Abstract
Background
Recent research suggests that the microbiota affects susceptibility to both respiratory tract infections (RTIs) and gastrointestinal infections (GIIs). In order to optimize global treatment options, it is important to characterize microbiota profiles across different niches and geographic/socioeconomic areas where RTI and GII prevalences are high.
Methods
We performed 16S sequencing of nasopharyngeal swabs from 209 Venezuelan Amerindian children aged 6 weeks–59 months who were participating in a 13-valent pneumococcal conjugate vaccine (PCV13) study. Using random forest models, differential abundance testing, and regression analysis, we determined whether specific bacteria were associated with RTIs or GIIs and variation in PCV13 response.
Results
Microbiota compositions differed between children with or without RTIs (P = .018) or GIIs (P = .001). Several species were associated with the absence of infections. Some of these health-associated bacteria are also observed in developed regions, such as Corynebacterium (log2(fold change [FC]) = 3.30 for RTIs and log2(FC) = 1.71 for GIIs), while others are not commonly observed in developed regions, such as Acinetobacter (log2(FC) = 2.82 and log2(FC) = 5.06, respectively). Klebsiella spp. presence was associated with both RTIs (log2(FC) = 5.48) and GIIs (log2(FC) = 7.20).
Conclusions
The nasopharyngeal microbiota of rural Venezuelan children included several bacteria that thrive in tropical humid climates. Interestingly, nasopharyngeal microbiota composition not only differed in children with an RTI but also in those with a GII, which suggests a reciprocal interplay between the 2 environments. Knowledge of region-specific microbiota patterns enables tailoring of preventive and therapeutic approaches.
Keywords: respiratory microbiota, children, rural, infections
The nasopharyngeal microbiota of rural Venezuelan children included several bacteria that thrive in tropical humid climates. Nasopharyngeal microbiota composition not only differed in children with a respiratory infection but also in those with a gastrointestinal infection.
Despite progress in the reduction of global childhood mortality over the past 2 decades, respiratory tract infections (RTIs) and gastrointestinal infections (GIIs) remain the leading causes of death in children aged <5 years worldwide, particularly in regions marked by socioeconomic inequality, poor sanitation, inadequate nutrition, and low public health services coverage [1]. When it comes to treatment, medical practitioners apply a “one microbe one disease” model. However, recent studies suggest that the microbiota (the collective abundance of bacterial species that inhabit the body) influences susceptibility to and outcome of both GIIs and RTIs [2–4]. In this context, the composition of the gut microbiota has received considerable attention due to its significant role in mucosal barrier function and healthy immune maturation in childhood [4]. Likewise, however, recent studies have revealed the important role of the respiratory microbiota in maintaining respiratory health [5]. While many observational studies and trials have assessed the influence of the intestinal microbiota on respiratory conditions [6–8], the reverse is much less studied.
Venezuela is a South American country with a rich cultural display and a large variation in the level of urbanization between regions. Warao Amerindians inhabit the Orinoco Delta and are the second largest indigenous group in Venezuela. Around one-third of Warao children die before the age of 12 years; the majority of deaths (97%) occur before age 5 years. RTIs and GIIs are the 2 most common causes of mortality in these children (18% and 63%, respectively), as previously described [9–11]. Although the respiratory microbiota has been increasingly recognized as a mediator of disease susceptibility over the past decade, most microbiota studies have been carried out in Westernized populations. A comprehensive understanding of the link between RTIs and GIIs and the composition of the microbiota in vulnerable rural populations is necessary to develop region-specific preventive and therapeutic strategies.
Further, an association between the gut microbiota and a range of vaccine responses has been described [12]. Immunization currently prevents between 2 million and 3 million deaths due to infectious diseases every year [13]. However, most vaccines do not convey 100% protection, and several factors can influence the strength of the immune responses mounted upon vaccination in children, such as age, season, and nutritional status [14]. The 13-valent pneumococcal conjugate vaccine (PCV13) was integrated in the Venezuelan national immunization program in 2014 due to high carriage rates of Streptococcus pneumoniae [10]. However, studies showed that protective antibody levels after PCV13 introduction in vulnerable indigenous children differed from those observed in studies in healthy nonnative children [15, 16]. Understanding the potential links between the respiratory microbiota and PCV13 response will help to address this discrepancy.
In this study, we used 16S sequencing of nasopharyngeal samples of indigenous Venezuelan children in order to characterize the nasopharyngeal microbiota of children living in rural conditions, assess the association between the nasopharyngeal microbiota and the presence of RTIs and GIIs, and study the relationship between the prevaccination nasopharyngeal microbiota and postvaccination PCV13 antibody titers. Because early-life microbial dysbiosis has been linked to subsequent susceptibility to infectious diseases [17], we studied only children aged <5 years.
METHODS
Study Population
We conducted a cross-sectional study to examine microbiota community profiles and the association between bacterial taxa and RTIs/GIIs combined with a prospective cohort study to examine the relationship between nasopharyngeal microbiota profiles at inclusion and antibody levels in serum after completion of a primary PCV13 vaccination series. Microbiota samples were collected between 23 May 2012 and 7 July 2012 from children aged <5 years who participated in a PCV13 study [16] in the Venezuelan Orinoco River Delta. Further details about the study population can be found in the Appendix. The Instituto de Biomedicina Ethical Committee (Caracas, Venezuela), the Delta Amacuro Indigenous Health Office, and community leaders approved the study.
Data Collection and Sampling
A team of physicians and medical students approached children from 9 Warao communities. The presence of an RTI at the time of sampling was defined as symptoms and signs of an upper (rhinorrhea, pharyngitis, sinusitis) and/or lower (bronchitis, bronchiolitis, pneumonia) respiratory tract infection according to previously described methods [9]. The presence of a GII at the time of sampling was defined as current diarrhea with or without other symptoms involving the gastrointestinal tract (such as vomiting or abdominal distension). Nasopharyngeal samples were obtained before administration of the first dose of the PCV13 vaccine using a flexible swab (Copan, Italia) in skim milk tryptone glucose glycerol medium [18]. Swabs were subjected to 1 freeze–thaw cycle to allow 300 µL to be sent to our laboratories for further processing [19].
