ABSTRACT
The vertical structuring of methanotrophic communities and its genetic controllers remain understudied in the water columns of oxygen-stratified lakes. Therefore, we used 16S rRNA gene sequencing to study the vertical stratification patterns of methanotrophs in two boreal lakes, Lake Kuivajärvi and Lake Lovojärvi. Furthermore, metagenomic analyses were performed to assess the genomic characteristics of methanotrophs in Lovojärvi and the previously studied Lake Alinen Mustajärvi. The methanotroph communities were vertically structured along the oxygen gradient. Alphaproteobacterial methanotrophs preferred oxic water layers, while Methylococcales methanotrophs, consisting of putative novel genera and species, thrived, especially at and below the oxic-anoxic interface and showed distinct depth variation patterns, which were not completely predictable by their taxonomic classification. Instead, genomic differences among Methylococcales methanotrophs explained their variable vertical depth patterns. Genes in clusters of orthologous groups (COG) categories L (replication, recombination and repair) and S (function unknown) were relatively high in metagenome-assembled genomes representing Methylococcales clearly thriving below the oxic-anoxic interface, suggesting genetic adaptations for increased stress tolerance enabling living in the hypoxic/anoxic conditions. By contrast, genes in COG category N (cell motility) were relatively high in metagenome-assembled genomes of Methylococcales thriving at the oxic-anoxic interface, which suggests genetic adaptations for increased motility at the vertically fluctuating oxic-anoxic interface.
Keywords: methanotroph, Methylococcales, vertical structuring, 16S rRNA, metagenomics, genetic potential
Genetic differences lead to variable vertical stratification patterns of methanotrophs in water columns of oxygen-stratified lakes.
INTRODUCTION
The concentration of atmospheric methane (CH4), a critical greenhouse gas, has increased substantially since industrialization, with the current net emissions approximating 550–600 Tg/year (Saunois et al. 2020). Of the total emissions, ~60% and 40% stem from anthropogenic and natural sources, respectively (Saunois et al. 2020). Although lakes occupy only a small part (3.7%) of the global non-glaciated land area, their CH4 emissions are estimated to be very high, 6–24% of the total natural release (Bastviken et al. 2004, 2011). The numerous lakes and ponds in the northern boreal and arctic zones with annual emissions of ∼16.5 Tg (∼7% of natural release) are especially significant components of the global CH4 budget (Wik et al. 2016). However, these emissions are partly balanced by microbial CH4 oxidation processes, which act as a filter substantially decreasing CH4 escape from ecosystems (Hanson and Hanson 1996; Blees et al. 2014; Cabrol et al. 2020). As the composition of the methane-oxidizing community is linked to CH4 oxidation rates in lakes (Reis et al. 2020), knowledge about the factors affecting the community structure of CH4-oxidizing microbes in lakes, and especially in lakes of the northern boreal zone, is essential for estimating and modeling global CH4 budgets. In microbial CH4 oxidation, CH4 can be consumed through aerobic oxidation by methanotrophic bacteria (MOB), which use O2 as an electron acceptor (EA) (Hanson and Hanson 1996; Milucka et al. 2015), or through anaerobic oxidation (i.e. anaerobic oxidation of methane [AOM]) by anaerobic methanotrophic (ANME) archaea, which use alternative EAs (e.g NO3−, SO42−, Mn4+/Fe3+ minerals, humic acids) (Beal, House and Orphan 2009; Knittel and Boetius 2009; Haroon et al. 2013; Ettwig et al. 2016; Scheller et al. 2016). In addition, bacteria belonging to the genus Candidatus Methylomirabilis drive AOM by gaining oxygen for the oxidation of CH4 under anoxia using the nitric oxide dismutase enzyme (Ettwig et al. 2010).
Many lakes have oxygen-stratified water columns, and a substantial part of CH4 oxidation takes place at and below the oxic-anoxic interface in these lakes (Blees et al. 2014; Oswald et al. 2016; Rissanen et al. 2018). In most of the studied lakes, water column CH4 oxidation is attributed dominantly to aerobic gammaproteobacterial MOB of the order Methylococcales, which particularly thrive in CH4-rich, hypoxic and anoxic water column layers, while aerobic alphaproteobacterial MOB (several genera/species within the families Methylocystaceae and Beijerinckiaceae) are more important in the CH4-poor, oxic surface layers (Eller, Känel and Krüger 2005; Milucka et al. 2015; Oswald et al. 2015; Kallistova et al. 2018; Cabrol et al. 2020; Reis et al. 2020). There is also vertical depth pattern variation among members of Methylococcales along the oxygen-methane counter gradient, as shown by a recent study of oxygen-stratified lakes from temperate area (Mayr et al. 2020b). Hence, MOB have different life strategies enabling them to predominate under different conditions (Ho et al. 2013). Furthermore, members of uncultivated Methylococcales taxon, CABC2E06, occurred mainly in the upper parts of the water column, under oxygen-rich and methane-deficient conditions, in temperate lakes, agreeing with vertical distribution of one of the Methylobacter-related species inhabiting a water column of a boreal lake (Rissanen et al. 2018; Mayr et al. 2020b). By contrast, a recent study of arctic lakes showed that both CABC2E06 (in the study it was named as an unclassified taxon) and Methylobacter-related taxa preferred lower, anoxic water layers (Cabrol et al. 2020). These results indicate that there are differences between lakes in the vertical variation patterns of Methylococcales MOB and that they are not predictable by their taxonomic classification. Thus, there is a need for additional studies on vertical stratification patterns of lake water column methanotrophs, especially from boreal areas, from where such studies are rare (Rissanen et al. 2018). Besides marker gene amplicon data (16S rRNA and pmoA gene) (Cabrol et al. 2020; Mayr et al. 2020b), data on the functional potential of Methylococcales, via analysis of metagenome-assembled genomes (MAGs), would provide further explanations for their vertical variation patterns (Oswald et al. 2017; Rissanen et al. 2018; van Grinsven et al. 2020).
Below the oxic-anoxic interface, Methylococcales may gain oxygen for aerobic CH4 oxidation through episodic oxygen intrusions from the oxic surface waters or through oxygenic photosynthesis (Blees et al. 2014; Kirf et al. 2014; Milucka et al. 2015; Kallistova et al. 2018). However, metagenomic and pure culture studies suggest that Methylococcales also have specific adaptations for living in hypoxic and anoxic conditions, via usage of high-affinity cytochromes (for aerobic respiration in low O2 conditions) and/or bacteriohemerythrin (to scavenge O2) and/or via driving fermentation or anaerobic respiration, i.e. denitrification or extracellular electron transfer to metal minerals and organic EAs (Kalyuzhnaya et al. 2013; Kits, Klotz and Stein 2015; Oswald et al. 2017; Rissanen et al. 2018; Smith and Wrighton 2019; van Grinsven et al. 2020; Zheng et al. 2020). A specific hypoxia stress response mechanism was also recently suggested to explain why Methylobacter-related MOB thrive in hypoxic ecosystems (Yu et al. 2020). Differences in genetic potential for these various microaerobic and anaerobic metabolism functions potentially explain the differences in vertical distribution patterns between members of Methylococcales. Our previous study from the water column of a boreal Lake Alinen Mustajärvi actually hinted towards that direction, as a MAG of Methylococcales MOB, which thrived in aerobic water layers, did not harbor denitrification genes, whereas the MAGs of other Methylococcales, which were most abundant below the oxic-anoxic interface, encoded denitrification enzymes (Rissanen et al. 2018). Differences in the genetic potential to fix N2 could also determine the vertical stratification patterns of Methylococcales, as availability of inorganic nitrogen is usually very low in surface layers but increases towards deeper layers in O2-stratified lakes (e.g. Rissanen et al. 2018). Besides focusing on specific genes, general genetic variations, i.e. the proportion of genes classified into different, broad functional categories, could also be compared among MAGs of lake water column Methylococcales.
The aim of this study was to gain more knowledge of the vertical stratification patterns of methanotrophs in water columns of oxygen-stratified lakes. We specifically focused on the rarely studied boreal lakes, as they are very important components of the global CH4 budget (Wik et al. 2016). Therefore, by using high-throughput 16S rRNA gene amplicon sequencing, we studied the bacterial and archaeal communities in water columns of two different types of shallow, boreal, O2-stratified lakes, (i) seasonally O2-stratified Lake Kuivajärvi and (ii) permanently O2-stratified Lake Lovojärvi. Furthermore, we assessed the potential genomic differences explaining the vertical stratification patterns of different members of Methylococcales in Lovojärvi and the previously studied boreal Lake Alinen Mustajärvi (see Rissanen et al. 2018) by analysis of MAGs constructed from shotgun metagenomic data.
MATERIALS AND METHODS
Study lakes
The study lakes, Lake Kuivajärvi (61° 50’N, 24° 17'E; area = 0.62 km2, max. depth = 13 m) and Lake Lovojärvi (61° 04’N, 25° 02’E; area = 0.054 km2, max. depth = 17.5 m), are small humic lakes (dissolved organic carbon: ∼13 mg L−1 in Kuivajärvi and ∼14 mg L−1 in Lovojärvi) located in southern Finland. Their catchment areas (Kuivajärvi: 9.4 km2; Lovojärvi: 7.2 km2) consist of managed forests as well as peatland and agricultural land. Each year both lakes are frozen for ~5 months. Kuivajärvi is dimictic with complete turnover of the water mass occurring immediately after ice-out and in the autumn (Heiskanen et al. 2015). By contrast, Lovojärvi is meromictic without complete water column turnover, which is due to its morphometry and sheltered position as well as chemical stratification due to historical anthropogenic effects, the soaking of flax and hemp in the Iron Age (Tolonen et al. 1976; Saarnisto, Huttunen and Tolonen 1977; Simola 1979; Hakala 2004). Due to the differing stratification patterns, the water column of Lovojärvi is always O2-stratified, while, in Kuivajärvi, O2 stratification and hypoxia/anoxia takes place only during late summer.
Sampling and analyses of physicochemical data
The lakes were sampled at their deepest points in September 2015 for Lovojärvi and September 2016 for Kuivajärvi. Vertical profiles of several physico-chemical variables as well as samples for bacterial and archaeal community composition were collected from both lakes at varying depth intervals, as summarized in Table S1 (supplementary data). Data from Lake Kuivajärvi for four sampling occasions from May to September 2016 were already presented in a previous paper (Saarela et al. 2020). However, in this paper, data are presented for September to allow for easier comparison with the microbial data. Dissolved O2 and temperature (°C) were measured with a YSI ProODO (optical dissolved oxygen) field meter (Yellow Springs Instruments, Yellow Springs, OH, USA; accuracy ± 0.2°C, ± 3 µmol O2 L−1 or ± 1% of reading) in both lakes. The pH and oxidation-reduction potential (ORP) were measured from samples taken with a 2 dm3 Limnos water sampler (length 30 cm) (Limnos Ltd., Turku, Finland) for Kuivajärvi and a 1.2 dm3 Limnos water sampler (length 20 cm) for Lovojärvi using SenTix 41 pH and SenTix ORP electrodes (Xylem Inc., Weilheim, Germany) connected to WTW ProfiLine pH 3110 (Xylem Inc., Weilheim, Germany). The samples for the analysis of microbial communities and background variables, i.e. concentrations of SO42−, combined NO3−+NO2−, NH4+, CO2 and CH4, as well as 13C/12C (i.e. δ13C) of CH4 and dissolved inorganic carbon (DIC), were also collected using Limnos samplers. The concentration and stable isotope analyses are described in detail in Supplementary Methods.
