dis 18, 25–32, doi: 10.1007/s00384-002-0415-6 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Maier E, Anderson RC & Roy NC Live Faecalibacterium prausnitzii Does Not Enhance Epithelial Barrier Integrity in an Apical Anaerobic Co-Culture Model of the Large Intestine. Nutrients 9, doi: 10.3390/nu9121349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulluwishewa D et al. Live Faecalibacterium prausnitzii in an apical anaerobic model of the intestinal epithelial barrier. Cell Microbiol 17, 226–240, doi: 10.1111/cmi.12360 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Maier E, Anderson RC, Altermann E & Roy NC Live Faecalibacterium prausnitzii induces greater TLR2 and TLR2/6 activation than the dead bacterium in an apical anaerobic co-culture system. Cellular Microbiology 20, doi:ARTN e1280510.1111/cmi.12805 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Peng L, Li ZR, Green RS, Holzman IR & Lin J Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr 139, 1619–1625, doi: 10.3945/jn.109.104638 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki N et al. Development of a scalable co-culture system for gut anaerobes and human colon epithelium. Gastroenterology, doi: 10.1053/j.gastro.2020.03.021. [DOI] [PubMed] [Google Scholar]
- 27.Fofanova TY et al. A novel human enteroid-anaerobe co-culture system to study microbial-host interaction under physiological hypoxia. bioRxiv, 555755, doi: 10.1101/555755 (2019). [DOI] [Google Scholar]
- 28.Kim HJ, Li H, Collins JJ & Ingber DE Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proceedings of the National Academy of Sciences 113, E7–E15, doi: 10.1073/pnas.1522193112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jalili-Firoozinezhad S et al. Complex human gut microbiome cultured in anaerobic human intestine chips. Nature Biomedical Engineering, 421404, doi: 10.1101/421404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin W et al. A Robust Longitudinal Co-culture of Obligate Anaerobic Gut Microbiome With Human Intestinal Epithelium in an Anoxic-Oxic Interface-on-a-Chip. Front Bioeng Biotechnol 7, 13, doi: 10.3389/fbioe.2019.00013 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah P et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nature Communications 7, 11535–11535, doi: 10.1038/ncomms11535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HJ, Huh D, Hamilton G & Ingber DE Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174, doi: 10.1039/c2lc40074j (2012). [DOI] [PubMed] [Google Scholar]
- 33.Kozuka K et al. Development and Characterization of a Human and Mouse Intestinal Epithelial Cell Monolayer Platform. Stem Cell Reports 9, 1976–1990, doi: 10.1016/j.stemcr.2017.10.013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cook CD et al. Local remodeling of synthetic extracellular matrix microenvironments by co-cultured endometrial epithelial and stromal cells enables long-term dynamic physiological function. Integr Biol (Camb) 9, 271–289, doi: 10.1039/c6ib00245e (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foster KA, Oster CG, Mayer MM, Avery ML & Audus KL Characterization of the A549 Cell Line as a Type II Pulmonary Epithelial Cell Model for Drug Metabolism. Experimental Cell Research 243, 359–366, doi: 10.1006/excr.1998.4172 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Fahey JV, Schaefer TM, Channon JY & Wira CR Secretion of cytokines and chemokines by polarized human epithelial cells from the female reproductive tract. Hum Reprod 20, 1439–1446, doi: 10.1093/humrep/deh806 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Kelly CJ et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 17, 662–671, doi: 10.1016/j.chom.2015.03.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inman W et al. Design, modeling and fabrication of a constant flow pneumatic micropump. Journal of Micromechanics and Microengineering 17, 891–899, doi: 10.1088/0960-1317/17/5/007 (2007). [DOI] [Google Scholar]
- 39.Edington CD et al. Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies. Sci Rep 8, 4530, doi: 10.1038/s41598-018-22749-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter E, Janich S, Roessler BJ, Hilfinger JM & Amidon GL HT29-MTX/Caco-2 Cocultures as an in Vitro Model for the Intestinal Epithelium: In Vitro–in Vivo Correlation with Permeability Data from Rats and Humans. Journal of Pharmaceutical Sciences 85, 1070–1076, doi: 10.1021/js960110x (1996). [DOI] [PubMed] [Google Scholar]
