Abstract
Bovine whey IgG enriched fraction contains antibodies against various human bacterial pathogens. It contains antibodies against some viral antigens, including human respiratory syncytial virus and influenza virus. We investigated whether the IgG enriched fraction has cross-reactivity with IgG antibodies against SARS-CoV-2 spike (S) and nucleocapsid (N) proteins. The full-length and partial-length SARS-CoV-2 S, N, a recombinant protein of the receptor binding domain (RBD) and nine peptides covering the receptor binding motif (RBM) of S were prepared. Direct enzyme-linked immunosorbent assays were conducted using these recombinant proteins and peptides as coating antigens and revealed the IgG enriched fraction contained antibodies against partial-length S [amino acids (aa) 177–512, 288–512, 348–578, 387–516 and 408–664], full-length N (aa 1–419) and partial-length N (aa 1–120, 111–220, 1–220 and 210–419), two RBD peptides, covering aa 427–446 and 502–520 of S, and recombinant RBD of S. These results indicate IgG enriched fraction contains antibodies against SARS-CoV-2.
1. Introduction
Bovine milk products contain IgG against several human bacterial pathogens, and rotavirus that cause gastrointestinal tract infections (Ulfman, Leusen, Savelkoul, Warner, & van Neerven, 2018), and bovine IgG was reported to bind to human respiratory syncytial virus (RSV) and influenza virus (Hartog et al., 2014). Bovine colostrum preparations obtained from cows immunised with antigens of several human gastrointestinal tract infections was called “hyperimmunised milk” (Golay, Ferrara, Felber, & Schneider, 1990). It is characterised by high antibody activities against specified pathogens. Clinical trials demonstrated that immune cow colostrum shortened the duration of gastrointestinal tract infections (Ulfman et al., 2018). Second-generation milk products obtained from colostrum derived from healthy non-immunised pasture fed cows provided immunity against Salmonella infection in calves (Funatogawa et al., 2002; Griffiths, 1969; Royal, Robinson, & Duganzich, 1986). An immunoglobulin preparation from non-immunised cows contained high levels of antibodies and neutralising activity against verotoxin of Escherichia coli O157:H7 (Funatogawa et al., 2002; Lissner, Schmidt, & Karch, 1996). Several reports that indicate bovine IgG has antibodies against various bacterial antigens and activates the human immune system to repel pathogens have been reviewed (Ulfman et al., 2018). Bovine IgG fraction was reported to protect mice against food-borne infections with enterohaemorrhagic E. coli O157:H7 and Salmonella enterica serovar Enteritidis (Funatogawa et al., 2019). This IgG fraction partially protected mice against respiratory tract infection with Mycobacterium avium (Funatogawa, Tada, Kuwahara-arai, Kirikae, & Takahashi, 2019). However, it is unclear whether bovine IgG recognises human viral pathogens except for rotavirus, RSV, and influenza virus.
The novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been causing a coronavirus disease (COVID-19) pandemic since 2019 (WHO, 2019). Sequence data analysis of human coronaviruses, including SARS-CoV-2 suggests they have an animal origin, especially bat (Cui, Li, & Shi, 2019). Here, we report IgG antibodies against SARS-CoV-2 spike protein (S) in bovine whey IgG rich fraction prepared from healthy non-immunised pasture fed cows in New Zealand.
2. Materials and methods
2.1. Construction and purification of recombinant SARS-CoV-2 spike protein (S) and nucleocapsid protein (N)
A partial-length of SARS-CoV-2 S gene (2055 bp) and the full-length of SARS-CoV-2 N gene (1260 bp) were synthesised based on SARS-CoV-2 isolate 2019-nCoV WHU01, complete genome (accession no. MN988668). Five sequences of SARS-CoV-2 S gene (529–1536 bp, 862–1536 bp, 1042–1734 bp, 1159–1548 bp and 1222–1992 bp), corresponding to amino acid (aa) 177–512, 288–512, 348–578, 387–516 and 408–664, and five sequences of SARS-CoV-2 N gene (1–360 bp, 330–660 bp, 1–660 bp, 628–1260 bp and 1–1260 bp), corresponding to amino acid (aa) 1–120, 111–220, 1–220, 210–419 and 1–419, were cloned into pET28a expression vectors (Novagen, Inc., USA) using primers listed in Table 1 . The constructed plasmids were used to transform E. coli BL21-CodonPlus (DE3)-RIP (Agilent Technologies, USA). Recombinant SARS-CoV-2 proteins were purified using Ni-NTA agarose, according to the manufacturer's instructions (Qiagen, Germany), and dissolved at 3 mg mL−1 in phosphate-buffered saline (PBS). A recombinant protein covering the RBD of SARS-CoV-2 spike protein was purchased from Sino Biological Inc, USA. For use in direct enzyme-linked immunosorbent assays (ELISAs), these recombinant proteins were diluted to 10 μg mL−1 in PBS-0.1% Tween 20 and used as coating antigens.