Both swabs and serum samples were refrigerated at 4°C for ≤3 days before being transported to a –20°C freezer and, within 4 weeks, to a –70°C freezer. Pneumococcal serotype-specific serum antibody concentrations were determined using a fluorescent bead-based multiplex immunoassay as described previously [20].
High-throughput Sequencing of Nasopharyngeal Samples
Bacterial DNA was isolated and polymerase chain reaction amplicon libraries of the 16S ribosomal RNA gene were generated by amplifying the V4 hypervariable region as previously described [3]. The pooled libraries were sequenced on the Illumina MiSeq platform (Illumina Inc, San Diego, CA). See the Appendix for a detailed description of laboratory procedures and bioinformatic processing, including validated methods to identify and remove possible contaminants (Supplementary Tables 1 and 2) [21]. Sequence data and R scripts are available from the Dryad data repository (doi: 10.5061/dryad.h44j0zpfq).
Statistical Analyses
All analyses were performed in R version 3.5.0. To identify bacterial community clusters, we used hierarchical clustering based on Bray-Curtis dissimilarity. Clusters were named after the most abundant taxa within a cluster. We determined whether microbiota compositions differed between children with or without RTIs or GIIs by calculating differences in alpha diversity between groups and by permutational multivariate analysis of variance (PERMANOVA). We tested whether certain bacterial taxa were associated with an RTI or GII using random forest and differential abundance testing using metagenomeSeq [22]. We used the mean of serotype-specific log-transformed pneumococcal antibody levels as a readout for pneumococcal vaccine response. To study the relationship between prevaccination microbiota profiles and postvaccination antibody levels, we performed a combination of random forest and linear regression analyses. P values were corrected using the Benjamini-Hochberg method to account for multiple testing. A detailed description of statistical analyses can be found in the Appendix.
RESULTS
A total of 191 children were included in the analyses. Characteristics of the study population are displayed in Table 1. A total of 36% of children showed signs of chronic malnourishment. An RTI (with upper and/or lower respiratory tract infection symptoms) was present in 54% of children, while 15% showed symptoms of a GII, with 15 children (8%) suffering from both an RTI and a GII. For microbiota analysis, we generated 2 953 950 reads (median, 10 375 reads per sample; range, 107–74 630), which were grouped into 121 operational taxonomic units (OTUs).
Table 1.
Characteristics of the Study Population
| Characteristics | Patients |
|---|---|
| Sex, n (%) | |
| Male | 87 (46) |
| Female | 104 (54) |
| Age, median (interquartile range), months | 15 (7–21) |
| Nutritional status categories, n (%)a | |
| Well-nourished (HAZ ≥ −2 SD) | 118 (62) |
| Stunted (HAZ < −2 SD) | 69 (36) |
| Feeding, n (%) | |
| Breastfed at the moment of sampling | 150 (79) |
| Not breastfed at the moment of sampling | 41 (21) |
| Presence of infectious illnesses, n (%) | |
| Respiratory infection | 103 (54) |
| Gastrointestinal infection | 28 (15) |
| Antibiotic use at or 1 week prior to sampling, n (%) | 15 (8) |
| Community, n (%) | |
| Araguabisi | 14 (7) |
| Araguaimujo | 33 (17) |
| Arature | 21 (11) |
| Bonoina | 20 (10) |
| Guayaboroina | 8 (4) |
| Ibaruma | 30 (16) |
| Jobure de Curiapo | 33 (17) |
| Merejina | 9 (5) |
| Winikina | 23 (12) |
Abbreviations: HAZ, height-for-age; SD, standard deviation.
aFor the calculation of nutritional status, 4 children were excluded because of probable measurement errors.
Microbial Community Composition of Rural Venezuelan Amerindian Children
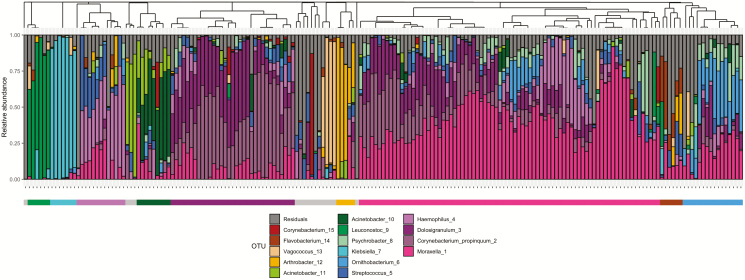
The most abundant bacteria in our study population were Moraxella (1) with a mean relative abundance of 22.5%, followed by Corynebacterium propinquum (2) (12.8%), Dolosigranulum (3) (11.3%), Haemophilus (4) (8.1%), and Streptococcus (5) (4.6%). We observed 15 clusters that represented different community profiles, with 9 clusters containing 5 children or more (Figure 1). The largest cluster (n = 80 children) was dominated by Moraxella (1) with a within-cluster mean relative abundance of 41.2%, followed by the second largest cluster (n = 33 children), which was dominated by a combination of C. propinquum (2) (39.4%) and Dolosigranulum (3) (29.1%). The remaining smaller clusters were dominated by Ornithobacterium (6) (n = 16 children, 26.6%), Haemophilus (4) (n = 13 children, 40.5%), Acinetobacter (10) (n = 9 children, 44.7%), Klebsiella (7) (n = 7 children, 91.4%), Flavobacterium (14) (n = 6 children, 22.3%), Leuconostoc (9) (n = 6 children, 78.0%), and Arthrobacter (12) (n = 5 children, 60.0%). Children in the Klebsiella- and Leuconostoc-dominated clusters were significantly more likely to have a GII than children in any other cluster (Fisher exact test: Klebsiella (7), P = .01, odds ratio [OR] = 8.7, 95% confidence interval [CI] = 1.4 to 63.3 and Leuconostoc (9), P = .042, OR = 6.3, 95% CI = .8 to 50.0), and there was a trend toward children in the Acinetobacter-dominated (10) cluster to be less likely to have an RTI (P = .083, OR = 0.2, 95% CI = .03 to 1.25).
