Abstract
Background
While severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection presents with mild or no symptoms in most cases, a significant number of patients become critically ill. Remdesivir has been approved for the treatment of coronavirus disease 2019 (COVID-19) in several countries, but its use as monotherapy has not substantially lowered mortality rates. Because agents from traditional Chinese medicine (TCM) have been successfully utilized to treat pandemic and endemic diseases, we designed the current study to identify novel anti-SARS-CoV-2 agents from TCM.
Methods
We initially used an antivirus-induced cell death assay to screen a panel of herbal extracts. The inhibition of the viral infection step was investigated through a time-of-drug-addition assay, whereas a plaque reduction assay was carried out to validate the antiviral activity. Direct interaction of the candidate TCM compound with viral particles was assessed using a viral inactivation assay. Finally, the potential synergistic efficacy of remdesivir and the TCM compound was examined with a combination assay.
Results
The herbal medicine Perilla leaf extract (PLE, approval number 022427 issued by the Ministry of Health and Welfare, Taiwan) had EC50 of 0.12 ± 0.06 mg/mL against SARS-CoV-2 in Vero E6 cells – with a selectivity index of 40.65. Non-cytotoxic PLE concentrations were capable of blocking viral RNA and protein synthesis. In addition, they significantly decreased virus-induced cytokine release and viral protein/RNA levels in the human lung epithelial cell line Calu-3. PLE inhibited viral replication by inactivating the virion and showed additive-to-synergistic efficacy against SARS-CoV-2 when used in combination with remdesivir.
Conclusion
Our results demonstrate for the first time that PLE is capable of inhibiting SARS-CoV-2 replication by inactivating the virion. Our data may prompt additional investigation on the clinical usefulness of PLE for preventing or treating COVID-19.
Keywords: COVID-19, Perilla frutescens (L.) Britt, SARS-CoV-2, Traditional Chinese medicine, Coronavirus, Zisu
At a glance of commentary
Scientific background on the subject
Over the last two decades, respiratory illnesses caused by coronavirus infections have compromised individual health, burdened health-care systems, and caused substantial economic and well-being losses. Unfortunately, effective prevention and treatment have been hampered by the absence of specific pharmacological interventions. In this scenario, new drugs derived from Chinese herbs have been actively researched in the ongoing quest to tackle the COVID-19 pandemic.
What this study adds to the field
Perilla leaf extract (PLE) inhibits the initial viral entry step by inactivating the SARS-CoV-2 virion. Additionally, it significantly decreases virus-induced cytokine release and viral protein/RNA levels in the human lung epithelial cell line Calu-3. PLE combined with remdesivir shows additive-to-synergistic efficacy against SARS-CoV-2. Altogether, our results provide empirical evidence that PLE may be part of the therapeutic armamentarium against COVID-19.
Severeacute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19) [1]. SARS-CoV-2 is a positive-sense, single-stranded RNA virus containing a canonical set of four major structural proteins. These include the spike (S), membrane (M), and envelope (E) proteins localized to the membrane envelope and the nucleocapsid (N) protein found in the ribonucleoprotein core. Similar to the SARS-CoV that caused a disease outbreak in 2003, SARS-CoV-2 uses its spike protein to interact with its cellular receptor, angiotensin-converting enzyme 2, for host cell entry [2,3]. Four other human coronaviruses (HCoVs) are typical causative agents of common colds. These include HCoV-OC43 and HCoV-229E that are responsible for ≤30% of all upper respiratory tract infections [4]. While SARS-CoV-2 infection presents with mild to no symptoms in most cases, a significant number of patients become critically ill and require hospitalization and intensive care. As of October 28, 2020, there were >44 million confirmed cases of COVID-19 and 1,172,000 COVID-19-related deaths worldwide. Notably, SARS-CoV-2 infections can elicit cytokine storms characterized by massively increased plasma concentrations of inflammatory cytokines and chemokines (i.e., IL-6, IFN-γ, TNF-α, MCP1, and CXCL10) [[5], [6], [7], [8]].
While vaccination programs against SARS-CoV-2 are currently ongoing, the long-term safety of available vaccines remains a major concern. In this scenario, there is an urgent need for both repurposed drugs and agents from traditional Chinese medicine (TCM) to contain the spread of this highly contagious disease.
The repurposed antiviral remdesivir – which was originally developed and administered as an Ebola polymerase inhibitor – is currently the only antiviral agent approved for COVID-19 therapy. While it reduces hospitalization length, it does not significantly lower COVID-19 mortality rates [9]. Agents from TCM have been frequently prescribed to prevent or treat the SARS-CoV outbreak in 2003. Medication records from antiquity disclosed that certain TCMs can be clinically useful to treat respiratory infections. In previous SARS-CoV outbreaks, several provinces in China issued TCM programs based on the principles tonifying qi, protecting against external pathogens, dispersing wind, discharging heat, and resolving dampness. The most frequently used herbs – prescribed either separately or in combination – included Radix Astragali (Huangqi), Radix Glycyrrhizae (Gancao), Radix Saposhnikoviae (Fangfeng), Rhizoma Atractylodis Macrocephalae (Baizhu), Lonicerae japonicae Flos (Jinyinhua), and Fructus forsythia (Lianqiao).
Perilla (Perilla frutescens L. Britt) is commonly known as perilla, Korean perilla, beefsteak plant, purple mint, perilla mint, Chinese basil, Zisu (in China), and Shiso (in Japan). Various parts of the perilla plant have different ethnopharmacological effects. While the leaves are used to tonify stomach function, discharge heat, and improve healthy qi, and seeds decrease qi, resolve phlegm, relieve cough and asthma, and alleviate constipation [10]. In the current study, we investigated the mechanisms underlying the anti-SARS-CoV-2 activity of perilla leaf extract (PLE) using standardized herbal preparations and identified PLE as a virucidal agent that directly inactivates the viral particle.
Materials and methods
Cell lines and viruses
Vero E6, human hepatocellular carcinoma (Huh7), human rhabdomyosarcoma (RD), and Calu-3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco, BRL, Gaithersburg, MD, USA) supplemented with 10% (w/v) fetal bovine serum (FBS). Madin–Darby canine kidney (MDCK) cells were cultured in DMEM containing 10% FBS, 2 mM l-glutamine (Gibco), 100 fold dilution nonessential amino acid mixture (Gibco), 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Gibco). Cell lines were obtained from the Chang Gung Memorial Hospital and passages were <25 in all cases. The SARS-CoV-2 strain CDC-4 (CGMH-CGU-01; GISAID accession number EPI_ISL_411915 and NCBI accession number MT192759) was provided by the Taiwan Center for Disease Control [11] and amplified in Vero E6 cells. HCoV-229 was obtained from the Research Center for Emerging Viral Infections of the Chang Gung University (Kweishan, Taoyuan, Taiwan) and amplified in Huh7 cells. The viral titer was determined by a plaque assay. SARS-CoV-2 was handled either in a biosafety level three or a biosafety level four laboratory.
