Competitive interactions between capping protein, twinfilin, and actin drive actin filament uncapping and recycling.
Abstract
Uncapping of actin filaments is essential for driving polymerization and depolymerization dynamics from capping protein–associated filaments; however, the mechanisms of uncapping leading to rapid disassembly are unknown. Here, we elucidated the x-ray crystal structure of the actin/twinfilin/capping protein complex to address the mechanisms of twinfilin uncapping of actin filaments. The twinfilin/capping protein complex binds to two G-actin subunits in an orientation that resembles the actin filament barbed end. This suggests an unanticipated mechanism by which twinfilin disrupts the stable capping of actin filaments by inducing a G-actin conformation in the two terminal actin subunits. Furthermore, twinfilin disorders critical actin-capping protein interactions, which will assist in the dissociation of capping protein, and may promote filament uncapping through a second mechanism involving V-1 competition for an actin-binding surface on capping protein. The extensive interactions with capping protein indicate that the evolutionary conserved role of twinfilin is to uncap actin filaments.
INTRODUCTION
Numerous cellular processes, such as morphogenesis, migration, cytokinesis, endocytosis, and memory, rely on rapid reorganization of actin cytoskeletal networks (1–3). Coordinated local assembly and disassembly of actin filaments generate force and structure, which is harnessed to drive these specific functions. A number of actin-regulating proteins control the assembly, disassembly, and organization of the filament networks (4, 5). Among the key evolutionarily conserved actin regulators are capping protein (CP) and the actin depolymerization factor homology (ADF-H) domain family of proteins, which include ADF/cofilins and twinfilin (6–10). While primitive, functional ADF-H domain proteins are found in Asgard archaea (7, 9), CP and twinfilin have only been found, and are ubiquitous, in eukaryotes (9, 10). Thus, CP and twinfilin likely arose during eukaryogenesis and were present in the last eukaryotic common ancestor (LECA). The architecture of twinfilin is unique, composed of two ADF-H domains connected by a short linker and followed by a conserved C-terminal tail (11). ADF-H proteins generally regulate cytoskeletal reorganization by accelerating the disassembly of actin filaments (6, 12, 13). Twinfilin-1 is ubiquitously expressed in almost all tissue types in mammals, where it regulates actin dynamics through mechanisms involving interactions with actin monomers, actin filaments, and CP (11, 14). The biological outputs from twinfilin regulation of actin dynamics include cell motility and synaptic endocytosis (15).
The reported in vitro roles of twinfilin in actin dynamics are numerous, diverse, and somewhat contradictory. Twinfilin binds and sequesters adenosine diphosphate (ADP)–actin monomers with high affinity, inhibiting nucleotide exchange and preventing assembly into filaments (11, 14, 16). Twinfilin also interacts directly with actin filament barbed ends, blocking filament elongation, suggestive of a capping activity (17–19). Recent studies have also demonstrated that twinfilin accelerates depolymerization of actin filament barbed ends containing ADP-actin subunits (20, 21), and at low pH, twinfilin can sever filaments (22). In addition, twinfilin interacts strongly with heterodimeric CP through interactions that include those involving the conserved twinfilin C-terminal tail (23). CP is a heterodimer that binds to the barbed ends of actin filaments to prevent actin subunit exchange (24–26). Although twinfilin binds to CP with high affinity, its exact biological role in promoting CP capping or uncapping is debated (27, 28). X-ray structural studies of twinfilin have been limited to single ADF-H domains, which show high structural conservation, and both domains bind actin monomers (19). CP’s interactions with the actin filaments or dynactin filaments are resolved to 23- and 3.4-Å resolution, respectively, via cryo–electron microscopy (cryo-EM) (29, 30). However, the molecular mechanism by which twinfilin interacts with CP at the actin filament barbed ends is unknown. Here, we address the role of twinfilin in uncapping of actin filaments by elucidating the x-ray structure of the twinfilin/CP/actin complex.
RESULTS
The crystal structure of the twinfilin/CP/actin complex
Previous biochemical data have shown that twinfilin’s interaction with CP-capped actin filaments protects CP from displacement by CARMIL, suggesting a stable interaction between twinfilin, CP, and barbed-end actin subunits (27). We therefore used purified twinfilin-1 (human), heterodimeric CP (mouse CapZα1/β2, henceforth CP), and skeletal muscle actin (rabbit) to reconstitute the complex between twinfilin, CP, and actin monomers. The twinfilin, CP, and actin complex was highly stable in gel filtration chromatography, and this complex was used to prepare protein crystals suitable for structure determination by x-ray crystallography at 3.2-Å resolution (fig. S1).
The complex consists of two ADP-bound actin subunits, one subunit each of the heterodimeric CP (CPα1 and CPβ2) and one full-length twinfilin-1 (Fig. 1, A and B). In the structure, CP adopts its canonical mushroom-shaped architecture consisting of a cap and stalk and interacts with the barbed-end faces of two G-actin subunits via the top surface of the mushroom cap (Fig. 1). The two actin subunits are in structurally similar conformations and adopt a typical G-actin fold consisting of four subdomains. The actin subunits do not show the subunit flattening and twisting associated with the G-to-F actin transition (31). The deoxyribonuclease I binding loops in the two actin subunits are disordered (fig. S2, A and B). The relative orientation of the two actin subunits resembles that of barbed-end actin subunits from the F-actin cryo-EM structure (fig. S2, C to E) (32). The actin subunits are arranged by the short pitch helical filament relationship, similar to two actin subunits across a filament. They do not adopt the relative positioning of two longitudinally related subunits in a single strand. Twinfilin adopts an elongated architecture in which the two ADF-H domains each bind one actin subunit, with ADF-H domain 2 (D2) binding to the terminal actin subunit, relative to a filament barbed end. The linker connecting the ADF-H domains, which includes an α-helix, extends across the upper surface (cap) of the mushroom-shaped CP and also contacts both actin subunits (Fig. 1, A and B). The twinfilin C-terminal tail extends from D2 and wraps around the stalk of the CP β-subunit, which is located opposite to the actin-binding interface, below the CP cap (Fig. 1, C and D).
Fig. 1. The twinfilin/CP/actin complex.