DNA extraction
Samples for molecular microbiological studies were collected in filter units (pore size 0.22 μm, Millipore®, Sterivex, Darmstadt, Germany), from 500 mL of lake water per sample, after pre-filtration through a plankton net (mesh size 25 μm). The filter units were stored frozen (at -80°C) before further steps. Half of the frozen filter material was used for DNA extraction with PowerSoil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA). DNA concentration was measured using a Qubit 2.0 Fluorometer and a dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Unfortunately, filter units from depths of 8.9 m, 9.4 m and 9.9 m of Lovojärvi had to be discarded from microbial analyses due to freezer malfunction.
16S rRNA gene amplicon and shotgun metagenomic sequencing
PCR of 16S rRNA genes and amplicon sequencing took place commercially at FISABIO (Valencia, Spain; http://www.fisabio-ngs.com/en/introduction/). For each sample, the V4 region of the bacterial and archaeal 16S rRNA genes was targeted using primer pair 515FB/806RB (Apprill et al. 2015; Parada, Needham and Fuhrman 2016). To gain more information on the diversity of methanotrophic bacteria and archaea, the V1-V2 region of bacterial and V3-V4 region of archaeal 16S rRNA gene were also targeted using primer pairs 27F/338R and 340F/806R, respectively, for part of the samples (Takai and Horikoshi 2000; Gantner et al. 2011) (see Table S1). PCR, library preparation and sequencing (Illumina MiSeq, Illumina, San Diego, CA, USA) took place as previously described (Myllykangas et al. 2020), except that, in PCR reactions, ~5 ng of DNA were used, and the annealing temperature was 53°C for primer pair 27F/338R. For shotgun metagenomic analysis of the Lovojärvi samples, the library preparation and sequencing (Illumina NovaSeq) were performed as previously described (Buck et al. 2020).
Bioinformatic analyses of 16S rRNA gene data
The sequencing facility, FISABIO, performed the quality assessment of the raw data and the merging of the paired end reads. Quality assessment (i.e. removing of too short and low quality reads) was conducted using prinseq-lite with the following parameters: min_length: 50; trim_qual_right: 30; trim_qual_type: mean; trim_qual_window: 20 (Schmieder and Edwards 2011). The paired end reads passing the quality check were joined using the FLASH program by applying default parameters (Magoč and Salzberg 2011).
Mothur was used in subsequent sequence analyses (Schloss et al. 2009), unless reported otherwise. The sequences were aligned using Silva reference alignment (Release 132). Chimeric sequences, identified using VSEARCH (Rognes et al. 2016), were removed from each library and a preclustering algorithm (Huse et al. 2010) was used to reduce the effect of sequencing errors. Sequences were assigned taxonomies with a naïve Bayesian classifier (bootstrap cutoff value 80%) (Wang et al. 2007), using the Silva database (Release 132), which was supplemented with a partial 16S rRNA gene sequence from Candidatus Methyloumidiphilus alinensis, a novel MOB belonging to order Methylococcales, which dominated the water column MOB community in a small Finnish boreal lake, Lake Alinen-Mustajärvi (Rissanen et al. 2018). Thereafter, sequences classified as chloroplast, mitochondria and eukaryota were removed from each library, while in addition, archaeal sequences were removed from the bacterial V1-V2 library and bacterial sequences from the archaeal V3-V4 library.
Sequences were clustered into operational taxonomic units (OTUs) at a 97% similarity level. Singleton OTUs (OTUs with only one sequence) were removed, and the data were then normalized by subsampling to the same size, which was 87173, 203287 and 2224 for prokaryotic (i.e. bacterial and archaeal) V4, bacterial V1-V2 and archaeal V3-V4 libraries, respectively. Good coverage was >0.96 in each library confirming that sequence variation was adequately covered.
This study focused on known aerobic and anaerobic methanotrophic bacterial and archaeal taxa. OTUs belonging to the order Methylococcales in prokaryotic V4 and bacterial V1-V2–libraries were further classified via blastn search (against the National Center for Biotechnology Information [NCBI] nr-database] (Altschul et al. 1990), as well as via phylogenetic tree analysis of the representative sequences of OTUs, which were performed using the maximum likelihood algorithm (generalized time reversible model) with 100 bootstraps in Mega 6.0 (Tamura et al. 2013). The Methylococcales OTUs from V4 and V1-V2 library sets were paired (determined to represent the same OTU) by their coherent depth variation (by correlation analysis) and position in the phylogenetic tree. The Methylococcales OTUs in both study lakes were also classified into groups based on their vertical stratification patterns using hierarchical clustering (UPGMA method, dissimilarity metric: correlation) in PAST (version 3) (Hammer, Harper and Ryan 2001).
Bioinformatic analysis of shotgun metagenomic data
Trimming of the raw data, assembly of the quality-controlled reads, as well as binning of the assembled contigs into metagenomic bins, are explained in detail in Buck et al. (2020). In addition, Kaiju was used for taxonomic classification of the trimmed reads against the NCBI nr-database (Menzel, Ng and Krogh 2016). Genes of the obtained bins were predicted and annotated using Prokka (version 1.13.3) (Seemann 2014). Prokaryotic completeness and redundancy of the bins were computed using CheckM (version 1.0.13) (Parks et al. 2015). Further, the bins were clustered into metagenomic OTUs if the average nucleotide identity (ANI) between them was above 95%, as computed with fastANI (version 1.3) (Jain et al. 2018). Bins were taxonomically annotated in a two-step process utilizing GTDB-Tk (version 102 with database release 89) and SourMASH´s lowest common ancestor database (version 1.0) and classifier (Brown and Irber 2016; Parks et al. 2018), as described in Buck et al. (2020). Thereafter, all bins, i.e. MAGs, which were affiliated to Methylococcales and had completeness >50% and contamination <10%, were chosen for further analyses.
The phylogenetic positions of MAGs affiliated to Methylococcales were specifically analyzed using PhyloPhlAn2 (Segata et al. 2013). Furthermore, ANIs and average amino acid identities between MAGs and reference genomes were calculated using tools at http://enve-omics.ce.gatech.edu/ (Goris et al. 2007; Rodriguez-R L 2014). Prokka annotations of the MAGs were screened for genes encoding enzymes involved in methane/methanol oxidation, nitrogen (N2) fixation, NOx− reduction (denitrification), hydroxylamine detoxification, microaerobic metabolism (bacteriohemerythrin and high-affinity oxidases) and fermentation (Campbell et al. 2011; Kalyuzhnaya et al. 2013; Kits, Klotz and Stein 2015; Smith and Wrighton 2019). MAGs were also analyzed using FeGenie for annotation of genes encoding enzymes involved in extracellular electron transfer (EET) (Garber et al. 2020). In addition, blastP was used to search similarities to genes coding for the type 1 secretion system (T1SS), which, based on comparative transcriptomics, was recently suggested to be involved in the EET of Methylomonas sp. LW13 (Altschul et al. 1990, 1997; Zheng et al. 2020). BlastP was also used (i) to confirm the presence of pmoC gene coding for gamma subunit of particulate methane monooxygenase, as it was not detected by Prokka, (ii) to analyze whether the detected methanol-dehydrogenase genes consist of mxaF or xoxF genes coding for calcium- and lanthanide-dependent enzymes, respectively, (iii) to detect genes coding for nitric oxide dismutase (nod), (iv) to detect genes involved in the recently described nitric oxide-mediated hypoxia stress response mechanism of Methylobacter sp. (Yu et al. 2020) and (v) to confirm the presence of genes coding for enzymes involved in hydroxylamine detoxification (Altschul et al. 1990, 1997). The predicted protein sequences of MAGs were also classified into the 26 functional categories of clusters of orthologous groups (COG) system using the online version of the eggNOG-mapper at http://eggnog-mapper.embl.de/ (accessed 15 October 2020) (Huerta-Cepas et al. 2017, 2019) This was also carried out for the four Methylococcales MAGs of the recently studied Lake Alinen Mustajärvi: MAG 10 (AMbin10; NCBI GenBank assembly accession GCA_003 242 955.1), MAG 126 (AMbin126; GCA_003 584 875.1), MAG 140 (AMbin140; GCA_003 584 865.1) and MAG 149 (AMbin149; GCA_003 584 895.1) (Rissanen et al. 2018).
Furthermore, pmoA sequences, coding for beta subunit of particulate methane monooxygenase, of the MAGs were subjected to phylogenetic tree analyses using the neighbor joining method (Jones-Taylor-Thornton model) with 500 bootstraps in Mega 6.0 (Tamura et al. 2013). The MAGs and 16S rRNA gene-based OTUs were also paired by their coherent depth variation (by correlation analysis of their relative abundances) and position in the PhyloPhlAn2 and phylogenetic trees. The MAGs of Lovojärvi as well as the aforementioned MAGs of Alinen Mustajärvi were also classified into groups based on their vertical stratification patterns using hierarchical clustering, as explained above.
Statistical analyses
The dependency of vertical stratification patterns of MAGs on their gene content was studied using one-way permutational multivariate analysis of variance (PerMANOVA) by testing differences in gene content of MAGs between groups representing different vertical stratification patterns (determined in UPGMA clustering, see above). In addition, Mantel´s test was used to analyze the correlation between the depth variation patterns and the gene content as well as the genome diagnostics values (i.e. genome completeness, contamination, genome size and corrected genome size [using completeness]) of MAGs. Furthermore, non-metric multidimensional scaling was used to visualize the differences in the gene content of the MAGs. The data for genetic potential for microaerobic and anaerobic metabolism were coded as presence/absence matrix of genes coding for the following 17 enzyme classes (coding genes in brackets) that were found in at least one MAG: periplasmic nitrate reductase (presence confirmed if napA and/or napB gene(s) are present), membrane-bound nitrate reductase (presence of narG, narH, narI and/or narJ), cd1-type nitrite reductase (nirS), Cu-type nitrite reductase (aniA), nitric oxide reductase (presence of norB and/or norC), decaheme c-type cytochrome (MtoA), decaheme-associated outer membrane protein (MtrB), bacteriohemerythrin (McHr), cytochrome bd ubiquinol oxidase (cydA, i.e. high-affinity oxidase), acetate kinase (ackA), phosphate acetyltransferase (pta), nad-reducing hydrogenase (presence of hoxF, hoxG, hoxH and/or hoxY), succinate dehydrogenase (presence of sdhA, sdhB, sdhC and/or sdhD), NAD(P)-dependent malic enzyme (sfcA), fumarate hydratase class II (fumC), 3-hydroxyacyl-CoA dehydrogenase (phbA) and malate dehydrogenase (mdh). In addition, the presence/absence of a gene encoding nitrogenase iron protein (nifH) was studied for N2 fixation potential. In calculation of the pairwise dissimilarity matrices, both Bray-Curtis and Jaccard dissimilarity metrics were used for the presence/absence data, Euclidean distance was used for genome diagnostics values as well as for the data on general genetic potential, i.e. the relative abundance of genes belonging to each COG category, and correlation-based metrics were used for the vertical depth patterns of MAGs. The analyses were conducted in PAST (version 3) (Hammer, Harper and Ryan 2001).