Reagent preparation
PLE (Cat. number 6216; approval number 022427 issued by the Ministry of Health and Welfare, Taiwan) was purchased – along with other herbal powders listed in [Table 1] – from SunTen Pharmaceutical Co. Ltd. (Taipei, Taiwan). The plant name was checked against http://www.theplantlist.org and verified by the Brion Research Institute of Taiwan. PLE (25 mg/mL stock solution) was sterilized using a 0.22-μm filter and maintained in small aliquots at −80 °C. The voucher specimen used in the present study was deposited in the herbarium of the Chang Gung University (Kweishan, Taoyuan, Taiwan). The results of high-performance liquid chromatography analysis are shown in Figure S1. The chromatograms detected the presence of rosmarinic acid – which serves as the constituent marker of PLE according to government guidelines.
Table 1.
Antiviral activity screening of herbal medicines in vitro.
| Herbal extracta | Final concentration | Antiviral activityb | Toxicity |
|---|---|---|---|
| 荊防敗毒散Ching-fang-pai-tu-san | 1.25 mg/mL | – | – |
| 柴葛解肌湯Chai-ge-jie-ji-tang. | – | – | – |
| 川芎茶調散Chuan-xiong-cha-tiao-san | 1.25 mg/mL | – | – |
| 麻杏石甘湯Ma-xing-shi-gan-tang | 1.25 mg/mL | partial | – |
| 葛根湯Ko-Ken Tang | 1.25 mg/mL | – | – |
| 廣藿香Pogostemon cablin (Blanco) Benth | 1.25 mg/mL | – | – |
| 菊花Chrysanthemi Flos | 1.25 mg/mL | – | – |
| 黃蘗Phellodendri Cortex | 1.25 mg/mL | – | – |
| 丁香Caryophylli Flos | 1.25 mg/mL | – | + |
| 紫蘇葉Perillae Folium | 1.25 mg/mL | + | – |
| 薑黃 Curcumae Longae Rhizoma | 1.25 mg/mL | – | – |
| 骨碎補Davallia mariesii Rhizoma (water-butanol extract) | 0.125 mg/mL | – | + |
| 白芍Paeoniae Alba Radix | 0.125 mg/mL | – | + |
| 苦丁茶Ilex Kaushue | 1.25 mg/mL | – | + |
| 赤勺Paeoniae Rubra Radix (ethanol extract) | 0.125 mg/mL | – | – |
| 牡丹皮Moutan Radicis Cortex | 0.125 mg/mL | – | + |
| 牛筋草Eleusine indica (Goosegrass) | 0.125 mg/mL | – | – |
| 龍牙草Agrimonia pilosa (Hairyvein agrimonia) | 0.125 mg/mL | – | + |
Water extract unless otherwise described in parenthesis.
“-” indicates no activity or no cytotoxicity; “+” indicates protection or cytotoxicity.
Cytotoxicity and cytopathic effect (CPE) assay
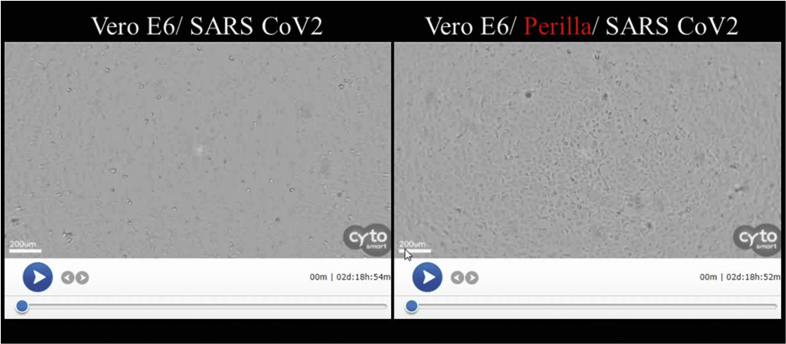
A 96-well tissue culture plate was seeded with Vero E6 cells (2 × 104/well) and incubated at 37 °C for 16–20 h under 5% CO2. Cell viability was measured following incubation with PLE for 72 h and subsequent staining with 5 mg/mL methyl thiazolyl tetrazolium (MTT). Thereafter, the crystal product formazan was dissolved in dimethyl sulfoxide (200 μL/well), and the absorbance (OD570nm) of each well was measured using a microplate reader. CC50 represents the cytotoxicity concentration at which 50% cell death occurs [12]. Live imaging of virus-induced CPE was recorded using CytoSMART Lux2 (Scintica Instrumentation Inc., Maastricht, the Netherlands).
EC50 determination
Vero E6 cells (2 × 104/well) plated in 96-well plates were inoculated with virus at a multiplicity of infection (MOI) of 2.5 × 10−4 plaque forming unit (PFU)/cell (which is equivalent to the 9TCID50, i.e., the median tissue culture infective dose). Cells were maintained in E2 (DMEM containing 2% [v/v] FBS) with different concentrations of PLE. After incubation at 37 °C under 5% CO2 for 72 h, cell viability was determined using the MTT assay. EC50 was defined as the PLE concentration required to inhibit 50% CPE. Remdesivir (MedChemExpress, Monmouth Junction, NJ, USA) was the positive control for each EC50 determination. Mock infection was used as negative control.
Time-of-drug-addition assay
PLE (1.25 mg/mL) was incubated with Vero E6 cells in six-well plates (1 mL/well) between −3 h post-infection (p.i.) and 0 h p.i., between −1 h p.i. and 0 h p.i., between 0 h p.i. and 24 h p.i., and between −3 h and 24 h p.i. at 37 °C. The virus was absorbed to cells between −1 h p.i. and 0 h p.i. at an MOI of 0.01. Cell lysates from each treatment were harvested at 24 h p.i. for quantitative reverse transcription polymerase chain reaction (qRT-PCR) and Western blot analyses.
Plaque assay and plaque reduction assays
For plaque assay, Vero E6 cells (4 × 105/well) were inoculated with serially diluted SARS-CoV-2 (0.5 mL/well) in 12-well plates for 1 h at 37 °C under occasional shaking. Cells were washed twice with phosphate-buffered saline. After the addition of 1.4% (v/v) methylcellulose in E2 (1.5 mL), cells were incubated for 3 days. Upon fixation with 4% (v/v) paraformaldehyde (PFA) for 1 h at room temperature, cells were stained with 0.5% (w/v) crystal violet for 30 min at room temperature. For plaque reduction assay, Vero E6 cells inoculated with SARS-CoV-2 (approximately 90 PFU/well; 0.5 mL/well) in presence of PLE for 1 h at 37 °C under occasional shaking. After washing, 1.5 mL of 1.4% (v/v) methylcellulose in E2 containing PLE was added to cells – which were subsequently incubated for 3 days.
RNA extraction and qRT-PCR analysis
Total RNA from Vero E6 or Calu-3 cells was extracted using TRIzol reagent (Thermo Fisher Scientific Inc, MA, USA) according to the manufacturer's instructions. Equal amounts of total RNA were reverse transcribed using the M-MLV reverse transcriptase system (Thermo Fisher Scientific Inc.) using random primers. Subsequently, qRT-PCR was performed with a QuantStudio3 RT-qPCR system (Applied Biosystems, Foster City, CA, USA) using primer pairs specific for the E gene. GAPDH was used as the internal control. The primer sequences are listed in Table S1, and the relative mRNA expression was calculated using the 2−△△Ct method [13].