Table 1.
List of primers used in this study.
| Name | Sequence (5′→3′) |
|---|---|
| SARS-CoV-2-S(529)-F | ATGGATCCATGGACCTTGAAGGAAAACAGG |
| SARS-CoV-2-S(862)-F | ATGGATCCATGGCTGTAGACTGTGCACTTG |
| SARS-CoV-2-S(1042)-F | ATGGATCCATGGCATCTGTTTATGCTTGG |
| SARS-CoV-2-S(1159)-F | ATGGATCCATGTTAAATGATCTCTGCTTTA |
| SARS-CoV-2-S(1222)-F | ATGGATCCATGAGACAAATCGCTCCAGGGC |
| SARS-CoV-2-S(1536)-R | ATGTCGACCTATACTACTACTCTGTATGGT |
| SARS-CoV-2-S(1548)-R | ATGTCGACCTATTCAAAAGAAAGTACTACT |
| SARS-CoV-2-S(1734)-R | ATGTCGACTTAATCACGGACAGCATCAGTA |
| SARS-CoV-2-S(1992)-R | ATGTCGACTTATATGTCACACTCATATGAG |
| SARS-CoV-2-N(1)-F | ATGGATCCATGTCTGATAATGGACCCCAAA |
| SARS-CoV-2-N(330)-F | ATGGATCCATGTTCTATTACCTAGGAACTG |
| SARS-CoV-2-N(628)-F | ATGGATCCATGGCTGGCAATGGCGGTGATG |
| SARS-CoV-2-N(360)-R | ATGTCGACTTAGAGGGCAGTTTCACCACCT |
| SARS-CoV-2-N(660)-R | ATGTCGACTTAAGCAAGAGCAGCATCACCG |
| SARS-CoV-2-N(1260)-R | ATGTCGACTTAGGCCTGAGTTGAGTCAGCA |
2.2. Peptides of SARS-CoV-2 S
Nine peptides of SARS-CoV-2 S, corresponding to aa 382–401, 397–416, 427–446, 442–461, 457–476, 472–491, 487–506 and 502–520, were synthesised by Eurofins Genetics Inc, Japan. These peptides were dissolved at 3 mg mL−1 in distilled water free of endotoxin (LONZA, Switzerland) and diluted to 10 μg mL−1 in PBS containing 0.1% Tween 20 for use as coating antigens in ELISAs and as inhibitors in competitive inhibition ELISA.
2.3. Bovine whey IgG enriched fraction
Bovine whey IgG enriched fraction (IgG30+; Aotearoa Co., Japan) was obtained from milk of pasture fed, non-immunised healthy New Zealand cows by New Zealand Dairy Group by centrifugation to remove fat and condensation of immunoglobulin. Two lots of the IgG enriched fraction prepared in November 2018 and August 2019 were used. The cows were not administered insecticidal drugs, antibiotics, or growth hormones. The powder included (w/w); 84.7% protein, <8.4% fat, 7.0 mg g−1 lactoferrin and 327.7 mg g−1 immunoglobulin.