Sequence data accession numbers
Raw sequences for 16S rRNA gene data are deposited in NCBI's short read archive under project number PRJNA516525, while shotgun metagenomic data are deposited in the European Nucleotide Archive (ENA) under project number PRJEB38681. Specific accession numbers for each library are shown in Table S1. The assembled shotgun metagenomic data are found under accession number ERZ1426841, where the contigs are named as ‘>bin_XXXX: contig_number/total_contig_count’, where XXXX denotes the MAG number the contig belongs to (e.g. bin_0959 denotes MAG 959). In addition, contig ‘ <bin-unbinned:52212/55684’ was later found to belong to MAG 959. Four MAGs with the lowest contamination % were also given direct ENA accession numbers (Table 1).
Table 1.
Medium and high quality metagenome-assembled genomes (MAGs) affiliated to Methylococcales in Lake Lovojärvi, their completeness, contamination, size, corrected size (using completeness values), taxonomic cluster and grouping based on their vertical distribution pattern, as well as their genes encoding enzymes involved in methane oxidation, methanol oxidation, N2 fixation and hydroxylamine (NH2OH) detoxification. Medium and high quality are defined as completeness >50% and contamination <10%, and, as completeness >90% and contamination <5%, respectively. Four MAGs with the lowest contamination % were also given direct ENA accession numbers, which are also shown. The analysis for depth distribution grouping is presented in Fig. S5C, while the procedure for taxonomic classification is explained in detail in Supplementary Results and Discussion, Table S2 and Figs.S3, S4, S6 and S7. Full names of the enzymes are provided in the table footnotes.
| MAG (accession) | Compl. (%) | Contam. (%) | Size (corrected) (Mb) | Taxonomic cluster | Vertical distribution group | CH4 → CH3OH | CH3OH → CH2O | N2–fixat. | NH2OH detox. |
|---|---|---|---|---|---|---|---|---|---|
| MAG 1608 (ERS4666687) | 98.6 | 1.0 | 3.3 (3.4) | 1 (Methylobacter sp.) | 1A | pmoCAB | xoxF | ||
| MAG 2248 | 88.4 | 5.7 | 2.6 (3.0) | 3 (CABC2E06) | 1A | xoxF | |||
| MAG 2500 | 94.8 | 5.8 | 2.9 (3.1) | 1 (Methylobacter sp.) | 1A | mmoC | xoxF | nifH | haoAB |
| MAG 3581 (ERS4666738) | 65.2 | 1.8 | 2.8 (4.2) | 4 (Methylovulum/Methylosoma-like) | 1B | nifH | |||
| MAG 4043 (ERS4666746) | 93.7 | 2.0 | 2.8 (3.0) | 3 (CABC2E06) | 1B | pmoCAB | xoxF | nifH | |
| MAG 2066 | 89.5 | 6.8 | 3.2 (3.5) | 1 (Methylobacter sp.) | 2A | pmoC | |||
| MAG 2588 | 75.9 | 7.4 | 3.4 (4.4) | 2 (Lake Crenothrix) | 2A | pmoCA; smmoXYBZDC | xoxF | ||
| MAG 2715 | 88.9 | 5.4 | 2.4 (2.7) | 3 (CABC2E06) | 2A | pmoCAB | xoxF | ||
| MAG 1620 | 97.9 | 6.6 | 5.1 (5.2) | 1 (Methylobacter sp.) | 2B | pmoCAB | xoxF | nifH | |
| MAG 959 (ERS4666674) | 95.4 | 3.8 | 5.8 (6.1) | 5 (Ca. Methyloumidiphilus sp.) | 2B | pmoCAB (pxmABC) | xoxF | nifH | haoA; cytL |
pmoCAB/smmoXYBZDC = particulate/soluble methane monooxygenase, pxmABC = sequence-divergent particulate methane monooxygenase, xoxF = lanthanide-dependent methanol-dehydrogenase, nifH = nitrogenase iron protein, haoAB = hydroxylamine oxidoreductase and cytL = cytochrome P460
RESULTS
Physico-chemical conditions in the water column of the study lakes
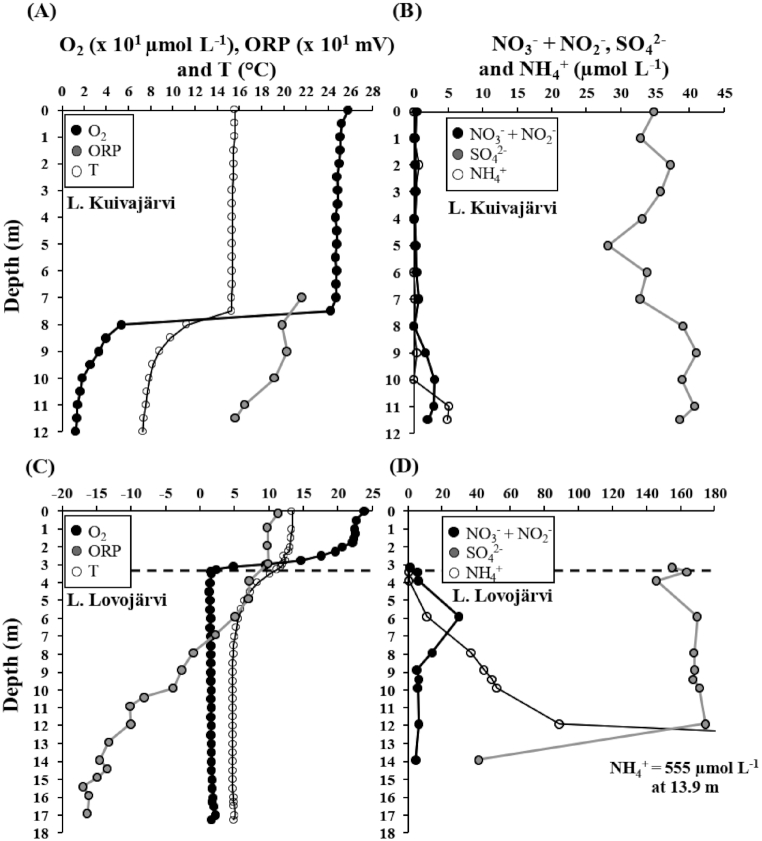
O2-stratification developed in Kuivajärvi during September, while Lovojärvi was permanently O2-stratified. Thus, both lakes had temperature- and O2-stratified water columns during the time of sampling (Fig. 1A and C). In this study, the oxic-anoxic interface was determined as the depth, where on-site measured O2 levels reached the detection limit of the optical sensor (i.e. when O2 approached steady levels). It was located at ∼3.3 m water depth in Lovojärvi, while, in Kuivajärvi, it was located virtually at the sediment surface, yet the water below 10 m depth was hypoxic (<20 µmol L−1, Fig. 1A and C). Due to the relatively high detection limit and accuracy of the O2 sensor, it is acknowledged that the actual oxic-anoxic interface was potentially located deeper in Lovojärvi. Lake water columns were slightly acidic: pH varied from 5.9 to 7.0 and from 6.3 to 6.9 in Kuivajärvi and Lovojärvi, respectively (Table S1). ORP was higher in Kuivajärvi than in Lovojärvi and decreased with depth in both lakes (Fig. 1A and C). NH4+ and NOx− (i.e. combined NO3− + NO2−) were generally higher in Lovojärvi (Fig. 1B and D). In both lakes, NH4+ increased in deeper water layers, while NOx− had a peak (i.e. showed a vertical increase followed by a decrease) in the water columns. In Kuivajärvi, the NOx− peak was at 10 m depth in the hypoxic water layer, while in Lovojärvi it was ~2.5 m below the oxic-anoxic interface at ∼6 m water depth (Fig. 1B and D). Sulfate concentration was higher in Lovojärvi than in Kuivajärvi and decreased drastically at deeper depths, from ∼12-14 m depth, in Lovojärvi but not in Kuivajärvi (Fig. 1B and D).
Figure 1.
Water column depth profiles of (A) O2, oxidation-reduction potential (ORP) and temperature (T) and (B) NO3−+ NO2−, SO42− and NH4+ for Lake Kuivajärvi, and (C) O2, ORP and T and (D) NO3−+ NO2−, SO42− and NH4+ for Lake Lovojärvi. The oxic-anoxic interface is defined with a black vertical dashed line in Lovojärvi (C and D) at ∼3.3 m depth. In Kuivajärvi (A and B), the oxic-anoxic interface was located so close to the sediment surface that the exact location was not possible to determine. For clarity, O2 and ORP are represented as x 101 µmol L−1 and x 101 mv, respectively (A and C), and NH4+ concentration of the deepest sample of Lovojärvi is not plotted (D).
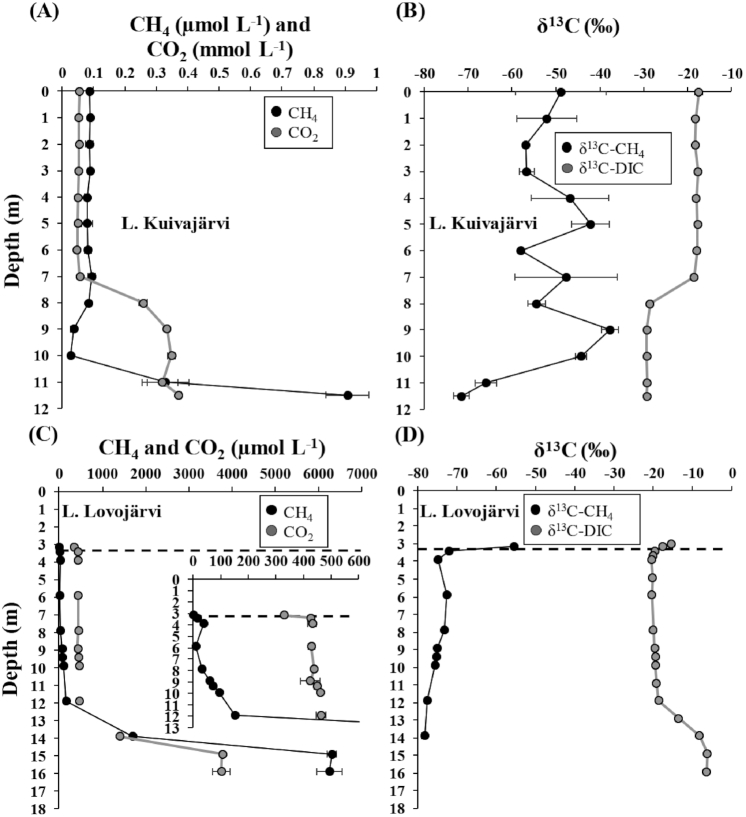
CH4 accumulated in deeper water layers of both lakes and its concentration was clearly higher in Lovojärvi (Fig. 2A and C). Vertically, δ13C of CH4 generally increased when CH4 concentration decreased. In particular, there was a zone of upward decreasing CH4 concentration and increasing δ13C of CH4 in hypoxic water layers between 11.5 m and 9 m depths in Kuivajärvi (Fig. 2A and B). In Lovojärvi, clearly below the oxic-anoxic interface between depths of 11.9 m and 5.9 m, there was a zone of slightly upward increasing δ13C of CH4 and decreasing CH4 concentration, while above 5.9 m and until the depth of 3.9 m, δ13C of CH4 again decreased and its concentration increased (Fig. 2C and D). However, above 3.9 m depth, in the zone below and around the oxic-anoxic interface in Lovojärvi, a drastic upward increase in δ13C of CH4 and decrease in its concentration took place (Fig. 2C and D).