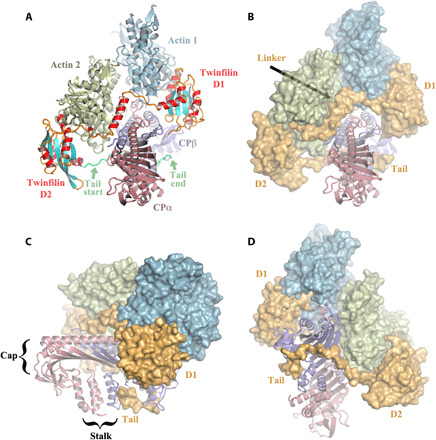
(A) Front view of the pentameric complex in cartoon representation. CP consists of two subunits, α-subunit (CPα) and β-subunit (CPβ). Twinfilin comprises two ADF-H domains (D1 and D2), a linker between D1 and D2 that includes a helix (residues 151 to 165), and a C-terminal tail (Tail; residues 316 to 342, the last eight amino acids are disordered). Actin subunit 1 is bound to twinfilin D1, and subunit 2 to twinfilin D2. The twinfilin secondary structure elements are colored differentially, helices in red, strands in cyan, and loops and extended regions in orange, with the Tail in lime green. (B) Front view [same as in (A)], in which the actin subunits and twinfilin are represented as surfaces. Twinfilin is shown entirely in orange. (C and D) Back and side views, respectively.
Binary interactions in the twinfilin/CP/actin complex
Each of the two twinfilin ADF-H domains binds to an actin subunit at analogous interfaces, providing basis for monomer sequestration (Fig. 2A). The individual ADF-H domains adopt similar architectures except for a difference in the conformation of the β-sheets. The β-3 and β-4 strands in D2 form a protrusive extension relative to that in ADF-H domain 1 (D1) (fig. S3, A to C). The key structural elements of the two ADF-H domains are highly conserved in mouse twinfilin and human twinfilin-2 (33). Despite twinfilin D2 having 10 times higher affinity for G-actin (16), its actin-binding interface is markedly similar to that of D1. Thus, the precise selection of residues in the two binding interfaces is likely to explain the differences in actin-binding affinity. Further, the α-helix in the linker between D1 and D2 loosely associates with the D2-bound actin subunit, and this likely strengthens D2 interaction with G-actin (Fig. 2A and fig. S4, A and C).
Fig. 2. Binary interactions in the twinfilin/CP/actin complex.
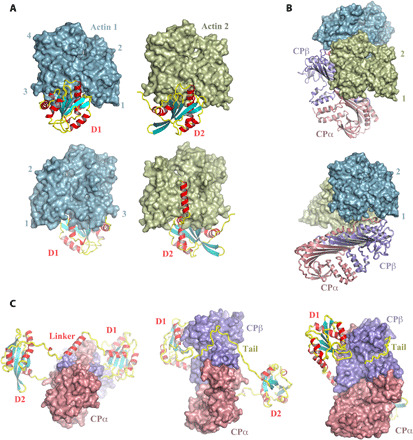
(A) Front and back views of actin bound to twinfilin D1 or twinfilin D2. (B) Two views of the CP interaction with actin, in which actin 1 and actin 2 are shown in similar orientations. (C) Three orientations of the CP/twinfilin interaction. Examples of the electron density of key features are shown in fig. S5 (H to K).
The CP actin-binding interface is located at the top surface of the mushroom cap (Fig. 2B). In the absence of twinfilin, CP interacts with the barbed-end protomers of a filament via C-terminal extensions to the α- and β-subunits, the α- and β-tentacles (24, 26, 29). In the twinfilin-bound structure, the CP β-tentacle is mostly disordered in the structure and, hence, has no direct contact with either of the actin subunits (fig. S4, D and E). The α-tentacle is partially ordered and forms an interaction with actin 1 (fig. S4F). Twinfilin binds CP through its C-terminal tail. This tail wraps around the stalk of CP β-subunit, a common binding site for filament uncapping proteins with CP interaction (CPI) motifs (Fig. 2C) (34). The ordered portion of the tail of twinfilin (residues 315 to 342) includes a basic stretch that interacts with the negatively charged groove between the CP β-subunit stalk and the underside of the mushroom-shaped cap (fig. S5, A to C). Furthermore, the complex reveals additional interactions between twinfilin and CP, beyond the C-terminal tail. First, the α-helix in the linker between D1 and D2 forms an interaction with CP that also involves the start of the β-tentacle (Fig. 2C and fig. S5, D to F). Second, twinfilin D1 forms a direct interface with the CP β-subunit (Fig. 2C and fig. S5G).
Comparison of the conformations of twinfilin tail–bound and CARMIL CPI–bound CP
The twinfilin tail-binding site on CP is distant from the actin-binding site, which is centered on the α-tentacle (Fig. 3, A and B). Comparison of the tail-binding site with the uncapping CPI motif from CARMIL shows an overlapping interaction on the underside of the CP β-subunit (34); however, the N termini of the two peptides take divergent paths around the CP stalk (Fig. 3A). The CARMIL CPI motif half encircles the CP stalk, with its N terminus making contact with the CP α-subunit stalk. By contrast, the N terminus of the twinfilin tail (residues 316 to 322) follows a straight path and does not form contacts with the CP α-subunit. This region (residues 316 to 322) is elongated yet ordered with clear electron density (Fig. 2C and fig. S5K), despite not being stabilized by interactions, suggesting that it may be under tension to extend the polypeptide chain into an ordered conformation. The overlapping interface provides a structural basis for the competition between the twinfilin tail and CARMIL CPI for CP binding, and twinfilin’s attenuation of CARMIL-mediated dissociation of CP from filament barbed ends (27).
Fig. 3. Conformations of CP.
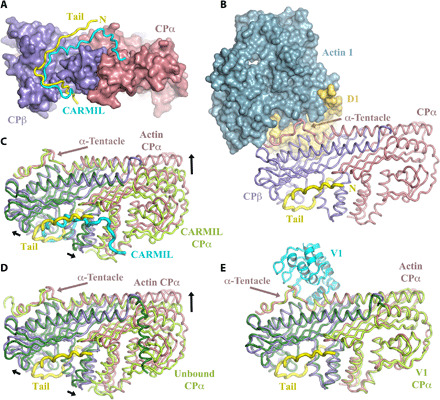
(A) Comparison of the twinfilin tail (yellow) binding site with CARMIL (cyan) on CP. Both CP-binding peptides run in the same direction, and N terminus of the twinfilin tail is labeled N. (B) The actin-binding site on the CP α-tentacle is distant from the twinfilin tail-binding site. (C and D) Structural superimposition of the β-subunits of CP reveals that the conformation of CP in the twinfilin/CP/actin complex is different to the CARMIL-bound and unbound conformations of CP. (E) Superimposition reveals that the conformation of CP in the twinfilin/CP/actin complex is similar to the V-1–bound conformation of CP. α- and β-subunits of CP are colored light green and dark green, respectively, for the CARMIL, unbound, and V-1 complexes. Black arrows indicate conformational changes in CP in adopting the twinfilin/CP/actin complex structure.