2.4. Direct enzyme-linked immunosorbent assays
Direct ELISAs, using a partial-length of recombinant SARS-CoV-2 S, the full-length recombinant SARS-CoV-2 N and a partial-length of recombinant SARS-CoV-2 N as coating antigens, were performed to investigate the presence of IgG antibodies with cross-reactivity against SARS-CoV-2 S and N in the IgG rich fraction. Triplicate 100 μL aliquots each containing either recombinant S or N in 0.05 m carbonate buffer (pH9.0) were placed in the wells of 96-well plates, incubated for 1 h at room temperature, then washed three times with PBS containing 0.1% Tween 20 between each of the following steps: blocking the wells with SuperBlock™ Blocking Buffer in PBS (Thermo Scientific, MA, USA) (150 μL well−1) for 30 min at room temperature; addition of 100 μL well−1 of IgG (0.003, 0.03, 0.3, 3 and 30 μg mL−1 in PBS containing 0.1% Tween 20) bovine IgG enriched fraction for 1 h at room temperature; addition of 100 μL well−1 of peroxidase-conjugated anti-bovine IgG (whole molecule) (1:10,000 dilution) (Sigma-Aldrich, St Louis, MO, USA) for 30 min at room temperature; addition of 50 μL TMB Peroxidase ELA Substrate Kit (Bio-Rad, Hercules, CA, USA) for 10 min. After 10 min incubation, 50 μL of 1 m sulphuric acid was added to stop the peroxidase reaction. Absorbance at 450 nm and 620 nm as reference were measured using infinite F50 microplate readers (TECAN, Switzerland).
Competitive inhibition ELISAs were performed by incubating the bovine IgG enriched fraction (0.3 μg mL−1) with one of the three synthesised peptides covering RBM of S (aa 382–401, 427–446 and 502–520) at concentrations of 0.001, 0.01, 0,1, 1, and 10 μg mL−1 in PBS containing 0.1% Tween 20 in 1.5-mL tubes overnight at 4 °C. The remaining free IgG against SARS-CoV-2 S was measured by direct ELISA, using plates coated with recombinant SARS-CoV-2 S (aa 288–512).
2.5. Endotoxin assay
Endotoxin concentrations in the solutions of the recombinant proteins and peptides were quantitatively measured using QCL-1000 Limulus Amebocyte Lysate, according to the manufacturer's instructions (LONZA, Switzerland). Solutions of recombinant proteins and peptide (3 mg mL−1 each) were diluted to 10 μg mL−1 in endotoxin-free water and endotoxin was measured.
3. Results and discussion
3.1. Recombinant SARS-CoV-2 S and N
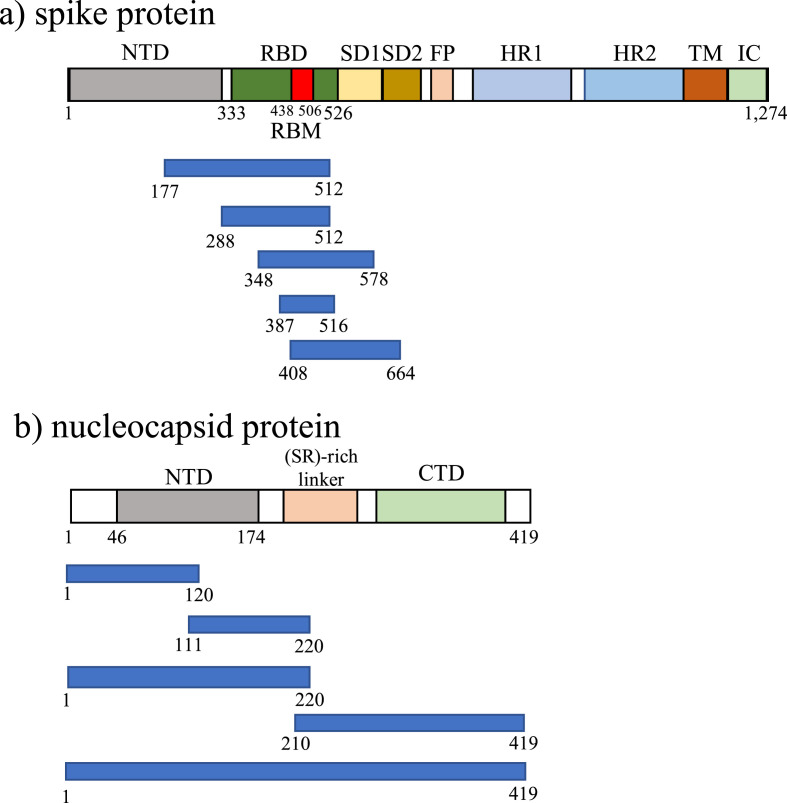
A total of 10 recombinant SARS-CoV-2 S and N, (five S and five N) samples were prepared by the expression system using E. coli BL21-CodonPlus (DE3)-RIP. As shown in Fig. 1 by the overall structures of SARS-CoV-2 S and N, SARS-CoV-2 S contains the N-terminal domain (NTD), receptor binding domain (RBD), subdomains (SDs), fusion peptide (FP), heptad repeats (HRs), transmembrane domain (TM) and intercellular domain (IC) (Lan et al., 2020), and SARS-CoV-2 N contains the N-terminal domain (NTD), (SR)-rich linker and C-terminal domain (CTD) (Kang et al., 2020). The regions of recombinant SARS-CoV-2 S (aa 177–512, 288–512, 348–578, 387–516 and 408–664) were selected from around RBD (aa 333–526), including receptor binding motif (RBM, aa 438–506), which was reported to interact with cell receptor ACE (Fig. 1a) (Lan et al., 2020). The full- and partial-lengths of recombinant SARS-CoV-2 N (aa 1–120, 111–220, 1–220, 210–419 and 1–419) were prepared (Fig. 1b) (Kang et al., 2020).