Figure 2.
Water column depth profiles of (A) CH4 and CO2 concentrations and (B) δ13C of CH4 and dissolved inorganic carbon (DIC) for Lake Kuivajärvi, and (C) CH4 and CO2 concentrations and (D) δ13C of CH4 and DIC for Lake Lovojärvi. For clarity, the depth profiles of CH4 and CO2 above 12 m depth in Lovojärvi are shown specifically in a subfigure (C). The oxic-anoxic interface is defined with a black vertical dashed line in Lovojärvi (C and D). The data are presented as average ± standard deviation (SD) of 2–3 replicates for Kuivajärvi (A and B) and of 2 replicates for Lovojärvi (C and D), except for δ13C of DIC for Lovojärvi that was only represented by 1 replicate per depth (D). In many cases, the error bars for SDs are narrower than the data symbols.
CO2 accumulated in deep layers of both lakes and its concentration was higher in Lovojärvi than in Kuivajärvi (Fig. 2A and C). The accumulation layer was located below 8 m depth in Kuivajärvi (Fig. 2A). In Lovojärvi, CO2 accumulated well below the oxic-anoxic interface, below 11.9 m depth. Above 11.9 m depth it was at stable levels up until the oxic-anoxic interface, above which it decreased slightly (Fig. 2C). Vertically, δ13C of DIC decreased when CO2 concentration increased in Kuivajärvi (Fig. 2A and B). In Lovojärvi, the slight upward decrease in CO2 concentration above the oxic-anoxic interface was accompanied with an increase in δ13C of DIC, while at the CO2 accumulation layer below 11.9 m depth, both the concentration of CO2 and δ13C of DIC increased (Fig. 2C and D).
Methanotrophic community in the study lakes
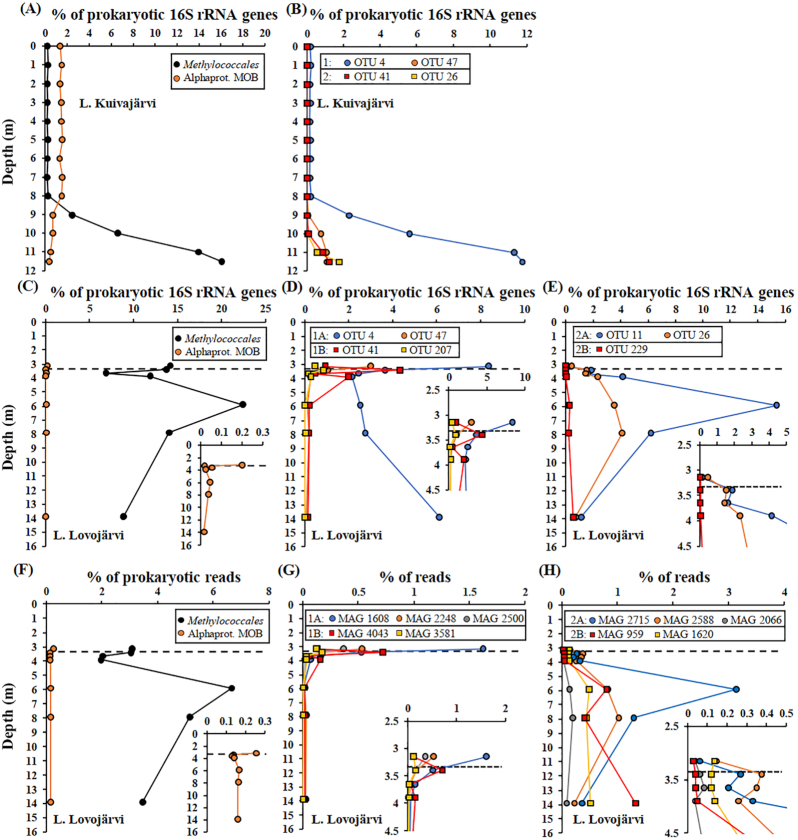
According to 16S rRNA gene amplicon sequencing, the methanotrophic community was dominated by gammaproteobacterial MOB, in the order Methylococcales, in the hypoxic water layers in both lakes, accounting for up to 16.1% and 22.4% of prokaryotic 16S rRNA gene amplicons in Kuivajärvi and Lovojärvi, respectively (Fig. 3A and C, Fig. S1A-D). Methylococcales had the highest relative abundance at the deepest water column layer in Kuivajärvi, whereas there were two Methylococcales peaks in Lovojärvi, one around the oxic-anoxic interface and another ~2.5 m below it (Fig. 3A and C). Other methanotrophs were rare, as alphaproteobacterial MOB were only up to 1.6% and 0.2% of prokaryotic 16S rRNA gene amplicons in Kuivajärvi and Lovojärvi, respectively, while verrucomicrobial MOB were less than 0.006% in both lakes, and anaerobic methanotrophic archaea, Ca. Methanoperedens (also known as ANME 2D), was less than 0.005% in Kuivajärvi but below the detection limit of prokaryotic 16S rRNA gene sequencing in Lovojärvi (Fig. 3A and C, Fig. S1A and C). Among archaea, which also generally were very rare, Ca. Methanoperedens had low relative abundance, up to 3.1% and 0.5% of archaeal 16S rRNA gene amplicons in Kuivajärvi and Lovojärvi, respectively (Fig. S1B and D). In Kuivajärvi, alphaproteobacterial MOB showed an opposite depth pattern compared with gammaproteobacterial MOB, being most abundant in upper well-oxygenated water layers and decreasing towards the deepest layers (Fig. 3A). Accordingly, they were at higher levels above than below the oxic-anoxic interface in Lovojärvi (Fig. 3C). Bacteria belonging to known anaerobically methane-oxidizing bacterial genus, Ca. Methylomirabilis sp., were not detected. Methanogens were also detected in both lakes and had a much higher relative abundance in Lovojärvi than in Kuivajärvi (Fig. S2).
Figure 3.
Water column depth profiles of relative abundance of (A) total Methylococcales and alphaproteobacterial MOBs, (B) four dominant operational taxonomic units (OTUs) of Methylococcales for Kuivajärvi, (C) total Methylococcales and alphaproteobacterial MOB and (D and E) seven dominant OTUs of Methylococcales for Lovojärvi based on 16S rRNA gene sequencing, as well as (F) total Methylococcales and alphaproteobacterial MOB and (G and H) metagenome-assembled genomes (MAGs) of Methylococcales for Lovojärvi, based on shotgun metagenomic sequencing. The OTUs (both lakes) and MAGs (Lovojärvi) are sorted according to their vertical distribution pattern into two major groups (groups 1 and 2), which are both sorted into two subgroups in Lovojärvi (subgroups 1A, 1B, 2A and 2B). See the text as well as Fig. S5 for further information on the grouping. The oxic-anoxic interface is defined with a black vertical dashed line in Lovojärvi (C-H). For clarity, depth profiles for Lovojärvi are also shown in subfigures (C-H).
The shotgun metagenomic analysis (reads classified by Kaiju) confirmed the results of the 16S rRNA gene amplicon analyses for Lovojärvi. Thus, Methylococcales dominated the methanotrophic communities, other methanotrophs were rare and Candidatus Methylomirabilis sp. were not detected (Fig. 3F, Fig. S1E). In addition, the depth distribution patterns of Methylococcales and alphaproteobacterial MOB were in accordance with those in the 16S rRNA gene dataset (Fig. 3F).
Vertical stratification patterns of different taxa of Methylococcales
Dominant Methylococcales OTUs (OTUs with maximum relative abundance >0.5% of prokaryotes at any depth) were defined for both lakes and assigned taxonomies, which is explained in detail in the Supplementary Data (Supplementary Results and Discussion; Table S2; Figs. S3 and S4). There were seven dominant OTUs (with the closest taxa in brackets) of which four, OTU 4 (Methylobacter), OTU 26 (Lake Crenothrix, i.e. MAG Crenothrix sp. D3), OTU 41 (CABC2E06 group) and OTU 47 (CABC2E06) were dominant in both lakes, whereas OTU 11 (CABC2E06), OTU 207 (Methylosoma) and OTU 229 (Ca. Methylomidiphilus alinensis) dominated only in Lovojärvi (Fig. 3B, D and E; Table S2; Figs. S3 and S4). Based on hierarchical clustering analysis of their vertical stratification patterns, the OTUs were divided into two major groups, groups 1 and 2, in both lakes, whereas, in Lovojärvi, both of these major groups were further divided into two subgroups, i.e. subgroups 1A, 1B, 2A and 2B (Fig. 3B, D and E; Fig. S5A and B). In Kuivajärvi, group 1 consisted of OTUs 4 and 47, which were found higher up in the water column, at depths of below 8 m and 9 m, respectively, than OTUs 26 and 41 in group 2, whose distribution was restricted to deeper depths, at depths of below 10 m (Fig. 3B). In Lovojärvi, group 1 consisted of OTUs that had their highest relative abundances near the oxic-anoxic interface, whereas OTUs in group 2 peaked clearly below the oxic-anoxic interface (Fig. 3D and E). OTUs 4 and 47 in subgroup 1A had their highest relative abundance just above the oxic-anoxic interface at depth 3.15 m, below which they decreased, however, OTU 4 again increased towards the deepest samples, while OTUs 41 and 207 belonging to subgroup 1B had their peak relative abundance just below the oxic-anoxic interface at depth 3.4 m and another peak slightly below that, at depth 3.9 m (Fig. 3D). By contrast, subgroup 2A consisted of OTUs 11 and 26, which both had a minor peak in their relative abundance just below the oxic-anoxic interface at depth 3.4 m, but which had their highest relative abundances deeper in the water column at depths of 5.9 and 7.9 m, respectively, while OTU 229 belonging to subgroup 2B had a minor peak at 5.9 m depth but had its highest relative abundance at the deepest sampled depth at 13.9 m (Fig. 3E).