- 41.Taketani M et al. Genetic circuit design automation for Bacteroides, applied to integrate signals from bile acid and antibiotic sensors. Nature Biotechnology, doi: 10.1038/s41587-020-0468-5 (2020). [DOI] [Google Scholar]
- 42.Cremer J et al. Effect of flow and peristaltic mixing on bacterial growth in a gut-like channel. Proceedings of the National Academy of Sciences of the United States of America 113, 11414–11419, doi: 10.1073/pnas.1601306113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khashab MA, Pickhardt PJ, Kim DH & Rex DK Colorectal anatomy in adults at computed tomography colonography: normal distribution and the effect of age, sex, and body mass index. Endoscopy 41, 674–678, doi: 10.1055/s-0029-1214899 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Schulze KS The imaging and modelling of the physical processes involved in digestion and absorption. Acta Physiol (Oxf) 213, 394–405, doi: 10.1111/apha.12407 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Varela E et al. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Alimentary Pharmacology & Therapeutics 38, 151–161, doi: 10.1111/apt.12365 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Machiels K et al. A decrease of the butyrate-producing species <em>Roseburia hominis</em> and <em>Faecalibacterium prausnitzii</em> defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275, doi: 10.1136/gutjnl-2013-304833 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Fujimoto T et al. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. Journal of Gastroenterology and Hepatology (Australia) 28, 613–619, doi: 10.1111/jgh.12073 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Quevrain E et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 65, 415–425, doi: 10.1136/gutjnl-2014-307649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao Y, Shen J & Ran ZH Association between faecalibacterium prausnitzii reduction and inflammatory bowel disease: A meta-analysis and systematic review of the literature. Gastroenterology Research and Practice 2014, 72725, doi: 10.1155/2014/872725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farhadi A, Banan ALI, Fields J & Keshavarzian ALI Intestinal barrier: An interface between health and disease. Journal of Gastroenterology and Hepatology 18, 479–497, doi: 10.1046/j.1440-1746.2003.03032.x (2003). [DOI] [PubMed] [Google Scholar]
- 51.Srinivasan B et al. TEER Measurement Techniques for In Vitro Barrier Model Systems. Journal of Laboratory Automation 20, 107–126, doi: 10.1177/2211068214561025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madden LR et al. Bioprinted 3D Primary Human Intestinal Tissues Model Aspects of Native Physiology and ADME/Tox Functions. iScience 2, 156–167, doi: 10.1016/j.isci.2018.03.015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fleischer D in Transport processes in pharmaceutical systems 163–200 (CRC Press, 1999). [Google Scholar]
- 54.Gurney MA, Laubitz D, Ghishan FK & Kiela PR Pathophysiology of Intestinal Na(+)/H(+) exchange. Cell Mol Gastroenterol Hepatol 3, 27–40, doi: 10.1016/j.jcmgh.2016.09.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johansson MEV, Larsson JMH & Hansson GC The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proceedings of the National Academy of Sciences of the United States of America 108 Suppl, 4659–4665, doi: 10.1073/pnas.1006451107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tramontano M et al. Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat Microbiol 3, 514–522, doi: 10.1038/s41564-018-0123-9 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Louis P & Flint HJ Formation of propionate and butyrate by the human colonic microbiota. Environmental Microbiology 19, 29–41, doi: 10.1111/1462-2920.13589 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Zhang J et al. Gut microbial beta-glucuronidase and glycerol/diol dehydratase activity contribute to dietary heterocyclic amine biotransformation. BMC Microbiology 19, 99, doi: 10.1186/s12866-019-1483-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sivaprakasam S, Bhutia YD, Yang S & Ganapathy V Short-Chain Fatty Acid Transporters: Role in Colonic Homeostasis. Compr Physiol 8, 299–314, doi: 10.1002/cphy.c170014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapecar M et al. Gut-Liver physiomimetics reveal paradoxical modulation of IBD-related inflammation by short-chain fatty acids. Cell Systems 10, 223–239, doi: 10.1016/j.cels.2020.02.