Western blot
Cells were harvested, treated with 3 × sodium dodecyl sulfate (SDS) sample buffer, denatured at 37 °C for 1 h, and subjected to 8% SDS-polyacrylamide gel electrophoresis. Proteins were immunoblotted onto polyvinylidene fluoride membranes and stained with mouse monoclonal anti-spike (GTX632604; GeneTex, Hsinchu, Taiwan, 1: 5000) or mouse monoclonal anti-nucleocapsid (GTX632269, GeneTex, 1: 2000) antibodies. Mouse monoclonal GAPDH (GTX627408, GeneTex, 1: 5000) was used as the loading control.
Viral particle inactivation assay
The virus (3000 PFUs) was incubated with increasing PLE concentrations in 500 μL DMEM for 30 min at 37 °C and 5% CO2. The reaction mixture was diluted 100-fold to reduce the antiviral effect of PLE. After treatment, the residual viral titer was measured using a plaque assay.
Cytokine analysis
Calu-3 cells (5 × 105/well) were seeded in six-well plates for 3–4 days before infection with SARS-CoV-2 (MOI = 0.01) for 1 h. The virus was subsequently removed and replaced with E2. At 48 h p.i., cells were harvested for qRT-PCR and Western blot analyses. The cytokine primer sequences are shown in Table S1.
Immunofluorescence assays
Calu-3 cells (5 × 105/well) were seeded on a coverslip glass, which was set in a six-well plate. After infection with SARS-CoV-2 at a MOI of 0.01, cells were fixed with PFA at 48 h p.i. and stained with anti-spike antibodies (GeneTex). Fluorescence was examined using a confocal microscope (LSM780, Carl Zeiss AG, Oberkochen, Germany).
Evaluation of PLE activity in combination with remdesivir
Vero E6 cells were seeded in a six-well tissue culture plate. Several dilutions of both remdesivir (concentrations: 0, 2, and 4 μM) and PLE (concentrations: 0, 1.25, and 2.5 mg/mL) were subsequently tested in a checkerboard matrix. None of the tested combinations was found to induce cytotoxicity. The inhibitory efficacy was assessed by qRT-PCR at 24 h p.i. To evaluate drug–drug interactions, the combination index was calculated by using Highest Single Agent (HSA) reference model of Synergy Finder, version 2.0 [14]. Interactions between drugs were considered antagonistic, additive, or synergistic when the combination indices were < −10, −10 ≤ × ≤ 10, or >10, respectively.
Statistical analysis
Data were analyzed using GraphPad Prism v. 8.0 (GraphPad Software, La Jolla, CA, USA) and presented as means ± standard error of the mean. Differences between pairs of means were analyzed using two-sample t-test. All tests were two-sided and P value less than 0.05 was considered statistically significant.
Results
PLE exhibits anti-SAR-CoV-2 activity in Vero E6 cells
To identify herbal extracts with potential activity against SARS-CoV-2, we performed a primary antiviral screen of a panel of Chinese herbs known to eliminate dampness and reduce heat. To this end, an assay of these materials was carried out to measure the inhibition of virus-induced cell death [Table 1]. PLE had an EC50 of 0.12 ± 0.06 mg/mL, a CC50 of 4.64 0.16 mg/mL, and a selectivity index of ∼40.65 for SARS-CoV-2 inhibition in Vero E6 cells [Table 2]. PLE was also capable of inhibiting EV-A71 and influenza virus – suggesting a broad-spectrum inhibition capacity against RNA viruses. The antiviral activity was apparently highly specific. No HCoV-229E inhibition was evident at a PLE concentration of ≤1.25 mg/mL [Table 1]. Remdesivir (positive control) had an EC50 of 3.28 ± 1.78 μM. PLE-mediated protection against virus-induced cell death was monitored in parallel using a live imaging system (Figure S2). Virus-mediated CPE was evident at a MOI of 0.01. The cells aggregated and detached from the dish by 48 h p.i. (Figure S2, left panel). However, PLE (1.25 mg/mL) delayed the onset of CPE (Figure S2, right panel).
Table 2.
CC50 and EC50 of PLE in various cell lines and viruses.
| Cell line or virus strain | CC50 (mg/mL)a | EC50 (mg/mL)b | SIc |
|---|---|---|---|
| Vero E6 | 4.64 0.16 | ||
| Calu-3 | >5 | ||
| Huh7 | >1.25 | ||
| RD | 10.92 ± 0.47 | ||
| MDCK |
1.73 ± 0.39 |
||
| HCov-229E | >1.25 | – | |
| SARS-CoV-2 | 0.12 ± 0.06 | 40.65 | |
| EV-A71 | 0.04 ± 0.00 | 280 | |
| Influenza A/WSN/1933 | 1.09 ± 0.37 | 1.58 |
CC50: Drug concentration causing 50% cytotoxicity as determined by the MTT assay. Vero E6, Huh7, RD, and MDCK cells were incubated with PLE for 3 days, whereas Calu-3 cells for 2 days.
EC50: Concentration of compounds inhibiting 50% of the cytopathic effects caused by viral infection according to the MTT assay. EC50 is presented as mean ± standard deviation of two-three independent experiments. HCoV-229E and SARS-CoV-2 were assayed in Huh7 and Vero E6 cells, respectively.
Selectivity index (SI): CC50/EC50.