Figure 1.
Cumulative relative abundance of the 15 most abundant OTUs in the samples. Each bar represents an individual sample. The dendrogram represents similarity between samples based on Bray-Curtis dissimilarity. The bar below the plot represents sample clusters after hierarchical clustering. Abbreviation: OTU, operational taxonomic unit.
Association of Nasopharyngeal Microbiota Profiles With the Presence of Infections
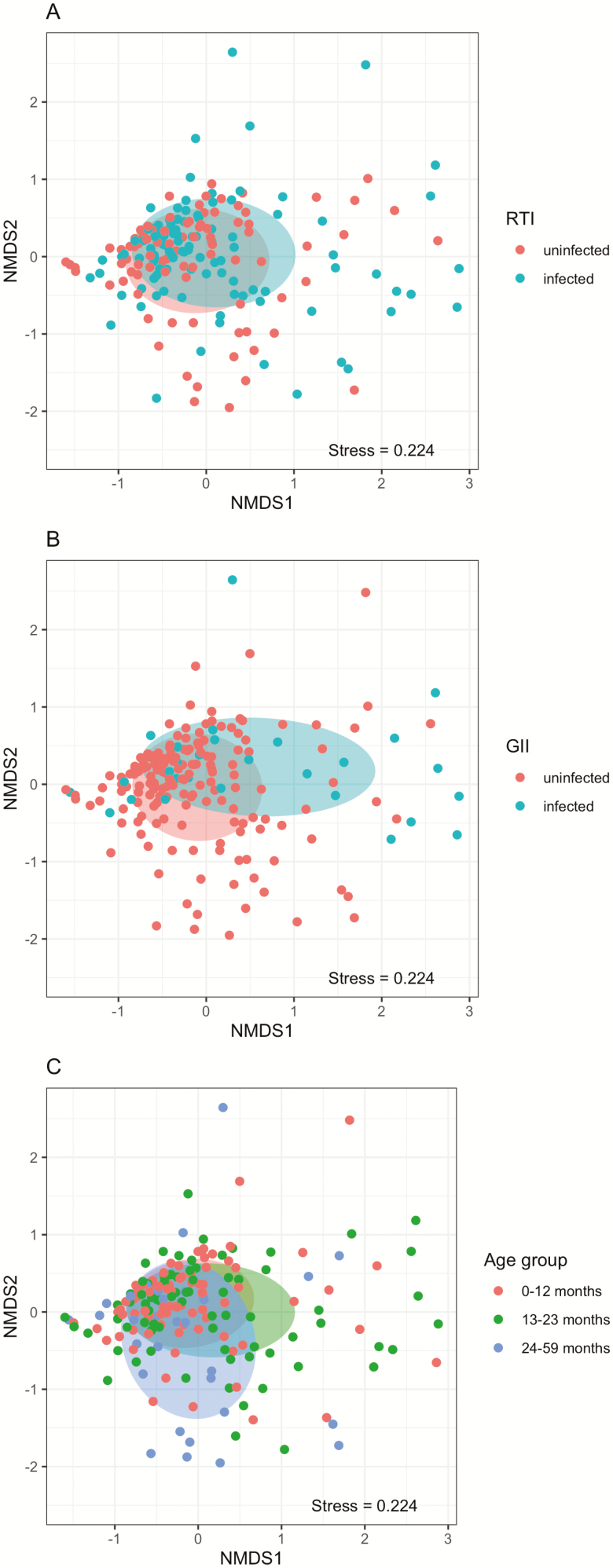
We found that microbiota profiles differed between children both with and without RTI (PERMANOVA: R2 = 1.2%, P = .018; Figure 2A) and between children with and without GII (R2 = 2.0%, P = .001; Figure 2B). Microbiota composition was further affected by age (R2 = 1.2, P = .018; Figure 2C, Supplementary Table 3). We also saw a difference in microbial compositions between children when we divided children into 4 categories based on their combined status for both RTIs and GIIs (Supplementary Figure 4).
Figure 2.
NMDS plots showing the difference in microbial community composition between (A) RTI groups, (B) GII groups, and (C) age groups. Abbreviations: GII, gastrointestinal infection; NMDS, non-metric dimensional scaling; RTI, respiratory tract infection.
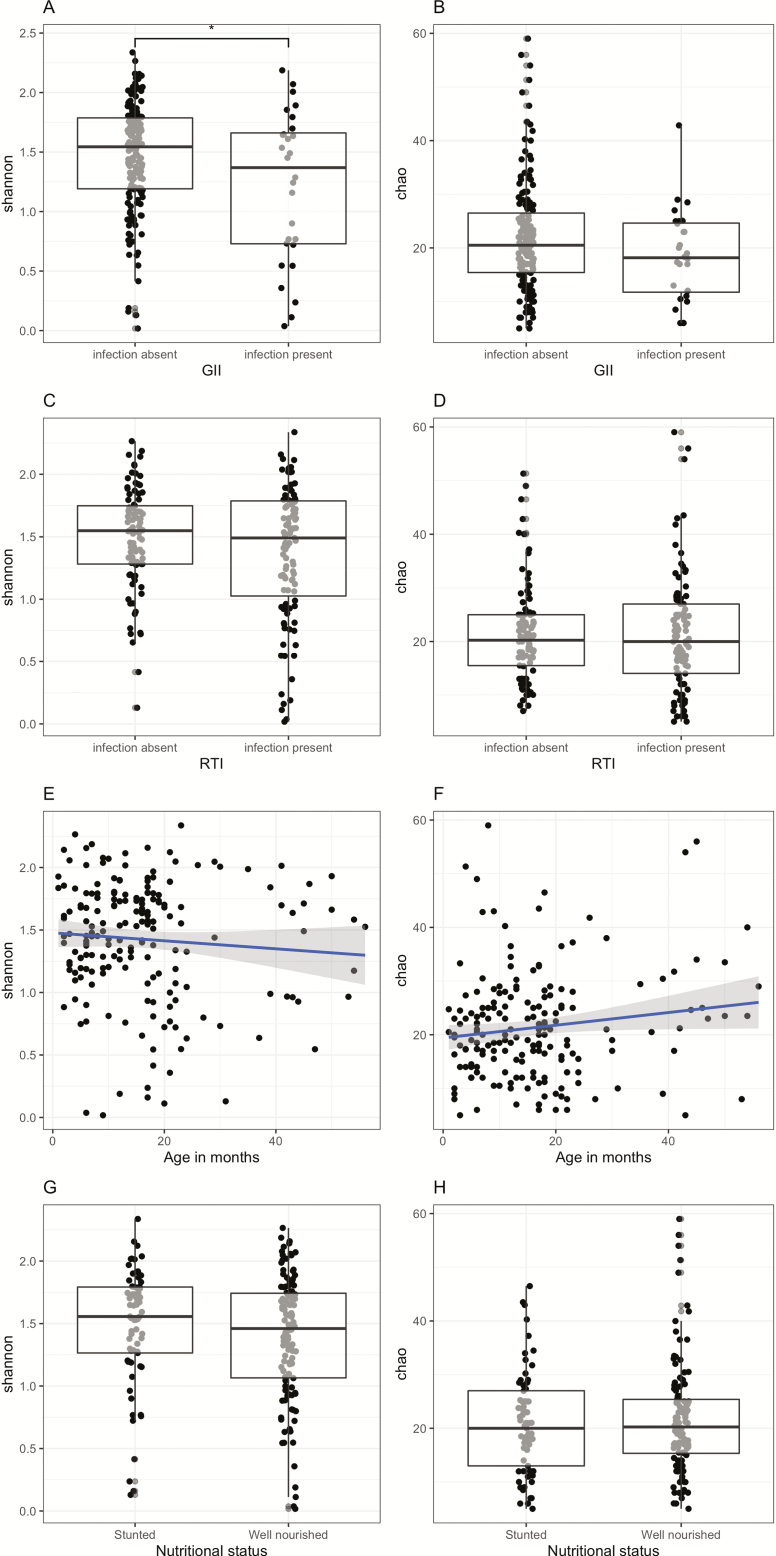
We measured alpha diversity to study the microbial evenness and richness within samples. Mean alpha diversity as measured using the Shannon diversity index (richness and evenness) and the Chao1 estimate of richness were 1.43 ± 0.48 standard deviation (SD) and 21.38 ± 10.00 SD, respectively. Children with a GII showed a lower Shannon index than children without a GII (linear model: F1,182 = 8.5, P = .004; Figure 3A, 3B), that is, their microbial profiles were less evenly distributed. Children with an RTI also showed a trend toward a lower Shannon index compared with children without an RTI (linear model: F1,182 = 3.40, P = .067; Figure 3C, 3D). In addition, age affected richness, with the Chao estimate (linear model: F1,182 = 3.59, P = .060) being lower in younger children compared with older children (Figure 3E, 3F). There were no differences in both of the measures of alpha diversity between stunted and well-nourished children (Figure 3G, 3H).
Figure 3.
Comparison of Shannon diversity (left panel) and Chao1 index (right panel) split by presence of GII (A and B), presence of RTI (C and D), across age (E and F), and nutritional status (G and H). Abbreviations: GII, gastrointestinal infection; RTI, respiratory tract infection.
Associations of Bacterial Taxa With the Presence of Infections
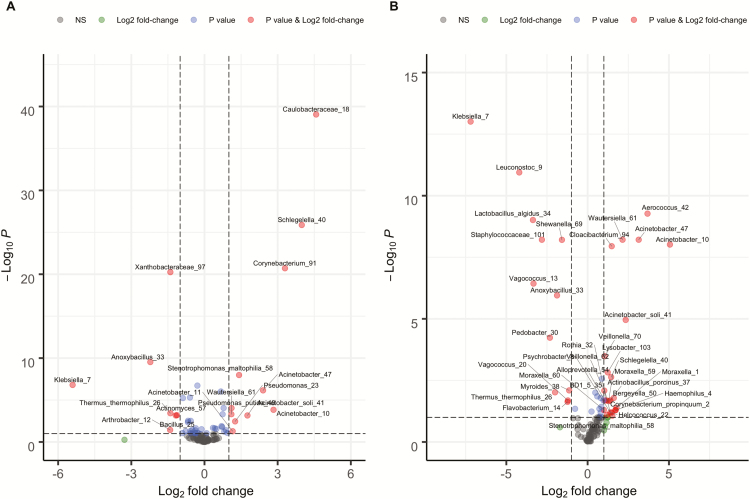
We found that 7 OTUs were more abundant in children with RTIs (Table 2, Figure 4A), with Klebsiella (7) (log2(fold change [FC]) = 5.48) and Anoxybacillus (33) (log2(FC) = 2.25) showing the highest fold changes. On the other hand, 13 OTUs were more abundant in children without RTIs, of which Caulobacteraceae (18) (log2(FC) = 4.58), Schlegelella (40) (log2(FC) = 3.99), Corynebacterium (91) (log2(FC) = 3.30), and Acinetobacter (10) (log2(FC) = 2.82) showed the highest fold changes (Table 2, Figure 4A). Random forest analysis identified an additional 2 OTUs that were associated with health (absence of RTIs), Corynebacterium propinquum (2) and Pseudomonas syringae (27) (Table 2). We identified 12 OTUs that were more abundant in children with a GII (Table 3, Figure 4B), with Klebsiella (7) again showing the highest fold change (log2(FC) = 7.20). Further, Klebsiella (7) was also the only OTU identified as discriminative between GII groups by random forest. Of the 23 OTUs that were more abundant in children without a GII, 3 Acinetobacter species were among those OTUs that showed a fold change of 2 or higher (Acinetobacter (10): log2(FC) = 5.06, Acinetobacter (47): log2(FC) = 3.14, Acinetobacter soli (41): log2(FC) = 2.34).
Table 2.
Bacteria Discriminative for Respiratory Tract Infection Groups as Identified by metagenomeSeq and Random Forest
| Operational Taxonomic Unit | Log Fold Change | P Value | Adjusted P Value | Methoda |
|---|---|---|---|---|
| Associated with disease (RTI) | ||||
| Bacillus (25) | −1.13 | .0001 | .0006 | Both |
| Klebsiella (7) | −5.48 | <.0001 | <.0001 | MGS |
| Anoxybacillus (33) | −2.25 | <.0001 | <.0001 | MGS |
| Thermus thermophilus (26) | −1.43 | .0003 | .0017 | MGS |
| Arthrobacter (12) | −1.41 | .0104 | .0359 | MGS |
| Xanthobacteraceae (97) | −1.40 | .0000 | <.0001 | MGS |
| Actinomyces (57) | −1.18 | .0001 | .0007 | MGS |
| Associated with health (absence of RTI) | ||||
| Caulobacteraceae (18) | 4.58 | <.0001 | <.0001 | Both |
| Schlegelella (40) | 3.99 | <.0001 | <.0001 | Both |
| Pseudomonas putida (49) | 1.10 | <.0001 | .0001 | Both |
| Corynebacterium (91) | 3.30 | <.0001 | <.0001 | MGS |
| Acinetobacter (10) | 2.82 | <.0001 | .0001 | MGS |
| Acinetobacter (47) | 2.38 | <.0001 | <.0001 | MGS |
| Acinetobacter soli (41) | 1.78 | .0001 | .0007 | MGS |
| Stenotrophomonas maltophilia (58) | 1.40 | <.0001 | <.0001 | MGS |
| Pseudomonas (23) | 1.25 | .0007 | .0034 | MGS |
| Acinetobacter (11) | 1.15 | <.0001 | <.0001 | MGS |
| Wautersiella (61) | 1.09 | .0001 | .0005 | MGS |
| Corynebacterium propinquum (2) | 0.50 | .2937 | .4581 | RFb |
| Pseudomonas syringae (27) | 0.12 | .5946 | .7027 | RF |
P values have been adjusted using Benjamini-Hochberg correction for multiple testing.
Abbreviation: MGS, metagenomeSeq; RF, random forest; RTI, respiratory tract infection; VSURF, variable selection using random forests.
aBoth methods adjusted for age.
bVariable selection by VSURF.
Figure 4.
Volcano plot depicting differential abundant operational taxonomic units (OTUs) for (A) respiratory tract infection groups and (B) gastrointestinal infection groups. Bacteria that are overrepresented during disease are depicted on the left (log2 fold change [FC] <0). Bacteria that are underrepresented during disease are depicted on the right (log2[FC]>0). Grey dots represent OTUs that were not differentially abundant; blue dots represent OTUs that were differentially abundant but with a log2(FC)<1; green dots represent OTUs that had a log2(FC)>1 but were not significantly differentially abundant; and red dots represent OTUs that were significantly differentially abundant and had a log2(FC)>1. Abbreviation: NS, non-significant.
Table 3.
Bacteria Discriminative for Gastrointestinal Infection Groups as Identified by metagenomeSeq and Random Forest
| Operational Taxonomic Unit | Log Fold Change | P Value | Adjusted P Value | Methoda |
|---|---|---|---|---|
| Associated with disease (GII) | ||||
| Klebsiella (7) | −7.20 | <.0001 | <.0001 | Both |
| Leuconostoc (9) | −4.20 | <.0001 | <.0001 | MGS |
| Lactobacillus algidus (34) | −3.36 | <.0001 | <.0001 | MGS |
| Vagococcus (13) | −3.33 | <.0001 | <.0001 | MGS |
| Staphylococcaceae (101) | −2.81 | <.0001 | <.0001 | MGS |
| Pedobacter (30) | −2.32 | <.0001 | .0001 | MGS |
| Thermus thermophilus (26) | −2.00 | .0017 | .0096 | MGS |
| Anoxybacillus (33) | −1.88 | <.0001 | <.0001 | MGS |
| Shewanella (69) | −1.58 | <.0001 | <.0001 | MGS |
| Flavobacterium (14) | −1.23 | .0065 | .0235 | MGS |
| Vagococcus (20) | −1.21 | .0052 | .0206 | MGS |
| Myroides (38) | −1.14 | .0013 | .0080 | MGS |
| Associated with health (absence of GII) | ||||
| Acinetobacter (10) | 5.06 | <.0001 | <.0001 | MGS |
| Aerococcus (42) | 3.68 | <.0001 | <.0001 | MGS |
| Acinetobacter (47) | 3.14 | <.0001 | <.0001 | MGS |
| Acinetobacter soli (41) | 2.34 | <.0001 | <.0001 | MGS |
| Wautersiella (61) | 2.15 | <.0001 | <.0001 | MGS |
| Moraxella (1) | 1.77 | .0120 | .0409 | MGS |
| Haemophilus (4) | 1.72 | .0172 | .0513 | MGS |
| Corynebacterium propinquum (2) | 1.71 | .0165 | .0502 | MGS |
| Bergeyella (50) | 1.61 | .0035 | .0164 | MGS |
| Psychrobacter (8) | 1.52 | .0333 | .0858 | MGS |
| Cloacibacterium (94) | 1.48 | <.0001 | .0000 | MGS |
| Helcococcus (22) | 1.48 | .0234 | .0634 | MGS |
| Schlegelella (40) | 1.45 | .0003 | .0023 | MGS |
| BD1 5 (35) | 1.39 | .0232 | .0634 | MGS |
| Actinobacillus porcinus (37) | 1.39 | .0051 | .0206 | MGS |
| Moraxella (59) | 1.29 | .0059 | .0220 | MGS |
| Lysobacter (103) | 1.23 | .0002 | .0015 | MGS |
| Stenotrophomonas maltophilia (58) | 1.19 | .0307 | .0808 | MGS |
| Rothia (32) | 1.16 | .0053 | .0206 | MGS |
| Veillonella (70) | 1.05 | <.0001 | .0003 | MGS |
| Alloprevotella (54) | 1.05 | .0053 | .0206 | MGS |
| Veillonella (62) | 1.03 | .0014 | .0082 | MGS |
| Moraxella (60) | 1.00 | .0162 | .0502 | MGS |
P values have been adjusted using Benjamini-Hochberg correction for multiple testing.
Abbreviations: GII, gastrointestinal infection; MGS, metagenomeSeq.
aBoth methods adjusted for age.
Association of Pre-PCV Microbiota Profiles With Post-PCV Antibody Levels
Postvaccination serum samples were collected at a median of 6.7 weeks (interquartile range, 6.3–6.9 weeks) and were available for 138/191 children. The remainder of the children were lost to follow-up, which is common in these nomadic communities. The random forest model that included OTUs from pre-PCV13 microbiota samples plus host characteristics explained 31.67% of the variance in PCV13 postvaccination response (Supplementary Figure 5). The top 15 covariates that were associated with post-PCV13 vaccine response in this model were used in the final mixed-effect model. Days to follow-up, increasing age, Arthrobacter (12), and Enhydrobacter (63) all had a significant negative effect on antibody levels (Table 4). Inclusion of mean log-transformed pre-vaccination antibody levels in the model did not affect these results.
Table 4.
Results from Linear Mixed Effect Model on Postvaccination Serum Antibody Levels for Each Covariate
| Model Covariate | Estimate (± Standard Error) | P Value |
|---|---|---|
| Days between vaccination and follow-up | −0.03 (0.01) | <.0001 *** |
| Age | −0.03 (0.01) | .0002*** |
| Nutritional status | ||
| Well-nourished vs stunted | −0.27 (0.15) | .08 |
| Breastfeeding | ||
| Breast-fed vs non-breastfed at the moment of sampling | −0.33 (0.21) | .13 |
| Enhydrobacter 63 | −0.24 (0.07) | .001** |
| Arthrobacter 12 | −0.19 (0.07) | .01* |
| Bacillus 25 | −0.17 (0.09) | .07 |
| Acinetobacter 11 | −0.08 (0.07) | .27 |
| Ornithobacterium 6 | −0.08 (0.07) | .30 |
| Moraxella 1 | −0.06 (0.08) | .43 |
| Pseudomonas syringae 27 | −0.06 (0.09) | .51 |
| Streptococcus 5 | −0.04 (0.08) | .65 |
| Sphingobacterium faecium 16 | −0.01 (0.08) | .90 |
| Haemophilus 4 | −0.004 (0.08) | .95 |
The estimate reflects the increase or decrease in log mean antibody concentration following 1 unit of change in the predictor variable. Bold values denote statistical significance at the P < 0.05 level.
***P < .001, **P < .01, *P < .05.
DISCUSSION
Here, we present an overview of nasopharyngeal microbiota profiles of indigenous Venezuelan children and associations with RTIs, GIIs, and post-PCV13 antibody levels.
The most abundant OTU in our cohort of Venezuelan Amerindian children was Moraxella, with 40% of children displaying a Moraxella-dominated profile. This finding is in line with those from other studies in both Western and tropical populations that included children of comparable age [17, 23–25]. Likewise, the top 5 most common genera in our study population (ie, Moraxella, Corynebacterium propinquum, Dolosigranulum, Haemophilus, and Streptococcus) are commonly observed in Western populations [2, 3, 17]. However, the top 15 most abundant OTUs in our study contained several bacteria that are not commonly observed in developed regions, including multiple Acinetobacter species. Interestingly, other studies performed in populations that reside in humid tropical regions also reported Acinetobacter presence in the upper respiratory tract of young children [25, 26], most likely because wet environments, such as moist soil, water plants, and seawater, are known environmental reservoirs for Acinetobacter [27]. This suggests a location-specific bacterial colonization pattern, which might have important implications for preventive and therapeutic strategies.
Microbial community composition differed significantly between RTI groups. For example, Corynebacterium was overrepresented in children without RTIs, similar to studies from Europe and the United States [17, 28, 29]. Interestingly, several of the bacteria that were associated with the absence of RTIs included environmental bacteria not commonly observed in developed regions such as Acinetobacter, Caulobacteraceae, and Pseudomonas. A recent study in young Fijian children residing on a tropical island also reported Pseudomonas to be abundantly present in the nasopharynx of healthy children [26]. Like Acinetobacter, Pseudomonas is present in watery environments, particularly in rivers during the period just before heavy rainfalls that lead to an influx of water into the river [30]. This is in line with the timing of sampling in our study, that is, in the beginning of the rainy season. Although the presence of these bacteria in nasopharyngeal samples could represent environmental contamination, our stringent decontamination procedures and the association of these environmental bacteria with health (absence of infections) suggest that they become part of the resident respiratory microflora in children in tropical watery areas.
Several bacterial taxa were overrepresented in children with RTIs, such as Klebsiella and Actinomyces. These bacteria are commonly found in immunocompromised patients or populations with chronic lung disease [31–33].The high prevalence of bronchiectasis caused by recurrent RTIs in indigenous children [34] might have led to alterations in microbiota profiles and facilitation of overgrowth by pathobionts. In addition, poverty, malnutrition, and infectious diseases may have weakened the children’s immune systems [35], thereby facilitating colonization by opportunistic bacteria. However, due to the cross-sectional design of our study, it was unknown whether host and environmental characteristics affected bacterial colonization or whether specific bacteria such as Klebsiella affected disease susceptibility. Longitudinal studies showed that the combination of environmental triggers and an aberrant nasopharyngeal microbial developmental trajectory may predispose children to RTIs [17, 36].
While previous studies also pointed toward the link between the respiratory tract microbiota and RTIs [17, 26, 36], disturbances related to GIIs have been much less studied. We observed significant differences in the upper respiratory tract microbiota of children with symptoms of a GII. In fact, the association between bacterial community composition and infection was stronger for GIIs compared with RTIs. While this suggests that respiratory microbiota patterns are related to gastrointestinal symptoms, most research up until this point has focused on the association between intestinal microbiota profiles and respiratory diseases. For example, in studies that included neonates, gut microbial dysbiosis in early life was associated with asthma at a later age [6, 7]. Our findings point toward a link between the respiratory microbiota and gastrointestinal disease. Gastroenteritis/diarrhea is the most common cause of mortality in our study population who live under precarious sanitary conditions, responsible for almost two-thirds of reported childhood deaths among live births [11]. If GIIs also negatively affect respiratory microbiota diversity in rural populations, this is worrisome since loss of diversity has been associated with respiratory disease severity and the overrepresentation of pathogenic bacteria [17, 37].
Vaccination against respiratory pathogens, including pneumococcal vaccination, has made an enormous contribution to global health [38]. The awareness of the immune-modulating effects of the human microbiota has begun to increase our understanding of its role in vaccine effectiveness [39]. However, again most studies focus on the gastrointestinal microbiota. One of the unique aspects of our cohort is that respiratory microbiota samples were collected before administration of the first PCV13 dose, while serum samples were collected after vaccination. An observed association between prevaccination bacterial taxa and postvaccination antibody response could thus point toward a causative effect of respiratory microbiota on PCV13 antibody response. However, we only observed a significant negative association between 2 bacterial taxa, Arthrobacter and Enhydrobacter, and antibody response, precluding firm conclusions on the role of the respiratory microbiota in modulating vaccine response. Interestingly, Arthrobacter presence dominated 1 of the 9 community profile clusters, underpinning the fact that this is a bacterial taxon to which rural Warao children are heavily exposed. In line with previous reports, we showed a significant effect of age on antibody response, with older children showing lower antibody levels, and a trend toward higher antibody responses in children compared with well nourished children [15].
In conclusion, this is the first explorative study of the nasopharyngeal microbiota of a large cohort of Venezuelan indigenous children living under extremely remote conditions. In addition to well-known colonizers of the upper respiratory tract, such as Moraxella and Streptococcus spp., less commonly observed Proteobacteria such as Klebsiella and Acinetobacter were part of the respiratory microbiota in these Amerindian children, possibly related to their living environment that may favor soil bacteria thriving in this humid area. From a public health perspective, the impact of RTIs and GIIs on morbidity and mortality in rural childhood populations is substantial. Our study contributes to a better understanding of the relationship between bacterial carriage and disease susceptibility, which has important implications for prevention and management of infectious diseases in vulnerable childhood populations.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the participating families and the field workers involved in the recruitment and sampling of children, in particular, Berenice del Nogal and the medical students of the Escuela de Medicina José Maria Vargas of the Universidad Central de Venezuela and Jochem Burghouts, Meyke Hermsen, Thor Küchler, Stèphan Kraai, and Marcella Overeem. The authors thank Irina Tcherniaeva and Kayleigh Arp for technical support and Dr P. C. J. L. Bruijning-Verhagen and Dr C. S. P. M. Uiterwaal for their critical assessment of the manuscript.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. The original study was supported by Pfizer Venezuela and the Fundacion para la Investigación en Micobacterias, Caracas, Venezuela. In addition, the study was partly supported by Integrated Microsystems for Biosensing (3E-01), FES0901:FES HTSM, a project of NanoNextNL. For the microbiota analyses, an European Society for Paediatric Infectious Diseases (ESPID) Small Grant Award was awarded to L. M. V.
Potential conflicts of interest. D. B. reports an unrestricted research grant from Nutricia, grants from MedImmune, and personal fees from Friesland Campina outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of under-5 mortality in 2000-15: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet 2016; 388:3027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Teo SM, Tang HHF, Mok D, et al. Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe 2018; 24:341–52.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Man WH, van Houten MA, Mérelle ME, et al. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir Med 2019; 7:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Libertucci J, Young VB. The role of the microbiota in infectious diseases. Nat Microbiol 2019; 4:35–45. [DOI] [PubMed] [Google Scholar]
- 5. Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 2017; 15:259–70. is J 2012; 31:255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arrieta M-C, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 2015; 7:307ra152. [DOI] [PubMed] [Google Scholar]
- 7. Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multi–sensitized atopy and T–cell differentiation. Nat Med 2016; 22:1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. King S, Glanville J, Sanders ME, Fitzgerald A, Varley D. Effectiveness of probiotics on the duration of illness in healthy children and adults who develop common acute respiratory infectious conditions: A systematic review and meta-analysis. Br J Nutr 2014; 112:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verhagen LM, Warris A, Hermans PWM, del Nogal B, de Groot R, de Waard JH. High Prevalence of Acute Respiratory Tract Infections Among Warao Amerindian Children in Venezuela in Relation to Low Immunization Coverage and Chronic Malnutrition. Pediatr Infect Dis J 2012; 31:255–62. [DOI] [PubMed] [Google Scholar]
- 10. Verhagen LM, Gómez-Castellano K, Snelders E, et al. Respiratory infections in Eñepa Amerindians are related to malnutrition and Streptococcus pneumoniae carriage. J Infect 2013; 67:273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Villalba JA, Liu Y, Alvarez MK, et al. Low child survival index in a multi-dimensionally poor Amerindian population in Venezuela. PLoS One 2013; 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zimmermann P, Curtis N. The influence of the intestinal microbiome on vaccine responses. Vaccine 2018; 36:4433–9. [DOI] [PubMed] [Google Scholar]
- 13. World Health Organization. Fact sheets. 10 facts on immunization March 2018. Available at: https://www.who.int/features/factfiles/immunization/en/. Accessed 27 December 2019.