CP can adopt two known conformations, which are likely to be in dynamic equilibrium in solution. The uncapping CPI motifs of CARMIL, CD2AP, and CKIP stabilize one conformation, actin-free conformation of CP (34). Superimposition of the complex with CARMIL-bound CP and unbound CP shows key structural changes that CP subunits undergo in adopting the twinfilin/CP/actin complex conformation (Fig. 3, C and D, and fig. S6). First, the CP β-subunit mushroom cap in this complex moves upward relative to the stalk, while the α-tentacle repositions to adopt the actin-bound conformation. Second, the α-subunit adjusts upward flattening the mushroom cap and adopts a similar conformation to that stabilized by V-1 or Arp1 (from the dynactin complex) (Fig. 3E and fig. S6) (30, 35). The binding site of V-1, a steric CP inhibitor, overlaps with that of actin in this complex (fig. S6J) (35). Therefore, the CP structure in the twinfilin/CP/actin complex represents the mushroom cap–bound conformational state, to either actin, Arp1, or V-1, while the CPI-bound structure of CP represents a stabilized mushroom cap–unbound conformation. Binding of the CARMIL CPI motif around the stem of CP locks CP in the mushroom cap–unbound conformation, which is less compatible with actin barbed-end interaction, leading to uncapping of the filament (34, 36). The inability of the twinfilin tail to induce a change in the CP conformation from mushroom cap–bound to unbound state suggests that, in the complex, actin binding dominates the CP conformation or the twinfilin tail does not stabilize the mushroom cap–unbound conformation. To distinguish between these possibilities, we tested for uncapping activity within the twinfilin tail in isolation. We performed pyrene-actin polymerization assays in which an increase in pyrene fluorescence reports on the efficiency of actin assembly from CP precapped actin filament seeds. The CARMIL CPI peptide, a positive control, displayed potent uncapping activity leading to polymerization from the CP precapped filament seeds, indicated by the increase in fluorescence (fig. S7). By contrast, the maltose-binding protein (MBP)–tagged twinfilin tail (residues Gln321-Asp350) had no detectable uncapping effect, showing a similar polymerization profile to the CP precapped filament seeds alone (fig. S7). Thus, in this assay, the twinfilin tail alone does not uncap CP-capped filaments, suggesting that it does not strongly stabilize the CP mushroom cap–unbound conformation.
Twinfilin influences CP interaction with actin barbed ends
The structure reveals additional interactions between twinfilin and CP that might influence the CP-binding mode to the actin filament barbed ends (fig. S5, D to G). The dynactin filament (consisting of Arp1 instead of actin) is structurally similar to the actin filament and is capped at its barbed end by CP (30). Comparison of the CP-binding modes to actin in the twinfilin/CP/actin complex and to Arp1 subunits reveals similar overall geometries (Fig. 4 and fig. S8). However, there are considerable differences in the binding of the CP α- and β-tentacles to their respective actin/Arp1 subunits. These differences arise from the presence of twinfilin, because the binding modes of the α- and β-tentacles are similar in the dynactin complex and in the 23-Å cryo-EM structure of the CP/actin filament (29, 30). The β-tentacle, which is bound to the terminal Arp1 subunit in the dynactin complex, is disordered in the twinfilin/CP/actin complex (Fig. 4A and fig. S8A). Structural superimposition indicates that the twinfilin linker obscures the β-tentacle binding site on actin in the actin/twinfilin/CP complex (Fig. 4A). In the dynactin complex, the α-tentacle is fully ordered and bound to Arp1. By contrast, the α-tentacle in the twinfilin/CP/actin complex only forms a partial interaction with actin (Fig. 4B and fig. S8, B and C). Superimposition reveals that the α-tentacle–binding site on actin is partially obstructed by twinfilin D1 (Fig. 4B). The binding of the tentacles to the actin protomers will also be influenced by the actin protomer conformation. A twinfilin-induced shift from an F-actin to G-actin conformation may aid dissociation of the tentacles.
Fig. 4. Comparison of the CP-binding mode to Arp1 in the dynactin complex (Arp1/CP) with the binding mode to actin in actin/twinfilin/CP complex.
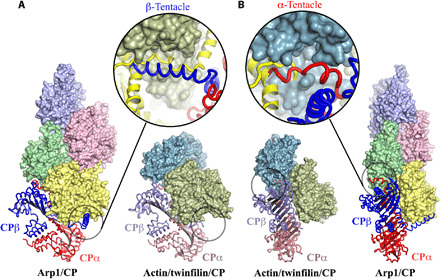
(A and B) Focus on the β- and α-tentacles, respectively, which are highlighted by black circles. The β-tentacle is disordered and unbound in the actin/twinfilin/CP complex, and the α-tentacle is partially bound and ordered, relative to the dynactin complex. Enlargements of the tentacle regions show the superimpositions of the CP (α- and β-subunits colored red and blue, respectively) from the dynactin complex on to actin (green and teal) and twinfilin (yellow) from the actin/twinfilin/CP complex. (A) Enlargement, the twinfilin linker (yellow) binds to the β-tentacle–binding site on actin 2 (green). (B) Enlargement, the N terminus of twinfilin D1 (yellow) occupies half of the α-tentacle–binding site on actin 1 (teal).
In the absence of twinfilin, the CP α-tentacle is a critical interaction with barbed end of a filament, while the β-tentacle offers a second important actin-binding interface to stabilize capping (36). The obstruction of the CP tentacles by twinfilin in the actin/twinfilin/CP complex will destabilize the CP interaction, leading to weakened CP affinity for actin filament barbed ends. Thus, the CP-binding mode in the dynactin complex represents fully bound CP, conferring strong and stable capping activity, while the CP-binding mode in the actin/twinfilin/CP complex represents a weak and unstable capping activity. This unstable interaction state may be targeted by other regulatory factors, such as the CP-sequestering protein V-1. We used the pyrene-actin polymerization assay to test whether V-1 can enhance CP uncapping in the presence of twinfilin (28). Addition of twinfilin into CP precapped actin filament seeds mixed with pyrene-labeled monomers and profilin did not accelerate polymerization, indicating either a lack of uncapping or uncapping followed by recapping (Fig. 5). Addition of V-1 alone displayed partial polymerization attributable to CP sequestration (Fig. 5). However, the presence of both twinfilin and V-1 accelerated polymerization to near the level of uncapped actin filament seeds, indicating that the cooperative activities of twinfilin and V-1 can induce dissociation of CP from barbed ends and prevent recapping by CP (Fig. 5). This indicates that twinfilin is able to remove CP from filaments; however, the CP-sequestering protein V-1 is required to prevent recapping of filaments. We propose that the high concentration of profilin (2.8 μM), relative to twinfilin (1 μM), used in this assay was sufficient to competitively remove actin from the CP/twinfilin complex and allow recapping in the absence of V-1. The requirement of both the uncapping agent twinfilin and CP-sequestering protein V-1 to observe robust uncapping in this in vitro assay partially explains some of the disparities in the reported activities of twinfilin. Uncapping has been difficult to observe in many assays in which recapping has not been excluded.
Fig. 5. V-1 uncaps filament barbed ends in the presence of twinfilin.
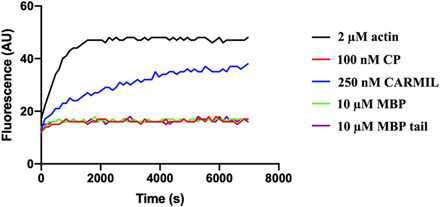
Pyrene-actin polymerization assay showing uncapping of F-actin seeds by V-1 in the presence of twinfilin. Unlabeled F-actin seeds (0.5 μM) were capped with 100 nM CP and subsequently polymerized in either 2 μM actin (10% pyrene)/2.8 μM profilin (red) or 2 μM actin (10% pyrene)/2.8 μM profilin supplemented with either 1 μM twinfilin (yellow), 5 μM V-1 (purple), or a mixture of 1 μM twinfilin and 5 μM V-1 (blue). As a control, F-actin seeds were polymerized in the absence of CP (black). As shown in the profile, the presence of both twinfilin and V-1 induces accelerated polymerization of precapped F-actin seeds (blue) to almost the same level as the positive control (black). By contrast, twinfilin alone (yellow) induces very low level of polymerization, similar to CP alone (red), while V-1 alone (purple) only induces minimal polymerization. AU, arbitrary units.
DISCUSSION
This analysis of the actin/twinfilin/CP complex reveals the structural basis by which twinfilin interacts with actin and with CP, and provides insight into molecular mechanisms for the regulation of CP in actin filament barbed-end dynamics. The unanticipated geometry in which twinfilin binds to two actin subunits in a pseudo-filament barbed-end orientation has major implications for its possible mechanisms of action. Previously proposed mechanisms for twinfilin’s activities assumed that twinfilin contacts two longitudinally related actin subunits at the barbed end of an actin filament (17, 18, 21, 27, 28). However, the structure presented here unambiguously identifies that the two actin subunits are laterally related. Twinfilin binds across the two strands of an actin filament, rather than down the side of a single strand in the filament. We discuss the ramifications of this binding orientation for actin filament regulation.
The primary biological role of twinfilin has not been settled. Mammalian twinfilin-1 was shown to preferentially localize to regions of the cell enriched for F-actin (14). Loss of twinfilin-1 in B16 cells leads to their inability to generate lamellipodia (27). In vitro and cell-based experiments have led to multiple proposed activities. Twinfilin’s actin-related functions have been variously described as follows: an actin monomer-sequestering protein that forms a 1:1 complex with actin that prevents actin polymerization and inhibits nucleotide exchange in actin (11, 14); a CP-interacting and phosphatidylinositol 4,5-bisphosphate (PIP2)–interacting protein (37); an actin monomer-shuttling protein between the pointed and barbed ends of filaments (38); a protein that does not affect CP-capping activity, nor does CP affect twinfilin’s monomer binding function (rather, twinfilin localizes actin monomers to sites of assembly through interaction with CP) (23); a barbed-end actin filament CP (17); an actin filament-severing protein (22); a protein that speeds up actin depolymerization at both ends of the actin filament in conjunction with cyclase-associated protein (CAP) (20); a protein that enhances CP actin filament capping, which competes with CARMIL for CP binding (27); and an actin filament CP-uncapping protein (28). The actin/twinfilin/CP complex provides the structural basis to reassess these activities.
In the complex, twinfilin forms extensive interactions with CP and with two actin protomers, which adopt a pseudo-actin filament barbed-end orientation relative to each other. These multiple interactions indicate that twinfilin’s primary and evolutionary conserved role, throughout eukaryotes, is to regulate CP capping at the barbed end of actin filaments. In yeast, the CP:twinfilin ratio is estimated to be 2.5:1 (37, 39), which indicates that the actin-related cellular activities of twinfilin will be dominated by the CP:twinfilin complex, rather than twinfilin acting alone on filaments. Similarly, the molar abundance of mammalian CP:twinfilin-1 ratio was determined as 2:1 (27). Twinfilin has been shown to strongly interact with actin filament barbed ends and with CP, dissociation constants (Kd) of 13 and 50 nM, respectively (17, 27). However, these affinities are at least one order of magnitude weaker than the CP affinity for actin filament barbed ends [Kd = 0.1 to 1.0 nM (40)]. The actin/twinfilin/CP complex structure demonstrates that twinfilin disrupts CP-actin interfaces, sterically competing with the CP α- and β-tentacles in binding to actin, indicating that twinfilin’s principal role is to destabilize CP capping (Fig. 6A). We discuss the possible mechanisms that lead to uncapping.
Fig. 6. Models of the filament uncapping.
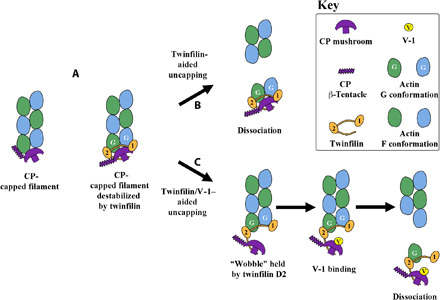
(A) Cartoon comparison of CP and CP/twinfilin association at the barbed end of a filament. (B) Twinfilin-aided uncapping is a result of twinfilin ADF-H domains inducing G-actin–like conformations in the terminal two actin protomers, weakening actin:actin interactions in the filament, leading to the dissociation of the complex. (C) Twinfilin/V-1–aided uncapping requires space for V-1 to reach its binding site on CP via a wobble state of the twinfilin-bound complex. Once V-1 is bound, CP is unable to reassociate with actin. In vitro effects of twinfilin alone on actin filaments and comparisons of uncapping models in the absence of twinfilin are shown in fig. S9.