Of the total 19 MAGs affiliated to Methylococcales, 10 were at least of medium quality and were thus considered further (Table 1). The MAGs did not contain 16S rRNA genes. However, they were paired with the 16S rRNA gene OTUs based on covarying depth patterns, and also assigned taxonomies based on phylogenomic (PhyloPhlAn2) as well as phylogenetic tree analyses (pmoA and 16S rRNA genes), as explained in detail in the Supplementary Data (Supplementary Results and Discussion; Table S2; Figs. S3, S4, S6 and S7). The MAGs were classified into five taxonomic clusters: cluster 1 belonging to Methylobacter sp.; cluster 2, which putatively represented a novel genus and was named as ‘Lake Crenothrix’; cluster 3, which putatively represented a novel genus and was named CABC2E06 (the name was based on 16S rRNA classification); cluster 4 belonging to either Methylovulum or Methylosoma and hence named as ‘Methylovulum/Methylosoma-like’; and cluster 5 belonging to Candidatus Methyloumidiphilus sp. (Table 1; Supplementary Results and Discussion; Table S2; Fig. S3, S4, S6 and S7). Based on their vertical stratification patterns, the MAGs belonged to two major groups (1 and 2) and four subgroups (1A, 1B, 2A and 2B), agreeing with the results on 16S rRNA gene OTUs (Fig. 3D, E, G and H, Fig. S5B and C). In comparison with OTU data, it was observed that OTU 4 (Methylobacter sp.), which thrived both above the oxic-anoxic interface as well as in the deepest sampled layers, very likely represented micro-diversity, which was resolved by MAGs 1608 and 1620 (both Methylobacter sp.) (Fig. 3D, G and H, Supplementary Results and Discussion).
Genomic potential of Methylococcales methanotrophs based on MAG data
MAGs 2248, 2500, 3581 and 2066 did not contain full gene sets essential for CH4 activation to methanol, i.e. those coding for particulate (pmoCAB) or soluble methane monooxygenase (mmoXYBZDC) (Table 1). These genes probably belonged to short contigs (<2500 base pairs (bp)) that were discarded before binning. By contrast, other MAGs coded for CH4 activation (Table 1). Most MAGs coded for methanol dehydrogenase, suggesting potential to oxidize methanol (Table 1). Genes encoding detoxification of hydroxylamine, a toxic product of methane monooxygenase-catalyzed NH4+ oxidation, were only found in two MAGs, while nifH gene encoding N2 fixation was found in five MAGs (Table 1). The presence/absence of nifH gene did not explain the observed depth variation patterns of the MAGs (correlation-based distance matrix) (Mantel´s test; Bray-Curtis: r = -0.02, P = 0.36, Jaccard: r = -0.06, P = 0.65).
The MAGs contained genes coding for micro-aerobic and anaerobic metabolism, i.e. denitrification (reduction of NO3− to N2O), EET, fermentation and ability to drive microaerobic respiration (high-affinity oxidase: cydA) and scavenge O2 (bacteriohemerythrin: McHr) in hypoxic conditions (Table 2). Genes encoding nitric oxide dismutase or a putative EET system, T1SS, were not found. Genes in the gene cluster encoding the NO-dependent hypoxia stress response mechanism, consisting of an anaerobic NO reductase transcriptional regulator (norR), a hybrid cluster protein (hcp), a cognate flavodoxin reductase (hcr), an oxygen and nitric oxide-sensing protein (hst) and hemerythrin (hmr), were present in only one MAG (MAG 2500), wherein the gene cluster was, however, incomplete (only norR, hcp and hcr). Based on visual observation of the presence/absence of genes encoding microaerobic and anaerobic metabolism (Table 2 and Fig. S8A) as well as ordination analysis (Fig. 4A), Mantel´s (P > 0.05, Table S3) and PerMANOVA tests (P > 0.05, Table S4), differences in the content of genes encoding microaerobic and anaerobic metabolism did not explain the differences in the depth variation patterns among MAGs.
Table 2.
Medium and high quality MAGs affiliated to Methylococcales in Lake Lovojärvi as well as their genes encoding enzymes involved in microaerobic (i.e. oxygen scavenging and high-affinity oxidases) and anaerobic metabolism (i.e. NOx− reduction [denitrification], extracellular electron transfer [EET, i.e. electron transfer to Fe3+ and Mn4+ minerals and organic EAs] and fermentation) in Lovojärvi. The MAGs are ordered based on their vertical distribution pattern group (see Table 1). Full names of the enzymes are provided in table footnote1.
| MAG | Vertical distribution group | NO3− → NO2− | NO2− → NO | NO → N2O | EET genes | Fermentation | Microaerobic metabolism |
|---|---|---|---|---|---|---|---|
| MAG 1608 | 1A | narGHIJ | aniA | MtoA; MtrB2 | hoxFGHY; sdhABCD; mdh | cydA | |
| MAG 2248 | 1A | nirS | norBC | MtoA; MtrB | ackA; hoxFH; sdhABC; fumC | McHr; cydA | |
| MAG 2500 | 1A | MtoA; MtrB | hoxFGHY; sdhABC; sfcA; mdh | McHr; cydA | |||
| MAG 3581 | 1B | narGH | aniA | norC | MtoA; MtrB | hoxGHY; sdhABC; fumC; sfcA | McHr |
| MAG 4043 | 1B | napAB | nirS | norBC | MtoA; MtrB | ackA; hoxFGHY; sdhABC; mdh | McHr; cydA |
| MAG 2066 | 2A | norBC | hoxH; sdhABD; mdh | cydA | |||
| MAG 2588 | 2A | aniA | norBC | MtoA; MtrB | phbA; pta; hoxFG; sdhABCD | McHr | |
| MAG 2715 | 2A | napAB | nirS | MtoA; MtrB | ackA; sdhAB; sfcA; mdh | McHr; cydA | |
| MAG 1620 | 2B | narGHIJ | nirS; aniA | MtoA; MtrB | ackA; hoxFGH; sdhABCD; sfcA; mdh | McHr; cydA | |
| MAG 959 | 2B | narHIJ | aniA | norBC | MtoA; MtrB | pta; hoxFGHY; sdhABCD; fumC; mdh | McHr; cydA |
1) napAB = periplasmic nitrate reducatse, narGHIJ = membrane-bound nitrate reductase, nirS = cd1-type nitrite reductase, aniA = Cu-type nitrite reductases, norBC = nitric oxide reductase, MtoA = A decaheme c-type cytochrome, MtrB = decaheme-associated outer membrane protein, ackA = acetate kinase, sdhABCD = succinate dehydrogenase, sfcA = NAD(P)-dependent malic enzyme, mdh = malate dehydrogenase, phbA = 3-hydroxyacyl-CoA dehydrogenase, pta = phosphate acetyltransferase, hoxFGHY = NAD-reducing hydrogenase, fumC = fumarate hydratase class II, McHr = bacteriohemerythrin, cydA = cytochrome bd ubiquinol oxidase
2) MtoA is classified as ‘probable iron oxidation and possible iron reduction’ while MtrB is classified as ‘dissimilatory iron reduction’ or ‘probable iron reduction’ by FeGenie (Garber et al. 2020)
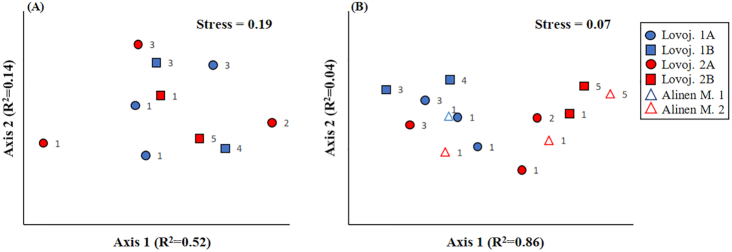
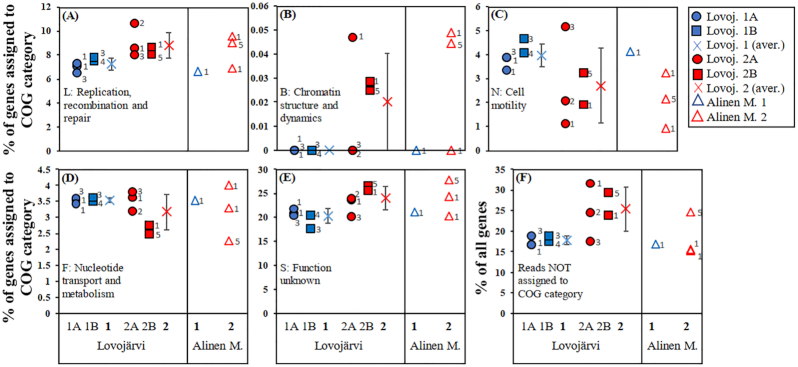
Figure 4.
Results of two-dimensional non-metric multidimensional scaling analysis on genetic differences between MAGs of Methylococcales based on (A) Bray-Curtis distance matrix on the presence/absence data of genes coding for microaerobic and anaerobic metabolism (see information on the genes in Materials and Methods and in Fig. S8A) in MAGs of Lake Lovojärvi and (B) Euclidean distance matrix on relative abundance of genes in different clusters of orthologous group (COG) categories (see the category list in Fig. S8B) in MAGs of Lake Lovojärvi and previously studied Lake Alinen Mustajärvi (see Rissanen et al. 2018). The MAGs are sorted according to their vertical distribution patterns into two major groups (1 and 2), which are both sorted into two subgroups in Lake Lovojärvi (subgroups 1A, 1B, 2A and 2B). See the text as well as Fig. S5 for further information on the grouping. The number at the right side of each symbol denotes the phylogenomic cluster the MAG belongs to (PhyloPhlAn analysis, Fig. S6). The presence/absence data of genes coding for microaerobic and anaerobic metabolism were also analyzed using Jaccard distance matrix with similar results as in panel A (data not shown).
The genes of the MAGs were also classified into functional (COG) categories to assess whether general genetic differences could explain the differences in their vertical stratification patterns (Fig. S8B). Of the genes, 68–83% were classified into COG categories, while the rest remained unclassified. Ordination analysis of the relative abundance of genes in different COG categories suggested that there were general genetic differences between the major groups and subgroups of depth distribution patterns of MAGs (Fig. 4B), which was supported by PerMANOVA analysis (P < 0.05, Table S5) as well as by Mantel´s test (P < 0.05, Table S6). The genes in the four Methylococcales-MAGs of the previously studied Lake Alinen Mustajärvi were also added into the analysis for comparison (75–85% could be classified as COG categories) (Fig. S8B) (Rissanen et al. 2018). These MAGs were also grouped into two groups, i.e. group 1 including MAG 126, which clearly thrived above the oxic-anoxic interface, and group 2 including MAGs 10, 140 and 149, which clearly thrived below the oxic-anoxic interface (Fig. S5D). Ordination analysis suggested that there were also general genetic differences between groups for MAGs of Alinen Mustajärvi (Fig. 4B). Due to low sample size, PerMANOVA analysis of MAGs of Alinen Mustajärvi was not possible. However, PerMANOVA analysis pooling MAGs of both Lovojärvi and Alinen Mustajärvi showed significant genetic differences between the depth distribution groups 1 and 2 (P < 0.05, Table S5).