Fig. 1.
Overall topology of (a) SARS-CoV-2 spike protein (S) and five regions of recombinant SARS-CoV-2 S and (b) SARS-CoV-2 nucleocapsid protein (N) and regions of recombinant SARS-CoV-2 N. NTD, N-terminal domain; CTD, C-terminal domain; RBD, receptor binding domain; RDM, receptor binding motif; SD1, subdomain 1; SD2, subdomain 2; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane region; IC, intracellular domain.
3.2. IgG enriched fraction containing antibodies against SARS-CoV-2
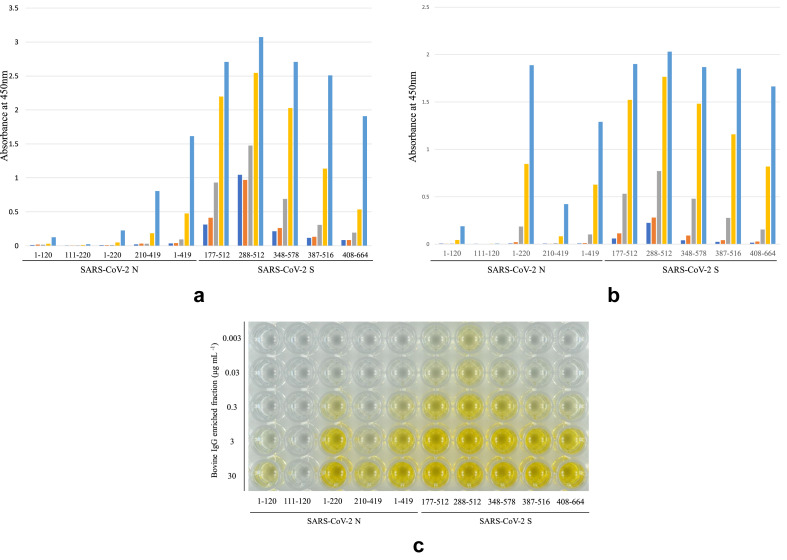
Reactivity against SARS-CoV-2 S was higher than that against SARS-CoV-2 N (Fig. 2 ). The IgG enriched fraction strongly recognised the 288–512 region compared with other regions of SARS-CoV-2 S (Fig. 2). The fraction recognised the full-length (aa 1–419) more than other partial-lengths of SARS-CoV-2 N (Fig. 2). There were few differences in reactivities against recombinant proteins from SARS-CoV-2 S and N among the two lots tested, except for reactivities against SARS-CoV-2 N (aa 1–220), i.e., the lot prepared in 2018 have more reactivity against SARS-CoV-2 N (aa 1–220) than that in 2019 (Fig. 2). The results of ELISA indicate that the bovine IgG enriched fraction contained IgG antibodies against SARS-CoV-2 S and N.
Fig. 2.