The ADP-bound actin protomer conformations in the actin/twinfilin/CP complex are very similar to that in the cofilin-decorated actin filament (fig. S2B) (41). Cofilin, which consists of a single ADF-H domain, severs actin filaments toward the pointed end of a section of filament decorated with cofilin, by inducing a G-actin–like conformation in the actin protomers, which are not stable as a filament (13, 41). We hypothesize that the orientation of the two ADF-H twinfilin domains, in binding across the filament, allows twinfilin to induce a G-actin–like conformation in the final two barbed-end actin protomers. This conformation will destabilize these two terminal actin subunits, leading to them being “severed” from the end of the filament, by an analogous mechanism to cofilin severing (13, 41). Thus, we propose that the two twinfilin ADF domains induce severing at the boundary between ADF-H–bound and ADF-H–free portions of F-actin, dissociating CP, twinfilin, and the two terminal actin subunits as a complex (Figs. 1A and 6, A and B). Immediate reassociation of the actin/twinfilin/CP complex with the filament would be unfavorable because the actin subunits in the complex are held in the ADP-bound G-actin state, and nucleotide exchange is inhibited by the twinfilin ADF-H domains (11).
We propose that the actin/twinfilin/CP complex will then undergo a process of recycling. Twinfilin strongly binds to ADP-bound actin monomers (Kd ~ 40 to 50 nM), and this affinity is unaffected by the presence of CP (16, 23). For comparison, CAP, thymosin-β4, profilin, and cofilin are characterized in their affinities for ADP-bound actin monomers by Kd of 20 nM, 80 to 100 nM, 0.17 μM, and 0.4 μM, respectively [reviewed in (42)]. The two ADP-bound actin subunits, from the actin/twinfilin/CP complex, may then be converted to adenosine triphosphate (ATP)–bound actin by the actions of CAP, due to the high affinity of CAP for ADP-bound actin (20, 21, 33, 43). Subsequently, the ATP-bound actin monomers will be released to profilin, and possibly to thymosin-β4, due to their superior affinities for ATP-bound actin relative to twinfilin, CAP, and cofilin, 0.1, 0.1 to 4.0, 0.5, 1.9, and 6 μM (16, 23, 42), respectively. This will replenish the polymerization-competent pool of actin monomers (42). Because CAP also binds to the pointed ends of actin filaments (33), association of the actin/twinfilin/CP complex with the pointed end bound CAP may provide a mechanism to spatially separate the twinfilin/CP complex away from elongating actin filament barbed ends close to the membrane.
The fate of the twinfilin/CP complex may then be several-fold. First, the twinfilin/CP complex may encounter a free barbed end of a filament and undergo a further round of transient binding followed by dissociation of the terminal actin protomers. Thus, the twinfilin/CP complex may depolymerize the filament in two subunit cycles, maintaining transient capping while depolymerizing the filament. Second, twinfilin may competitively dissociate from CP in favor of CP-capped filaments due to the superior affinity for filament ends relative to CP alone, Kd of 13 and 50 nM, respectively (17, 27). This mechanism assumes that twinfilin affinity for the CP-capped filament is also high; however, the value is unknown and possibly difficult to measure due to the uncapping mechanism. Last, the twinfilin/CP complex may be dissociated at the membrane by PIP2 or by competition with CPI proteins, such as CARMIL (27, 37).
The structure of the actin/twinfilin/CP complex suggests that a second possible mechanism of filament uncapping may operate in the presence of V-1. In this mechanism, the twinfilin/CP complex “wobbles” (44) at the barbed end of a filament, remaining attached through the high-affinity actin-binding site on twinfilin D2, while twinfilin D1 dissociates from actin (Fig. 6, A and C) (16). The extended portion of the twinfilin tail (residues 316 to 322, between the end of D2 and the first tail residue with significant contact with CP), which we hypothesize is under tension, may aid the wobble process. There is homology between twinfilin and CARMIL in this region (317-HAHKQSFAKP-326 in twinfilin-1 and 981-KLEHFTKLRP-990 in CARMIL1) (27), which may allow the twinfilin tail to associate more intimately with CP in a manner similar to the CARMIL CPI interaction with CP (Fig. 3A) (34). Association of this region of twinfilin with CP would hold CP away from the actin subunit, to which it was previously bound via the α-tentacle, thus freeing the actin-binding site on the α-tentacle to bind to V-1, and preventing the α-tentacle from reassociating with actin. The partially bound complex would then dissociate as twinfilin/CP/V-1 bound to a single actin, with the high-affinity twinfilin D2 inducing a G-actin–like conformation in the actin subunit to destabilize its association with the filament (16). Flow-cell total internal reflection fluorescence microscopy observations of CP uncapping by twinfilin detected enhanced uncapping in the presence of V-1 (28). This suggests that V-1 may play an active role in twinfilin-aided uncapping as hypothesized in this mechanism (Fig. 6, A and C).
In summary, the actin/twinfilin/CP complex structure clarifies the possible mechanisms by which twinfilin operates as a diffusing uncapping agent. The majority of the in vitro observations outlined above can be rationalized in terms of twinfilin’s proposed role in transforming CP from a strong capping agent into a transiently capping depolymerization complex, which aids in the recycling of actin monomers and is sensitive to the presence of V-1. In the bulk cytoplasm, the outcome of twinfilin’s uncapping activities is likely to be actin depolymerization while maintaining transient capping. This role contrasts starkly to the CPI-containing proteins, which are target-bound uncapping agents that are hypothesized to engender actin polymerization for membrane remodeling (45, 46). The location of uncapping, in either actin assembly- or disassembly-rich regions of a cell, will determine whether filament polymerization or depolymerization will be the product of the uncapping. Any twinfilin-induced uncapping at a membrane may therefore result in the opposite activity, actin polymerization. Because twinfilin/CP-bound filaments, relative to CP-bound filaments, have enhanced barbed-end filament dynamics, we propose that cells are able to differentially regulate these actin filament barbed-end binding states to dictate the lifetimes of individual actin filaments. The proposed mechanism of depolymerization while maintaining transient filament capping adds to the evidence that actin filament ends are highly controlled and are rarely free in mammalian cells. During depolymerization, filament barbed ends are controlled by twinfilin/CP and filament pointed ends by CAP (33). During polymerization modes, the filament barbed ends can be regulated by formins (47), or alternatively by the VASP family proteins (48), following either ARP2/3 nucleation or filament uncapping at the membrane (34, 40, 49, 50). In each case, the filament end is protected.