- 14. Zimmermann P, Curtis N. Factors that influence the immune response to vaccination. Clin Microbiol Rev 2019; 32:e00084–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Verhagen LM, Hermsen M, Rivera-Olivero I, et al. Stunting correlates with high salivary and serum antibody levels after 13-valent pneumococcal conjugate vaccination of Venezuelan Amerindian children. Vaccine 2016; 34:2312–20. [DOI] [PubMed] [Google Scholar]
- 16. Verhagen LM, Rivera-Olivero IA, Hermsen M, et al. Introduction of the 13-valent pneumococcal conjugate vaccine in an isolated pneumococcal vaccine-naïve indigenous population. Eur Respir J 2016; 48:1492–6. [DOI] [PubMed] [Google Scholar]
- 17. Bosch AATM, de Steenhuijsen Piters WAA, van Houten MA, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am J Respir Crit Care Med 2017; 196:1582–90. [DOI] [PubMed] [Google Scholar]
- 18. O’Brien KL, Bronsdon MA, Dagan R, et al. Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J Clin Microbiol 2001; 39:1021–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Biesbroek G, Tsivtsivadze E, Sanders EA, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 2014; 190:1283–92. [DOI] [PubMed] [Google Scholar]
- 20. Elberse KE, Tcherniaeva I, Berbers GA, Schouls LM. Optimization and application of a multiplex bead-based assay to quantify serotype-specific IgG against Streptococcus pneumoniae polysaccharides: response to the booster vaccine after immunization with the pneumococcal 7-valent conjugate vaccine. Clin Vaccine Immunol 2010; 17:674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018; 6:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paulson JN, Stine OC, Bravo HC, Pop M. Robust methods for differential abundance analysis in marker gene surveys. Nat Methods 2013; 10:1200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cardenas PA, Cooper PJ, Cox MJ, et al. Upper airways microbiota in antibiotic-naïve wheezing and healthy infants from the tropics of rural Ecuador. PLoS One 2012; 7:e46803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kelly MS, Surette MG, Smieja M, et al. Pneumococcal colonization and the nasopharyngeal microbiota of children in Botswana. Pediatr Infect Dis J 2018; 37:1176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H, Dai W, Feng X, et al. Microbiota composition in upper respiratory tracts of healthy children in Shenzhen, China, differed with respiratory sites and ages. Biomed Res Int 2018; 2018:6515670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boelsen LK, Dunne EM, Mika M, et al. The association between pneumococcal vaccination, ethnicity, and the nasopharyngeal microbiota of children in Fiji. Microbiome 2019; 7:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 2017; 30:409–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Man WH, van Dongen TMA, Venekamp RP, et al. Respiratory microbiota predicts clinical disease course of acute otorrhea in children with tympanostomy tubes. Pediatr Infect Dis J 2019; 38:e116–25. [DOI] [PubMed] [Google Scholar]
- 29. Mccauley K, Durack J, Valladares R, et al. Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J Allergy Clin Immunol 2019; 144:1187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cannon MV, Craine J, Hester J, et al. Dynamic microbial populations along the Cuyahoga River. PLoS One 2017; 12:e0186290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Béder T, Sales RK, Oliveira EP, Costa FM, Colares PFB, Costa AN. Pulmonary actinomycosis in Brazil: a retrospective case series. Eur Respir J 2018; 52(Suppl 62). [Google Scholar]
- 32. Cribbs SK, Uppal K, Li S, et al. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome 2016; 4:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu W, Yu J, Ai Q, Liu D, Song C, Li L. Increased constituent ratios of Klebsiella sp., Acinetobacter sp., and Streptococcus sp. and a decrease in microflora diversity may be indicators of ventilator-associated pneumonia: a prospective study in the respiratory tracts of neonates. PLoS One 2014; 9:e87504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Basnayake TL, Morgan LC, Chang AB. The global burden of respiratory infections in indigenous children and adults: a review. Respirology 2017; 22:1518–28. [DOI] [PubMed] [Google Scholar]
- 35. Rytter MJ, Kolte L, Briend A, Friis H, Christensen VB. The immune system in children with malnutrition— a systematic review. PLoS One 2014; 9:e105017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Man WH, Clerc M, de Steenhuijsen Piters WAA, et al. Loss of microbial topography between oral and nasopharyngeal microbiota and development of respiratory infections early in life. Am J Respir Crit Care Med 2019; 200:760–70. [DOI] [PubMed] [Google Scholar]
- 37. Edouard S, Million M, Bachar D, et al. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur J Clin Microbiol Infect Dis 2018; 37:1725–33. [DOI] [PubMed] [Google Scholar]
- 38. Izurieta P, Bahety P, Adegbola R, Clarke C, Hoet B. Public health impact of pneumococcal conjugate vaccine infant immunization programs: assessment of invasive pneumococcal disease burden and serotype distribution. Expert Rev Vaccines 2018; 17:479–93. [DOI] [PubMed] [Google Scholar]
- 39. Harris VC. The significance of the intestinal microbiome for vaccinology: from correlations to therapeutic applications. Drugs 2018; 78:1063–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Advisory Committee on Immunization Practices. Licensure of a 13-valent pneumococcal conjugate vaccine (PCV13) and recommendations for use among children. MMWR Morb Mortal Wkly Rep 2010; 59:258–61. [PubMed] [Google Scholar]
- 41. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013; 79:5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Joshi NA, Fass JN. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software] 2011. https://github.com/najoshi/sickle.
- 43. Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: PAired-eND Assembler for Illumina sequences http://emboss.sourceforge.net/apps/release/6.2/emboss/. Accessed 15 May 2019. [DOI] [PMC free article] [PubMed]
- 44. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011; 27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yilmaz P, Parfrey LW, Yarza P, et al. The SILVA and ‘“All-species Living Tree Project (LTP)”’ taxonomic frameworks. Nucleic Acids Res 2014; 42:D643–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Subramanian S, Huq S, Yatsunenko T, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014; 510:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Satzke C, Turner P, Virolainen-Julkunen A, et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 2013; 32:165–79. [DOI] [PubMed] [Google Scholar]
- 48. Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014; 12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oksanen J, Blanchet FG, Friendly M, et al. Package ‘vegan’ — Community Ecology Package 2019. https://github.com/vegandevs/vegan.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.