PLE may target the early stage of viral infection cycle
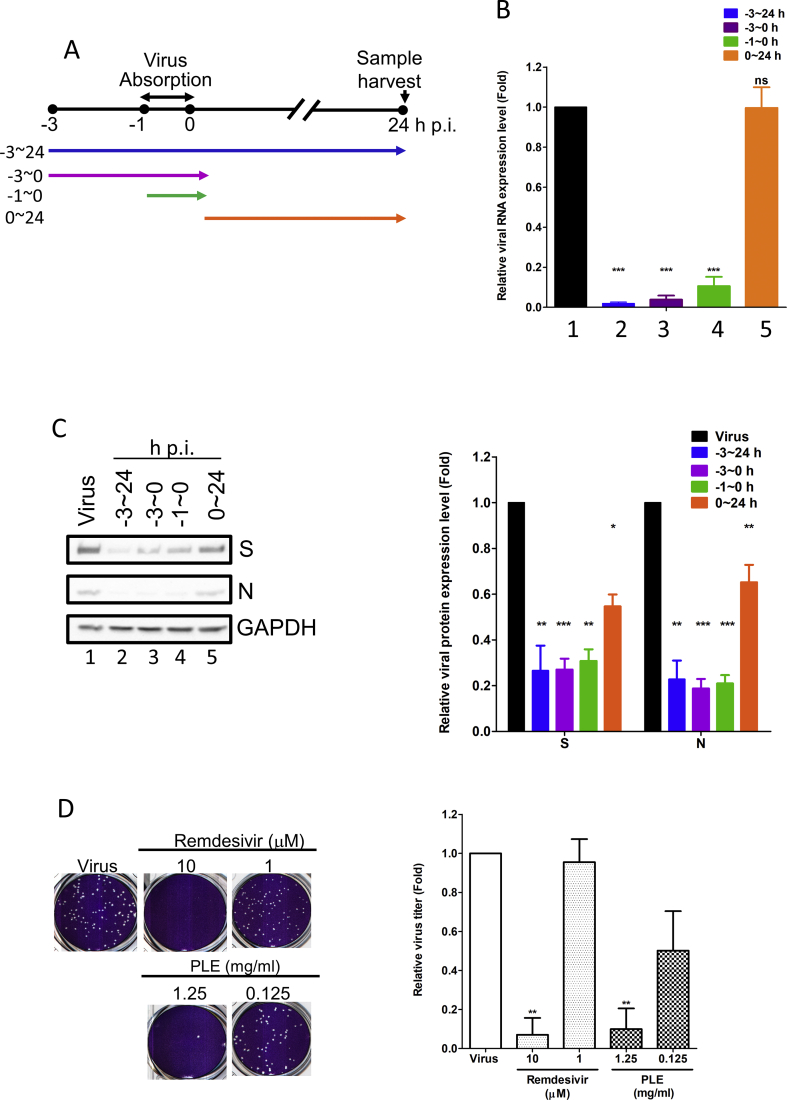
We assessed the time course of drug addition to identify the stage at which PLE inhibits the viral replication cycle [Fig. 1A]. Compared with the mock-treated control, the addition of PLE markedly inhibited viral RNA and protein syntheses at treatment course from −3 h p.i. to 24 h p.i. [lanes 2; Fig. 1B,C]. PLE showed a stronger inhibitory activity when it was added at earlier time points (from −3 h to 0 h p.i. and from −1 h to 0 h p.i.) than when it was added between 0 h p.i. and 24 h p.i. [Fig. 1B,C]. These findings indicate that PLE may play a role in blocking viral entry. PLE treatment inhibited SARS-CoV-2 plaque formation in a dose-dependent manner [Fig. 1D]. In addition, we found that the antiviral activity of PLE (1.25 mg/mL) was similar to that of remdesivir (10 μM; positive control) as shown by the extent of inhibition of plaque formation [Fig. 1D]. The foregoing data strongly support the anti-SARS-CoV-2 activity of PLE.
Fig. 1.
Treatment with PLE inhibits SARS-CoV-2 at early stages of replication. (A) Schematic representation of the time-of-addition assay. (B–C) Vero E6 cells were infected with SARS-CoV-2 at a MOI of 0.01. Subsequently, PLE (1.25 mg/mL) was added at the following time points: before virus entry (between −3 and 0 h p.i.), during virus absorption (−1−0 h p.i.), and following virus adsorption (0–24 h p.i.). Infected cells were collectively harvested at 24 h p.i.; viral RNA synthesis and viral protein expression were analyzed with qPCR (B) and western blotting (C), respectively. (B) Expression levels of viral RNA were initially normalized to GAPDH mRNA at each experimental condition. Moreover, the ratio measured in PLE-treated cells was normalized to the RNA level of virus control (arbitrarily set to 1). (C) The intensity of SARS-CoV-2 spike protein (S) and nucleocapsid (N) expression was normalized to GAPDH. Moreover, the ratio measured in PLE-treated cells was normalized to the protein level of virus control (arbitrarily set to 1). N = 3. (D) The results of the plaque reduction assay revealed that SARS-CoV-2 infectivity was diminished after exposure of Vero E6 cells to PLE. SARS-CoV-2 was pre-incubated with various concentrations of PLE or remdesivir before its addition to Vero E6 cells for the plaque assay. The number of plaques was calculated and normalized to that of virus control (arbitrarily set to 1). Data in bar charts are expressed as means ± standard error of the mean from at least two independent experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005; ns = not significant.
PLE inhibits virus-induced proinflammatory cytokine expression
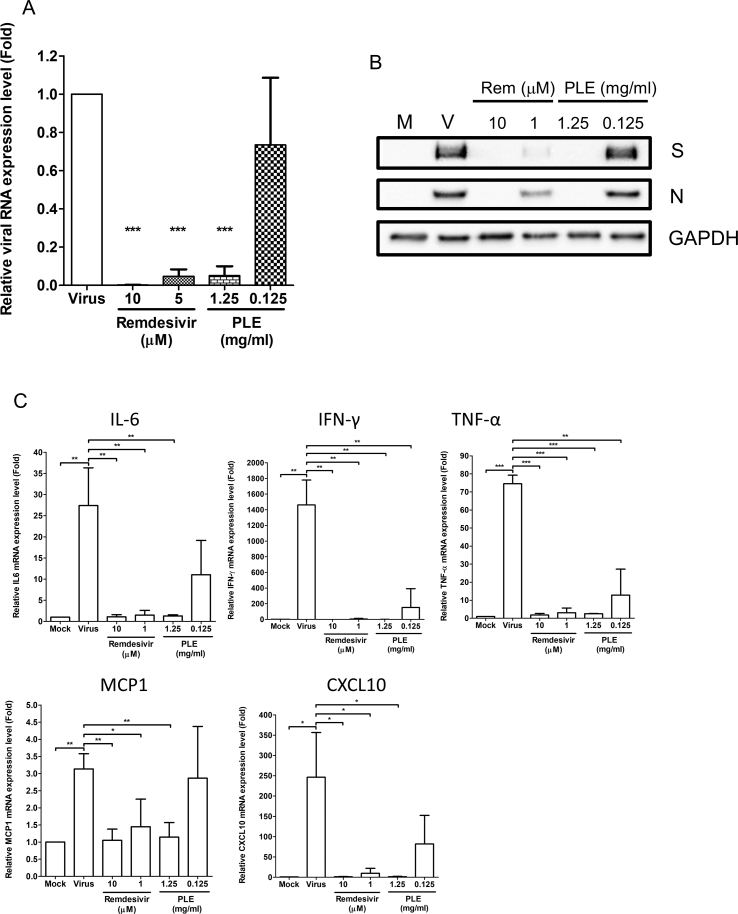
The primary targets of SARS-CoV-2 are the epithelial cells in the airways and lungs. Therefore, the antiviral efficiency of PLE was assessed in the human lung alveolar cell line Calu-3, which is highly susceptible to viral infection [15]. As expected, SARS-CoV-2 infection was robust in Calu-3 cells. Viral RNA and protein syntheses were markedly increased by 48 h p.i. but were inhibited by both remdesivir and PLE in a dose-dependent manner [Fig. 2A,B]. Dramatic changes in cytokine levels are associated with COVID-19 progression. For this reason, we examined the effects of PLE on cytokine and chemokine expression induced by SARS-CoV-2 in Calu-3 cells. The mRNA levels of CXCL10, IL-6, TNF-α, IFN-γ, and MCP1 were compared between PLE- and mock-treated Calu3 cells [Fig. 2C]. Relative cytokine/chemokine mRNA levels were increased several fold upon viral infection [Fig. 2C]. Similar to remdesivir, 1.25 mg/mL PLE significantly decreased the relative cytokine/chemokine mRNA levels. The viral protein/RNA synthesis was highly positively correlated with cytokine levels, indicating that cytokine inhibition is closely associated with PLE-mediated reduction in viral replication [Fig. 2A–C].