The single COG categories correlating with the depth distribution variations were further identified using Mantel´s test (Table S6). For Lovojärvi MAGs, the pairwise differences in depth variation patterns correlated significantly with the pairwise differences in the relative abundance of genes of COG categories L (replication, recombination and repair), B (chromatin structure and dynamics), N (cell motility), F (nucleotide transport and metabolism) and S (function unknown) (P < 0.05, Table S6, Fig. 5). Expectedly, genes encoding COG class B, which is generally associated with eukaryotic organisms, were rare (0–0.05% of genes with COG classification) and were not considered further (Fig. 5B). Furthermore, the pairwise differences in depth variation patterns correlated significantly with the pairwise differences in the proportion of genes that remained unclassified into COG categories (P < 0.05), as well as with the pairwise differences in genome sizes (P < 0.05), but not with the corrected genome sizes (corrected using completeness percentage) (Table S6). Such analysis was not performed for the MAGs of Alinen Mustajärvi due to a too low number of MAGs. However, in both Lovojärvi and Alinen Mustajärvi, the relative abundance of genes encoding replication, recombination and repair (L), as well as unknown functions (S), were generally higher, while those encoding cell motility (N) were generally lower in MAGs, which had their peak relative abundances clearly below the oxic-anoxic interface (depth distribution group 2), than in MAGs, which thrived higher up in the water column closer to the oxic-anoxic interface (depth distribution group 1) (Fig. 5A, C and E). Furthermore, the relative abundance of genes encoding nucleotide transport and metabolism (F) were lower in MAGs thriving in the deepest studied water column layers (subgroup 2B) than in other MAGs (subgroups 1A, 1B and 2A) in Lovojärvi and a similar tendency was observed for two out of three MAGs in depth distribution group 2 compared with the one MAG in group 1 for Alinen Mustajärvi (Fig. 5D). The proportion of genes not assigned to COG categories was also generally higher in MAGs of depth distribution group 2 than group 1 for Lovojärvi, but such a pattern was not observed for Alinen Mustajärvi (Fig. 5F). Notably, MAG 2715 in Lovojärvi, representing depth distribution group 2A, had genetic content more similar to MAGs thriving higher up in the water column close to the oxic-anoxic interface (Figs 4B and 5).
Figure 5.
Proportion of genes of MAGs of Lake Lovojärvi and previously studied Lake Alinen Mustajärvi in the following COG categories: (A) L: replication, recombination and repair, (B) B: chromatin structure and dynamics, (C) N: cell motility, (D) F: nucleotide transport and metabolism and (E) S: function unknown (% of genes assigned to COG categories), as well as (F) the proportion of genes not assigned to a COG category (% of all genes). These variables correlated with the vertical distribution patterns of MAGs of Lake Lovojärvi (Mantel´s test, P < 0.05; see Table S6). The MAGs are sorted according to their vertical distribution patterns into two major groups (1 and 2), which are both sorted into two subgroups in Lake Lovojärvi (subgroups 1A, 1B, 2A and 2B). Besides values for single MAGs in each subgroup, average (±SD) for the two major groups are also shown for Lake Lovojärvi. See the text as well as Fig. S5 for further information on the grouping. The number at the right side of each symbol denotes the phylogenomic cluster the MAG belongs to (PhyloPhlAn analysis, Fig. S6).
DISCUSSION
Methane dynamics
As microbial CH4 oxidation fractionates against the heavier isotope, enriching the residual CH4 in 13C (Whiticar 1999), the concurrent upward decrease in CH4 concentration and increase in its δ13C indicate zones of active CH4 oxidation (Blees et al. 2014; Oswald et al. 2016). In Kuivajärvi, CH4 oxidation was dominantly found in the hypoxic zone, i.e. between 11.5 m and 9 m (Fig. 2A and B), which agrees with the measured rates of potential CH4 oxidation (Saarela et al. 2020). In Lovojärvi, δ13C and concentration data suggested that CH4 oxidation took place around the oxic-anoxic interface as well as deeper in the hypoxic and anoxic water column between 11.9 m and 5.9 m; however, confirmation of CH4 oxidation at the deeper zone would require additional analyses, as the change in δ13C of CH4 in the deeper zone was actually very slight (Fig. 2C and D). Changes in δ13C of DIC did not show signals of CH4 oxidation, as they were possibly masked by the isotopic signals from other substrates and processes than CH4 and CH4 oxidation (Fig. 2B and D).
Methanotrophic community structure
The dominance of Methylococcales, especially in the layers at and below the oxic-anoxic interface, where isotopic data indicated CH4 oxidation, agrees with previous studies from arctic, boreal and temperate lakes (Blees et al. 2014; Oswald et al. 2015, 2016, 2017; Crevecoeur et al. 2017; Rissanen et al. 2018; Cabrol et al. 2020). (Meta)genomic data from this and previous studies as well as previous experimental data strongly suggests that besides aerobic methanotrophy taking place in oxic water layers and being partly enabled through oxygenic photosynthesis below the oxic-anoxic interface, Methylococcales in lakes may drive microaerobic metabolism, anaerobic respiration (denitrification and EET) and/or fermentation during times of oxygen depletion, yet besides anaerobic respiration, EET mechanisms can also act as electron sinks (Kalyuzhnaya et al. 2013; Kits, Klotz and Stein 2015; Oswald et al. 2016, 2017; Graf et al. 2018; Smith et al. 2018; He et al. 2019; Knief 2019; Smith and Wrighton 2019; van Grinsven et al. 2020; Zheng et al. 2020). However, besides denitrification pathway, nitrite and nitric oxide reduction can also be part of the hydroxylamine detoxification mechanism in methanotrophs (Campbell et al. 2011), while Yu et al. (2020) proposed that the main role of the denitrification pathway in Methylococcales may not be anaerobic respiration but production of nitric oxide, which acts as a signaling molecule in response to hypoxia. However, our results suggest the presence of denitrification potential as a means for anaerobic respiration, as the genes coding for hydroxylamine detoxification, as well as the nitric oxide-dependent hypoxia stress response mechanism, were rare in the MAGs (Table 1).
In contrast to deep, temperate, permanently O2-stratified lakes, Lake Zug and Lake Lugano (max. depth ≥∼200 m), where bacteria belonging to Ca. Methylomirabilis were abundant (Graf et al. 2018; Mayr et al. 2020b), the results of this study also further highlight the very low abundance of anaerobic methanotrophs in the water columns of shallow lakes (max. depth ≤∼20 m) (Oswald et al. 2015; Rissanen et al. 2018; Cabrol et al. 2020; Mayr et al. 2020b). The discrepancy between shallow and deep lakes is most likely due to environmental instability caused by day-night variations in light penetration-driven oxygenic photosynthesis taking place in the seemingly anoxic waters of shallow lakes but not in deep lakes with dark conditions, preventing the establishment of populations of slow-growing anaerobic methanotrophs in shallow lakes (Oswald et al. 2015; Rissanen et al. 2018; Cabrol et al. 2020; Mayr et al. 2020b). Penetration of photosynthetically active radiation (PAR) is not known for Lovojärvi, but it can be roughly estimated according to Karhunen et al. (2013) using the previously measured water color values, from 60 to 105 mg Pt L−1 (Hakala 2004; unpublished data from Lammi Biological Station). According to these estimates, the depth with minimum PAR allowing for oxygenic photosynthesis, i.e. ∼0.1 µmol photons m−2 s−1 (Gibson 1985; Brand et al. 2016), extended well below the oxic-anoxic interface, between depths of ∼7.5 and ∼5.5 m, during a bright summer day in Lovojärvi (surface PAR = ∼1400 µmol photons m−2 s−1). Besides light dynamics, the seasonal stratification pattern also contributes to the environmental instability decreasing the population sizes of anaerobic methanotrophs in shallow lakes; however, respective data do not exist from seasonally stratified deep lake basins (Mayr et al. 2020b). Recent studies suggest that methanogens may also drive AOM (Bar-Or et al. 2017; Yu et al. 2021). Indeed, the relative abundance of methanogens was higher in Lovojärvi, where isotopic data indicated CH4 oxidation below the oxic-anoxic interface, than in Kuivajärvi (Fig. S2). Unfortunately, the role of water column methanogens (i.e. driving either methanogenesis or AOM) is impossible to assess using the current dataset. Hence, further studies on the potential AOM activity of lake water column methanogens are well justified.
Vertical stratification patterns of MOB
The degree of vertical structuring and diversity (based on the number of dominant OTUs) of MOB community was higher in Lovojärvi than in Kuivajärvi, which was due to wider physicochemical gradients, especially those of O2 and CH4, which are key drivers of water column MOB community structure, as well as due to higher environmental stability, in the constantly O2-stratified Lovojärvi than in the seasonally stratified Kuivajärvi (Figs. 1A, C, 2A, C and 3) (Reis et al. 2020; Mayr et al. 2020b). In contrast to Methylococcales, which thrived in hypoxic/anoxic water layers, alphaproteobacterial MOB had increased relative abundance in the upper water layers in both lakes. There were also similarities in the observed stratification patterns of Methylococcales between the study lakes, with OTUs 4 (Methylobacter sp.) and 47 (CABC2E06) thriving just above the oxic-anoxic interface in Lovojärvi, and accordingly, extending higher in the water column than other OTUs in Kuivajärvi (Fig. 3B and D). Furthermore, OTUs 26 (Lake Crenothrix) and 41 (CABC2E06) thrived below the oxic-anoxic interface in Lovojärvi, agreeing with their distribution in Kuivajärvi, where they were limited to deeper layers than OTUs 4 and 47 (Fig. 3B, D and E). O2 was recently suggested to drive the distributional differences between alphaproteobacterial MOB and Methylococcales, while both CH4 and O2 were considered to be the major factors affecting the vertical stratification of Methylococcales in lake water columns (Reis et al. 2020; Mayr et al. 2020b). Given the similarities in the vertical structuring and high proportion of shared dominant Methylococcales OTUs (i.e. 4/7 OTUs, representing 3/5 taxa) between the study lakes, which differ over 40-fold in their CH4 concentration but not much in their O2 concentrations, it can be suggested that O2 gradient was more important in structuring the MOB communities than CH4 in the study lakes (Figs. 1A, C, 2A, C, 3A and C). In addition to abiotic factors such as O2, biotic factors, i.e. microbial interactions, were also very recently shown to affect the structure of methanotrophic communities, which, however, was beyond the scope of this study (Guggenheim et al. 2020).
Our results also further highlight that the depth distribution patterns of different members of Methylococcales are not taxon-dependent (Rissanen et al. 2018; Cabrol et al. 2020; Mayr et al. 2020b). In contrast to temperate lakes, where CABC2E06 thrived at the high-O2 and low-CH4 conditions, members of CABC2E06 (OTU 11/MAG 2715) were detected to dominate the MOB community also clearly below the oxic-anoxic interface in Lovojärvi, agreeing with results from arctic lakes (Fig. 3E and H) (Cabrol et al. 2020; Mayr et al. 2020b). Furthermore, Methylobacter-related MOB were detected to thrive both above and clearly below the oxic-anoxic interface in Lovojärvi (OTU 4, MAG 1608 and MAG 1620) and previously studied Alinen Mustajärvi (MAG 126, MAG 140 and MAG 149), which disagrees with results from arctic lakes, where Methylobacter sp. thrived only in the deep, anoxic, CH4-rich water layers (Rissanen et al. 2018; Cabrol et al. 2020). In addition, in contrast to Lovojärvi, where Methylosoma/Methylovulum-like MOB were abundant, Methylosoma-related MOB had only marginal abundance during the O2-stratification period, yet thrived during the lake overturn, in the water column of L. Rotsee (Mayr et al.2020a, 2020b).