Bovine IgG enriched fraction containing IgG against SARS-CoV-2 assessed by direct enzyme-linked immunosorbent assays (ELISA) using a partial-length of recombinant SARS-CoV-2 S (aa 177–512, 288–512, 348–578, 387–516 and 408–664), full-recombinant SARS-CoV-2 N (aa 1–419) and partial-length of recombinant SARS-CoV-2 N (aa 1–120, 111–220, 1–220 and 210–419) (Fig. 1) as coating antigens. Two different lots of bovine IgG enriched fraction prepared in 2019 and 2018 were used (2a and 2b, respectively):  , 0.003 μg mL−1;
, 0.003 μg mL−1;  , 0.03 μg mL−1;
, 0.03 μg mL−1;  , 0.3 μg mL−1;
, 0.3 μg mL−1;  , 3 μg mL−1;
, 3 μg mL−1;  , 30 μg mL−1. A picture of a representative ELISA result is shown in 2c.
, 30 μg mL−1. A picture of a representative ELISA result is shown in 2c.
3.3. Epitope mapping of RBD in SARS-CoV-2 spike protein
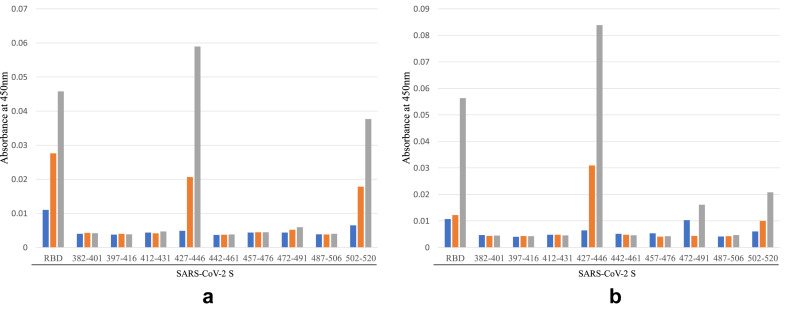
ELISA using a recombinant RBD protein revealed that the IgG enriched fraction contained antibodies against the RBD in the S protein of SARS-CoV-2 (Fig. 3 ). ELISA using peptides covering the RBD revealed that this fraction contained IgG antibodies against two peptides (aa 427–446 and 502–520), indicating that the IgG enriched fraction contained antibodies against two epitopes of the RBD in S protein (Fig. 3).
Fig. 3.
Determination of epitopes by direct ELISA using nine peptides of SARS-CoV-2 S protein, corresponding to aa 382–401, 397–416, 427–446, 442–461, 457–476, 472–491, 487–506 and 502–520, with plates coated with a recombinant protein covering the RBD of SARS-CoV-2 S protein. Two lots of bovine IgG enriched fraction prepared in 2019 and 2018 (3a and 3b, respectively), were tested:  , 0.3 μg mL−1;
, 0.3 μg mL−1;  , 3 μg mL−1;
, 3 μg mL−1;  , 30 μg mL−1.
, 30 μg mL−1.
3.4. Competitive inhibition ELISA
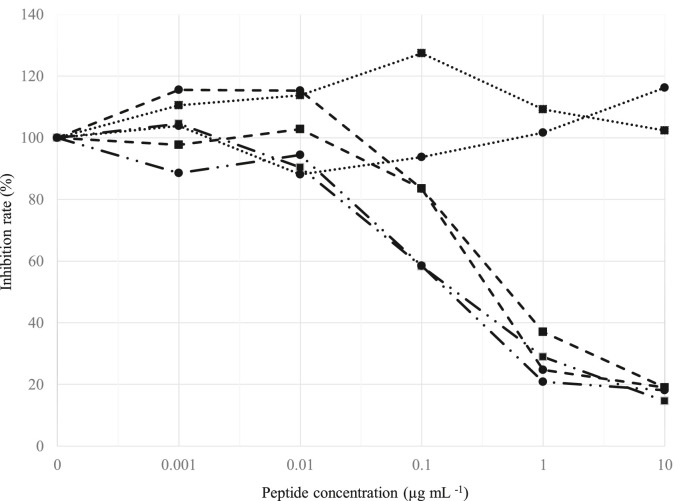
The ability of two peptides (aa 427–446 and 502–520) to inhibit the binding of IgG antibodies present in the enriched fraction with the S protein of SARS-CoV-2 was evaluated by competitive inhibition ELISA. Both peptides dose-dependently inhibited the binding of two lots of bovine IgG enriched fraction with the S protein of SARS-CoV-2 (aa 288–512) (Fig. 4 ). The 50% inhibitory concentrations (ID50s) of peptide (aa 502–520) (0.09 and 0.11 μg mL−1, respectively) were slightly lower than those of peptide (aa 427–446) (0.20 and 0.24 μg mL−1, respectively) (Fig. 4). A third peptide (aa 382–401), with which the IgG enriched fraction did not react, was unable to inhibit the binding of the two lots in ranges of 0.001–10 μg mL−1 (Fig. 4).