Fig. 2.
PLE inhibits SARS-CoV-2 replication in Calu-3 cells. The expression of viral RNA and proteins (A–B) and cytokine mRNA (C) in Calu-3 cells was inhibited by PLE. Calu-3 cells were infected with SARS-CoV-2 in presence of various concentrations of PLE with remdesivir serving as positive control. Upon cell harvesting, RNA and viral protein quantification was performed with qRT-PCR (A) and western blotting (B), respectively. (A, C) Expression levels of viral or cytokine RNA were initially normalized to GAPDH mRNA. Moreover, the ratio measured in PLE-treated cells was normalized to the RNA level of virus control (arbitrarily set to 1). Data are expressed as means ± standard error of the mean from at least three independent experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005.
PLE inactivates viral particles and blocks their entry into host cells
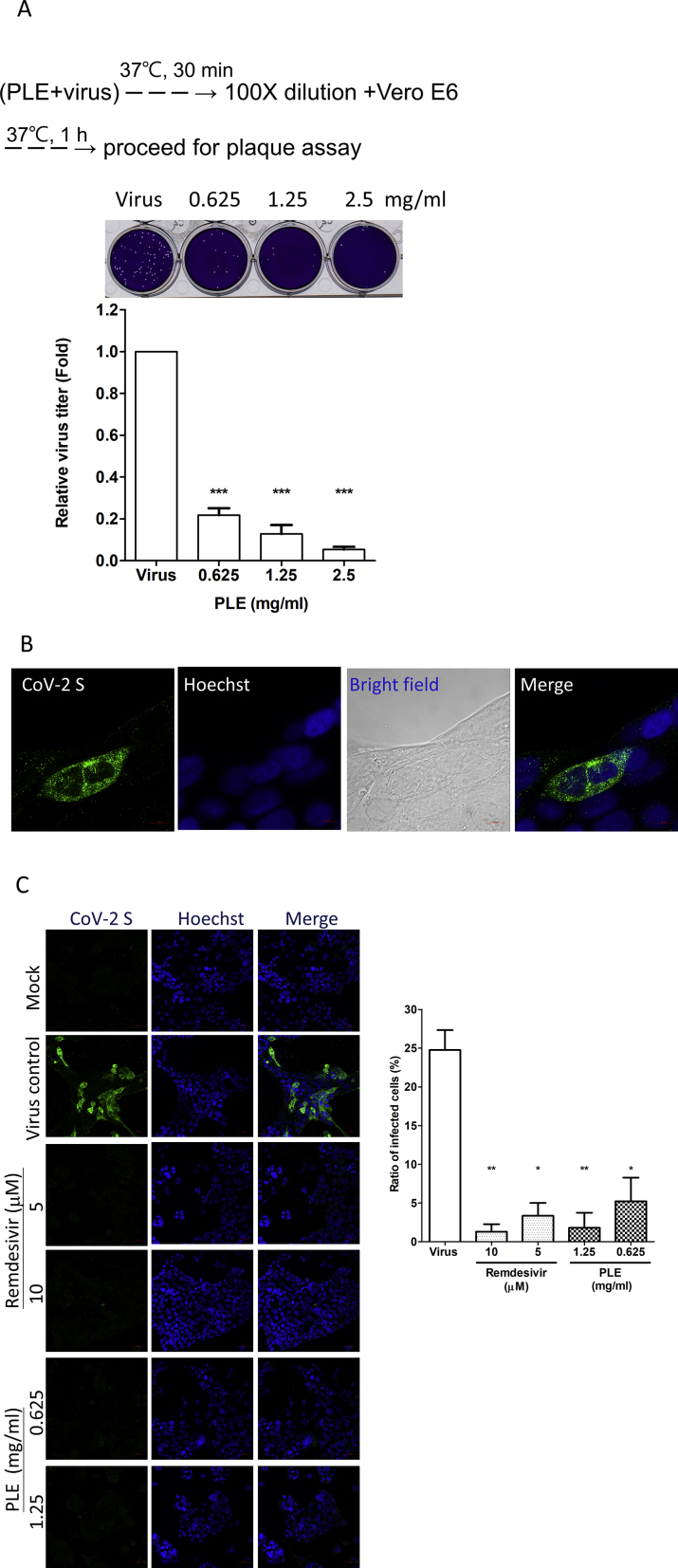
We observed that PLE inhibited the initial step of viral replication in a time-of-addition assay [Fig. 1]. We thus performed a viral inactivation assay to determine whether PLE can directly target the virion. Viruses subjected to PLE treatment showed significant dose-dependent decrease in their relative titers. Thus, PLE directly targets the virus and has virucidal efficacy [Fig. 3A]. We used immunofluorescence staining to evaluate the reduction in viral protein synthesis in infected cells treated with PLE [Fig. 3B,C]. [Fig. 3B] (bright field) shows Calu-3 cells clustered in a characteristic multinuclear structure. The immunofluorescence foci of the viral spike protein were evenly distributed in the cytoplasm at the site of viral replication. As shown in [Fig. 3C], the intensity of viral spike protein (left panels) and the ratio of infected cells (right panel) were markedly reduced in both remdesivir- and PLE-treated Calu-3 cells. Hoechst staining demonstrated that the observed reduction in immunofluorescent foci was not attributable to the absence of cells. While these morphological data are consistent with the Western blot results shown in [Fig. 2], they corroborate the hypothesis that PLE treatment inhibits viral protein synthesis and viral replication by inhibiting viral entry.
Fig. 3.
SARS-CoV-2 was inactivated by PLE. (A) The virus stock was pre-treated with increasing concentrations of PLE, and the remaining viral titers were subsequently determined using a plaque assay carried out in Vero E6 cells. The number of plaques for PLE-treated viruses was normalized to that of the virus control (arbitrarily set to 1). Data are expressed as means ± standard error of the mean from at least three independent experiments. (B–C) Confocal immunofluorescence microscopy revealed that PLE treatment reduced viral protein synthesis in Calu-3 cells. Cells treated with or without remdesivir and PLE were infected with SARS-CoV-2 at a MOI of 0.01. Cells were harvested at 48 h p.i. for confocal microscopy using the anti-S antibodies as indicated. Fluorescence images of S protein subcellular distribution in SARS-CoV-2-infected Calu-3 cells obtained either in absence (B) or presence (C) of inhibitors. Magnifications of objective lenses: 100 × (B) and 20 × (C). Nuclei were stained with a Hoechst dye. The transmitted light in the bright field revealed the overall morphology of Calu-3 cells. The bar chart in the right panel illustrates the ratios of infected cells under different experimental conditions (magnification of objective lens: 20 × ). For each condition, the ratio of spike-positive cells was calculated in two independent experiments from >200 cells in randomly selected fields. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005.