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cummings JH, Pomare EW, Branch WJ, Naylor CPE & Macfarlane GT Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28, 1221–1227, doi: 10.1136/gut.28.10.1221 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cummings J, Pomare EW, Branch WJ, Naylor CP & Macfarlane GT Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28, 1221–1227 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiefer J, Beyer-Sehlmeyer G & Pool-Zobel BL Mixtures of SCFA, composed according to physiologically available concentrations in the gut lumen, modulate histone acetylation in human HT29 colon cancer cells. Br J Nutr 96, 803–810, doi: 10.1017/bjn20061948 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Roediger WEW et al. Luminal Ions and Short Chain Fatty-Acids as Markers of Functional-Activity of the Mucosa in Ulcerative-Colitis. J Clin Pathol 35, 323–326, doi:DOI 10.1136/jcp.35.3.323 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamamoto K et al. Fluid shear stress induces differentiation of Flk-1-positive embryonic stem cells into vascular endothelial cells in vitro. Am J Physiol Heart Circ Physiol 288, H1915–1924, doi: 10.1152/ajpheart.00956.2004 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Parada Venegas D et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol 10, 277, doi: 10.3389/fimmu.2019.00277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mootha VK et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273, doi: 10.1038/ng1180 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Hung JH, Yang TH, Hu Z, Weng Z & DeLisi C Gene set enrichment analysis: performance evaluation and usage guidelines. Brief Bioinform 13, 281–291, doi: 10.1093/bib/bbr049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bertoli C, Skotheim JM & de Bruin RA Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol 14, 518–528, doi: 10.1038/nrm3629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laoukili J et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol 7, 126–U134, doi: 10.1038/ncb1217 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Roche KC et al. SOX9 Maintains Reserve Stem Cells and Preserves Radioresistance in Mouse Small Intestine. Gastroenterology 149, 1553–+, doi: 10.1053/j.gastro.2015.07.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li J et al. Tetrandrine inhibits colon carcinoma HT-29 cells growth via the Bcl-2/Caspase 3/PARP pathway and G1/S phase. Biosci Rep 39, doi: 10.1042/BSR20182109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wells V & Mallucci L Identification of an autocrine negative growth factor: Mouse β-galactoside-binding protein is a cytostatic factor and cell growth regulator. Cell 64, 91–97, doi: 10.1016/0092-8674(91)90211-G (1991). [DOI] [PubMed] [Google Scholar]
- 75.Wierstra I & Alves J FOXM1, a typical proliferation-associated transcription factor. Biol Chem 388, 1257–1274, doi:Doi 10.1515/Bc.2007.159 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Kormish JD, Sinner D & Zorn AM Interactions Between SOX Factors and Wnt/beta-Catenin Signaling in Development and Disease. Dev Dynam 239, 56–68, doi: 10.1002/dvdy.22046 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mah AT, Yan KS & Kuo CJ Wnt pathway regulation of intestinal stem cells. J Physiol 594, 4837–4847, doi: 10.1113/JP271754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Byndloss MX et al. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575, doi: 10.1126/science.aam9949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rivera-Chavez F et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host and Microbe 19, 443–454, doi: 10.1016/j.chom.2016.03.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iyer NV et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev 12, 149–162, doi: 10.1101/gad.12.2.149 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ratcliffe P et al. Update on hypoxia-inducible factors and hydroxylases in oxygen regulatory pathways: from physiology to therapeutics. Hypoxia (Auckl) 5, 11–20, doi: 10.2147/HP.S127042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schofield CJ & Ratcliffe PJ Signalling hypoxia by HIF hydroxylases. Biochem Biophys Res Commun 338, 617–626, doi: 10.1016/j.bbrc.2005.08.111 (2005). [DOI] [PubMed] [Google Scholar]