Fig. 4.
Competitive inhibition ELISA of the bovine IgG enriched fraction (IgG 0.3 μg mL−1), incubated with one of three peptides of S protein of SARS-CoV-2 ( , aa 382–401;
, aa 382–401;  , aa 427–446;
, aa 427–446;  , aa 502–520) at concentrations 0.001, 0.01, 0,1, 1, and 10 μg mL−1, the remaining free IgG against the S protein of SARS-CoV-2 was assayed by direct ELISA, using plates coated with the peptide corresponding to aa 288–512 of S protein of SARS-CoV-2. Two lots of bovine IgG enriched fraction prepared in 2019 (
, aa 502–520) at concentrations 0.001, 0.01, 0,1, 1, and 10 μg mL−1, the remaining free IgG against the S protein of SARS-CoV-2 was assayed by direct ELISA, using plates coated with the peptide corresponding to aa 288–512 of S protein of SARS-CoV-2. Two lots of bovine IgG enriched fraction prepared in 2019 ( ) and 2018 (
) and 2018 ( ) were tested.
) were tested.
3.5. Endotoxin levels
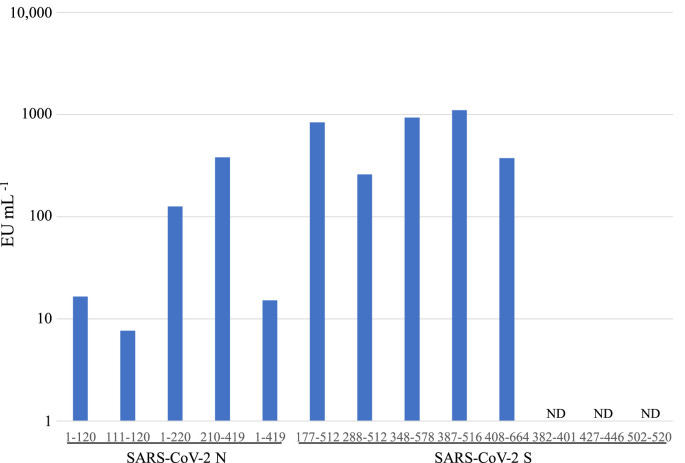
The solutions of all the recombinant SARS-CoV-2 S and N proteins (10 μg mL−1) were found to be contaminated with significant concentrations of endotoxin, ranging from 7.7 to 1108 EU mL−1 (0.77–110.8 EU μg−1) (Fig. 5 ). In contrast, endotoxin was not detected in any of the solutions of the three SARS-CoV-2 S peptides (aa 382–401, 427–446 and 502–520) tested (Fig. 5). These results together, with those of direct and indirect ELISA indicated that bovine IgG directly and specifically binds to the two peptides (aa 427–446 and 502–520), although we could not exclude that possibility that endotoxin disrupted the binding of bovine IgG to recombinant SARS-CoV-2 S and N proteins.
Fig. 5.
Endotoxin levels in solutions of the recombinant proteins and peptides measured quantitatively using QCL-1000 Limulus Amebocyte Lysate; each solution contained 10 μg of recombinant protein or peptide (ND, not detected).
3.6. Bovine IgG enriched fraction
It is unlikely these pasture-fed healthy New Zealand cows were exposed to or naturally immunised against SARS-CoV-2. The two lots of bovine IgG enriched fraction investigated in this study were prepared in November 2018 and August 2019, respectively, which predate the emergence of SARS-CoV-2 in December 2019 in China. But it cannot be excluded that the cows had been exposed to some unknown coronavirus that share the immunogenicity of SARS-CoV-2.
The present study showed that the bovine IgG enriched fraction binds specifically to the RBD of SARS-CoV-2 S protein, indicating that the IgG enriched fraction would have potential to neutralise SARS-CoV-2. SARS-CoV-2 S forms homotrimers and uses angiotensin-converting enzyme 2 (ACE2) to enter cell (Walls et al., 2020).