Synergy between PLE and remdesivir
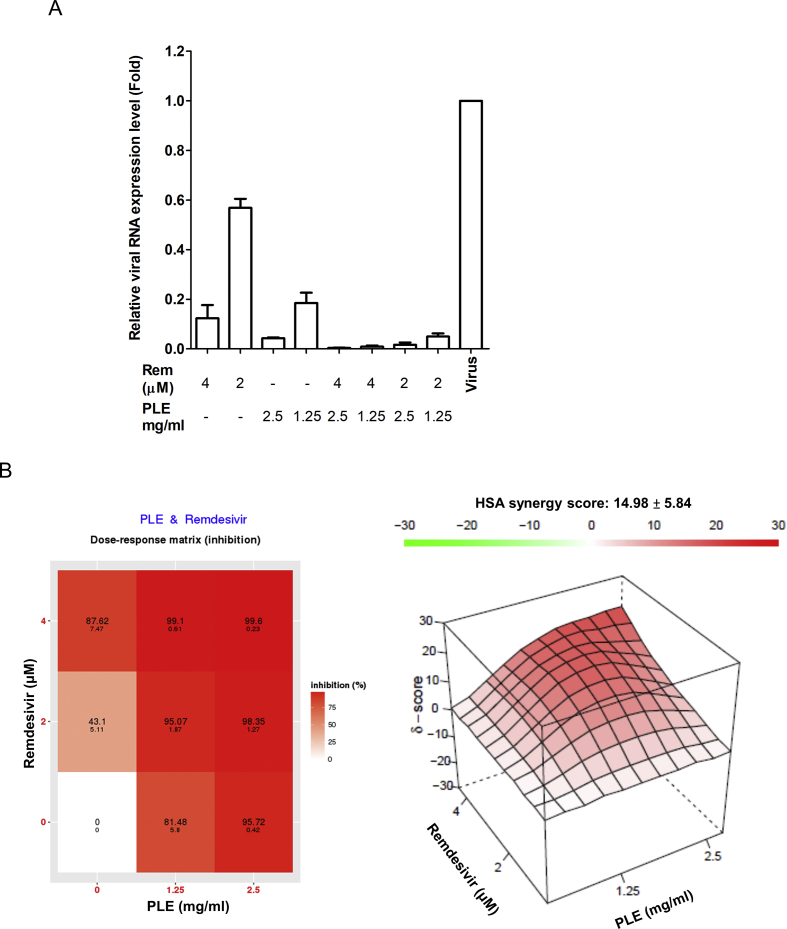
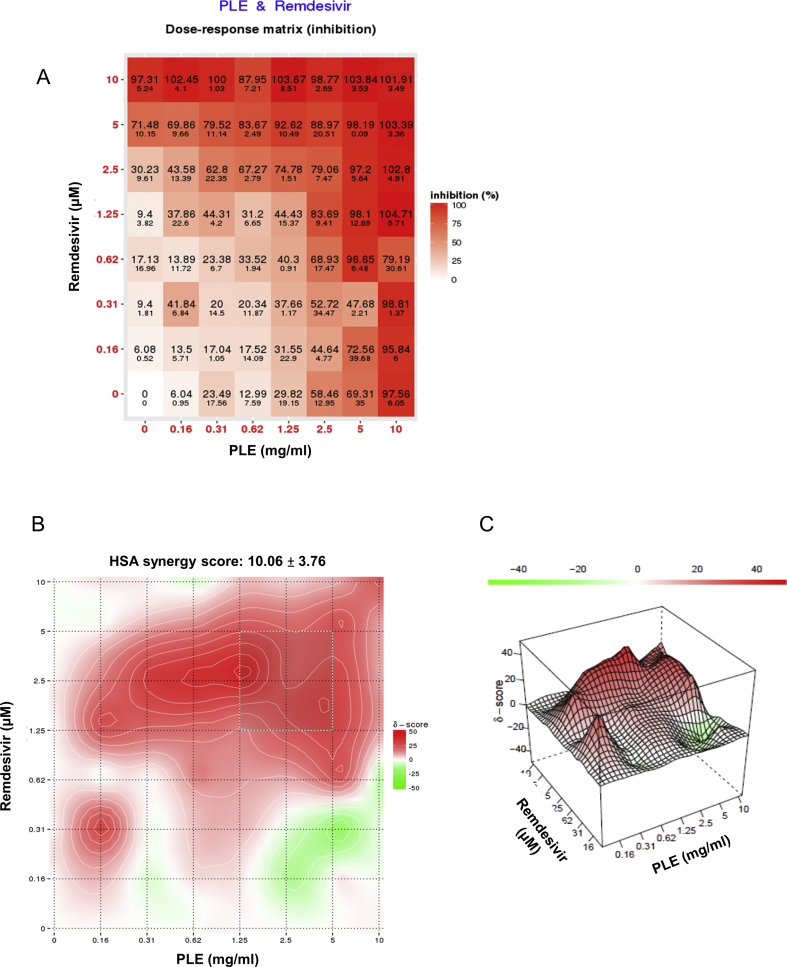
While remdesivir does not reduce COVID-19-related mortality, clinical trials have shown improvements in terms of recovery time. Hence, it is necessary to combine this drug with other therapeutic agents characterized by different modes of action. We assessed drug–drug interactions between PLE and remdesivir using a cell-based assay. We used qRT-PCR to evaluate the inhibition of viral RNA synthesis by drug combinations [Fig. 4A]. The results of data analysis obtained using SynergyFinder version 2 are presented in [Fig. 4B]. The combination of PLE and remdesivir had a synergistic score of 14.98 ± 5.84, indicating an additive-to-synergistic effect of PLE when combined with remdesivir [Fig. 4B]. We also used MTT staining to evaluate the inhibitory efficacy on cell viability by drug combinations (Figure S3). A similar additive-to-synergistic effect was evident (synergistic score: 10.06 ± 3.76). Taken together, these results provide a strong rationale for clinical trials investigating the potential utility of combinatory PLE and remdesivir treatment.
Fig. 4.
Antiviral activity of PLE and remdesivir used in combination (A). Vero E6 cells were infected with SARS-CoV-2 with serial dilutions of PLE in combination with remdesivir and subsequently harvested at 24 h p.i. to quantify viral RNA loads by qRT-PCR. Expression levels of viral RNA were initially normalized to GAPDH mRNA. The ratio of drug-treated cells was subsequently normalized to the RNA level of virus control (arbitrarily set to 1). (B) The ratio of drug inhibition elicited by the combination of PLE and remdesivir was calculated with the HSA reference synergy model available in the SynergyFinder software (version 2). The graphs illustrate average results from two independent experiments carried out in duplicate.