- 83.Schofield CJ & Ratcliffe PJ Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol 5, 343–354, doi: 10.1038/nrm1366 (2004). [DOI] [PubMed] [Google Scholar]
- 84.Taylor CT & Colgan SP Hypoxia and gastrointestinal disease. J Mol Med (Berl) 85, 1295–1300, doi: 10.1007/s00109-007-0277-z (2007). [DOI] [PubMed] [Google Scholar]
- 85.Manalo DJ et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105, 659–669, doi: 10.1182/blood-2004-07-2958 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Elvidge GP et al. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J Biol Chem 281, 15215–15226, doi: 10.1074/jbc.M511408200 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Fachi JL et al. Butyrate Protects Mice from Clostridium difficile-Induced Colitis through an HIF-1-Dependent Mechanism. Cell Rep 27, 750–761.e757, doi: 10.1016/j.celrep.2019.03.054 (2019). [DOI] [PubMed] [Google Scholar]
- 88.Hamer HM et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27, 104–119, doi: 10.1111/j.1365-2036.2007.03562.x (2008). [DOI] [PubMed] [Google Scholar]
- 89.Zhou L et al. Faecalibacterium prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm Bowel Dis 24, 1926–1940, doi: 10.1093/ibd/izy182 (2018). [DOI] [PubMed] [Google Scholar]
- 90.Miquel S et al. Faecalibacterium prausnitzii and human intestinal health. Current Opinion in Microbiology 16, 255–261, doi: 10.1016/j.mib.2013.06.003 (2013). [DOI] [PubMed] [Google Scholar]
- 91.Liu T, Zhang L, Joo D & Sun SC NF-kappaB signaling in inflammation. Signal Transduct Target Ther 2, 17023, doi: 10.1038/sigtrans.2017.23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawasaki T & Kawai T Toll-like receptor signaling pathways. Front Immunol 5, 461, doi: 10.3389/fimmu.2014.00461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rabiei N et al. Induction effects of Faecalibacterium prausnitzii and its extracellular vesicles on toll-like receptor signaling pathway gene expression and cytokine level in human intestinal epithelial cells. Cytokine 121, 154718, doi: 10.1016/j.cyto.2019.05.005 (2019). [DOI] [PubMed] [Google Scholar]
- 94.Kawai T & Akira S Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med 13, 460–469, doi: 10.1016/j.molmed.2007.09.002 (2007). [DOI] [PubMed] [Google Scholar]
- 95.Shahbazi S et al. Impact of novel N-aryl piperamide NO donors on NF-kappaB translocation in neuroinflammation: rational drug-designing synthesis and biological evaluation. Innate Immun 24, 24–39, doi: 10.1177/1753425917740727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Della Ragione F et al. Genes modulated by histone acetylation as new effectors of butyrate activity. FEBS Letters 499, 199–204, doi: 10.1016/s0014-5793(01)02539-x (2001). [DOI] [PubMed] [Google Scholar]
- 97.Wilson AJ et al. Apoptotic sensitivity of colon cancer cells to histone deacetylase inhibitors is mediated by an Sp1/Sp3-activated transcriptional program involving immediate-early gene induction. Cancer Res 70, 609–620, doi: 10.1158/0008-5472.CAN-09-2327 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Donohoe DR et al. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell 48, 612–626, doi: 10.1016/j.molcel.2012.08.033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kazemi Sefat NA et al. Sodium Butyrate as a Histone Deacetylase Inhibitor Affects Toll-Like Receptor 4 Expression in Colorectal Cancer Cell Lines. Immunol Invest 48, 759–769, doi: 10.1080/08820139.2019.1595643 (2019). [DOI] [PubMed] [Google Scholar]
- 100.Zhai S et al. Dietary butyrate suppresses inflammation through modulating gut microbiota in high-fat diet-fed mice. FEMS Microbiol Lett 366, fnz153, doi: 10.1093/femsle/fnz153 (2019). [DOI] [PubMed] [Google Scholar]
- 101.Liu J et al. Sodium Butyrate Inhibits the Inflammation of Lipopolysaccharide-Induced Acute Lung Injury in Mice by Regulating the Toll-Like Receptor 4/Nuclear Factor kappaB Signaling Pathway. J Agric Food Chem 67, 1674–1682, doi: 10.1021/acs.jafc.8b06359 (2019). [DOI] [PubMed] [Google Scholar]
- 102.Inan MS et al. The luminal short-chain fatty acid butyrate modulates NF-kappaB activity in a human colonic epithelial cell line. Gastroenterology 118, 724–734 (2000). [DOI] [PubMed] [Google Scholar]