MATERIALS AND METHODS
Protein expression and purification
The gene sequences encoding full-length human twinfilin-1 and profilin-1 were codon-optimized for Escherichia coli, synthesized (GenScript), and cloned into a pSY5 vector that includes an N-terminal eight-histidine tag followed by a human rhinovirus 3C protease cleavage site (51). The mouse full-length CP (α1/β2) construct was provided by P. Lappalainen (University of Helsinki, Finland). The construct was in the pRSFDuet-1 vector designed for coexpression of two interacting target proteins and contains an N-terminal six-histidine tag on the α-subunit (52).
All constructs encoding proteins of interest were transformed and recombinantly expressed in phage-resistant E. coli strain BL21 Star (DE3) (New England Biolabs). Fresh LB medium supplemented with either ampicillin (pSY5) or kanamycin (pRSFDuet-1) was inoculated with respective overnight cultures and shaken at 37°C until the cell density reached OD600 (optical density at 600 nm) ~ 0.6. Cells were induced for protein expression with 0.25 mM isopropyl-β-d-thiogalactopyranoside at 16°C overnight. Cells were harvested via centrifugation at 4000g at 4°C for 1 hour, and the pellets were resuspended in 50 ml of His-binding buffer [50 mM tris-HCl (pH 7.5), 500 mM NaCl, 20 mM imidazole, and one protease inhibitor tablet]. The cells were lysed by sonication with a Vibra-Cell ultrasonic processor and clarified by centrifugation at 19,000 rpm in SS-34 rotor using an RC 5C Plus centrifuge (Sorvall) at 4°C for 1 hour followed by filtration through a 0.45-μm Minisart syringe filter (Sartorius). The proteins were purified on an ÄKTAxpress system (GE Healthcare) by affinity chromatography using 5 ml of HisTrap FF column with or without (CP) on-column cleavage of the His-tag with human rhinovirus 3C protease. Proteins purified with the His-tag were eluted in buffer containing 50 mM tris-HCl (pH 7.5), 500 mM NaCl, and 500 mM imidazole. The eluted proteins were concentrated to 5 ml and subjected to size exclusion chromatography (SEC) in a Superdex 75 pg column (GE Healthcare) preequilibrated with buffer containing 50 mM tris-HCl (pH 7.5) and 150 mM NaCl. All purified proteins were verified by SDS–polyacrylamide gel electrophoresis (PAGE) before being snap-frozen and stored at −80°C.
MBP-tagged twinfilin tail constituting residues Gln321-Asp350 was polymerase chain reaction (PCR)–amplified from full-length human twinfilin-1 expression plasmid and inserted into pSY7 vector, which incorporates a cleavable N-terminal histidine and MBP tag. MBP control was prepared by expressing pSY7 vector alone in BL21(DE3) E. coli. Both MBP-tagged twinfilin tail and MBP were purified by affinity chromatography using 1 ml of HisTrap FF column with on-column digestion with human rhinovirus 3C protease to remove the histidine tag. The proteins were further purified by gel filtration chromatography with a Superdex 75 pg column (GE Healthcare) equilibrated with 50 mM tris-HCl (pH 9) and 150 mM NaCl.
Human V-1 (pGEX-6P-3 vector) was expressed as a glutathione S-transferase (GST) fusion E. coli BL21 Star (DE3) and affinity-purified on an ÄKTAxpress system (GE Healthcare) by loading cleared lysate onto 1 ml of GSTrap FF Column (GE Healthcare) equilibrated with 50 mM tris-HCl (pH 7.4), 150 mM NaCl, and 1 mM dithiothreitol (DTT). GST tag was removed by on-column digestion with human rhinovirus 3C protease overnight followed by further purification by gel filtration chromatography. CARMIL CP–binding region (CBR) was expressed and purified as previously described (34).
Preparation of the actin, twinfilin, and CP complex
Rabbit skeletal muscle actin was purified from skeletal muscle acetone powder (Pel-Freez) (53, 54). The protein was subjected to a final SEC using HiLoad Superdex 200 on an ÄKTA Prime system. The purity of G-actin was assessed by SDS-PAGE, and concentration was determined by measuring OD at 290 nm. The protein complex was prepared by mixing human twinfilin-1, mouse CP α1β2, and rabbit actin in buffer A [2 mM tris-HCl (pH 7.5), 0.2 mM ATP, 0.5 mM DTT, 1 mM NaN3, and 0.1 mM CaCl2] at a molar ratio of 1:1:2.5. The mixture was incubated on ice for 10 min to allow complex formation and then purified by SEC (HiLoad 16/60 Superdex 200 preequilibrated with buffer A) on an ÄKTAxpress system (GE Healthcare). Fractions corresponding to ultraviolet absorption peak were analyzed by SDS-PAGE gel to verify complex formation.
Crystallization and crystal optimization
The fractions corresponding to the peak from SEC were pooled together, concentrated with Vivaspin 20 MWCO 10,000 concentrator (Sartorius) to approximately 20 mg/ml, and subjected to commercial crystallization screens. Crystallization screens were set up as sitting drops in three-drop Intelli-Plate 96 (Hampton Research) in 2:1, 1:1, and 1:2 ratios consisting of protein solution and precipitant and stored in room temperature (25°C). Crystal hits were observed in a number of conditions in the JBScreen Classic HTS I screen after 3 days. One condition [12% polyethylene glycol (PEG) 8000, 10% glycerol, and 500 mM potassium chloride] produced the best shaped crystals composed of thin rods. A seed bead kit (Hampton Research) was used to generate seed stocks of protein crystals for further optimization. Crystals were set up using the hanging-drop vapor diffusion method in 2-μl drops of 1:1 ratio consisting of protein solution and seed stock in the crystallization condition (12% PEG 8000, 10% glycerol, and 500 mM potassium chloride).
Crystal data processing and structure determination
Harvested crystals were soaked in 25% glycerol before being carefully fished with cryoloops and flash-frozen in liquid nitrogen for subsequent data collection at the National Synchrotron Radiation Research Center, Taiwan. Indexing, scaling, and merging of datasets were carried out in HKL2000 (55). The crystal structure twinfilin/CP/actin complex was solved at a resolution of 3.2 Å by molecular replacement using Phaser (56), sequentially searching for one copy of mouse twinfilin-1 D2 (3DAW) (19) and one copy of chicken CP heterodimer (3AA7) (35) followed by a second copy of mouse twinfilin-1 D2 (3DAW) (19). The resultant model was rebuilt by hand, and the structure was subjected to several rounds of refinement using Phenix (56) and further manual rebuilding in COOT (57). The crystal data collection and structure refinement statistics are presented in table S1.