Discussion
COVID-19 has threatened human health on an unprecedented scale owing to the lack of specific anti-viral treatments. Recently, emergency use authorization has been granted to SARS-CoV-2 mRNA vaccines based on new technologies. Both Pfizer and Moderna vaccines – which have been developed on a nucleoside modified mRNA platform – induce immunogenicity to a stabilized membrane anchored spike protein. In addition, Regeneron antibodies (REGN-COV2) consist of an antibody cocktail comprising casirivimab and imdevimab – two monoclonal antibodies that are specifically directed against the spike protein of SARS-CoV-2. Pfizer and Moderna vaccines have been shown to confer a greater than 90 percent protection. REGN-COV2 induced a significant reduction in viral loads and mitigated clinical symptoms in patients with COVID-19; thus, it might act as a therapeutic substitute for the naturally-occurring immune response. However, the safety and duration of immunity conferred by these vaccines have not yet been fully elucidated. Additionally, children under the age of 12 years, African Americans, and immunocompromised patients were underrepresented in currently available trials. In this scenario, there is an urgent need to identify novel antiviral agents with satisfactory efficacy against this virus. In this study, we empirically demonstrated that PLE has potent anti-SARS-CoV-2 activity. Specifically, our results provide evidence that it blocks viral entry into the host cell by directly inactivating the virion. It also inhibits viral RNA and protein syntheses, as well as virus-induced cytokine production. Viral entry inhibitors offer several therapeutic advantages. Accordingly, they may directly attenuate or destroy the virus and can be relatively less cytotoxic than agents with other mechanisms of action as they need not permeate the host cell. In addition, they may also display a potential prophylactic efficacy. Intriguingly, PLE can be formulated not only into agonists but also as external preparations such as hand sanitizers and environmental disinfectants.
Health authorities in various provinces of mainland China have officially recommended the use of herbal formulas to control the spread of COVID-19 in the general population. The main functions of these herbal formulas are to strengthen qi, expel external pathogens, disperse wind, and discharge heat. To this end, a total of 54 herbs have been listed, and PLE was the sixth most common extract in all formulas [16]. However, the exact role of PLE in such herbal formulations when used for directly targeting the virus remains unknown. The TCM Huoxiang Zhengqi capsules/Shui contain PLE and other herbs have been administered to dissipate cold and eliminate dampness. Few studies have described the use of these capsules to treat patients with COVID-19 during the medical observation period [[17], [18], [19]]. A clinical trial of Huoxiang Zhengqi capsules combined with Western medicines for treating patients with COVID-19 reported an improved therapeutic efficacy [20]. As PLE is a component of this formulation, it might be responsible for its anti-SARS-CoV-2 activity. We tested PLE derived from different batches and commercial suppliers and found that all of them displayed a similar anti-SARS-CoV-2 activity. Taken together, these findings clearly indicate that all of these formulations contain the same pharmacologically active principle(s) and attempts to identify and quantify them are currently ongoing.
Dramatic relative changes in cytokine levels have been observed in patients with severe COVID-19 [[5], [6], [7], [8]]. The occurrence of a hyperinflammatory state and the accumulation of monocytes/macrophages and neutrophils in lung tissues are typical features of severe disease [6]. In this context, anti-inflammatory inhibitors such as IL-1 and IL-6 blockers can be therapeutically useful [21]. Expectedly, we observed elevated levels of different proinflammatory cytokines and chemokines (i.e., TNF-α, IFN-γ, IL-6, CCL-2/MCP1, and CXCL10/IP-10) in Calu-3 cells infected with SARS-CoV-2. However, PLE treatment inhibited all of these proinflammatory factors and alleviated the cytokine storm induced by SARS-CoV-2. It is therefore plausible that PLE could act as an anti-inflammatory inhibitor. Nevertheless, this hypothesis requires additional in vivo investigations.
Synergistic or additive drug combinations can reduce the required dose of each individual drug, mitigate toxicity and adverse reactions, and enhance overall therapeutic efficacy. The combination of compounds with at least two different antiviral mechanisms may also mitigate the risk of inducing drug-resistant viral mutants [22]. Certain drug combinations administered for treating HIV and HCV infections have shown remarkable therapeutic efficacy. Thus, remdesivir should be combined with other drugs to improve the overall survival rate of patients with COVID-19. Several attempts are currently ongoing to address this issue. Based on laboratory data and the results of a small randomized trial [23], the National Institute of Allergy and Infectious Diseases (USA) conducted the Adaptive COVID-19 Treatment Trial 3 to evaluate the safety and efficacy of a combination of remdesivir and IFN-1β in hospitalized patients with confirmed SARS-CoV-2 infection and lung involvement. Here, we demonstrate that the combination of PLE and remdesivir showed additive-to-synergistic efficacy against SARS-CoV-2 in Vero-E6 cells. Because PLE has been widely used in Asian countries to treat patients with various viral infections, our findings should prompt additional clinical investigations of this extract as an adjunct to remdesivir in patients with COVID-19.
Conclusion
In this study, we have shown that PLE is capable of preventing SARS-CoV-2 entry into host cells. Our findings may pave the way to the clinical use of this extract for preventing and/or treating COVID-19. Notably, perilla has a widespread distribution and large amounts of its herbal extracts can easily be obtained from intensive farming. While the exact active ingredients of PLE have not been elucidated yet, clinically useful preparations can easily be obtained following standard protocols and formulated in oral supplements or medicated topical products. Finally, the use of PLE can theoretically alleviate the ongoing shortage of prescription drugs such as remdesivir.
Authors’ contribution
W.-F. Tang, H.-P. Tsai, Y.-H. Chang, T.-Y. Chang, C.-F. Hsieh, C.-Y. Lin, G.-H. Lin, Y.-L. Chen, J.-R. Jheng, P.-C. Liu, C.-M. Yang, and Y.-F. Chin performed the experiments and conducted data analysis. H.-P. Tsai, Y.-H. Chang, T.-Y. Chang, P.-C. Liu C.-M. Yang, C.-C. Chen, Y.-F. Chin, J.-H. Kau, Y.-J. Hung, and P.-S. Hsieh were in charge of high-security experiments. W.-F. Tang and J.-T. Horng designed the study, analyzed and interpreted data, and drafted the paper. All authors have read and approved the final manuscript.
Funding
This research was funded by the Chang Gung Memorial Hospital, Taoyuan, Taiwan (BMRP416, CMRPD1G0301-3, CMRPD1F0581-3, and CMRPD1K0241-2), the Ministry of Science and Technology of Taiwan (106-2320-B-182-004-MY3, 106-2811-B-182-011, 106-2632-B-182-001, 107-2811-B-182-512, 108-2320-B-182-039-, 109-2320-B-182-026 -MY3, 109-2327-B-182-002, and 109-2327-B-182-003- to J.-T. Horng and 109-2327-B-016 -002 – to C.-C. Chen), the Research Centre for Emerging Viral Infections from The Featured Areas Research Centre Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan, and the Ministry of Science and Technology, Taiwan (MOST 109-2634-F-182-001).
Availability of data and materials
The analytical methods and all study materials will be made available upon publication of the study. The original data set can be obtained from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Not applicable.
Conflicts of interest
The authors declare they have no actual or potential competing financial interests.