- 103.Xiao T et al. Butyrate upregulates the TLR4 expression and the phosphorylation of MAPKs and NK-κB in colon cancer cell in vitro. Oncol Lett 16, 4439–4447, doi: 10.3892/ol.2018.9201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Davie JR Inhibition of histone deacetylase activity by butyrate. J Nutr 133, 2485S–2493S, doi: 10.1093/jn/133.7.2485S (2003). [DOI] [PubMed] [Google Scholar]
- 105.Kyrylenko S, Kyrylenko O, Suuronen T & Salminen A Differential regulation of the Sir2 histone deacetylase gene family by inhibitors of class I and II histone deacetylases. Cell Mol Life Sci 60, 1990–1997, doi: 10.1007/s00018-003-3090-z (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spurling CC et al. HDAC3 overexpression and colon cancer cell proliferation and differentiation. Mol Carcinog 47, 137–147, doi: 10.1002/mc.20373 (2008). [DOI] [PubMed] [Google Scholar]
- 107.Dangond F & Gullans SR Differential expression of human histone deacetylase mRNAs in response to immune cell apoptosis induction by trichostatin A and butyrate. Biochem Biophys Res Commun 247, 833–837, doi: 10.1006/bbrc.1998.8891 (1998). [DOI] [PubMed] [Google Scholar]
- 108.Patel BM Sodium Butyrate Controls Cardiac Hypertrophy in Experimental Models of Rats. Cardiovasc Toxicol 18, 1–8, doi: 10.1007/s12012-017-9406-2 (2018). [DOI] [PubMed] [Google Scholar]
- 109.Lee C et al. Sodium butyrate inhibits the NF-kappa B signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int Immunopharmacol 51, 47–56, doi: 10.1016/j.intimp.2017.07.023 (2017). [DOI] [PubMed] [Google Scholar]
- 110.Wang Y et al. In Vitro Generation of Mouse Colon Crypts. ACS Biomater Sci Eng 3, 2502–2513, doi: 10.1021/acsbiomaterials.7b00368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kurtz CB et al. An engineered <em>E. coli</em> Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Science Translational Medicine 11, eaau7975, doi: 10.1126/scitranslmed.aau7975 (2019). [DOI] [PubMed] [Google Scholar]
- 112.Yu EW et al. Fecal microbiota transplantation for the improvement of metabolism in obesity: The FMT-TRIM double-blind placebo-controlled pilot trial. PLOS Medicine 17, e1003051, doi: 10.1371/journal.pmed.1003051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cermak N, Datta MS & Conwill A Rapid, Inexpensive Measurement of Synthetic Bacterial Community Composition by Sanger Sequencing of Amplicon Mixtures. iScience 23, 100915, doi: 10.1016/j.isci.2020.100915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frazier SB, Roodhouse KA, Hourcade DE & Zhang L The quantification of glycosaminoglycans: a comparison of HPLC, carbazole, and alcian blue methods. Open glycoscience 1, 31, doi: 10.2174/1875398100801010031 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Han J, Lin K, Sequeira C & Borchers CH An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography-tandem mass spectrometry. Analytica Chimica Acta 854, 86–94, doi: 10.1016/j.aca.2014.11.015 (2015). [DOI] [PubMed] [Google Scholar]
- 116.Dobin A et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kramer A, Green J, Pollard J Jr. & Tugendreich S Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530, doi: 10.1093/bioinformatics/btt703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Parameters used for the simulation of flow pattern and oxygen distribution. Related to Figure 2 and Figure S1.
Table S2. Absolute amount of SCFA under Static, GuMI-NB, and GuMI-FP conditions. Related to Figure 2 and Table 1.
Table S4. IPA revealing upstream regulators in GuMI-FP vs GuMI-NB cells. Related to Figure 5.
Table S5. (A) Composition of all media used for colon organoids and monolayers. Related to STAR Methods Experimental Model and Subject Details, and Key Resource Table. (B) TaqMan probes used for RT-qPCR. Related to STAR Methods Method Details, and Key Resource Table.
Table S3. (A) The list of genes that are significantly changed in GuMI-NB compared with Static cells. Related to Figure 3 and Figure S3. (B) The list of genes that are significantly changed in GuMI-FP compared with GuMI-NB. Related to Figure 3 and Figure S3. (C) The list of core genes in the cell differentiation gene set enrichment analysis. Related to Figure 3. (D) IPA revealing upstream regulators in GuMI-NB compared with Static cells. Related to Figure 3.
Data Availability Statement
Raw RNAseq data of the colon epithelial donor could not be included due to sharing restrictions but can be shared upon request to the Lead Contact (Linda G. Griffith, griff@mit.edu).