Pyrene-actin polymerization assays
Pyrene-actin polymerization assays to monitor filament uncapping were performed in 100-μl reactions containing 2 μM rabbit skeletal muscle G-actin (10% pyrene–labeled), 0.5 μM F-actin seeds, 2.8 μM profilin (to prevent pointed-end polymerization), 100 nM CP, and variable concentrations of the test proteins. In all pyrene-actin polymerization assays, the final concentrations of twinfilin, CP, CARMIL, V-1, MBP-tagged tail, and MBP were set at 1 μM, 100 nM, 250 nM, 5 μM, 10 μM, and 10 μM, respectively. The components were mixed in buffer A [2 mM tris-HCl (pH 7.5), 0.2 mM ATP, 0.5 mM DTT, 1 mM NaN3, and 0.1 mM CaCl2] to a final volume of 70 μl in a black flat-bottom 96-well plate (Corning).
To prepare actin filament seed stocks, 5 μM G-actin was polymerized for 1 hour at room temperature, after which the filaments were mechanically sheared by repeatedly passing the F-actin solution through a 0.7-mm-diameter needle for 1 min. Thereafter, 10 μl of the F-actin seed stock was mixed with 10 μl of 1 μM CP and left for 5 min to allow barbed-end capping of the F-actin seeds. As a control, the F-actin seed stock was mixed with buffer A, without CP. Then, 20 μl of this precapped F-seed stock, or control, was added to pyrene-actin polymerization mixture and actin polymerization initiated by addition of 10 μl of 10× KMEI buffer (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, and 100 mM imidazole, pH 7.4) in a total reaction volume of 100 μl. The pyrene fluorescence intensities were monitored immediately on a Safire2 fluorimeter plate reader (Tecan) with excitation and emission wavelengths set at 365 and 407 nm, respectively.
Acknowledgments
We thank B. Xue for technical advice, S. Takeda and B. Xue for valuable discussions, P. Lappalainen (University of Helsinki, Finland) and J. Cooper (Washington University School of Medicine, USA) for constructs, and the experimental facility and the technical services provided by the Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology and the National Synchrotron Radiation Research Center, a national user facility supported by the Ministry of Science and Technology, Taiwan. Funding: This work was supported by Biomedical Research Council of A*STAR under the Singapore International Graduate Award (SINGA) scholarship and NMRC OFIRG/0027/2016, and by Human Frontiers Science Program award RGP0028/2018. Author contributions: D.M.M. and R.C.R. conceived experiments, analyzed data, and wrote the paper. D.M.M. performed experiments. R.C.R. and E.M. supervised the work. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under the accession code 7CCC. All other data are available from the corresponding author upon reasonable request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/5/eabd5271/DC1
REFERENCES AND NOTES
- 1.Mooren O. L., Galletta B. J., Cooper J. A., Roles for actin assembly in endocytosis. Annu. Rev. Biochem. 81, 661–686 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Pollard T. D., Cooper J. A., Actin, a central player in cell shape and movement. Science 326, 1208–1212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudy J. W., Actin dynamics and the evolution of the memory trace. Brain Res. 1621, 17–28 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Pollard T. D., Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8, a018226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siripala A. D., Welch M. D., SnapShot: Actin regulators I. Cell 128, 626 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Poukkula M., Kremneva E., Serlachius M., Lappalainen P., Actin-depolymerizing factor homology domain: A conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton (Hoboken) 68, 471–490 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Akıl C., Tran L. T., Orhant-Prioux M., Baskaran Y., Manser E., Blanchoin L., Robinson R. C., Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea. Proc. Natl. Acad. Sci. U.S.A. 117, 19904–19913 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunning P. W., Ghoshdastider U., Whitaker S., Popp D., Robinson R. C., The evolution of compositionally and functionally distinct actin filaments. J. Cell Sci. 128, 2009–2019 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Akıl C., Kitaoku Y., Tran L. T., Liebl D., Choe H., Muengsaen D., Suginta W., Schulte A., Robinson R. C., Mythical origins of the actin cytoskeleton. Curr. Opin. Cell Biol. 68, 55–63 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Rivero F., Cvrcková F., Origins and evolution of the actin cytoskeleton. Adv. Exp. Med. Biol. 607, 97–110 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Goode B. L., Drubin D. G., Lappalainen P., Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer-sequestering protein. J. Cell Biol. 142, 723–733 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ono S., Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol. 258, 1–82 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Elam W. A., Kang H., De la Cruz E. M., Biophysics of actin filament severing by cofilin. FEBS Lett. 587, 1215–1219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vartiainen M., Ojala P. J., Auvinen P., Peranen J., Lappalainen P., Mouse A6/twinfilin is an actin monomer-binding protein that localizes to the regions of rapid actin dynamics. Mol. Cell. Biol. 20, 1772–1783 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D., Zhang L., Zhao G., Wahlstrom G., Heino T. I., Chen J., Zhang Y. Q., Drosophila twinfilin is required for cell migration and synaptic endocytosis. J. Cell Sci. 123, 1546–1556 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Ojala P. J., Paavilainen V. O., Vartiainen M. K., Tuma R., Weeds A. G., Lappalainen P., The two ADF-H domains of twinfilin play functionally distinct roles in interactions with actin monomers. Mol. Biol. Cell 13, 3811–3821 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helfer E., Nevalainen E. M., Naumanen P., Romero S., Didry D., Pantaloni D., Lappalainen P., Carlier M. F., Mammalian twinfilin sequesters ADP-G-actin and caps filament barbed ends: Implications in motility. EMBO J. 25, 1184–1195 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paavilainen V. O., Hellman M., Helfer E., Bovellan M., Annila A., Carlier M. F., Permi P., Lappalainen P., Structural basis and evolutionary origin of actin filament capping by twinfilin. Proc. Natl. Acad. Sci. U.S.A. 104, 3113–3118 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paavilainen V. O., Oksanen E., Goldman A., Lappalainen P., Structure of the actin-depolymerizing factor homology domain in complex with actin. J. Cell Biol. 182, 51–59 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston A. B., Collins A., Goode B. L., High-speed depolymerization at actin filament ends jointly catalysed by Twinfilin and Srv2/CAP. Nat. Cell Biol. 17, 1504–1511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilton D. M., Aguilar R. M., Johnston A. B., Goode B. L., Species-specific functions of twinfilin in actin filament depolymerization. J. Mol. Biol. 430, 3323–3336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moseley J. B., Okada K., Balcer H. I., Kovar D. R., Pollard T. D., Goode B. L., Twinfilin is an actin-filament-severing protein and promotes rapid turnover