Genetic factors controlling the vertical stratification patterns of Methylococcales methanotrophs
Based on the MAG data, variations in the genetic potential for N2 fixation or microaerobic and anaerobic metabolism did not explain the variable depth distribution patterns of Methylococcales in Lovojärvi, which contrast with the results on the denitrification potential of Methylococcales from the previously studied Lake Alinen Mustajärvi (Rissanen et al. 2018). This suggests, that in contrast to Alinen Mustajärvi, Methylococcales thriving in the oxic water layer in Lovojärvi benefitted from having genetic potential for microaerobic and anaerobic metabolism, which could be due to differences in fluctuations of the oxic-anoxic interface between the lakes. Although previous studies suggest that the oxic-anoxic interface was slowly moving downwards in both lakes during the time of sampling (in September) due to the autumn-time extension of the mixed layer (Mutyaba 2012; Nykänen et al. 2014), there can still be differences in short-term (hourly-daily) fluctuations in the vertical location of the oxic-anoxic interface between the lakes, which is impossible to assess with the existing data. In any case, these results suggest that lakes differ in how genetic potential for microaerobic and anaerobic metabolism explains the depth distribution patterns of Methylococcales, which warrants further studies. However, the results could also represent a sampling artifact, as the whole oxic water column was sampled in Alinen Mustajärvi, while only one oxic depth layer was sampled in Lovojärvi, potentially leaving some MOB thriving in the oxic layers of Lovojärvi unnoticed (Rissanen et al. 2018). Hence, future studies should also adequately cover the oxic water column layers.
In contrast to genetic potential for N2 fixation and microaerobic/anaerobic metabolism, the general genetic differences among Methylococcales, analyzed as the proportion of MAG genes in different COG categories, were related to their variable depth distribution patterns, both in Lovojärvi and Alinen Mustajärvi (Figs 4B and 5, Tables S5 and S6). The genetic differences were especially clear between Methylococcales thriving at (just above and below) and clearly below the oxic-anoxic interface in Lovojärvi, and between those thriving above and clearly below the oxic-anoxic interface in Alinen Mustajärvi (Figs 4B and 5). Despite it not being possible to determine the exact functional roles of the genes belonging to the COG categories without experimental confirmation, some suggestions for the possible reasons underlying the relationship between genetic and depth distribution differences of Methylococcales can be made. The higher proportion of genes encoding replication, recombination and repair in Methylococcales thriving clearly below than at/above the oxic-anoxic interface in both lakes is very likely related to higher stress tolerance adaptations (Ma et al. 2015; Le et al. 2016), which could be due to more stressful conditions, for example, the absence/very limited availability of O2, decreased ORP, presence of sulfide (as seen in Alinen Mustajärvi; see Rissanen et al. 2018) and accumulation of methane-derived extracellular metabolites to potentially stressful/inhibitory levels (e.g. formate and volatile fatty acids; Kalyuzhnaya et al. 2013), in deeper, hypoxic/anoxic water column layers than in upper, oxic water column layers (Fig. 5A). The generally higher proportion of genes encoding unknown functions (in both lakes) and of unknown genes (i.e. not assigned to any COG category, in Lovojärvi), in genomes of Methylococcales thriving clearly below than in those at/above the oxic-anoxic interface, also suggest that diverse, currently uncharacterized genetic potential is needed to cope with the harsh conditions of the hypoxic/anoxic water layers (Fig. 5E and F). The contrasting pattern in the proportion of genes encoding cell motility probably reflects higher cell motility activity by Methylococcales thriving at/above than clearly below the oxic-anoxic interface (Fig. 5C). This is most likely due to more pronounced vertical fluctuations in physicochemical conditions in the upper than in the lower water layers (Fig. 5C). MOB thriving close to the oxic-anoxic interface may have been specifically adapted to living in such conditions. Hence, increased genetic investment into cell motility may enable them to actively move alongside with changes in the vertical location of the oxic-anoxic interface. Interestingly, MAG 2715 of the CABC2E06 group, which thrived clearly below the oxic-anoxic interface (depth distribution group 2A), had more similar general genetic content with the two other CABC2E06 representatives (MAGs 2248 and 4043), which peaked at the oxic-anoxic interface (Figs 4B and 5). The relative abundance of MAG 2715 actually peaked 2 m higher in the water column, at 5.9 m depth, than the two other MAGs (MAGs 2066 and 2588) in the same depth distribution group (Fig. 3H). Actually, it peaked at depths where light levels were estimated to sustain oxygenic photosynthesis (i.e. above 7.5 m depth, see above). Furthermore, the representative 16S rRNA gene OTU for MAG 2715, OTU 11 was absent in Kuivajärvi, where it was too dark for oxygenic photosynthesis in the CH4 oxidation layers (Fig. 3B, D and E). This, together with the relatively high proportion of genes encoding cell motility in its genome, suggests that MOB represented by MAG 2715 are specifically adapted to living below the oxic-anoxic interface in water column layers, where (micro)aerobic CH4 oxidation is supported by oxygenic photosynthesis, and that it is adapted to changing its vertical position along with changes in light and O2 levels (Fig. 5C).
CONCLUSIONS
We showed that Methylococcales and alphaproteobacterial MOB as well as different taxa (i.e. OTUs and MAGs representing putative novel genera and species) within Methylococcales vary in their vertical stratification patterns in the water columns of boreal, oxygen-stratified lakes. Together with previous results from temperate lakes, this confirms that the vertical structuring of MOB communities is a general phenomenon in the water columns of oxygen-stratified lakes. However, the vertical stratification patterns of the Methylococcales methanotrophs were not completely predictable by their taxonomic classification. Instead, for the first time, we provide evidence suggesting that the variable depth distribution patterns of MOB are governed by their genetic differences. The role of genetic potential for microaerobic and anaerobic metabolism in explaining the distribution of MOB along the O2 gradient remained unclear due to inconsistent data between the study lakes. Yet the results suggested that there was genetic adaptation for increased stress tolerance in MOB thriving in the harsh conditions of the hypoxic and anoxic water column layers clearly below the oxic-anoxic interface, and for increased cell motility in MOB thriving at the vertically fluctuating oxic-anoxic interface. We acknowledge that these results are based on analyses of genetic potential, thus requiring further confirmation, for example, via experiments with lake MOB isolates. However, in any case, the results highlight, that in addition to physicochemical conditions (especially O2) and microbial interactions, genetic adaptation must be considered as a driving factor of vertical structuring of MOB communities in water columns of oxygen-stratified lakes, and potentially also in other oxygen-stratified methane-containing ecosystems.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Xavier Prieto Mollar (MARUM, University of Bremen) for laboratory support. In addition, Lammi Biological Station and Hyytiälä Forestry Field Station are acknowledged for their support in field and laboratory work. Furthermore, we thank the reviewers and editor for their valuable comments.
Contributor Information
Antti J Rissanen, Faculty of Engineering and Natural Sciences, Tampere University, Korkeakoulunkatu 6, FI-33720, Tampere, Finland.
Taija Saarela, Department of Environmental and Biological Sciences, University of Eastern Finland, Yliopistonranta 1 E, FI-70210, Kuopio, Finland.
Helena Jäntti, Department of Environmental and Biological Sciences, University of Eastern Finland, Yliopistonranta 1 E, FI-70210, Kuopio, Finland.
Moritz Buck, Department of Ecology and Genetics/Limnology, Uppsala University, Norbyvägen 18D, SE-75236, Uppsala, Sweden; Department of Aquatic Sciences and Assessment, Swedish University of Agricultural Sciences, box 7050, SE-75007, Uppsala, Sweden.
Sari Peura, Department of Forest Mycology and Plant Pathology, Science for Life Laboratory, Swedish University of Agricultural Sciences, Almas allé 5, SE-75651, Uppsala, Sweden.
Sanni L Aalto, Department of Environmental and Biological Sciences, University of Eastern Finland, Yliopistonranta 1 E, FI-70210, Kuopio, Finland; Department of Biological and Environmental Sciences, University of Jyväskylä, Survontie 9 C, FI-40014, Jyväskylä, Finland.
Anne Ojala, Ecosystems and Environment Research Programme, Faculty of Biological and Environmental Sciences, University of Helsinki, Viikinkaari 1, FI-00014, Helsinki, Finland; Institute of Atmospheric and Earth System Research (INAR)/Forest Sciences, Faculty of Agriculture and Forestry, University of Helsinki, Viikinkaari 1, FI-00014, Helsinki, Finland.
Jukka Pumpanen, Department of Environmental and Biological Sciences, University of Eastern Finland, Yliopistonranta 1 E, FI-70210, Kuopio, Finland.
Marja Tiirola, Department of Biological and Environmental Sciences, University of Jyväskylä, Survontie 9 C, FI-40014, Jyväskylä, Finland.
Marcus Elvert, MARUM - Center for Marine Environmental Sciences & Faculty of Geosciences, University of Bremen, Leobener Str. 8, D-28359, Bremen, Germany.
Hannu Nykänen, Department of Environmental and Biological Sciences, University of Eastern Finland, Yliopistonranta 1 E, FI-70210, Kuopio, Finland.
FUNDING
This study was supported by the Kone Foundation (grant no. 201803224) for AJR, Olvi-säätiö (grant no. 201720037), Maa-ja Vesitekniikan tuki ry (grant no. 34348), the University of Eastern Finland Doctoral Programme in Environmental Physics, Health and Biology (EPHB) and Water JPI ERA-NET Cofund WaterWorks2017 and Academy of Finland (project no. 326818) for TS, the Deutsche Forschungsgemeinschaft through the cluster of Excellence EXC 309 ‘The Ocean in the Earth system’ (project no. 49926684) for ME, Academy of Finland (project no. 286642 for AJR, project no. 275127 for HJ, project no. 310302 for SLA, and project nos. 136455 and 140964 for HN), and European Research Council (ERC) CoG project no. 615146 for MT. The authors also acknowledge the Academy of Finland Centre of Excellence (project nos. 272041, 118780 and 307331) and ARCTICFIRE-project (project no. 286685) funded by Academy of Finland for JP. In addition, the authors acknowledge the University of Eastern Finland Water Research Programme funded by Olvi-säätiö, the Jenny and Antti Wihuri Foundation and the Saastamoinen Foundation for HJ. The shotgun metagenomic sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project uppstore2018116 and SNIC2017/1–616. Tampere University provided funding for the open access fees.
Conflict of interest
None declared.