Acknowledgements
The authors are grateful to Scintica Instrumentation Inc. for the kind gift of the CytoSMART Lux2 instrument. The authors thank Drs. Shin-Ru Shih, Pei-Wen Hsieh, and Tsong-Long Hwang for their constructive suggestions. We acknowledge the help of Dr. Ming-Chung Lee (Brion Research Institute) in botanical identification experiments. We also indebted to the Institute of Preventive Medicine, National Defense Medical Center, for providing biosafety level three and four laboratory in which SARS-CoV-2 was handled.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2021.01.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
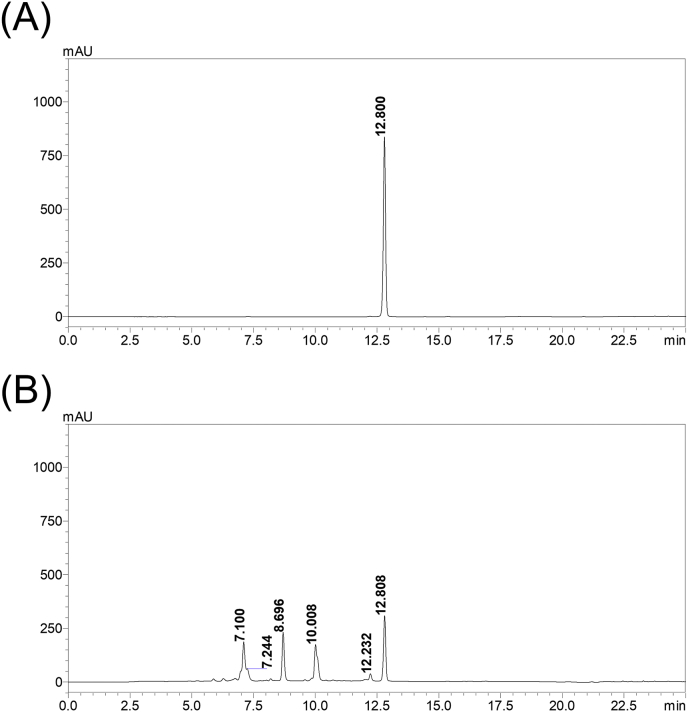
Figure S1. PLE fingerprint analysis. PLE and rosmarinic acid were analyzed using a Shimadzu Nexera-I LC-2040C-3D HPLC system (Kyoto, Japan) with a Cosmosil 5C18-MS-II HPLC column (4.6 × 250 mm; Nacalai Tesque, Inc., Kyoto, Japan). PLE (50 mg/mL) and rosmarinic acid (5 μg/mL) were filtered through a 0.22-μm filter before analysis. Solvents consisted of 0.1% formic acid aqueous solution and acetonitrile. An acetonitrile gradient elution was carried out at the following conditions: 15−40%, 0−18 min; 40−100%, 18−20 min; 100%, 20−25 min, 100−15%, 25−35 min. The flow rate was 1 mL/min, whereas the detection wavelength and the injection volume were 330 nm and 20 μL, respectively. (A) Rosmarinic acid was detected at a retention time of 12.800 min; (B) Rosmarinic acid in PLE was detected at a retention time of 12.808 min.
figs2.
Figure S2. Video clip of the cytopathic effect assay obtained using a real-time CytoSmart Lux2 imaging system. The filming was carried out over a 48-h timeframe. Vero-E6 cells were infected with SARS-CoV-2 at a MOI 0.01 either in the presence or absence of PLE.
figs3.
Figure S3. Antiviral activity of PLE and remdesivir as observed in the combination assay. Vero E6 cells were infected with SARS-CoV-2 in the presence of serial dilutions of PLE combined with remdesivir. Cells were subsequently harvested at 72 h p.i. and cell viability was quantified with the MTT assay. The absorbance of drug-treated cells was normalized to mock-infected cells (arbitrarily set to 1). The ratio of drug inhibition elicited by the combination of PLE and remdesivir was calculated with the HSA reference synergy model available in the SynergyFinder software (version 2). The dose-response plot (A) as well as the two-dimensional (B) and three-dimensional (C) synergy maps are reported. Results should be considered representative of those obtained from two independent experiments carried out in duplicate.
figs4.

figs5.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masters P.S., Perlman S. Coronaviridae. In: David M., Knipe P.M.H., Martin Malcolm A., Griffin Diane E., Lamb Robert A., Bernard Roizman, Straus Stephen E., editors. Fields virology. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 825–858. [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rochwerg B., Agarwal A., Zeng L., Leo Y.S., Appiah J.A., Agoritsas T. Remdesivir for severe covid-19: a clinical practice guideline. BMJ. 2020;370:m2924. doi: 10.1136/bmj.m2924. [DOI] [PubMed] [Google Scholar]
- 10.Ahmed H.M. Ethnomedicinal, phytochemical and pharmacological investigations of perilla frutescens (L.) Britt. Molecules. 2018;24:102. doi: 10.3390/molecules24010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong Y.N., Tsao K.C., Hsiao M.J., Huang C.G., Huang P.N., Huang P.W. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg Microb Infect. 2020;9:1457–1466. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C.W., Leu Y.L., Horng J.T. Daphne Genkwa sieb. Et zucc. Water-soluble extracts act on enterovirus 71 by inhibiting viral entry. Viruses. 2012;4:539–556. doi: 10.3390/v4040539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Ianevski A., He L., Aittokallio T., Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics. 2017;33:2413–2415. doi: 10.1093/bioinformatics/btx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo H., Tang Q.L., Shang Y.X., Liang S.B., Yang M., Robinson N. Can Chinese medicine Be used for prevention of corona virus disease 2019 (COVID-19)? A review of historical classics, research evidence and current prevention programs. Chin J Integr Med. 2020;26:243–250. doi: 10.1007/s11655-020-3192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Y.Y., Zhao M.R., Shi B., Song Z.H., Zhou S.P., He Y. Application of integrative therapy protocols for treatment of coronavirus disease 2019 (COVID-19) Chin Tradit Herb Drugs. 2020;4:878–882. [Google Scholar]
- 18.General Office of National Health Committee Notice on the issuance of guidelines of diagnosis and treatment for 2019-nCoV infected pneumonia. 2000. http://bgs.satcm.gov.cn/zhengcewenjian/2020-02-06/12847.html./ [accessed 6 February 2020]
- 19.Zhu Y.G., Deng Z.W., Liu L.H., Liu X.H., Li X.Z., Chen W.H. Compilation of drug information for the diagnosis and treatment of COVID-19 (version 1) Cent S Pharm. 2020;18:1–14. [Google Scholar]
- 20.Xiao M., Tian J., Zhou Y., Xu X., Min X., Lv Y. Efficacy of Huoxiang Zhengqi dropping pills and Lianhua Qingwen granules in treatment of COVID-19: a randomized controlled trial. Pharmacol Res. 2020;161:105126. doi: 10.1016/j.phrs.2020.105126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu L., Zhang H., Zhan M., Jiang J., Yin H., Dauphars D.J. Preventing mortality in COVID-19 patients: which cytokine to target in a raging storm? Front Cell Dev Biol. 2020;8:677. doi: 10.3389/fcell.2020.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsiodras S., Mooney J.D., Hatzakis A. Role of combination antiviral therapy in pandemic influenza and stockpiling implications. BMJ. 2007;334:293–294. doi: 10.1136/bmj.39105.428981.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahmani H., Davoudi-Monfared E., Nourian A., Khalili H., Hajizadeh N., Jalalabadi N.Z. Interferon beta-1b in treatment of severe COVID-19: a randomized clinical trial. Int Immunopharm. 2020;88:106903. doi: 10.1016/j.intimp.2020.106903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analytical methods and all study materials will be made available upon publication of the study. The original data set can be obtained from the corresponding author upon reasonable request.