of actin structures in vivo. J. Cell Sci. 119, 1547–1557 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Falck S., Paavilainen V. O., Wear M. A., Grossmann J. G., Cooper J. A., Lappalainen P., Biological role and structural mechanism of twinfilin-capping protein interaction. EMBO J. 23, 3010–3019 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wear M. A., Yamashita A., Kim K., Maeda Y., Cooper J. A., How capping protein binds the barbed end of the actin filament. Curr. Biol. 13, 1531–1537 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Cooper J. A., Sept D., New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol. 267, 183–206 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita A., Maeda K., Maeda Y., Crystal structure of CapZ: Structural basis for actin filament barbed end capping. EMBO J. 22, 1529–1538 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston A. B., Hilton D. M., McConnell P., Johnson B., Harris M. T., Simone A., Amarasinghe G. K., Cooper J. A., Goode B. L., A novel mode of capping protein-regulation by twinfilin. eLife 7, e41313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.M. Hakala, H. Wioland, M. Tolonen, A. Jegou, G. Romet-Lemonne, P. Lappalainen, Twinfilin uncaps filament barbed ends to promote turnover of lamellipodial actin networks. bioRxiv 864769 [Preprint]. 4 December 2019. 10.1101/864769. [DOI] [PubMed]
- 29.Narita A., Takeda S., Yamashita A., Maeda Y., Structural basis of actin filament capping at the barbed-end: A cryo-electron microscopy study. EMBO J. 25, 5626–5633 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urnavicius L., Lau C. K., Elshenawy M. M., Morales-Rios E., Motz C., Yildiz A., Carter A. P., Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554, 202–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oda T., Iwasa M., Aihara T., Maeda Y., Narita A., The nature of the globular- to fibrous-actin transition. Nature 457, 441–445 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Chou S. Z., Pollard T. D., Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. U.S.A. 116, 4265–4274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotila T., Wioland H., Enkavi G., Kogan K., Vattulainen I., Jégou A., Romet-Lemonne G., Lappalainen P., Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin. Nat. Commun. 10, 5320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez-Valladares M., Kim T., Kannan B., Tung A., Aguda A. H., Larsson M., Cooper J. A., Robinson R. C., Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat. Struct. Mol. Biol. 17, 497–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeda S., Minakata S., Koike R., Kawahata I., Narita A., Kitazawa M., Ota M., Yamakuni T., Maeda Y., Nitanai Y., Two distinct mechanisms for actin capping protein regulation—Steric and allosteric inhibition. PLOS Biol. 8, e1000416 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim T., Ravilious G. E., Sept D., Cooper J. A., Mechanism for CARMIL protein inhibition of heterodimeric actin-capping protein. J. Biol. Chem. 287, 15251–15262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmgren S., Ojala P. J., Wear M. A., Cooper J. A., Lappalainen P., Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J. Cell Biol. 155, 251–260 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmgren S., Vartiainen M., Lappalainen P., Twinfilin, a molecular mailman for actin monomers. J. Cell Sci. 115, 881–886 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Kim K., Yamashita A., Wear M. A., Maéda Y., Cooper J. A., Capping protein binding to actin in yeast: Biochemical mechanism and physiological relevance. J. Cell Biol. 164, 567–580 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schafer D. A., Jennings P. B., Cooper J. A., Dynamics of capping protein and actin assembly in vitro: Uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135, 169–179 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka K., Takeda S., Mitsuoka K., Oda T., Kimura-Sakiyama C., Maeda Y., Narita A., Structural basis for cofilin binding and actin filament disassembly. Nat. Commun. 9, 1860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue B., Robinson R. C., Guardians of the actin monomer. Eur. J. Cell Biol. 92, 316–332 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Kotila T., Kogan K., Enkavi G., Guo S., Vattulainen I., Goode B. L., Lappalainen P., Structural basis of actin monomer re-charging by cyclase-associated protein. Nat. Commun. 9, 1892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattacharya N., Ghosh S., Sept D., Cooper J. A., Binding of myotrophin/V-1 to actin-capping protein: Implications for how capping protein binds to the filament barbed end. J. Biol. Chem. 281, 31021–31030 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A. I. Fokin, V. David, K. Oguievetskaia, E. Derivery, C. E. Stone, L. Cao, N. Rocques, N. Molinie, V. Henriot, M. Aumont-Nicaise, M.-V. Hinckelmann, F. Saudou, C. Le Clainche, A. P. Carter, G. Romet-Lemonne, A. M. Gautreau, The Arp1/11 minifilament of dynactin primes the endosomal Arp2/3 complex. bioRxiv 2020.05.29.123372 [Preprint]. 29 May 2020. 10.1101/2020.05.29.123372. [DOI]
- 46.Lanier M. H., McConnell P., Cooper J. A., Cell migration and invadopodia formation require a membrane-binding domain of CARMIL2. J. Biol. Chem. 291, 1076–1091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zigmond S. H., Evangelista M., Boone C., Yang C., Dar A. C., Sicheri F., Forkey J., Pring M., Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13, 1820–1823 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Hansen S. D., Mullins R. D., VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 191, 571–584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welch M. D., Iwamatsu A., Mitchison T. J., Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature 385, 265–269 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Janmey P. A., Chaponnier C., Lind S. E., Zaner K. S., Stossel T. P., Yin H. L., Interactions of gelsolin and gelsolin-actin complexes with actin. Effects of calcium on actin nucleation, filament severing, and end blocking. Biochemistry 24, 3714–3723 (1985). [DOI] [PubMed] [Google Scholar]
- 51.Nag S., Ma Q., Wang H., Chumnarnsilpa S., Lee W. L., Larsson M., Kannan B., Hernandez-Valladares M., Burtnick L. D., Robinson R. C., Ca2+ binding by domain 2 plays a critical role in the activation and stabilization of gelsolin. Proc. Natl. Acad. Sci. U.S.A. 106, 13713–13718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soeno Y., Abe H., Kimura S., Maruyama K., Obinata T., Generation of functional beta-actinin (CapZ) in an E. coli expression system. J. Muscle Res. Cell Motil. 19, 639–646 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Wang H., Robinson R. C., Burtnick L. D., The structure of native G-actin. Cytoskeleton (Hoboken) 67, 456–465 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Spudich J. A., Watt S., The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 (1971). [PubMed] [Google Scholar]
- 55.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nag S., Larsson M., Robinson R. C., Burtnick L. D., Gelsolin: The tail of a molecular gymnast. Cytoskeleton (Hoboken) 70, 360–384 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/5/eabd5271/DC1