REFERENCES
- Altschul SF, Gish W, Miller W et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apprill A, McNally S, Parsons R et al. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. 2015;75:129–37. [Google Scholar]
- Bar-Or I, Elvert M, Eckert W et al. Iron-coupled anaerobic oxidation of methane performed by a mixed bacterial-archaeal community based on poorly reactive minerals. Environ Sci Technol. 2017;51:12293–301. [DOI] [PubMed] [Google Scholar]
- Bastviken D, Cole J, Pace M et al. Methane emissions from lakes: Dependence of lake characteristics, two regional assessments, and a global estimate. Glob Biogeochem Cycles. 2004;18:GB4009. [Google Scholar]
- Bastviken D, Tranvik LJ, Downing JA et al. Freshwater methane emissions offset the continental carbon sink. Science. 2011;331:50. [DOI] [PubMed] [Google Scholar]
- Beal EJ, House CH, Orphan VJ.. Manganese- and iron-dependent marine methane oxidation. Science. 2009;325:184–7. [DOI] [PubMed] [Google Scholar]
- Blees J, Niemann H, Wenk CB et al. Micro-aerobic bacterial methane oxidation in the chemocline and anoxic water column of deep south-Alpine Lake Lugano (Switzerland). Limnol Oceanogr. 2014;59:311–24. [Google Scholar]
- Brand A, Bruderer H, Oswald K et al. Oxygenic primary production below the oxycline and its importance for redox dynamics. Aquat Sci. 2016;78:727–41. [Google Scholar]
- Brown C., Irber L.. sourmash: a library for MinHash sketching of DNA. J Open Source Softw. 2016;1:27. [Google Scholar]
- Buck M, Garcia SL, Fernandez Vidal L et al. Comprehensive dataset of shotgun metagenomes from stratified freshwater lakes and ponds. bioRxiv. 2020, DOI: https://doi.org/10.1101/2020.11.12.379446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrol L, Thalasso F, Gandois L et al. Anaerobic oxidation of methane and associated microbiome in anoxic water of Northwestern Siberian lakes. Sci Total Environ. 2020;736:139588. [DOI] [PubMed] [Google Scholar]
- Campbell MA, Nyerges G, Kozlowski JA et al. Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol Lett. 2011;322:82–9. [DOI] [PubMed] [Google Scholar]
- Crevecoeur S, Vincent WF, Comte J et al. Diversity and potential activity of methanotrophs in high methane-emitting permafrost thaw ponds. PLoS One. 2017;12:e0188223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eller G, Känel L, Krüger M.. Cooccurrence of aerobic and anaerobic methane oxidation in the water column of Lake Plußsee. Appl Environ Microbiol. 2005;71:8925–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–8. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, Zhu B, Speth D et al. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci. 2016;113:12792–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner S, Andersson AF, Alonso-Sáez L et al. Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J Microbiol Methods. 2011;84:12–8. [DOI] [PubMed] [Google Scholar]
- Garber AI, Nealson KH, Okamoto A et al. FeGenie: A comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front Microbiol. 2020;11:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CE. Growth rate, maintenance energy and pigmentation of planktonic Cyanophyta during one-hour light: Dark cycles. Br Phycol J. 1985;20:155–61. [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA et al. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. [DOI] [PubMed] [Google Scholar]
- Graf JS, Mayr MJ, Marchant HK et al. Bloom of a denitrifying methanotroph, ‘Candidatus Methylomirabilis limnetica’, in a deep stratified lake. Environ Microbiol. 2018;20:2598–614. [DOI] [PubMed] [Google Scholar]
- Guggenheim C, Freimann R, Mayr MJ et al. Environmental and microbial interactions shape methane-oxidizing bacterial communities in a stratified lake. Front Microbiol. 2020;11:2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakala A. Meromixis as a part of lake evolution; observations and a revised classification of true meromictic lakes in Finland. Boreal Environ Res. 2004;9:37–53. [Google Scholar]
- Hammer O, Harper DA., Ryan P.. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:1–9. [Google Scholar]
- Hanson RS, Hanson TE.. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon MF, Hu S, Shi Y et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;500:567–70. [DOI] [PubMed] [Google Scholar]
- Heiskanen JJ, Mammarella I, Ojala A et al. Effects of water clarity on lake stratification and lake-atmosphere heat exchange. J Geophys Res Atmos. 2015;120:7412–28. [Google Scholar]
- He S, Lau MP, Linz AM et al. Extracellular electron transfer may be an overlooked contribution to pelagic respiration in humic-rich freshwater lakes. mSphere. 2019;4:e00436–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A, Kerckhof F-M, Luke C et al. Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies. Environ Microbiol Rep. 2013;5:335–45. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Forslund K, Coelho LP et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34:2115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG et al. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C, Rodriguez-R LM, Phillippy AM et al. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallistova A, Kadnikov V, Rusanov I et al. Microbial communities involved in aerobic and anaerobic methane cycling in a meromictic ferruginous subarctic lake. Aquat Microb Ecol. 2018;82:1–18. [Google Scholar]
- Kalyuzhnaya MG, Yang S, Rozova ON et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun. 2013;4:2785. [DOI] [PubMed] [Google Scholar]
- Karhunen J, Arvola L, Peura S et al. Green sulphur bacteria as a component of the photosynthetic plankton community in small dimictic humic lakes with an anoxic hypolimnion. Aquat Microb Ecol. 2013;68:267–72. [Google Scholar]
- Kirf MK, Dinkel C, Schubert CJ et al. Submicromolar oxygen profiles at the oxic–anoxic boundary of temperate lakes. Aquat Geochemistry. 2014;20:39–57. [Google Scholar]
- Kits KD, Klotz MG, Stein LY.. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ Microbiol. 2015;17:3219–32. [DOI] [PubMed] [Google Scholar]
- Knief C. Diversity of methane cycling microorganisms in soils and their relation to oxygen. Curr Issues Mol Biol. 2019;33:23–56. [DOI] [PubMed] [Google Scholar]
- Knittel K, Boetius A.. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol. 2009;63:311–34. [DOI] [PubMed] [Google Scholar]
- Le PT, Makhalanyane TP, Guerrero LD et al. Comparative metagenomic analysis reveals mechanisms for stress response in hypoliths from extreme hyperarid deserts. Genome Biol Evol. 2016;8:2737–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Lyu X-F, Zha T et al. Reconstructed metagenomes reveal changes of microbial functional profiling during PAHs degradation along a rice (Oryza sativa) rhizosphere gradient. J Appl Microbiol. 2015;118:890–900. [DOI] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL.. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MJ, Zimmermann M, Dey J et al. Growth and rapid succession of methanotrophs effectively limit methane release during lake overturn. Commun Biol. 2020a;3:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MJ, Zimmermann M, Guggenheim C et al. Niche partitioning of methane-oxidizing bacteria along the oxygen–methane counter gradient of stratified lakes. ISME J. 2020b;14:274–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel P, Ng KL, Krogh A.. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7:11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milucka J, Kirf M, Lu L et al. Methane oxidation coupled to oxygenic photosynthesis in anoxic waters. ISME J. 2015;9:1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutyaba C. A stable isotope study of the hydrological and carbon cycle in meromictic lake, Lovojärvi. Master of Science thesis, University of Jyväskylä, Jyväskylä, Finland, 2012. [Google Scholar]
- Myllykangas J-P, Rissanen AJ, Hietanen S et al. Influence of electron acceptor availability and microbial community structure on sedimentary methane oxidation in a boreal estuary. Biogeochemistry. 2020;148:291–309. [Google Scholar]
- Nykänen H, Peura S, Kankaala P et al. Recycling and fluxes of carbon gases in a stratified boreal lake following experimental carbon addition. Biogeosciences Discuss. 2014;2014:16447–95. [Google Scholar]
- Oswald K, Graf JS, Littmann S et al. Crenothrix are major methane consumers in stratified lakes. ISME J. 2017;11:2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald K, Milucka J, Brand A et al. Aerobic gammaproteobacterial methanotrophs mitigate methane emissions from oxic and anoxic lake waters. Limnol Oceanogr. 2016;61:S101–18. [Google Scholar]
- Oswald K, Milucka J, Brand A et al. Light-dependent aerobic methane oxidation reduces methane emissions from seasonally stratified lakes. PLoS One. 2015;10:e0132574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada AE, Needham DM, Fuhrman JA.. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14. [DOI] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Waite DW et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. [DOI] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT et al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis PCJ, Thottathil SD, Ruiz-González C et al. Niche separation within aerobic methanotrophic bacteria across lakes and its link to methane oxidation rates. Environ Microbiol. 2020;22:738–51. [DOI] [PubMed] [Google Scholar]
- Rissanen AJ, Saarenheimo J, Tiirola M et al. Gammaproteobacterial methanotrophs dominate methanotrophy in aerobic and anaerobic layers of boreal lake waters. Aquat Microb Ecol. 2018;81:257–76. [Google Scholar]
- Rodriguez-R L KK. Bypassing cultivation to identify bacterial species. Microbe Mag. 2014;3:111–8. [Google Scholar]
- Rognes T, Flouri T, Nichols B et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarela T, Rissanen AJ, Ojala A et al. CH4 oxidation in a boreal lake during the development of hypolimnetic hypoxia. Aquat Sci. 2020;82:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarnisto M, Huttunen P, Tolonen K.. Annual lamination of sediments in Lake Lovojärvi, southern Finland, during the past 600 years. Ann Bot Fenn. 1977;14:35–45. [Google Scholar]
- Saunois M, Stavert AR, Poulter B et al. The global methane budget 2000–2017. Earth Syst Sci Data. 2020;12:1561–623. [Google Scholar]
- Scheller S, Yu H, Chadwick GL et al. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science. 2016;351:703–7. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R.. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. [DOI] [PubMed] [Google Scholar]
- Segata N, Börnigen D, Morgan XC et al. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun. 2013;4:2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola H. Micro-stratigraphy of sediment laminations deposited in a chemically stratifying eutrophic lake during the years 1913–1976. Holarct Ecol. 1979;2:160–8. [Google Scholar]
- Smith GJ, Angle JC, Solden LM et al. Members of the genus Methylobacter are inferred to account for the majority of aerobic methane oxidation in oxic soils from a freshwater wetland. MBio. 2018;9:e00815–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Wrighton KC.. Metagenomic approaches unearth methanotroph phylogenetic and metabolic diversity. Curr Issues Mol Biol. 2019;33:57–84. [DOI] [PubMed] [Google Scholar]
- Takai K, Horikoshi K.. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol. 2000;66:5066–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D et al. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen K, Tolonen M, Honkasalo L et al. Esihistoriallisen ja historiallisen maankäytön vaikutuksesta Lammin Lampellonjärven kehitykseen. Luonnon Tutkija. 1976;80:1–15.(in Finnish with English abstract). [Google Scholar]
- van Grinsven S, Sinninghe Damsté JS, Abdala Asbun A et al. Methane oxidation in anoxic lake water stimulated by nitrate and sulfate addition. Environ Microbiol. 2020;22:766–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM et al. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiticar MJ. Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem Geol. 1999;161:291–314. [Google Scholar]
- Wik M, Varner RK, Anthony KW et al. Climate-sensitive northern lakes and ponds are critical components of methane release. Nat Geosci. 2016;9:99–105. [Google Scholar]
- Yu L, He D, Zhang E et al. Electricity from anaerobic methane oxidation by a single methanogenic archaeon Methanosarcina barkeri. Chem Eng J. 2021;405:126691. [Google Scholar]
- Yu Z, Pesesky M, Zhang L et al. A complex interplay between nitric oxide, quorum sensing, and the unique secondary metabolite tundrenone constitutes the hypoxia response in Methylobacter. mSystems. 2020;5:e00770–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wang H, Liu Y et al. Methane-dependent mineral reduction by aerobic methanotrophs under hypoxia. Environ Sci Technol Lett. 2020;7:606–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.