Abstract
Photosynthetic light-harvesting complexes (LHCs) of higher plants, moss, and green algae can undergo dynamic conformational transitions, which have been correlated to their ability to adapt to fluctuations in the light environment. Herein, we demonstrate the application of solid-state NMR spectroscopy on native, heterogeneous thylakoid membranes of Chlamydomonas reinhardtii (Cr) and on Cr light-harvesting complex II (LHCII) in thylakoid lipid bilayers to detect LHCII conformational dynamics in its native membrane environment. We show that membrane-reconstituted LHCII contains selective sites that undergo fast, large-amplitude motions, including the phytol tails of two chlorophylls. Protein plasticity is also observed in the N-terminal stromal loop and in protein fragments facing the lumen, involving sites that stabilize the xanthophyll-cycle carotenoid violaxanthin and the two luteins. The results report on the intrinsic flexibility of LHCII pigment-protein complexes in a membrane environment, revealing putative sites for conformational switching. In thylakoid membranes, fast dynamics of protein and pigment sites is significantly reduced, which suggests that in their native organelle membranes, LHCII complexes are locked in specific conformational states.
Significance
Photosynthetic light-harvesting complexes (LHCs) undergo dynamic conformational transitions that regulate the capacity of the light-harvesting antenna. We demonstrate the application of solid-state NMR spectroscopy to investigate the structural dynamics of LHCII, the most abundant LHC of plants and algae, in native membranes. Selective dynamic protein and pigment residues are identified that are putative sites for a conformational switch.
Introduction
Multipigment-protein complexes in plants, moss, and photosynthetic algae perform delicate photophysical and chemical tasks. To maintain a homeostatic balance in fluctuating sunlight conditions, these processes are tightly regulated by dynamic membrane responses to prevent photodamage (1,2). The most abundant light-harvesting complexes (LHCs), the peripheral antenna complexes LHCII, have been studied extensively for their molecular role in light-harvesting regulation. Through Raman spectroscopy of the LHCII xanthophylls, it was revealed that isolated LHCIIs can undergo conformational changes in vitro and that they respond to excess light by undergoing similar conformational changes inside chloroplast organelle membranes called thylakoids (3). Fluorescence studies have shown that individual LHCIIs have the capacity to switch between light-harvesting and photoprotective, excitation-quenched states (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12). This concept has led to fluctuating-antenna models for describing excitation energy migration over a two-dimensional (2D) protein lattice (13,14). The LHCII crystal structures show selective sites with high B-factors. The structural dynamics of LHCII has been characterized with use of electron paramagnetic resonance spin labeling combined with molecular modeling (MD) (4,10,15,16). An MD study on monomeric LHCII in lipid membrane was also applied to correlate protein fluctuations to functional changes in pigment-pigment interactions (9). Those studies, however, do not capture the structural elements of native chloroplast membranes. In thylakoid membranes, many LHCIIs are held in specific arrangements within LHCII-photosystem-II supercomplexes (17). The surrounding lipids and free xanthophylls that are present in thylakoid membranes further control the conformational dynamics of the membrane-embedded proteins (18). Furthermore, in living plants or algae, reversible phosphorylation takes place during state transitions that involve membrane rearrangements and a redistribution of LHCII over photosystem I and photosystem II (19).
The LHCII trimer complexes are isomers consisting of different pigment-bound polypeptides with overlapping sequences. The different polypeptides have been found to have distinct roles in photoprotection and state transitions (20,21), suggesting that molecular recognition plays a role in LHCII regulatory functions. Phosphorylation will change the local structure of LHCII and may modify specific recognition sites (22). The phosphorylation sites are located in the N-terminus of LHCII, which is not resolved in the crystal structures (10,15). Protein-associated galactosyl lipids further have shown to play a role in stabilizing LHCII complexes and their interactions with partner proteins (23) as well as in controlling the supramolecular membrane architectures (24). Little is known about the functional roles of selective thylakoid lipids and how their dynamic interactions may contribute to the regulation of light harvesting.
Until now, no structure-based methods have been presented to experimentally probe the dynamics of LHCII inside a native lipid membrane environment. We, here, apply solid-state NMR spectroscopy to study the conformational dynamics of LHCII in lipid bilayers and in native thylakoid membranes. Solid-state NMR is a powerful tool for atomistic detection of membrane proteins, and in some studies, NMR signals of membrane proteins could even be detected in native membrane or cellular environments by using overexpression of the target proteins in prokaryotic and eukaryotic host-expression systems (25, 26, 27, 28, 29, 30, 31). In the following, we recorded NMR spectra of isolated LHCIIs that were reconstituted in lipid bilayers prepared from native-like lipid mixtures. In addition, we made use of the fact that LHCIIs are abundant in natural chloroplast membranes and performed NMR experiments directly on chloroplast thylakoid membranes containing LHCII that were isolated from wild-type U-13C Chlamydomonas reinhardtii (Cr) cells. Strong overlap is observed between the spectrum of LHCII proteoliposomes and that of the thylakoid membranes, which allowed us to assess the dynamics of LHCII in its native environment.
Materials and Methods
Biosynthetic isotope labeling of Cr cells and sample preparation
For the experiments on isolated LHCII trimers, Cr cells from strain cw15 were cultivated in Erlenmeyer flasks with liquid Tris-acetate phosphate (TAP) medium at 100 rpm agitation and 21°C in a growth chamber. For optimal labeling of the LHCII complexes, cells were grown under mixotrophic conditions under dim light to minimize the uptake of 12CO2 from the air. Continuous illumination was provided from cool-white fluorescent lamps under low (<25 μmol photons m−2 s−1), photosynthetically active radiation (400–700 nm). The TAP medium (32) used to grow labeled cells was prepared using 13C-labeled sodium acetate and 15N-labeled ammonium chloride (Sigma-Aldrich, Zwijndrecht, the Netherlands). Cultures in labeled medium were set up starting from an optical density at 750 nm equal to 0.1 and were grown until OD750 = 1. Three rounds of cultivation in labeled medium were performed to ensure more than 95% labeling of the cells with 13C and 15N atoms. Thylakoid membranes were then isolated under previously described conditions (33). After the isolation, Cr thylakoids were resuspended in buffer (50 mM Hepes-KOH (pH 7.5) and 5 mM MgCl2 with 50% glycerol). For isolation of the LHCII fractions, thylakoid membranes corresponding to 3 mg/mL of total chlorophylls (Chls), according to the optical density at 680 nm, were washed with 50 mM EDTA and solubilized for 20 min on ice in 3 mL of final 1.2% n-dodecyl α-D-maltoside (α-DM) in 10 mM Hepes (pH 7.5) after vortexing for 1 min. The solubilized samples were centrifuged at 15,000 × g for 30 min to eliminate any insolubilized material, and the supernatant with the photosynthetic complexes was then fractionated by ultracentrifugation in a 0–1-M sucrose gradient containing 0.06% α-DM and 10 mM Hepes (pH 7.5) at 141,000 × g for 40 h at 4°C. The green fraction (see Document S1. Figs. S1–S15 and Tables S1–S4, Document S2. Article plus Supporting Material D) corresponding to LHCII proteins was harvested with a syringe, and the Chl concentration was adjusted to 2 mg/mL with a buffer (50 mM Hepes and 5 mM MgCl2 (pH 7.5)). LHCII proteins solubilized in α-DM were reconstituted in lipid membranes whose composition mimics the native thylakoid membrane (47% monogalactosyldiacylglycerol (MGDG), 12% sulfoquinovosyldiacylglycerol (SQDG), 14% phosphatidylglycerol (PG), and 27% digalactosyldiacylglycerol (DGDG)) with a protein/lipid molar ratio of 1:55 according to the method described in Crisafi and Pandit (34). The chosen protein/lipid ratio is in the range of native protein packing densities in thylakoid membranes, in which 70–80% of the membrane surface area is occupied with proteins (35).
For experiments on whole fresh thylakoid membranes, Cr cells were cultivated in TAP medium using 13C-labeled sodium acetate in a home-built photo chamber under continuous illumination with cool-white LEDs (∼50 μmol m−2 s−1). Cells were harvested in the exponential growth phase, centrifuged, and resuspended in 0.2 volumes of MgCl2 buffer (1 mM MgCl2, 0.1 M Hepes (pH 7.5)/KOH, and 10% sucrose) and were ruptured by sonication on a 2500-W sonicator set at 10%. The isolation of fresh thylakoids was performed according to Chua and Bennoun (36) with some modifications. This procedure differed from the steps described above for LHCII isolation by using sucrose gradient layers for purification of the thylakoid membranes to obtain more pure fractions. In the procedure, disrupted cells were overlaid with layers of sucrose (1 mL of 1.3 M, 1 mL of 0.5, and 0 M sucrose). The gradients were ultracentrifuged for 1 h at 4°C in an SW41 swing rotor at 24,000 rpm (100,000 × g). The thylakoid fraction was isolated from the dark-green sucrose band (see Fig. S2 A). With the used isolation procedure, membrane stacking is preserved.
For the LHCII NMR sample, 18 mL of LHCII in liposomes, containing ∼10 mg LHCII and 1.5 mg Chl (as determined by OD680 of the Chls), was pelleted by ultracentrifugation (223,000 × g, 4°C, 90 min) and transferred to a thin-wall, 3.2-mm solid-state NMR magic-angle spinning (MAS) rotor through centrifugation. For the thylakoid membrane NMR sample, 12 mL of freshly isolated thylakoid membrane containing 2 mg Chl and ∼10 times more in protein content was pelleted by ultracentrifugation (100,000 × g, 4°C, 45 min) and transferred to a thin-wall, 3.2-mm MAS rotor.
Gel electrophoresis
Coomassie-stained sodium-dodecyl-sulphate -polyacrylamide-gel-electrophoresis (SDS-PAGE) was performed using 15% Tris-glycine gels (37). Samples were solubilized with a solubilization buffer (4×) containing 30% glycerol, 125 mM Tris (pH 6.8), 0.1 M dithiothreitol, and 9% SDS.
Time-resolved and 77-K fluorescence spectroscopy
Time-resolved fluorescence measurements on U-13C-15N LHCII in α-DM and on the LHCII proteoliposomes were performed using a FluoTime 200 (PicoQuant, Berlin, Germany) time-correlated photon-counter spectrometer. Samples were held in a 1 × 1 cm quartz cuvette that was thermostated at 20°C and excited at 440 nm using a diode laser (PicoQuant), 440-nm excitation. Fluorescence decay traces were fitted with multiexponentials using a χ2 least-square fitting procedure. 77-K fluorescence measurements were performed using a Fluoromax 3 spectrophotometer (Horiba, Jobin-Yvon, France). The samples were diluted in 50 mM Hepes and 5 mM MgCl2 buffer and cooled in a nitrogen-bath cryostat to 77 K. Excitation was performed at 440 nm, and a bandwidth of 2 nm was used for selecting the excitation and emission wavelengths.
Solid-state NMR experiments
Solid-state NMR spectra of U-13C-15N LHCII in proteoliposomes and of 13C-enriched thylakoid membranes were recorded on an ultrahigh-field, 950-MHz 1H Larmor frequency spectrometer (Bruker BioSpin, Billerica, MA) equipped with a triple-channel (1H, 13C, and 15N), 3.2-mm MAS probe. Typical π/2 pulses were 3 μs for 1H, 5 μs for 13C, and 8 μs for 15N. The 1H/15N and 1H/13C cross-polarization (CP) (38) contact times were 800 μs and 1 ms, respectively, with a constant radio frequency field of 35 and 50 kHz on nitrogen and carbon, respectively, whereas the proton lock field was ramped linearly around the n = 1 Hartmann-Hahn condition (39). The 15N-13Ca SPECIFIC-CP transfer (40) was implemented with an optimized contact time of 4.2 ms with a constant spin lock field of 2.5 × νr applied on 15N, whereas the 13C field was ramped linearly (10% ramp) around 1.5 × νr. 1H decoupling during direct and indirect dimensions was performed using SPINAL64 (41) with ∼83-kHz irradiation. The presented 2D 13C-13C PARIS (42) spectra were collected with a mixing time of 30 ms at 17-kHz MAS at a set temperature of −18°C. The 2D NCA and NCACX experiments (43) were performed on the LHCII sample at a 14-kHz MAS frequency and a readout temperature of −18°C. For the NCACX experiment, a PARIS (13C,13C) mixing time of 50 ms was used. The J-coupling-based 2D 13C-13C INEPT-TOBSY (44,45) experiments were recorded at −3°C with a TOBSY mixing time of 6 ms at 14-kHz MAS. All spectra were processed using Bruker TopSpin software version 3.2 (Bruker BioSpin) with linear prediction and Fourier transformed in forward quadrature complex mode. Spectra were analyzed by Sparky version 3.114 (46) and MestReNova 11.0 (Mestrelab Research SL, Santiago de Compostela, Spain).
NMR chemical-shift prediction using a plant-LHCII homology model
Homology models of Cr LHCII were built using the SWISS model web server (47) based on the LHCII crystal structure of spinach and using the Lhcbm1 or Lhcbm2 sequence of Cr LHCII (48). The Lhcbm sequences and respective Protein Data Bank (PDB) models were used as input for SHIFTX2 (49) to predict the 13C and 15N chemical shifts. 13C-13C predicted correlation spectra for use in Sparky (46) were generated by FANDAS (50).
Results
A biochemical analysis of our thylakoid preparations (Figs. S1 and S2) shows that the LHCII trimers are the most abundant pigment-containing complexes in the thylakoid membranes, whereas the higher-weight bands in the SDS-PAGE contain the photosystems, ATP synthase, and cytochrome b6f complexes. Different molecular-weight bands are distinguished for LHCII because of the fact that the LHCII trimers are isomers consisting of different polypeptides of which the most abundant types are Lhcbm1, Lhcbm2/7 (Lhcbm2 and Lhcbm7 have identical mature peptide sequences), and Lhcbm3 (51). Our LHCII proteoliposomes contain the lipids MGDG, DGDG, SQDG, and the phospholipid PG at a protein/lipid ratio of 1:55 (mol/mol) to mimic the protein density and lipid composition of the native thylakoid membranes (52). The 77-K fluorescence spectrum of the proteoliposomes shows a prominent band at 700 nm (Fig. S3), which is characteristic for LHCII aggregates. Aggregation of LHCII in liposome membranes is associated with the formation of quenched states (34,53). Indeed, a fluorescence lifetime analysis gives an average lifetime of 0.7 ns for LHCII proteoliposomes compared with a lifetime of 3.5 ns for LHCII in α-DM detergent (Fig. S4; Table S1).
Two complementary types of solid-state NMR experiments were employed to distinguish rigid and highly dynamic molecular species and protein subdomains. In CP-based experiments, 1H-13C or 13C-15N magnetization is transferred via the dipolar interaction. Dipolar-based transfer experiments become inefficient for dynamic molecules or molecular segments that display submicrosecond motions because the dipolar couplings are averaged because of the fast overall or local motion. Such dynamic species remain visible in NMR experiments such as INEPT (10) and TOBSY (9), in which magnetization is transferred via J couplings. The combination of such J-based pulse sequences can hence be employed for screening very flexible parts of large biomolecules that undergo large-amplitude motions on picosecond-to-microsecond timescales (54). On the other hand, membrane proteins typically contain long transmembrane domains with limited flexibility, which are predominantly detected in dipolar-based experiments (54).
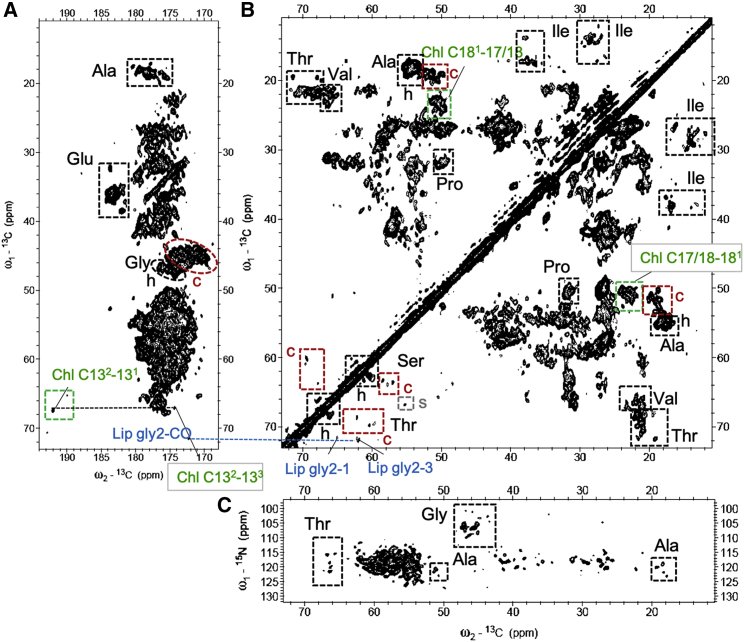
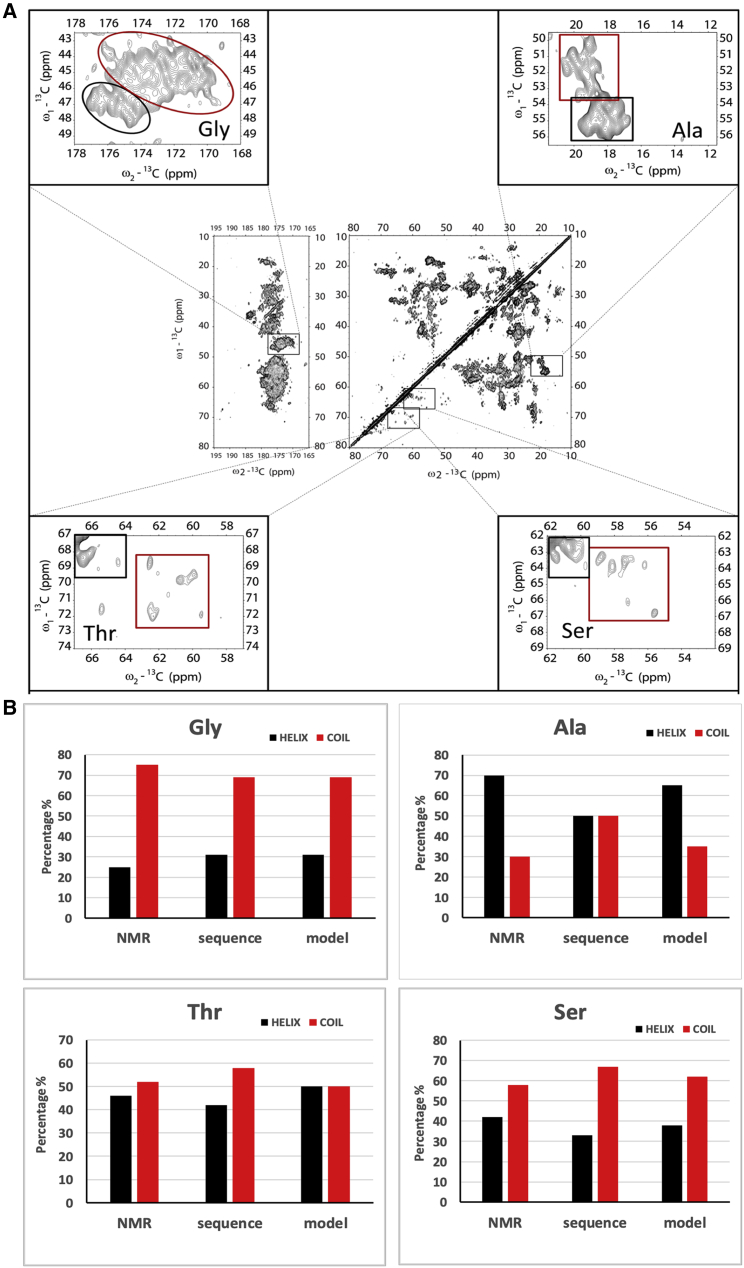
Dipolar-based 13C-13C (CC) and 15N-13C (NC) experiments of LHCII in proteoliposomes are shown in Fig. 1 for the protein regions, and the selective pigment and lipid regions are shown and discussed further down in Fig. 3. The CP-PARIS CC spectrum verifies that our sample contains pigment-protein complexes of which all components are uniformly labeled. For clarity, the full spectrum in the aliphatic region without annotations is also shown in Fig. S5. Resonance signals of specific amino acids, i.e., the Cα-Cβ correlations of threonine (Thr), serine (Ser), and alanine (Ala) and the Cα-C′ correlations of glycine (Gly) can be identified and classified as helix or coil based on their unique chemical-shift patterns and clear spectral separation of their Cα-Cβ (Thr, Ser, and Ala) or Cα-C′ (Gly) correlation signals in the CC spectrum. We use those amino acids as spectroscopic markers. To evaluate if their correlation patterns agree with the known secondary structure of LHCII, integrated NMR signal intensities in helix and coil regions of Thr, Ser, Ala, and Gly are presented in Fig. 2. The NMR-estimated coil and helix contributions are compared with the predicted helix and coil contents according to the amino acid sequence and according to a structural model that was build based on the plant-LHCII crystal structures (10,15) using the amino acid sequence of Lhcbm1, one of the most abundant polypeptides of Cr LHCII.
Figure 1.
NMR spectra of LHCII proteoliposomes. (A and B) Shown is the 13C-13C CP-PARIS spectrum in the aliphatic (A) and carbonyl (B) region; (C) Shown is the NC NCACX spectrum. Identified clusters of amino-acid types are indicated with boxes. Helix, coil, and strand correlations of Thr, Ser, Ala, and Gly are indicated in black, red, and gray in the 13C-13C spectrum. Chl and lipid correlations are indicated in green and blue. To see this figure in color, go online.
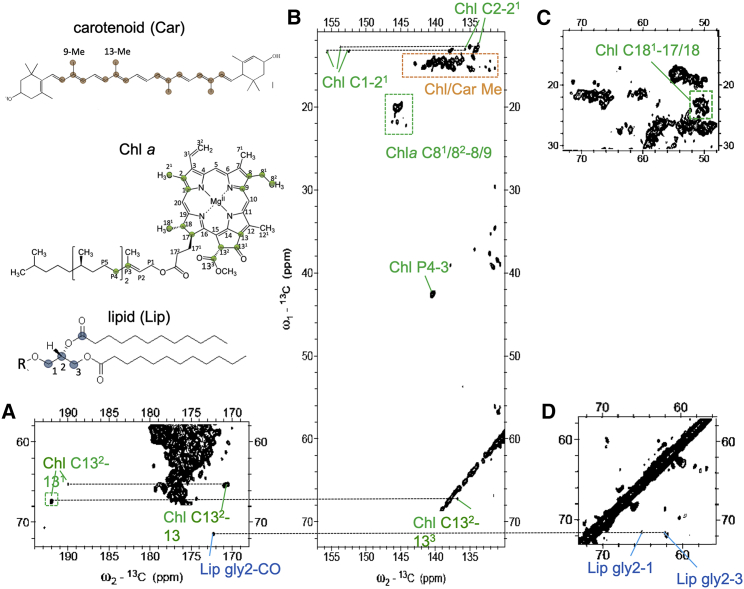
Figure 3.
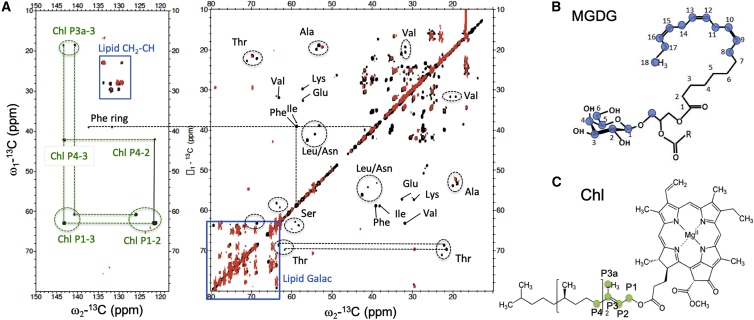
Pigment and lipid correlations in the PARIS 13C-13C NMR spectrum of LHCII proteoliposomes. Shown are the insets of the carbonyl (A), aromatic (B), and aliphatic (C and D) region, with identified chlorophyll (Chl), carotenoid, and lipid correlations indicated in green, orange, and blue. The structures of carotenoid (lutein), of Chl α and of a glyco- or phospholipid are drawn in the left side of the figure, with their attributed carbon atom types color coded. To see this figure in color, go online.
Figure 2.
NMR-estimated helix and coil percentages of Gly, Ala, Thr, and Ser residues. (A) Shown are the insets of the 13C-13C PARIS spectrum containing Gly, Ala, Thr, or Ser correlation signals. The helix and coil regions are indicated with black and red boxes, respectively. (B) Given are the helix and coil percentages according to the NMR, Lhcbm1 sequence, and Cr LHCII homology model. To see this figure in color, go online.
The structural model lacks the first 13 residues in the N-terminus of LHCII because this part is not resolved in the LHCII crystal structures and therefore has lower numbers of coil amino acids than the full Lhcbm1 sequence. Comparing the NMR-estimated helix and coil contents of Gly, of which the amino acid types are abundant in LHCII and evenly distributed over the Lhcbm1 sequence (see Fig. S6), the estimated fractions agree with those anticipated and confirm that the LHCII complexes have well-folded secondary structures. For Ser and Thr, the NMR-estimated contents also reasonably agree with the anticipated numbers. One Thr is at the edge of a 310-helix fragment on the luminal site and may adopt a coil conformation. For Ala, the NMR-estimated helix and coil contents agree with the model, but the estimated coil content is lower than anticipated based on the full sequence. The full sequence contains additional Ser, Thr, and Ala amino acids in the N-terminus (2 × S, 2 × T, and 3 × A). We suspect that the flexible N tail is not detected in the NMR CP-PARIS spectrum, in agreement with earlier work on 13C-15N-Arg Cr LHCII, in which the number of detected Arg signals matches with the anticipated number based on the structure if the N-terminal part was left out (55). No Gly amino acids are found in the N-terminus of Lhcbm1 so that the model and the full sequence contain the same helix and coil Gly contents. In Fig. S7, NMR-estimated helix and coil contents are also compared with those calculated from the structure of Lhcbm2/7 and to the model and full sequence of Lhcbm3. Lower agreement is observed for the Ala and Ser helix/coil contents than when compared with Lhcbm1 (model1), and the model of Lhcbm3 gives a better match than the full sequence. The data in Fig. S7 suggest that in our LHCII preparations, Lhcbm1 is dominant and, again, that signals from the N tail are not visible in the CP-PARIS spectrum.
Next to signals originating from the protein content, Chl and carotenoid signals can be identified in regions of the CC spectrum where no protein signals are expected. They are shown in Fig. 3, which shows enlarged regions of the LHCII CC spectrum where Chl, carotenoid, and lipid signals occur. Signals are identified from several Chl macrocycle ring carbons (C1, C2, C8, C9, C13, C131, C132, and C17/18) and their neighboring side-chain atoms (C21, C81, C82, C133, and C181). The C8, C9, C81, and C82 signals are specific for the side chain Chl a, whereas the other signals could be either from Chl a or b. In addition, signals are detected of atoms P3 and P4 in the Chl tail. Strong signals of carotenoids are observed that are easily distinguished from lipid signals owing to their correlations between the conjugated chain and methyl (Me) resonances of carotenoids, accumulating between 15 and 19 ppm (ω1) and 130–145 ppm (ω2) (Fig. 3 B). The Chl correlations of macrocycle ring atoms with side-chain atoms C71 and C121 also fall in this region. A single set of lipid glycerol carbon resonances is observed. It is well-known that each monomer unit of LHCII contains one PG lipid, which forms the ligand for Chl611 (nomenclature Liu et al. (15)). Based on their chemical shifts, we attribute the detected lipid resonances to the head carbons of LHCII-bound PG lipid molecules. From its appearance in a CP-based spectrum, it can be deduced that this lipid does not exchange on a typical NMR timescale (∼0.1 ms) and is a structural lipid.
We now compare the NMR spectrum of LHCII in liposomes with the NMR spectrum of thylakoid membranes containing LHCII. To assess the overall dynamics of our proteoliposome and thylakoid membrane sample preparations, we first compared the relative intensities of one-dimensional (1D) 13C MAS NMR spectra obtained through 1H-13C CP with those obtained through a 90° pulse with direct polarization (DP) of the carbons (Fig. S8). The spectra were recorded at −3°C to include NMR signals of mobile sites. For pure solids, the CP signal intensities will be ∼4× enhanced compared with the DP signal intensities based on the gyromagnetic ratio of 1H relative to 13C, whereas for dynamic molecules, the relative CP signal intensities will be lower because polarization transfer becomes less efficient. For both samples, the CP and DP signal intensities in the aliphatic region (0–60 ppm) that include the Cα, Cβ, and other amino acid side-chain atom signals are similar, whereas the DP spectrum is enhanced in the aromatic (110–130 ppm) and carbonyl (170–180 ppm) region. The additional peaks in the DP spectrum contain those of fatty acids and carbohydrate atoms from the galactosyl lipid heads. The relative CP and DP intensities indicate that the samples are not real solids and, rather, form gel-like states. From the similar results for the LHCII proteoliposome and the thylakoid sample, we conclude that conditions in terms of overall protein dynamics for the two samples are comparable.
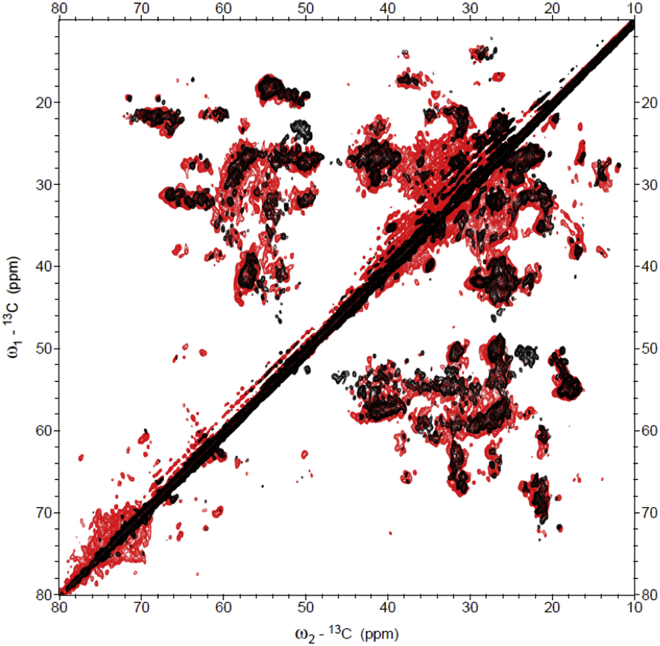
Fig. 4 shows the 2D 13C-13C CP-PARIS spectrum of LHCII (black) overlaid with the spectrum of thylakoid membranes (red) that was recorded and processed under identical conditions. Strong overlap of the two spectra demonstrates in agreement with the biochemical analysis that the most abundant signals that dominate the 2D 13C-13C thylakoid spectrum arise from LHCII. Contributions of other membrane components are visible as additional signals in the thylakoid spectrum compared with the overlaid LHCII spectrum. Strong resonance correlations accumulate between 70 and 80 ppm, which can be assigned to the galactosyl heads of thylakoid lipids. Fig. 5 show the overlaid spectra of LHCII in liposomes (black) and thylakoid membranes (red) focusing on the Ala (Fig. 5 A), Ser (Fig. 5 B), and Thr (Fig. 5 C) selective spectral regions. Fig. S9 shows the spectra in the aromatic region where Chl and carotenoid signals appear. Note that the pigment compositions and stoichiometric contents in the thylakoid membranes differ from those of isolated LHCII because the thylakoid contains additional photo-complexes with different pigment types and stoichiometries.
Figure 4.
NMR comparison of LHCII proteoliposomes and thylakoid membranes. Shown is the 13C-13C CP-PARIS spectrum of thylakoid membranes (red) with the LHCII spectrum (black) overlaid. To see this figure in color, go online.
Figure 5.
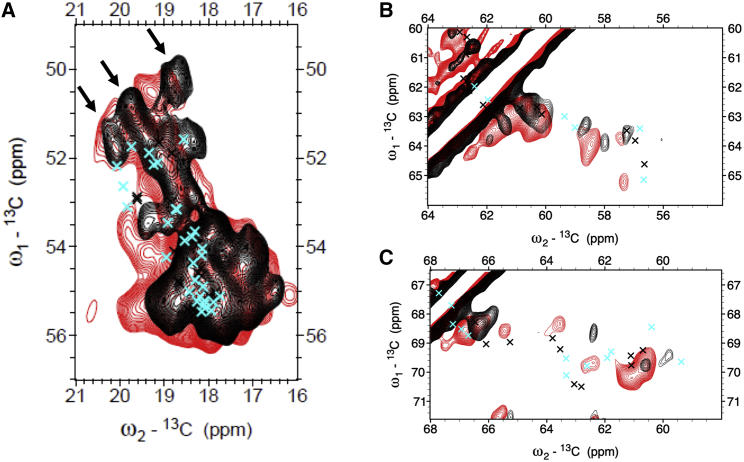
13C-13C CP-PARIS spectrum of thylakoid membranes (red) with the LHCII spectrum (black) overlaid in the Ala (A), Ser (B), and Thr (C) spectral regions. Chemical-shift predictions of Lhcbm1 (black crosses) and Lhcbm2/7 (cyan crosses) are overlaid. To see this figure in color, go online.
Representative 1D slices of the spectral regions are also shown (Fig. S9) to indicate the signal to noise levels at different ω1 frequencies. Although most of the Ala signals in the two spectra overlap, low agreement is seen comparing their Ser and Thr coil signals. The spectra in Fig. 5 are overlaid with predicted Cα and Cβ chemical-shift correlations that were generated from the Lhcbm1 and Lhcbm2/7 homology models using the program SHIFTX2 (49) and simulated as CC spectrum using the program FANDAS (50).
The full CC spectrum of LHCII with all the predictions overlaid can be found in Fig. S10, and an NC spectrum with overlaid predictions can be found in Fig. S11. Overall, the pattern of experimental correlations matches well with the predictions. However, in the selective regions, we observe several deviations. Remarkably, three strong Ala peaks appear in the coil region of the spectrum in Fig. 5 A (indicated by the arrows for the LHCII spectrum) that are better resolved in the proteoliposome spectra and Cα signals that are more upfield than any of the predicted peaks. In the Thr region (Fig. 5 C), several predicted signals are close to Cα-Cβ cross correlation signals in the thylakoid spectrum but not to any experimental peak in the LHCII proteoliposome spectrum. None of the evaluated residues are in van der Waals contact with pigment ligands, excluding direct contact interactions as the cause of chemical-shift anomalies. Looking at the pigment correlations, we note that the signals that we attributed to Chl C181-C17 or C181-18 correlations in the aliphatic region of the CC spectrum of LHCII proteoliposomes (see Fig. 3) are not visible in the spectrum of thylakoid membranes (Fig. S9). In contrast, Chl signals of ring carbons resonating in the region 130–145 ppm that correlate with the side-chain Me signals in the region 10–20 ppm appear in both spectra.
To explore the presence of dynamic protein sites, we proceeded with J-based (INEPT-TOBSY) experiments that are exclusively selective for molecules with strong dynamics and large-amplitude motions. To enhance the mobility and emphasize signals from very flexible regions, the INEPT-TOBSY experiments were carried out at and were recorded at a higher readout temperature of −3°C. Fig. 6 presents the 2D INEPT-TOBSY spectrum of the proteoliposomes (black) overlaid on the INEPT-TOBSY spectrum of thylakoid membranes (red). As expected by the fact that the majority of the protein correlations are detected in the dipolar-based spectra, only a limited set of protein signals are detected that we refer to as “J” amino acids. The overlaid spectra clearly show that many more J signals are detected in the INEPT-TOBSY spectrum of LHCII (black) than in the spectrum of thylakoid membranes (red). Because strong overlapping signals were observed in the dipolar-based correlation plots of LHCII and thylakoid membranes, the absence of J signals in the thylakoid spectrum cannot be attributed to a lower LHCII content in the thylakoid sample and demonstrate that the thylakoid-contained LHCIIs are lacking highly dynamic sites.
Figure 6.
J-analysis of LHCII proteoliposomes and thylakoid membranes. (A) Shown is the overlay of the INEPT-TOBSY spectrum of LHCII (black) and of thylakoid membranes (red). Resonance signals of Chl phytol chains, lipids, and Ala, Thr, Ser, and Phe amino acids are indicated. (B) Shown is the chemical structure of MGDG with assigned carbon atom types colored in blue. (C) Shown is the chemical structure of Chl with the assigned carbon atom types colored in green. To see this figure in color, go online.
We could assign the J residues in the LHCII spectrum to Ala, Thr, Ser, phenylalanine (Phe), Pro, Val, Lys, Ile, Glu, Asn, and Leu amino acid types (see Fig. S11 and Table S2 for the chemical-shift assignments). Based on the Cα and Cβ chemical shifts, the J amino acids can all be classified as having nonhelical structure. Because they represent amino acids that display fast and large-amplitude motions, we expect them to be located in the nonhelical stretches and in the N- and C-termini. Remarkably, whereas random coil correlations often overlap, here, distinct chemical-shift peaks are observed for J signals of same amino acid types. For instance, we see three distinct Ala Cα-Cβ correlation peaks in the spectrum. This indicates that the J amino acid containing segments are dynamical, but structured, elements. The INEPT-TOBSY spectrum of LHCII also contains two sets of Chl phytol chain signals (chemical-shift assignments in Table S3), revealing that two Chls have dynamic tails. In the thylakoid INEPT-TOBSY spectrum, no Chl correlations are observed.
The lipid signals in the NMR spectra of LHCII allow us to identify the nature of the LHCII-associated lipid molecules. The 13C lipid NMR signals must originate from the original thylakoid lipids that remained associated with the LHCII complex after purification because the reconstituted lipids are not isotope labeled. Several galactolipids are detected in the INEPT-TOBSY spectrum that can be assigned to MGDG or DGDG (56). Their chemical-shift assignments are presented in Table S4, and the connectivities are drawn in Fig. S12.
The lipid signals in the thylakoid spectra provide a unique molecular picture of lipid dynamics inside the thylakoid membrane. Lipid signals are detected both in the J-based and in the dipolar-based CC spectrum, indicating that the thylakoid membrane contains mobile lipids with strong dynamics and low segmental order as well as immobilized lipids with low dynamics, the notion of which is in agreement with our previous work (57). In the previous work, we tentatively assigned lipid signals from 1D 13C MAS NMR (58). Owing to the higher resolution and 2D spectra in this study, we now can distinguish different lipid types via differences in the sugar headgroups of MGDG, DGDG, and SQDG and make a connecting walk through the 13C-13C spectra to correlate CH-CH2 correlations. The thylakoid membrane contains lipids with high degree of unsaturation. Their CH-CH2 correlations involving double-bonded carbons exclusively appear in the INEPT spectrum and not in the CP-based spectrum, revealing that the unsaturated thylakoid lipid molecules have highly mobile tails.
Discussion
Dynamic sites in LHCII according to J-based NMR spectroscopy
The majority of NMR signals of LHCII proteoliposomes appear in the CP-based spectrum, whereas only a selective set of signals is seen in the J-based spectrum, verifying that the LHCII complexes are stabilized in the lipid membranes. The J-base INEPT-TOBSY spectra reveal selective rapid, large-amplitude motions on a picosecond-to-nanosecond timescale that occur for LHCII in liposomes but are lacking for LHCII in thylakoid membranes. According to the 77-K fluorescence spectrum and fluorescence lifetime analysis, the LHCIIs form aggregate states inside the liposome membranes. We conclude that despite their strong aggregation, selective sites in liposome-reconstituted LHCIIs have considerable dynamics compared with LHCIIs in a native thylakoid environment. The INEPT-TOBSY spectrum was collected at −3°C, whereas the CP spectrum was collected at −18°C. Because rigid amino acids that are detected in the CP-based spectrum at −18°C may become dynamic at −3°C, the CP-PARIS and INEPT-TOBSY spectrum of LHCII are overlaid in Fig. S13. One Ser J signal indeed overlaps with a correlation in the CP-based spectrum, and one Ala J signal overlaps, indicating that those two amino acids are rigid at −18°C and become dynamic at −3°C. Other J signals do not overlap with any of the protein signals seen in the CP-based spectrum and must arise from different amino acid residues. Those could be amino acids in the N-terminus as we suspect, supported by analysis of the integrated the helix and coil intensities and by earlier work (55) that amino acids in the N-terminus are not detected in a CP-based spectrum. The N-terminal domain part of Lhcbm1 is VEARRTVKPASKASTPD, which matches with many of the observed J residue types (3 × A, 3 × S, 3 × P, 3 × T, 1 or 2 L, and 1 × V, I, F, K, E, and N amino acid signals). For Lhcbm3, the equivalent stretch is KATGKKGTGKTAAKQAPASSG, whereas the sequence of Lhcbm2 in the N-terminal domain is only IA. The N-terminus could not be resolved in plant-LHCII crystal structures, which was ascribed to their nonuniform positioning in the crystals (10,15). To resolve the structure of this protein stretch, Fehr et al. modeled the N-terminal section based on distance mapping using electron paramagnetic resonance spin labels. They showed that the N-terminal domain is dynamic but does not have a random structure and covers only a restricted area above the superhelix in LHCII (16). This would in fact match with our notion that J amino acids form highly dynamical but structured elements. In a coarse-grained simulation of LHCII in thylakoid lipid bilayer, the N-terminus was shown to be preferentially located on top of the membrane-protein interface, enabling interaction with the lipid headgroups (59).
Inspection of the LHCII crystal structures shows that Chl605 and -606 (nomenclature according to Liu et al. (15)) are the only Chls in the structure with unresolved phytol chains, emphasizing their dynamics, whereas the other phytol chains are resolved and form tight interactions within the pigment-protein complex. The chains of Chl605 and -606 are oriented outward according to the orientation and positioning of their macrocycle rings in the structure and likely will not be motion limited by intracomplex interactions. In fact, Chl606 is ligated by a water molecule and could be very dynamic. In a coarse-grain model of LHCII in thylakoid lipid bilayer, the phytol tail ends of Chl605 and -606 undergo the largest fluctuations (59). We attribute the two sets of Chl phytol resonances in the INEPT-TOBSY spectrum to Chl606 and -605. The CP-PARIS spectrum shows correlations between P3 and P4, which are the two phytol atoms close to the Chl ring. No other phytol chain atom signals are observed in the CP-based spectrum. In previous studies we found that the dynamics of protein-bound chromophores often fall in an intermediate regime in which their 13 carbon signals are neither detected in CP- nor in INEPT-based NMR experiments (8). Indeed, additional cross correlation peaks of carbons along the Chl phytol chain are seen in a CC spectrum that is obtained by direct excitation (DP), as illustrated in Fig. S14.
The INEPT-TOBSY spectrum of LHCII proteoliposomes further reveals that dynamic membrane lipids are associated with LHCII. According to the 13C lipid signals, those must be from galactosyl lipids that were purified together with the LHCII complexes. Because of their visibility in the INEPT spectrum, we assume that they are not structural lipids but copurified annular lipids that in the original thylakoid membrane formed a shell around the LHCII complexes. In particular, MGDG lipids have been shown to interact with the LHCII complexes (59,60). Upon reconstitution of LHCII, those lipids may have exchanged with the liposome bulk lipids. In contrast, a PG lipid is observed in the CP-PARIS spectrum, consistent with the fact that this is a structural lipid that stabilizes the LHCII trimer conformation and does not exchange on NMR timescale. MD simulations of LHCII monomers embedded in a lipid bilayer performed by Liguori et al. predict that flexibility of the N-terminal stretch induces significant disorder of the PG molecule that ligates Chl611 and that is stabilized by a conserved Tyr (Y31 in the Lhcbm1 sequence) (9). This is not in agreement with what is observed in our experiments. In the dipolar-based NMR spectrum, the correlating set of PG lipid signals is well resolved (see Fig. 3), implying that this lipid has a steady conformation. The discrepancy may be explained by the fact that the MD simulations were performed on LHCII monomers, whereas our samples contained LHCII trimers. The PG structural lipid is located at the interface between the monomers and possibly forms a more rigid structure in trimeric LHCII.
Flexible sites in LHCII according to dipolar-based NMR spectroscopy
Comparing the CP-PARIS spectrum of LHCII proteoliposomes with the CP-PARIS spectrum of thylakoid membranes containing LHCII, poor overlap is observed in the Thr and Ser spectral coil regions, and the correlation peaks in the LHCII proteoliposome spectrum are weak. Yet, the integrated helix and coil Thr peak intensities of the LHCII proteoliposome spectrum match quite well with those anticipated. This discrepancy may be solved if we look at the predicted chemical-shift correlations for the Thr amino acids. The predicted peaks of T188 and T205, two amino acids that we counted as helical, whereas T205 is a 310 helix, fall into the coil region. According to the predictions generated using the Lhcbm1 model, the anticipated fraction of Thr signals in the coil region is 70%, which is significantly higher than the NMR-estimated percentage. Relying on the structure-based predictions, we arrive to the conclusion that—also for Thr amino acids outside the N terminus—the Thr coil signals are not fully resolved in the LHCII proteoliposome spectrum. In addition to the N-terminal loop, the C-terminus is predicted to be very dynamic for lipid-embedded LHCII trimers according to coarse-grain MD simulations (59). The flexible part of the C-terminus contains the stretch TKFTPQ (Lhcbm1) or TKFTPSA (Lhcbm2) involving the two Thr amino acids T213 and T216. One of the observed J amino acids is a Phe, whereas there are no Phe amino acids in the N-terminus. We therefore consider that some of the J amino acids could belong to the C-terminus.
To identify the NMR chemical-shift anomalies, we consider deviations between predicted (ω pred) and experimentally observed (ω exp) Cα-Cβ or NH-Cα protein backbone correlations significant if √[wω1 exp − ω1 pred)2 + (ω2 exp − ω2 pred)2] ≥ 1.2 ppm (61). In the LHCII proteoliposome spectrum, anomalies in the Ser and Thr coil regions are observed for the residues T16, T35, T188, T205, and S110 (in Lhcbm1) and T22, T38, S113, and T191 (in Lhcbm2) as shown in Fig. S15. Additional anomalies are seen in the NCA spectrum for V106, I111, and T213 and are shown in Fig. S11. For the highlighted amino acids, backbone chemical shifts either fall into a different region than predicted or are lacking in the LHCII spectrum. In contrast, in the thylakoid spectrum, matching correlation signals are found. In other words, coil correlation signals of LHCII in thylakoid membranes match with structure-based predictions, whereas selective coil signals of LHCII in proteoliposomes are lacking or fall into different regions, indicating conformational flexibility of those specific protein sites. Weak coil signals in the LHCII proteoliposome CP spectrum could be indicative of intermediate protein dynamics because thermal motions on a microsecond timescale lower the efficiency of CP. In addition, dynamic solvent-exposed residues may have multiple conformers at low temperature when their motions are frozen out, resulting in inhomogeneous line broadening or invisibility (62). Fig. 7 presents the amino acids with anomalous shifts and their location in the Lhcbm1 or Lhcbm2/7 sequences and Lhcbm1 structure. Anomalies are found for residues in the stromal loop, the luminal edge of helix C and helix A, and for residues close to the C-terminus. Indeed, a CG protein model of LHCII trimers predict that the C-terminus, stromal loop, and edges of the AC and BC loops have the highest B-factors (59).
Figure 7.
Protein sites with deviating NMR chemical shifts. Left: shown are amino acid sequences of Lhcbm1 and Lhcbm2 with deviating shift residues indicated. Right: shown is the homology model of Lhcbm1 highlighting Thr, Ser, and Ala residues with matching chemical shifts in blue and residues with deviating NMR chemical shifts in red (deviations in CC spectrum) or green (deviations in NC spectrum). To see this figure in color, go online.
In a previous study, we found that an arginine in the stromal loop adopts a different structure in aggregated LHCII than in frozen solutions of LHCII trimers in detergent micelles (55). The notion of a flexible stromal loop is consistent with the observations in this work, in which we find Thr chemical-shift anomalies in the same loop segment.
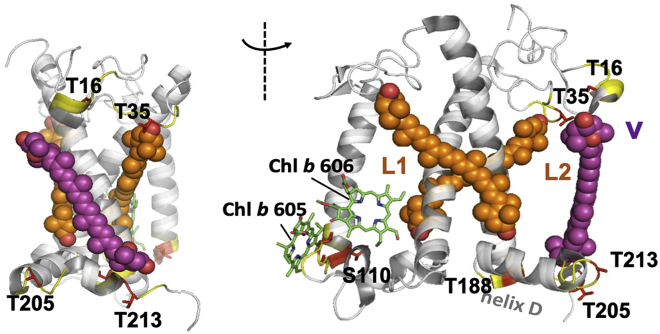
Several of the amino acids with anomalous shifts or weak signals are close to pigment-binding sites as illustrated in Fig. 8. We tentatively speculate that flexibility of the protein matrix at these spots could influence the pigment properties. It is known that the loop fragment containing T35 controls the position and orientation of L2 carotenoids and in the structure T35 is close to the headgroup of Lutein 2 (10,15). The geometries of LHCII carotenoids in the lutein pocket are found to tune light-harvesting efficiency (63), and structural changes of those sites could permit access to different dark states (64). At the lumen site of LHCII, S110, and I111 control the position and dynamics of Chl605 that is ligated to I111 and of Chl606, which are the Chl molecules with fast moving tails. T205 connects the amphipathic helix D to a 310 helix fragment at the C-terminus of LHCII containing T213, which, together with the stromal loop, stretch around T16, stabilizes the xanthophyll-cycle carotenoid violaxanthin. Plasticity of the V1 pocket could be a requirement for LHCII to bind and release carotenoids under influence of (de)-epoxidase enzymes during the xanthophyll cycle and explain why V1 carotenoids are loosely bound and easily removed during protein isolation.
Figure 8.
LHCII sites with deviating shifts and pigments in close proximity. Given are the side view (left) and front view (right) of spinach LHCII with Lutein 1 (L1), Lutein 2 (L2), and violaxanthin (V) represented as orange and purple spheres and Chl605 and -606 as green sticks. Deviating residues are highlighted in red, and adjacent residues (residue i ± 1, i ± 2) are colored yellow. To see this figure in color, go online.
Dynamics of LHCII in native thylakoid membranes
The NMR spectrum of thylakoid membranes containing LHCII provides us with a view on the dynamics of LHCII in a native membrane environment. We observe that fast, large-amplitude motions are suppressed for LHCII in native thylakoid membranes. The INEPT-TOBSY spectrum of the native thylakoid membranes lacks the protein J signals that are present in the LHCII proteoliposome spectrum and also lacks the signals of two mobile Chl tails. In the CP-PARIS spectrum of thylakoid membranes, coil signals of Thr have stronger intensities than in the LHCII proteoliposome spectrum. The Ser coil signals are also more intense and broadened, which could point to inhomogeneous broadening. From the comparable CP and DP spectral intensities in the 1D spectra, we conclude that the overall phase of matter conditions of the two samples are comparable, excluding a trivial conclusion that the thylakoid sample as a whole is more solid-like. To gain further insight, we repeated the CP-PARIS experiment for the thylakoid membrane sample at a readout temperature of −3°C (Fig. S16). To avoid high spinning of liquid-like contents, the MAS frequency was set to 14 kHz, matching with the MAS frequency at which the INEPT-TOBSY experiments were performed. At T = −3°C and 14 kHz conditions, the spectrum overall is better resolved. Thr and Ser coil correlation signals are clearly visible in the spectrum, and new Ser coil correlation signals actually emerge that can be classified as strand amino acids according to the spectral region they fall into. Those could be from dynamic water-exposed amino acids that have multiple conformers leading to coalescence and inhomogeneous broadening at low temperatures.
The cross correlation peaks that we attributed to Chl C17/18-C181 correlations in the CP-based spectrum of LHCII proteoliposomes are not visible in the thylakoid spectrum at −18°C, whereas Chl signals in other regions are observed in both. However, they do emerge in the thylakoid spectrum recorded at −3°C (Fig. S16). We presume that this has to do with differential dynamics of the Chls in proteoliposomes and in thylakoid membranes with respect to the 17, 18, and 181 macrocycle atoms, which form the ring site that is connected to the phytol chain. In an earlier study, we noted that the visibility of the LHCII Chl double-bonded ring atoms in CP-based NMR spectra were strongly dependent on temperature conditions and disappeared when the temperature was raised from −50 to −30°C (8). In this study, specific Chl ring signals are only observed when the temperature is raised from −18 to −3°C. This suggests that a complex relationship exists between the dynamics of the Chl macrocycle ring atoms and its visibility in CP experiments, which makes it hard to draw strong conclusions whether those Chl sites are more dynamic or more rigid for LHCII in thylakoid membranes than in LHCII in proteoliposomes.
Taken together, a picture emerges for the intrinsic dynamics in LHCII. The N-terminal tail and C-terminus of LHCII can undergo selective fast segmental motions and the connected stromal loop, the EC loop and luminal sites, involving sites that are close to carotenoid binding pockets, have conformational flexibility and may undergo thermal equilibrium motions on a microsecond timescale. In native thylakoid membranes, fast segmental motions are suppressed, and selective flexible sites have differential dynamics or adapt a conformation that is different from that of liposome-embedded LHCII.
To explain the differences, we consider the supramolecular interactions that LHCII complexes can have inside the thylakoid membranes. Thylakoid membranes form stacks that are stabilized by transversal salt bridges between the stromal sites of LHCIIs (the N-terminal site) (33) that could significantly reduce the dynamics of the N-terminus compared with the liposome-embedded LHCII, where the N-terminus will be exposed to the lipid-water interface. Phosphorylation of specific Thr in the N-terminus of LHCII may constrain dynamics of the N tail in thylakoid membranes and could modify the structure of the connected stromal loop.
Recent cryogenic electron microscopy (cryo-EM) structures of Cr PSII-LHCII supercomplexes show extensive interactions between LHCIIs and the monomeric complexes CP43, CP47, CP26, and CP29 and protein PsbW (65,66). Strongly bound LHCII (S-LHCII) in the supercomplex interacts with CP43, CP26, and the protein PsbW via the AC and EC loops. Loosely bound LHCII (N-LHCII) interacts with CP47 and PsbW via Chl611 on the stromal side and Chl614 and short helix D on the luminal side. Moderately bound LHCII (M-LHCII) and N-LHCII are tightly associated with CP29 at the stromal side via extensive interactions at the AC loop, Chl608, and neoxanthin and at the luminal side via Chl605 and the BC loop. Multiple interactions are observed among the LHCIIs via the amphipathic helix D that bring Chl605 and Chl606 of connected LHCIIs close together. The phytol chain of Chl605 that is lacking in LHCII crystal structures is partly resolved in the Cr LHCII-PSII structure. Interactions involving helix D could influence the conformation and dynamics of T188 and T205 at the lumen side, whereas the interactions involving the BC loop are close to V106, S110, and I111. With our chemical-shift analysis, we do not have suitable NMR reporters at the AC loop. The AC loop of Lhcbm1 contains one Ser (S147), one Ala (A153), and a number of Gly residues. Predicted Cα-Cβ correlations of Ser and Ala, however, fall in regions where multiple signals accumulate, and also, the Gly Cα-CO coil correlations are not dispersed. Whereas reduced dynamics of LHCII protein and pigment sites can be explained by their interactions in supercomplexes, thylakoid membranes also contain a pool of free LHCIIs. The thylakoid INEPT-TOBSY spectrum, which has excellent signal to noise, does not show weak signals from a subset of free LHCIIs. This suggests that also for free complexes, fast protein and pigment fluctuations are suppressed by the surroundings of the thylakoid environment.
The thylakoid INEPT-based CC spectrum contains only few protein signals, but multiple signals of unsaturated thylakoidal lipids are detected that are indicative of lipids with highly mobile tails. This could point to a phase separation between those dynamical lipids and rigid protein-rich membrane regions. Lipid polymorphism has been revealed in chloroplast thylakoid membranes, including the reversible formation of inverted hexagonal phase structures by the nonbilayer lipid MGDG (67).
Finally, our findings call to question whether or not spontaneous fluctuations of individual LHCIIs between different conformational states, as has been observed by single-molecule spectroscopy and has been suggested by MD simulations, do occur in vivo. The NMR experiments on whole thylakoid membranes containing LHCII are the first attempt, to the best of our knowledge, to obtain dynamic information of LHCII in a native thylakoid environment and under noncrystallizing conditions. Our NMR data results show that in a native setting in which the proteins are embedded inside thylakoidal membranes, fast large-amplitude protein and pigment fluctuations are suppressed and flexible sites, involved in stabilization of the carotenoids, are motion limited. A comparision of the spectra of thylakoid-contained and liposome-embedded LHCII suggest that the protein can be stabilized in somewhat different conformational states depending on its local environment. The thylakoid membrane, however, is not a static architecture, but in living organisms actively responds to changes in the light conditions. Thylakoid remodeling under light stress involves phosphorylation, membrane unstacking, supercomplex reorganization, and xanthophyll exchange, all of which may modify the protein landscape of LHCII and (temporarily) reduce the energetic transition barriers between distinct protein states. This work shows that by MAS NMR structural dynamics can be uncovered from proteins, pigment, and associated lipid components inside native, heterogeneous organelle membranes. The approach may open ways to explore how thylakoid plasticity controls the conformational states of the most abundant light-harvesting proteins, which are among the key players in photo regulatory processes.
Author Contributions
A.P. and F.A.-C. designed the research, with the input of T.M. and M.B. F.A.-C. and M.E.W. performed the experiments. G.P. and D.S. prepared the U-13C-LHCII complexes, and F.A.-C. performed the protein membrane reconstitution and prepared the U-13C-thylakoid membranes. F.A.-C. and A.P. wrote the manuscript, with input from all authors.
Acknowledgments
We thank Emanuela Crisafi for assistance with the liposome preparations and Dr. Lijin Tian for assistance with the fluorescence experiments and the Biophysics Department of the Vrije University Amsterdam for use of their fluorescence spectrometry equipment.
A.P. and F.A.-C. were financially supported by a CW-VIDI grant of the Netherlands Organization of Scientific Research under grant no. 723.012.103. M.E.W. is a recipient of a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship. This work was supported in part by uNMR-NL, a the-Netherlands-Organization-of-Scientific-Research-funded National Roadmap Large-Scale Facility of the Netherlands (grant number 184.032.207).
Editor: Timothy Cross.
Footnotes
Giorgio Perin’s present address is Department of Life Sciences, Imperial College London, SW72BB London, United Kingdom.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.11.2265.
Supporting Material
References
- 1.Kirchhoff H. Structural changes of the thylakoid membrane network induced by high light stress in plant chloroplasts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130225. doi: 10.1098/rstb.2013.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horton P., Ruban A. Molecular design of the photosystem II light-harvesting antenna: photosynthesis and photoprotection. J. Exp. Bot. 2005;56:365–373. doi: 10.1093/jxb/eri023. [DOI] [PubMed] [Google Scholar]
- 3.Ruban A.V., Berera R., van Grondelle R. Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature. 2007;450:575–578. doi: 10.1038/nature06262. [DOI] [PubMed] [Google Scholar]
- 4.Krüger T.P., Ilioaia C., van Grondelle R. Controlled disorder in plant light-harvesting complex II explains its photoprotective role. Biophys. J. 2012;102:2669–2676. doi: 10.1016/j.bpj.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holt N.E., Zigmantas D., Fleming G.R. Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science. 2005;307:433–436. doi: 10.1126/science.1105833. [DOI] [PubMed] [Google Scholar]
- 6.Robert B., Horton P., Ruban A.V. Insights into the molecular dynamics of plant light-harvesting proteins in vivo. Trends Plant Sci. 2004;9:385–390. doi: 10.1016/j.tplants.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Schlau-Cohen G.S., Yang H.-Y., Moerner W.E. Single-molecule identification of quenched and unquenched states of LHCII. J. Phys. Chem. Lett. 2015;6:860–867. doi: 10.1021/acs.jpclett.5b00034. [DOI] [PubMed] [Google Scholar]
- 8.Pandit A., Reus M., de Groot H.J. An NMR comparison of the light-harvesting complex II (LHCII) in active and photoprotective states reveals subtle changes in the chlorophyll a ground-state electronic structures. Biochim. Biophys. Acta. 2013;1827:738–744. doi: 10.1016/j.bbabio.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Liguori N., Periole X., Croce R. From light-harvesting to photoprotection: structural basis of the dynamic switch of the major antenna complex of plants (LHCII) Sci. Rep. 2015;5:15661. doi: 10.1038/srep15661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Standfuss J., Terwisscha van Scheltinga A.C., Kühlbrandt W. Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. EMBO J. 2005;24:919–928. doi: 10.1038/sj.emboj.7600585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papadatos S., Charalambous A.C., Daskalakis V. A pathway for protective quenching in antenna proteins of Photosystem II. Sci. Rep. 2017;7:2523. doi: 10.1038/s41598-017-02892-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miloslavina Y., Wehner A., Holzwarth A.R. Far-red fluorescence: a direct spectroscopic marker for LHCII oligomer formation in non-photochemical quenching. FEBS Lett. 2008;582:3625–3631. doi: 10.1016/j.febslet.2008.09.044. [DOI] [PubMed] [Google Scholar]
- 13.Chmeliov J., Trinkunas G., Valkunas L. Light harvesting in a fluctuating antenna. J. Am. Chem. Soc. 2014;136:8963–8972. doi: 10.1021/ja5027858. [DOI] [PubMed] [Google Scholar]
- 14.Chmeliov J., Gelzinis A., Valkunas L. The nature of self-regulation in photosynthetic light-harvesting antenna. Nat. Plants. 2016;2:16045. doi: 10.1038/nplants.2016.45. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z., Yan H., Chang W. Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 16.Fehr N., Dietz C., Paulsen H. Modeling of the N-terminal section and the lumenal loop of trimeric light harvesting complex II (LHCII) by using EPR. J. Biol. Chem. 2015;290:26007–26020. doi: 10.1074/jbc.M115.669804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X., Su X., Liu Z. Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature. 2016;534:69–74. doi: 10.1038/nature18020. [DOI] [PubMed] [Google Scholar]
- 18.Ruban A.V., Johnson M.P., Duffy C.D. The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta. 2012;1817:167–181. doi: 10.1016/j.bbabio.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 19.Tikkanen M., Grieco M., Aro E.M. Novel insights into plant light-harvesting complex II phosphorylation and ‘state transitions’. Trends Plant Sci. 2011;16:126–131. doi: 10.1016/j.tplants.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Ferrante P., Ballottari M., Bassi R. LHCBM1 and LHCBM2/7 polypeptides, components of major LHCII complex, have distinct functional roles in photosynthetic antenna system of Chlamydomonas reinhardtii. J. Biol. Chem. 2012;287:16276–16288. doi: 10.1074/jbc.M111.316729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietrzykowska M., Suorsa M., Jansson S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell. 2014;26:3646–3660. doi: 10.1105/tpc.114.127373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen J.F. Why we need to know the structure of phosphorylated chloroplast light-harvesting complex II. Physiol. Plant. 2017;161:28–44. doi: 10.1111/ppl.12577. [DOI] [PubMed] [Google Scholar]
- 23.Seiwert D., Witt H., Paulsen H. The non-bilayer lipid MGDG stabilizes the major light-harvesting complex (LHCII) against unfolding. Sci. Rep. 2017;7:5158. doi: 10.1038/s41598-017-05328-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seiwert D., Witt H., Paulsen H. The nonbilayer lipid MGDG and the major light-harvesting complex (LHCII) promote membrane stacking in supported lipid bilayers. Biochemistry. 2018;57:2278–2288. doi: 10.1021/acs.biochem.8b00118. [DOI] [PubMed] [Google Scholar]
- 25.Fu R., Wang X., Tian F. In situ structural characterization of a recombinant protein in native Escherichia coli membranes with solid-state magic-angle-spinning NMR. J. Am. Chem. Soc. 2011;133:12370–12373. doi: 10.1021/ja204062v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao Y., Qin H., Cross T.A. M2 proton channel structural validation from full-length protein samples in synthetic bilayers and E. coli membranes. Angew. Chem. Int.Engl. 2012;51:8383–8386. doi: 10.1002/anie.201204666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renault M., Pawsey S., Baldus M. Solid-state NMR spectroscopy on cellular preparations enhanced by dynamic nuclear polarization. Angew. Chem. Int.Engl. 2012;51:2998–3001. doi: 10.1002/anie.201105984. [DOI] [PubMed] [Google Scholar]
- 28.Shi P., Li D., Tian C. In situ 19F NMR studies of an E. coli membrane protein. Protein Sci. 2012;21:596–600. doi: 10.1002/pro.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward M.E., Wang S., Ladizhansky V. In situ structural studies of Anabaena sensory rhodopsin in the E. coli membrane. Biophys. J. 2015;108:1683–1696. doi: 10.1016/j.bpj.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan M., Narasimhan S., Baldus M. EGFR dynamics change during activation in native membranes as revealed by NMR. Cell. 2016;167:1241–1251.e11. doi: 10.1016/j.cell.2016.10.038. [DOI] [PubMed] [Google Scholar]
- 31.Baker L.A., Sinnige T., Grunewald K. Combined 1H-detected solid-state NMR spectroscopy and electron cryotomography to study membrane proteins across resolutions in native environments. Structure. 2018;26:161–170.e3. doi: 10.1016/j.str.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niyogi K.K., Bjorkman O., Grossman A.R. Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell. 1997;9:1369–1380. doi: 10.1105/tpc.9.8.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sculley M.J., Duniec J.T., Boardman N.K. The stacking of chloroplast thylakoids. Quantitative analysis of the balance of forces between thylakoid membranes of chloroplasts, and the role of divalent cations. Arch. Biochem. Biophys. 1980;201:339–346. doi: 10.1016/0003-9861(80)90519-6. [DOI] [PubMed] [Google Scholar]
- 34.Crisafi E., Pandit A. Disentangling protein and lipid interactions that control a molecular switch in photosynthetic light harvesting. Biochim. Biophys. Acta Biomembr. 2017;1859:40–47. doi: 10.1016/j.bbamem.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Kirchhoff H. Molecular crowding and order in photosynthetic membranes. Trends Plant Sci. 2008;13:201–207. doi: 10.1016/j.tplants.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Chua N.H., Bennoun P. Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: wild-type and mutant strains deficient in photosystem II reaction center. Proc. Natl. Acad. Sci. USA. 1975;72:2175–2179. doi: 10.1073/pnas.72.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Pinest A., Gibby M.G., Waugh J.S. Proton-enhanced NMR of dilute spins in solids. J. Chem. Phys. 1973;59:569–590. [Google Scholar]
- 39.Hartmann S.R., Hahn E.L. Nuclear double resonance in the rotating frame. Phys. Rev. 1962;128:2042–2053. [Google Scholar]
- 40.Baldus M., Petkova A.T., Griffin R.G. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol. Phys. 1998;95:1197–1207. [Google Scholar]
- 41.Fung B.M., Khitrin A.K., Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 42.Weingarth M., Demco D.E., Tekely P. Improved magnetization transfer in solid-state NMR with fast magic angle spinning. Chem. Phys. Lett. 2009;469:342–348. [Google Scholar]
- 43.Baldus M. Correlation experiments for assignment and structure elucidation of immobilized polypeptides under magic angle spinning. Prog. Nucl. Magn. Reson. Spectr. 2002;41:1–47. [Google Scholar]
- 44.Baldus M., Meier B.H. Total correlation spectroscopy in the solid state. The use of scalar couplings to determine the through-bond connectivity. J. Magn. Reson. A. 1996;121:65–69. [Google Scholar]
- 45.Morris G.A., Freeman R. Enhancement of nuclear magnetic resonance signals by polarization transfer. J. Am. Chem. Soc. 1979;101:760–762. [Google Scholar]
- 46.Goddard T. D. and D. G. Kneller. SPARKY: University of California, San Francisco, CA.
- 47.Arnold K., Bordoli L., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 48.Natali A., Croce R. Characterization of the major light-harvesting complexes (LHCBM) of the green alga Chlamydomonas reinhardtii. PLoS One. 2015;10:e0119211. doi: 10.1371/journal.pone.0119211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han B., Liu Y., Wishart D.S. SHIFTX2: significantly improved protein chemical shift prediction. J. Biomol. NMR. 2011;50:43–57. doi: 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gradmann S., Ader C., Baldus M. Rapid prediction of multi-dimensional NMR data sets. J. Biomol. NMR. 2012;54:377–387. doi: 10.1007/s10858-012-9681-y. [DOI] [PubMed] [Google Scholar]
- 51.Drop B., Webber-Birungi M., Croce R. Light-harvesting complex II (LHCII) and its supramolecular organization in Chlamydomonas reinhardtii. Biochim. Biophys. Acta. 2014;1837:63–72. doi: 10.1016/j.bbabio.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 52.Trémolières A. Glycerolipids: composition, biosynthesis and function in chlamydomonas. In: Rochaix J.D., Goldschmidt-Clermont M., Merchant S., editors. The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Springer; 1998. pp. 415–431. [Google Scholar]
- 53.Natali A., Gruber J.M., Croce R. Light-harvesting complexes (LHCs) cluster spontaneously in membrane environment leading to shortening of their excited state lifetimes. J. Biol. Chem. 2016;291:16730–16739. doi: 10.1074/jbc.M116.730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andronesi O.C., Becker S., Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J. Am. Chem. Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 55.Sunku K., de Groot H.J., Pandit A. Insights into the photoprotective switch of the major light-harvesting complex II (LHCII): a preserved core of arginine-glutamate interlocked helices complemented by adjustable loops. J. Biol. Chem. 2013;288:19796–19804. doi: 10.1074/jbc.M113.456111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Souza L.M., Iacomini M., Sassaki G.L. Glyco- and sphingophosphonolipids from the medusa Phyllorhiza punctata: NMR and ESI-MS/MS fingerprints. Chem. Phys. Lipids. 2007;145:85–96. doi: 10.1016/j.chemphyslip.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 57.Azadi Chegeni F., Perin G., Pandit A. Protein and lipid dynamics in photosynthetic thylakoid membranes investigated by in-situ solid-state NMR. Biochim. Biophys. Acta. 2016;1857:1849–1859. doi: 10.1016/j.bbabio.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 58.Azadi-Chegeni F., Schiphorst C., Pandit A. In vivo NMR as a tool for probing molecular structure and dynamics in intact Chlamydomonas reinhardtii cells. Photosynth. Res. 2018;135:227–237. doi: 10.1007/s11120-017-0412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thallmair S., Vainikka P.A., Marrink S.J. Lipid fingerprints and cofactor dynamics of light-harvesting complex II in different membranes. Biophys. J. 2019;116:1446–1455. doi: 10.1016/j.bpj.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaller S., Latowski D., Goss R. The main thylakoid membrane lipid monogalactosyldiacylglycerol (MGDG) promotes the de-epoxidation of violaxanthin associated with the light-harvesting complex of photosystem II (LHCII) Biochim. Biophys. Acta. 2010;1797:414–424. doi: 10.1016/j.bbabio.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 61.Seidel K., Etzkorn M., Baldus M. Comparative analysis of NMR chemical shift predictions for proteins in the solid phase. Solid State Nucl. Magn. Reson. 2009;35:235–242. doi: 10.1016/j.ssnmr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 62.Linden A.H., Franks W.T., Oschkinat H. Cryogenic temperature effects and resolution upon slow cooling of protein preparations in solid state NMR. J. Biomol. NMR. 2011;51:283–292. doi: 10.1007/s10858-011-9535-z. [DOI] [PubMed] [Google Scholar]
- 63.Son M., Pinnola A., Schlau-Cohen G.S. The electronic structure of lutein 2 is optimized for light harvesting in plants. Chem. 2019;5:575–584. [Google Scholar]
- 64.Liguori N., Xu P., Croce R. Different carotenoid conformations have distinct functions in light-harvesting regulation in plants. Nat. Commun. 2017;8:1994. doi: 10.1038/s41467-017-02239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen L., Huang Z., Zhang X. Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 2019;116:21246–21255. doi: 10.1073/pnas.1912462116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheng X., Watanabe A., Liu Z. Structural insight into light harvesting for photosystem II in green algae. Nat. Plants. 2019;5:1320–1330. doi: 10.1038/s41477-019-0543-4. [DOI] [PubMed] [Google Scholar]
- 67.Garab G., Ughy B., Lambrev P.H. Lipid polymorphism in chloroplast thylakoid membranes - as revealed by 31P-NMR and time-resolved merocyanine fluorescence spectroscopy. Sci. Rep. 2017;7:13343. doi: 10.1038/s41598-017-13574-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.