SARS-CoV-2 S murine polyclonal antibodies potently inhibited SARS-CoV-2 mediated entry into cells, indicating that cross-neutralising antibodies targeting conserved S epitopes may be induced by vaccination (Walls et al., 2020). A human monoclonal antibody bound a conserved epitope of spike RBD and neutralised SARS-CoV-2 (Wang et al., 2020).
SARS-CoV-2 N is an intercellular protein that has a multifunctional RNA-binding protein for viral RNA transcription and replication (Kang et al., 2020; Lan et al., 2020). SARS-CoV-2 N would have relatively lower antigenicity against cows. Accordingly, the bovine IgG enriched fraction may show less potent activity against SARS-CoV-2 N than SARS-CoV-2 S.
Author contributions
SO and NM created the research data. SO and TT wrote the draft of the manuscript. All authors read, made significant edits to the first version, and approved the final manuscript.
Conflict of interest
S. J. works for Kohjin Bio Co., Ltd.
M.T. works for Aotearoa Co., Ltd.
Acknowledgements
This study was supported by a grant from Japan Agency for Medical Research and Development, Japan (grant number 20he0622015h0001), a joint research fund from Kohjin Bio Co., Ltd and a joint research fund from Aotearoa Co., Ltd.
References
- Cui J., Li F., Shi Z. Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatogawa K., Ide T., Kirikae F., Saruta K., Nakano M., Kirikae T. Use of immunoglobulin enriched bovine colostrum against oral challenge with enterohaemorrhagic Escherichia coli O157:H7 in mice. Microbiology and Immunology. 2002;46:761–766. doi: 10.1111/j.1348-0421.2002.tb02761.x. [DOI] [PubMed] [Google Scholar]
- Funatogawa K., Tada T., Kuwahara-arai K., Kirikae T., Takahashi M. Enriched bovine IgG fraction prevents infections with enterohaemorrhagic Escherichia coli O157:H7, Salmonella enterica serovar Enteritidis, and Mycobacterium avium. Food Science and Nutrition. 2019;7:2726–2730. doi: 10.1002/fsn3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golay A., Ferrara J.M., Felber J.P., Schneider H. Cholesterol-lowering effect of skim milk from immunized cows in hypercholesterolemic patients. American Journal of Clinical Nutrition. 1990;52:1014–1019. doi: 10.1093/ajcn/52.6.1014. [DOI] [PubMed] [Google Scholar]
- Griffiths O.V. Colostral immunity against salmonella infection in calves. New Zealand Veterinary Journal. 1969;17 doi: 10.1080/00480169.1969.33779. [DOI] [PubMed] [Google Scholar]
- Hartog G.D., Jacobino S., Bont L., Cox L., Ulfman L.H., Leusen J.H.W., et al. Specificity and effector functions of human RSV-specific IgG from bovine milk. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain potential unique drug targeting sites. Acta Pharmaceutica Sinica B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure if the SARS-CoV-S spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lissner R., Schmidt H., Karch H. A standard immunoglobulin preparation produced from bovine colostra shows antibody reactivity and neutralization activity against Shiga-like toxins and EHEC-hemolysin of Escherichia coli O157:H7. Infection. 1996;24:378–383. doi: 10.1007/BF01716084. [DOI] [PubMed] [Google Scholar]
- Royal W.A., Robinson R.A., Duganzich D.M. Colostral immunity against salmonella infection in calves. New Zealand Veterinary Journal. 1986;16:141–145. doi: 10.1080/00480169.1968.33761. [DOI] [PubMed] [Google Scholar]
- Ulfman L.H., Leusen J.H.W., Savelkoul H.F.J., Warner J.O., van Neerven R.J.J. Effects of bovine immunoglobulins on immune function, allergy, and infection. Frontiers in Nutrition. 2018;5 doi: 10.3389/fnut.2018.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2020;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., Haperen R.V., Osterhaus A.D.M. A human monoclonal antibody blocking SARS-CoV-2 infection. Nature Communications. 2020;11 doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus disease 2019 (COVID-19) situation report. 2019. https://www.who.int/docs/default-source/coronaviruse/situation-reports Available from: