Abstract
Cell division control protein 42 homolog (Cdc42) protein, a Ras superfamily GTPase, regulates cellular activities, including cancer progression. Using all-atom molecular dynamics (MD) simulations and essential dynamic analysis, we investigated the structure and dynamics of the catalytic domains of GDP-bound (inactive) and GTP-bound (active) Cdc42 in solution. We discovered substantial differences in the dynamics of the inactive and active forms, particularly in the “insert region” (residues 122–135), which plays a role in Cdc42 activation and binding to effectors. The insert region has larger conformational flexibility in the GDP-bound Cdc42 than in the GTP-bound Cdc42. The G2 loop and switch I at the effector lobe of the catalytic domain exhibit large conformational changes in both the GDP- and the GTP-bound systems, but in the GTP-bound Cdc42, the switch I interactions with GTP are retained. Oncogenic mutations were identified in the Ras superfamily. In Cdc42, the G12V and Q61L mutations decrease the GTPase activity. We simulated these mutations in both GDP- and GTP-bound Cdc42. Although the overall structural organization is quite similar between the wild type and the mutants, there are small differences in the conformational dynamics, especially in the two switch regions. Taken together, the G12V and Q61L mutations may play a role similar to their K-Ras counterparts in nucleotide binding and activation. The conformational differences, which are mainly in the insert region and, to a lesser extent, in the switch regions flanking the nucleotide binding site, can shed light on binding and activation. We propose that the differences are due to a network of hydrogen bonds that gets disrupted when Cdc42 is bound to GDP, a disruption that does not exist in other Rho GTPases. The differences in the dynamics between the two Cdc42 states suggest that the inactive conformation has reduced ability to bind to effectors.
Significance
Cell division control protein 42 homolog (Cdc42), a Ras superfamily and Rho family protein, acts in cancer metastasis and progression. Like Ras proteins, it switches between active GTP-bound and inactive GDP-bound forms. Rho family members have a helical “insert region” that plays a role in binding and activation. We simulated the active and inactive Cdc42, as well as two mutants: G12V and Q61L. We found that the inactive (GDP-bound) and active (GTP-bound) systems differ mainly in the conformational dynamics of the insert region and, to a lesser extent, in the switch regions flanking the nucleotide binding site. The high fluctuations in Cdc42-GDP, but not Rac-GDP, can disrupt the insert region integrity, reducing the inactive conformation ability to bind to effectors.
Introduction
The cell division control protein 42 homolog (Cdc42) is a Rho family protein, a subgroup of the Ras superfamily, which regulates cellular activities, including cytoskeletal organization, gene expression, and transformation (1, 2, 3, 4, 5, 6, 7, 8). Superfamily members have been linked to multiple human cancers (9, 10, 11, 12, 13, 14, 15). These proteins act as molecular switches between active GTP-bound and inactive GDP-bound forms. The best-characterized members of the Rho family are RhoA, Rac1, and Cdc42. Cdc42 controls filopodia formation. Filopodia are actin-rich fingers that establish the direction of motility. Rac1 controls lamellipodia formation—actin-rich ruffles at the leading edge of the cell that initiate motion—and Rho controls the establishment of stress fibers whose formation results in the contractile force that moves the body of the cell behind the leading edge (3,16). Increasing evidence has validated Cdc42 as involved in cancer metastasis and progression and has shown it to be overexpressed in many types of cancer (17,18).
The 166-residue catalytic domain is conserved among the Ras isoforms and several other GTPases (19). The sequence includes several highly conserved regions related to binding of guanine nucleotides (GDP and GTP) and activation. These include the P-loop (residues 10–15), switch I (resides 30–38), and switch II (residues 60–76), which are responsible for the interaction with GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs). The switch regions undergo conformational changes upon guanosine activation and binding (20, 21, 22, 23, 24, 25, 26). A major distinguishing structural component between the Rho family members and the Ras isoforms is the “insert region” spanning residues 122–135 (27). The region contains an α-helix that replaces loop 8 of the Ras isoforms (Fig. 1). It is rich in charged amino acids (28). The insert region is not required for Rho kinase binding, but it plays a role in its activation (1). The insert region plays a role in many processes, including the activation of phospholipase D1 (PLD1) by Cdc42 and the activation of superoxide-producing nicotinamide adenine dinucleotide phosphate (NADPH) oxidases in Rac (29). The dedicator of cytokinesis (DOCK) family of atypical guanine nucleotide exchange factors activates Rac and/or Cdc42 through DOCK homology region 2 (DHR-2) (30). Molecular dynamics (MD) simulations of the dissociation mechanism of GDP from Cdc42 via DOCK9 revealed that Mg2+ influences the conformational change of switch I through residue Pro34, which functions as a “clasp” to control its flexibility (31). Upon activation, Rho-GTP interacts with downstream effector proteins through its switch I effector binding site (32,33). In the active GTP-bound state, Cdc42 and Rac can interact with the IQ motif-containing GAP2 (IQGAP2), and IQGAP interacts with α-catenin (34). However, the GDP-bound Cdc42 and Rac are unable to interact with IQGAP, inducing the β-catenin interaction and dissociating α-catenin from the complex.
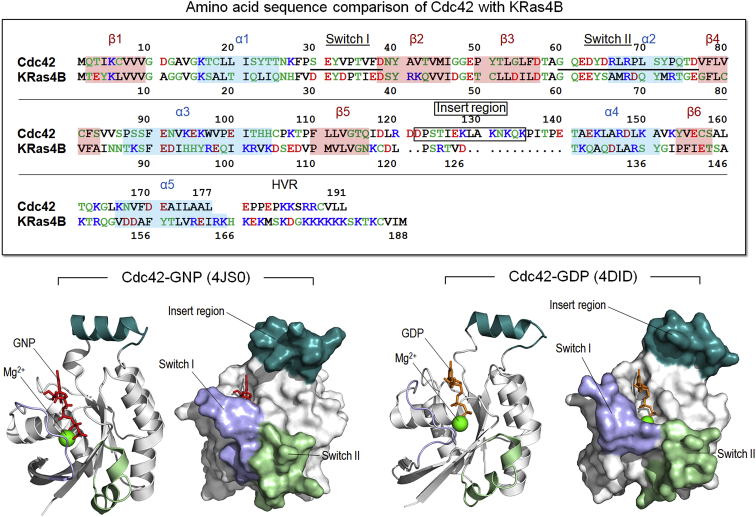
Figure 1.
The sequence of Cdc42 aligned with the sequence of K-Ras4B (top panel). The crystal structures of GNP-bound (PDB: 4JS0) and GDP-bound (PDB: 4DID) Cdc42 are shown (bottom panel). Although the sequence and most structural motifs are highly conserved, K-Ras4B is missing the insert region (residues 122–135), an α-helix rich in charged amino acids, which only appears in members of the Rho family. To see this figure in color, go online.
Several oncogenic mutations have been identified in the Ras superfamily (15,35, 36, 37, 38, 39, 40, 41, 42, 43, 44). Gly12, Ala13 (in Rho proteins) or Gly13 (in Ras proteins), and Gln61 are involved in GAP-assisted hydrolysis (45,46), which the mutants impair (47,48). K-Ras4B Gly12, Gly13, and Gln61 mutants decrease GAP-assisted hydrolysis rate drastically with respect to the wild type, but that of Q61L and G12A is still ∼15- to 25-fold higher than their intrinsic hydrolysis rates, which may indicate that a portion of the GAP-dependent catalytic mechanism is still somewhat functional in these mutants (47). Gln61 is involved in hydrogen bonding with a catalytic water molecule, Arg789 of GAP, and the GTP molecule, initiating a nucleophilic attack that hydrolyzes GTP. The Q61L mutation stabilizes the active GTP-bound form. Mutations in residues 12 and 13 disrupt the catalysis-favored arrangement of Gln61, as well as the hydrogen bonding of the backbone of residue 13 with GTP through steric hindrance. In KRAS-driven cancer, Gly12 mutations (89%) are predominant, followed by Gly13 (9%) and Gln61 (1%) mutations (49). Recent studies shed more light on the structural basis of the nucleotide (GDP or GTP) binding pockets of the wild type and mutants of the K-Ras4A and K-Ras4B proteins (48,50). The catalytic domain of GDP-bound K-Ras4A has a more exposed nucleotide binding pocket than K-Ras4B, and the dynamic fluctuations in the switch I and II regions also differ. The wild-type K-Ras4B-GTP exists in two (active and inactive) conformations (48). Mutations in Gly12 and Gly13 differentially elicit an inactive-to-active conformational transition in K-Ras4B-GTP. In K-Ras4B-GDP, the mutants expose the bound nucleotide, which facilitates the GDP-to-GTP exchange.
A recent study shed light on the dimerization of IQGAPs through interaction of the GAP-related domain 2 (GRD2) of IQGAP2 with the active GTP-bound Cdc42 (34,51). Importantly, the study revealed that the GTP-bound Rac1 displays only single-site binding to the GRD2 as opposed to Cdc42, which binds the GRD2 in two sites, indicating that only Cdc42 promotes IQGAP2 dimerization. More specifically, residues 25–38 (switch I) and 57–75 (switch II) are involved in contacts with GRD2, and Cdc42 binding promoted allosteric changes in the RasGAP site, providing a binding site for the second Cdc42 (34). Cdc42 insert region was essential for the binding. As a result of the differences in the sequence and structure of the insert region, Rac1 could bind only to the RasGAP site of apo-GRD2 and was unable to mediate IQGAP2 dimerization. Like the Ras isoforms, Cdc42 can bind to the membrane through the interactions of its positively charged hypervariable region (HVR) with lipids (52, 53, 54, 55, 56, 57). The C-terminal Cys gets a geranylgeranyl post-translational modification (PTM), which can be inserted into the membrane, anchoring the protein.
In this study, we aim to figure out the role of the insert and the switch regions in the active and inactive forms. We performed explicit MD simulations of the catalytic domain of Cdc42 in the active (GTP-bound) and inactive (GDP-bound) states. Additionally, we simulated the G12V and Q61L mutants of the two states to reveal their impact on both states. Being located on the P-loop and switch II regions, respectively, these two residues are involved in interaction with the active site. We found that the insert region exhibits much larger conformational flexibility in the GDP-bound state than in the GTP-bound state in both the wild type and mutant systems. This is interesting because the insert region does not interact directly with the binding site. The switch I and II regions undergo conformational changes in both cases, but to a lesser extent. We also show that the mutants affect the binding site exposure to the solvent and the interactions with other amino acids, Tyr32 and Thr35, near the binding site. This may indicate that the mutants play a similar role in GTP hydrolysis as in K-Ras4A and K-Ras4B and other Ras proteins (41,58, 59, 60, 61).
Materials and Methods
Preparation of models
The GDP-bound and GTP-bound Cdc42 proteins were modeled based on the crystal structures (Protein Data Bank, PDB: 4DID (chain A) and PDB: 4JS0 (chain A), respectively) (Fig. 1) and represented in full atomic level. We replaced the 4JS0 GNP molecule with GTP and removed the inhibitor bound to it. Missing side chains and hydrogen atoms were added using the CHARMM software (62). The G12V and Q61L mutants were modeled using CHARMM as well, using the wild-type system as a starting point. The simulations were conducted using the CHARMM force field (62). The solvent was represented explicitly using the TIP3P model. The charge of all potential titratable groups was fixed at values corresponding to neutral pH (i.e., all acidic and basic side chains were represented in their negatively and positively charged forms, respectively). We used a cubic simulation box with periodic boundary conditions and the nearest image convention. Atom pair cutoff distance was set at 14.0 Å to compute the van der Waals (vdW) interactions. To avoid discontinuities in the potential energy function, nonbonding energy terms were forced to slowly converge to zero by applying a smoothing factor from a distance of 12.0 Å. The nontruncated electrostatic potential was computed through particle mesh Ewald (PME) summations. We used a charge mesh with a grid thickness of one point per cubic Å. Na+ and Cl− ions were added to each system to neutralize the charge and simulate physiological conditions. Overall, the wild-type GDP-bound system had 74,197 atoms, and the wild-type GTP-bound system had 74,290 atoms.
Simulation protocol
All simulations were performed using the NAMD 2.13 software package (63) and carried out according to our previously published protocol (4,22,34,48,50,64). First, the potential energy was minimized by using 10,000 conjugate gradient steps. Then, the protein atoms were held fixed while the solvent was heated to a temperature of 310 K to ensure uniform distribution of the ions in solution. Next, the system was isothermally and isobarically equilibrated at 310 K and 1 bar (NPT conditions) to allow reaching infinite dilution conditions (i.e., water density close to 1 g/cm3) for 500 ps. Later, the solute was allowed to move, and the whole system was heated and equilibrated (50 ps) at the production temperature of 310 K and pressure of 1 bar. A numerical integration time step of 2 fs was used for all the simulations, and the nonbonded pair list was updated every five steps. In the production runs, the constant temperature of 310 K was maintained by the Langevin temperature control, and the pressure at 1 atm was sustained by the Nosé-Hoover Langevin piston pressure control. During the production runs, the trajectories were recorded every 50,000 steps (25 ps) for subsequent analysis, and in all cases, the production simulations were run for a period of 500 ns each, totaling 3 μs. The first 30 ns were removed from the analysis to ensure equilibration. The analysis was done using the VMD (65), ProDy (66), and MATLAB (The MathWorks, Natick, MA) software packages.
Results
Nucleotide-dependent conformational changes of the insert region
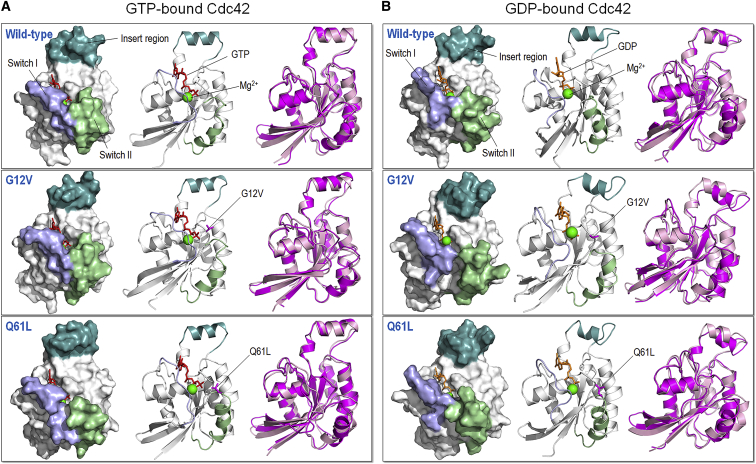
We performed explicit MD simulations on the catalytic domains of the wild-type Cdc42 (hereafter referred to as Cdc42) and the G12V and Q61L Cdc42 mutants (denoted as Cdc42G12V and Cdc42Q61L, respectively) in the GTP- and GDP-bound states. During the simulations, we observed that there are significant differences in the conformational flexibility of the insert region. The insert region maintains its helical structure in the GTP-bound systems (Fig. 2 A) but loses some of its helical conformation in the GDP-bound systems (Fig. 2 B). Although the crystal structures of Cdc42 exhibited a helical conformation of the insert region for both GTP- and GDP-bound systems, the GDP-bound crystal structure showed much higher B-factor values, especially in the insert region (Fig. S1). For Cdc42-GTP, the averaged root mean-square deviation (RMSD) of the insert region is 1.4 ± 0.2 Å, whereas it is 3.9 ± 0.8 Å for Cdc42-GDP. The overall RMSD of the entire system is also a little higher for the GDP-bound system, but the difference is smaller: 1.8 ± 0.2 Å vs. 2.4 ± 0.2 Å for the GTP- and GDP-bound systems, respectively. Similar results are observed in the mutants (see Table 1). The main loss of helical integrity in Cdc42-GDP starts in the beginning of the simulation. In Cdc42-GTP, the active regions consisting of the insert region, switch I, and switch II maintained closer distance to GTP than in the GDP-bound state (see structures in Fig. 2). The switch regions come close to the binding site to form interactions with GTP in the active site, whereas upon hydrolysis, the loops open to allow GDP to dissociate. This has been shown to be the case in K-Ras4B as well (48). The average distances between the Cα atom of the middle residue in the loop (Glu31) and the Pβ atom of the GTP/GDP molecule are 12.3 ± 0.4 and 14.0 ± 1.0 Å for the GTP- and GDP-bound Cdc42, respectively. The average distances between Gln61 (on switch II) and the GTP/GDP molecule are 9.3 ± 0.3 and 11.5 ± 1.0 Å for the GTP- and GDP-bound Cdc42, respectively.
Figure 2.
Snapshots depicting the final conformation in surface and cartoon representations, and superimposition of the final (magenta cartoon) and initial (pink cartoon) configurations of the wild type and two mutants of (A) the GTP-bound and (B) GDP-bound Cdc42. The insert region (light teal) loses its structural integrity in all the GDP-bound systems but not in the GTP-bound systems. To see this figure in color, go online.
Table 1.
The Averaged RMSD of the Entire Protein and the Insert Region (Residues 122–135)
| System | RMSD (Å) |
|
|---|---|---|
| Total | Insert Region | |
| Cdc42-GTP | 1.8 ± 0.2 | 1.4 ± 0.2 |
| Cdc42G12V-GTP | 1.8 ± 0.1 | 1.6 ± 0.2 |
| Cdc42Q61L-GTP | 2.2 ± 0.3 | 1.5 ± 0.2 |
| Cdc42-GDP | 2.4 ± 0.2 | 3.9 ± 0.8 |
| Cdc42G12V-GDP | 2.2 ± 0.2 | 3.3 ± 0.6 |
| Cdc42Q61L-GDP | 2.5 ± 0.2 | 3.5 ± 0.8 |
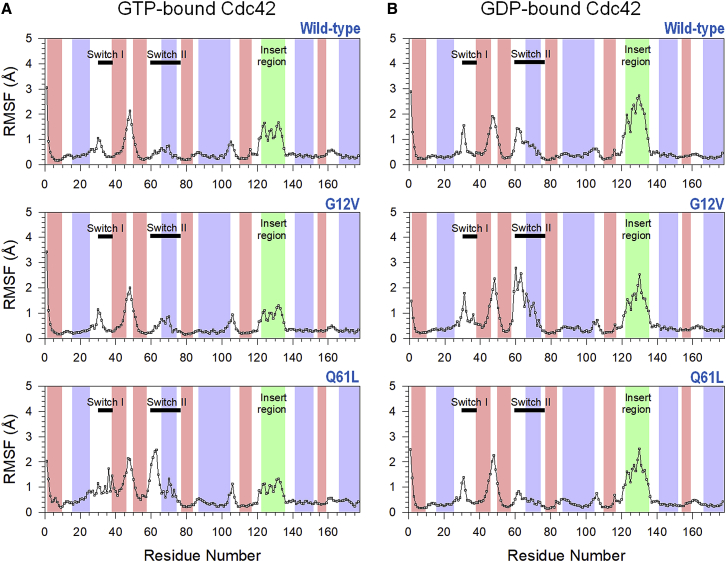
To monitor the fluctuations of individual residues in Cdc42, we measured the root mean-square fluctuations (RMSFs) for each amino acid in both the GTP- and GDP-bound systems. In general, the GTP-bound systems (Fig. 3 A) showed less fluctuations than the GDP-bound systems (Fig. 3 B). We observed that the insert region showed the greatest difference. The RMSF was higher in the GDP-bound systems in both the wild type and mutants, indicating much greater fluctuations for this region. In the GTP-bound systems, the two switch regions show lower RMSF than the insert region, except the Q61L mutant. In both the GTP- and GDP-bound Cdc42, the switch I and II regions exhibit RMSF peaks because both regions interact directly with the binding site. There are differences between the wild type and mutants, but they all follow the same trend. In the GTP-bound systems, the Q61L mutant exhibits higher fluctuations in the two switch regions—especially switch II—whereas the G12V mutant showed approximately the same mean fluctuations as the wild type. The Q61L mutant binding site residues, especially the mutated Leu61, interact with the nucleotide only intermittently, whereas the wild-type and the G12V mutant show more constant interactions, which indicates that the Q61L mutant may sample inactive conformations (see below). In the GDP-bound case, all the systems show high fluctuations in the insert regions despite the mutations being located elsewhere on the protein. In the other two regions that show higher fluctuations, corresponding approximately to the switch I and II regions, the G12V mutant exhibits the largest fluctuations, whereas the Q61L fluctuates a little less than the wild type.
Figure 3.
The root mean-square fluctuations (RMSFs) for (A) the GTP-bound and (B) GDP-bound Cdc42 as a function of the residue number. The thick black bars denote the locations of the switch I and switch II regions, and the light-green background corresponds to the insert region in each figure. The backgrounds with light red and blue denote the β-sheet and α-helical secondary structures of the protein, respectively. Although the two switch regions and the insert region exhibit the largest fluctuations in all systems, the insert region fluctuates much more in the GDP-bound systems. The switch II region fluctuates more in the Cdc42Q61L-GTP and Cdc42G12V-GDP systems. To see this figure in color, go online.
Essential dynamic analysis reveals different conformational dynamics between the GTP- and GDP-bound Cdc42
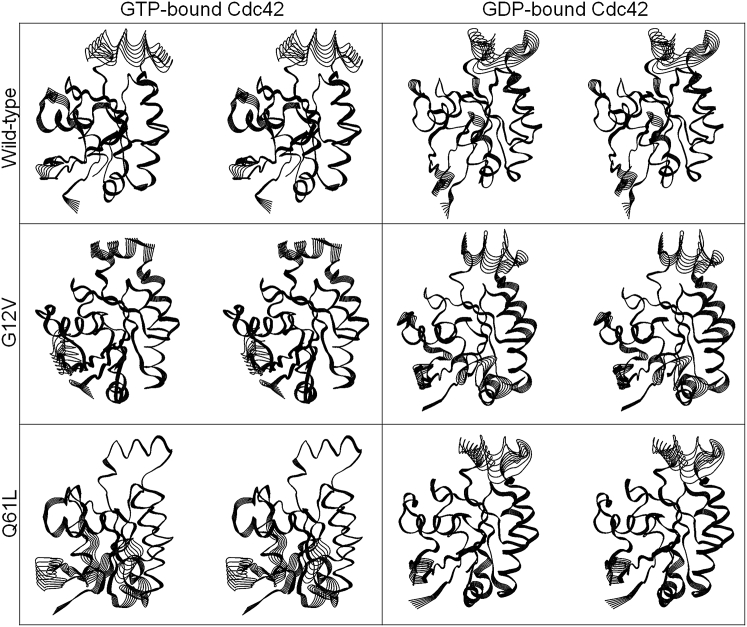
To measure how they differ in their essential modes of motion, we performed essential dynamics analysis on the six systems. The analysis included principal component analysis (PCA) and residue-residue cross correlations. The PCA analysis reveals major differences between the GTP- and GDP-bound systems, illustrating the directions of the main mode of motion for the six Cdc42 systems (Fig. 4). The main mode of motion is the lowest-frequency motion, corresponding to the global motion of the protein. For Cdc42-GTP, the largest motions are in the G2 loop, the region before the start of switch I. In the wild-type Cdc42-GTP, there is also a large motion in the insert region, even though the insert region itself does not lose its integrity but moves as a whole with respect to the rest of the protein. The regions with the largest motions are highly correlated with the largest RMSF values (Fig. 3). For Cdc42-GDP and the mutants, the largest motion is in the insert region.
Figure 4.
Superimpositions of protein motion at the main normal mode with the lowest-frequency motion in stereo pair view for the wild-type and mutant GTP-bound (left column) and wild-type and mutant GDP-bound (right column) Cdc42. The images show the full range of motion for each residue. The motion clearly shows the loss of insert region integrity in the GDP-bound systems and the conformational flexibility of the switch I and switch II regions. Although the insert region has a visible range of motion in the GTP-bound wild-type and G12V mutant, it still maintains its helical structure.
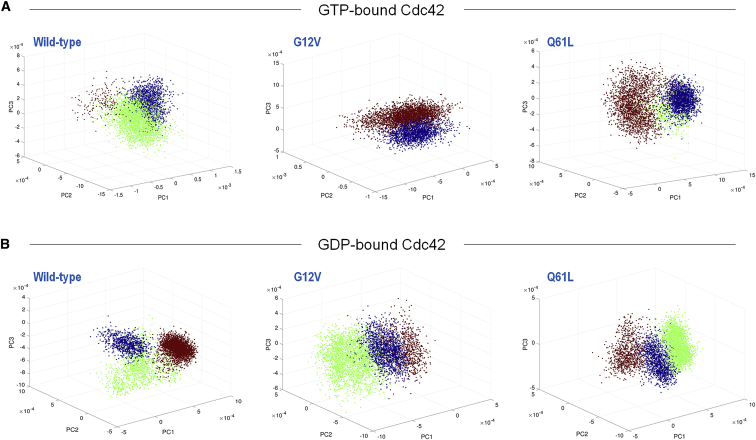
We calculated the PCA projection of the six systems. The projection of the first three principal components is presented for the wild-type and mutant systems in the GTP-bound (Fig. 5 A) and GDP-bound states (Fig. 5 B). In the projections, we clustered the principal components using linkage clustering, which allows us to select the desired number of clusters. We searched for between two and three distinct clusters shown in different colors; clusters 1, 2, and 3 are denoted as blue, green, and red, respectively. Searching for more than three clusters did not result in significantly sized clusters. In the case of CdcG12V-GTP, we obtained two clusters, and all the other systems produced three clusters. For Cdc42-GTP, cluster 1 appears in 28.1% of the conformations (around 100–250 and 350–450 ns of the simulation). Cluster 2 appears in ∼68.2% of the conformations and is dominant in the beginning and middle of the simulation. A small but distinct third cluster exists in only 3.5% of the conformations and appears toward the end of the simulation. The superimposition of representative structures from the clusters illustrates that they differ mainly in the configuration of the switch I region and in the position of the insert region with respect to the rest of the protein (Fig. S2). The G2 loop gets farther from the binding site. However, in all cases, the Thr35 and Tyr32 side chains face the GTP binding site, interacting with it. For Cdc42G12V-GTP, we obtained only two clusters. The difference between them is noticeable mostly in the switch I region. Cluster 2 appears in 67% of the conformations, and cluster 1 appears intermittently in the middle and is dominant in the end of the simulation. For Cdc42Q61L-GTP, cluster 1 appears mostly in 200–400 ns of the simulation and is responsible for 38% of the conformations. Cluster 2 appears in the end of the simulation and is responsible for 20% of the conformations. Cluster 3 appears in the first half of the simulation and is responsible for 42% of the conformations. As in the case of the wild type, the main difference between the three clusters is in the switch I region, where the loop gets farther from the binding site. In cluster 1, the Tyr32 side chain faces away from the GTP, resulting in loss of interaction toward the middle of the simulation. In the case of Cdc42-GDP, the clusters differ in the conformations of the insert region and the switch II loop. Clusters 1, 2, and 3 appear in 17, 25, and 58% of the conformations during the simulations. Cluster 1 appears in the first 80 ns of the simulation, and again toward the end. Then, cluster 2 is more common throughout 80–200 ns. Cluster 3 appears throughout the rest of the simulation (Fig. S2). It appears that the second principal component probably corresponds to the insert region and switch II conformational changes. Cluster 2 differs from the two other clusters in the conformation of the switch I region. On the PC projection, the main difference is along the first principal component. For Cdc42G12V-GDP, the clusters appear at 30, 52, and 18% of the conformations, respectively. Cluster 1 appears between 80 and 250 ns, cluster 2 dominates the middle and end of the simulation, and cluster 3 appears in the first 80 ns. For Cdc42Q61L-GDP, there are also three clusters. Cluster 1 differs from the other two mostly in switch I, but all clusters differ also in the conformation of switch II and the insert region. Cluster 1 appears in 50–200 ns of the simulations and in 26.5% of the conformations. Cluster 2 appears toward the middle and end, with a frequency of 56%. Cluster 3 appears mostly in the beginning, in 17.5% of the conformations. In all three clusters, the side chains of Thr35 and Tyr32 tend to face away from the GDP group. The main backbone difference between the clusters resides in the insert region conformation.
Figure 5.
The projection of the first three principal components, PC1, PC2, and PC3, for the (A) GTP-bound and (B) GDP-bound Cdc42. The PCA projection was subject to linkage clustering, and each cluster is shown in a different color. All the systems show three clusters, except the GTP-G12V mutant, which has two. Representatives of the clusters are shown in Fig. S2. To see this figure in color, go online.
We compared the most populated cluster of Cdc42-GTP with the crystal structure of the GTP-activated Cdc42 (PDB: 5CJP) bound to the GRD of IQGAP2 (Fig. S3). The Cdc42 conformation when bound to GRD2 is very similar to the most populated structure. The overall RMSD was 0.97 Å, including the position of the GTP, even though the simulation did not include the GRD or any other effector binding. Conversely, when a representative of the most populated cluster of Cdc42-GDP was superimposed on the same GRD-bound Cdc42, the RMSD was larger (1.34 Å). Most importantly, the insert region is positioned in a very different way toward the GRD. This further supports the hypothesis that the presence of GDP hinders the conformation of the insert region and, hence, the binding to effectors.
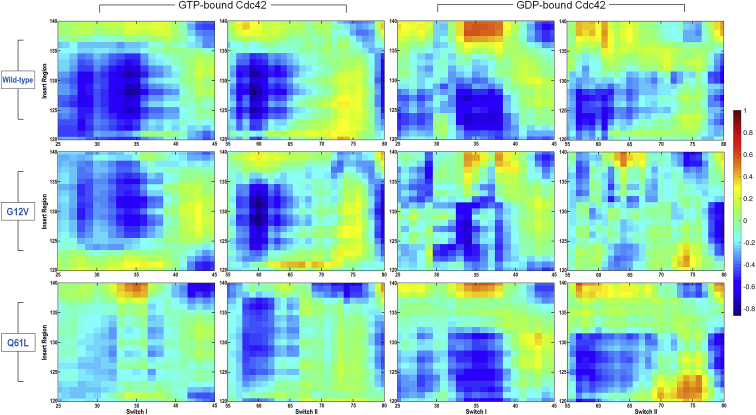
To observe how individual residue motion affects the protein conformation, the dynamic cross correlation map representing the covariance of residues in the insert region and two switch regions, as measured by the PCA analysis, were calculated (Fig. 6). The differences between the GTP-bound and GDP-bound systems are visible, but a closer focus gives us more insight into the important regions (between the switch I and insert regions, the switch II and insert regions, and the switch I and II regions (Fig. S4)). The most prominent correlation is a strong negative correlation between the switch I region and the insert regions of both GTP-bound and GDP-bound systems. Switch I undergoes the largest conformational changes in Cdc42-GTP, while the insert region is rather stable. In Cdc42-GDP, the insert region, which undergoes the largest conformational change throughout the simulation, shows a slightly weaker negative correlation with the two switch regions. There are several visible differences between the GTP-bound and GDP-bound systems, as well as between the wild type and mutants in each case. The Cdc42G12V-GTP behaves in a similar way to the wild type, but in the Cdc42Q61L-GTP, the mutations cause a stronger positive correlation in the motion of the two switches but a weak correlation between switch I and II and the insert region. In contrast, in the GDP-bound systems, the Cdc42Q61L-GDP shows a stronger positive correlation between the two switch regions (Fig. S4), similar to the Cdc42-GTP systems. Notice that even though the conformational change pattern is similar between the wild type and the mutants in the GDP-bound state (in all cases the insert region undergoes large-scale changes), there is a marked difference in the cross correlation pattern.
Figure 6.
Dynamic cross correlation map representing the covariance of residues between the switch I and insert region regions, and the switch II and insert region for the GTP-bound (left panels) and GDP-bound (right panels) Cdc42. The cross correlation values are depicted on a scale from blue to red. The figure shows big differences in the cross correlation between the GTP-bound and GDP-bound systems. In particular, the wild-type and G12V GTP-bound systems show stronger negative cross correlation between the insert region and the two switch regions (especially switch I). Weaker correlations are observed in the GTP-bound Q61L mutant. To see this figure in color, go online.
Interactions between CDC42 and the nucleotide around the binding site
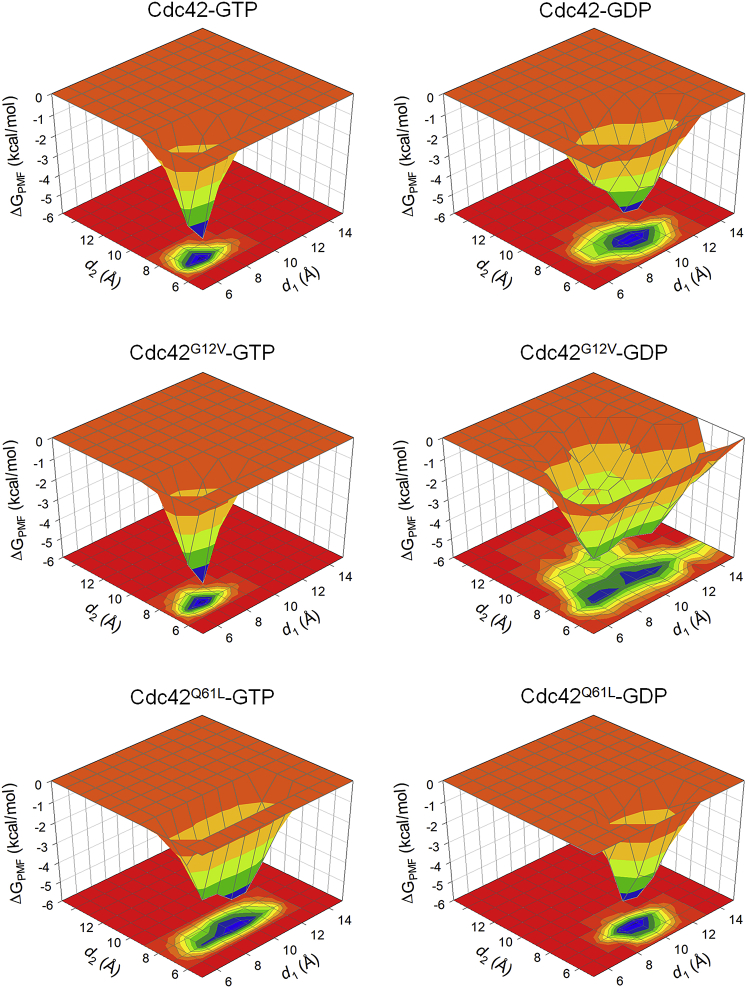
Several residues are involved in the binding of Ras superfamily proteins with the nucleotide—in particular, Gly12/Val12, Ala13, Tyr32, Thr35, and Gln61 (37,47). We calculated the potential of mean force (PMF) along the reaction coordinates d1, defined as the distance between the Cα atom of Gly60 and the Pβ atom (the second phosphorus) in the GTP/GDP molecule, and d2, defined as the distance between the Cα atom of Thr35 and the Pβ atom in the nucleotides (Fig. 7). The choice of distances was made to compare with the results obtained for K-Ras4B and K-Ras4A in our previous studies (40,48,50). The PMF profiles allow us to detect regions of energy minima and elucidate active and inactive states. In all GTP-bound systems, the distance between the Cα atom of Thr35 and the Pβ atom in the nucleotides was stable, as well as in the GDP-bound Q61L mutant. The main difference was in the distance of Gly60 from the Pβ atom, which was still stable in the GTP-bound systems but fluctuated in the GDP-bound systems, in particular the GDP-bound G12V mutant. In addition to these atoms, we also measured the distances from the Cα atom of Ala13, the OH group of Tyr32, the Oγ group of Thr35, and the Cδ atom of Gln61 (or Leu61 in the Q61L mutants) to the Pβ atom to monitor the side-chain orientation (Fig. S5). The distances are summarized in Table 2. As expected, the GDP-bound systems fluctuate more than the GTP-bound systems. In the GTP-bound systems, Ala13 and Thr35 show stable distances from the GTP molecule, indicating that the G2 loop and the switch I loop maintain close, consistent distance from the GTP. Tyr32 fluctuates throughout the simulation, but to a lesser extent than the GDP-bound system. The distances are usually between ∼6 and 8 Å, with a few spikes to ∼12–14 Å, indicating that, at times, the Tyr32 side chain faces away from the GTP. In both the wild-type and the G12V mutant of Cdc42-GTP, Gln61 maintained a rather stable distance of ∼8–10 Å from the nucleotide group. In the Q61L mutant, the distance of the CD1 atom of Leu61 was larger and less stable, indicating loss of interaction. In the GDP-bound systems, the largest fluctuations are observed with respect to the Tyr32 and, in the case of the G12V mutant and to a lesser extent the wild-type GDP, also with respect to the Thr35 interactions. The distances between these two residues and the GDP molecule fluctuate between ∼8 and ∼14–16 (for Thr35) and 22 Å (for Tyr 32) throughout the simulation. This shows that, for most of the simulation time, the two residues lose their interaction with the GDP, except for short periods. The distances are also larger than in the GTP-bound systems, which is consistent with the GDP-bound molecules having an open conformation. In the K-Ras4B simulations, we observed that several mutants also caused a near-total loss of switch I interactions with the GDP. The distance of Ala13 from the GDP is rather stable in all the GDP-bound systems, whereas Gln/Leu61 had a larger distance from the GDP than the GTP-bound systems. As in the K-Ras4B simulations (48), the distance between Thr35 and the GDP fluctuated.
Figure 7.
The potential of mean force (ΔGPMF) along the reaction coordinates d1 (defined by the distance from G60-Cα to GTP/GDP-Pβ atom) and d2 (defined by the distance from T35-Cα to GTP/GDP-Pβ). As seen, the distances are very stable in the GTP-bound systems and the GDP-bound Q61L mutant. Both d1 and d2 fluctuate more in the GDP-bound G12V mutant, and they fluctuate to a lesser extent in the GDP-bound wild type. To see this figure in color, go online.
Table 2.
The Averaged Distances (in Angstroms) of Selected Residues from the Pβ Atom of the GDP/GTP Molecule during the Simulation
| System | Thr35-Cα | Gly60-Cα | Ala13-Cα | Tyr32-OH | Gln/Leu61-CD |
|---|---|---|---|---|---|
| Cdc42-GTP | 7.2 ± 0.1 | 6.8 ± 0.3 | 4.0 ± 0.1 | 7.6 ± 1.5 | 9.4 ± 0.8 |
| Cdc42G12V-GTP | 7.2 ± 0.1 | 6.8 ± 0.3 | 4.0 ± 0.1 | 8.1 ± 2.2 | 10.1 ± 0.7 |
| Cdc42Q61L-GTP | 7.3 ± 0.2 | 9.2 ± 1.0 | 4.0 ± 0.1 | 8.0 ± 1.8 | 12.1 ± 1.5 |
| Cdc42-GDP | 6.9 ± 0.3 | 9.5 ± 0.7 | 4.0 ± 0.1 | 13.6 ± 2.9 | 10.5 ± 1.9 |
| Cdc42G12V-GDP | 7.4 ± 0.9 | 9.5 ± 1.8 | 3.9 ± 0.1 | 12.7 ± 2.3 | 10.5 ± 1.8 |
| Cdc42Q61L-GDP | 6.8 ± 0.2 | 9.7 ± 0.5 | 4.0 ± 0.1 | 12.7 ± 2.5 | 12.8 ± 1.5 |
Discussion
In this work, we used MD simulations and essential dynamics analysis to investigate the structural and dynamical properties of the GTP-bound (active) and GDP-bound (inactive) Cdc42. We found that the active and inactive systems differ significantly in their dynamical properties, especially in areas formerly characterized to be important for activation or for binding with the GRD of IQGAP2 (34,67). Specifically, we show that GDP-bound Cdc42 exhibits much larger conformational flexibility in the insert region, which is associated with activation as well as binding to effectors. The GDP-bound insert region is highly dynamic, exhibiting the unfolded/folded α-helical secondary structure during the simulation time (Fig. S6). This may suggest that the inactive form has reduced ability to bind to effectors, possibly as a result of allosteric effects taking place at the nucleotide binding site. The effect is through allosteric pathways, as the insert region does not interact directly with the binding site. Recently, it has been shown that order-disorder-mediated allosteric interactions are involved in many instances of protein activity, including the inhibition of the Rac1 GTPase (68). Recent work suggests that mutations may also have an effect on allosteric pathways (69,70).
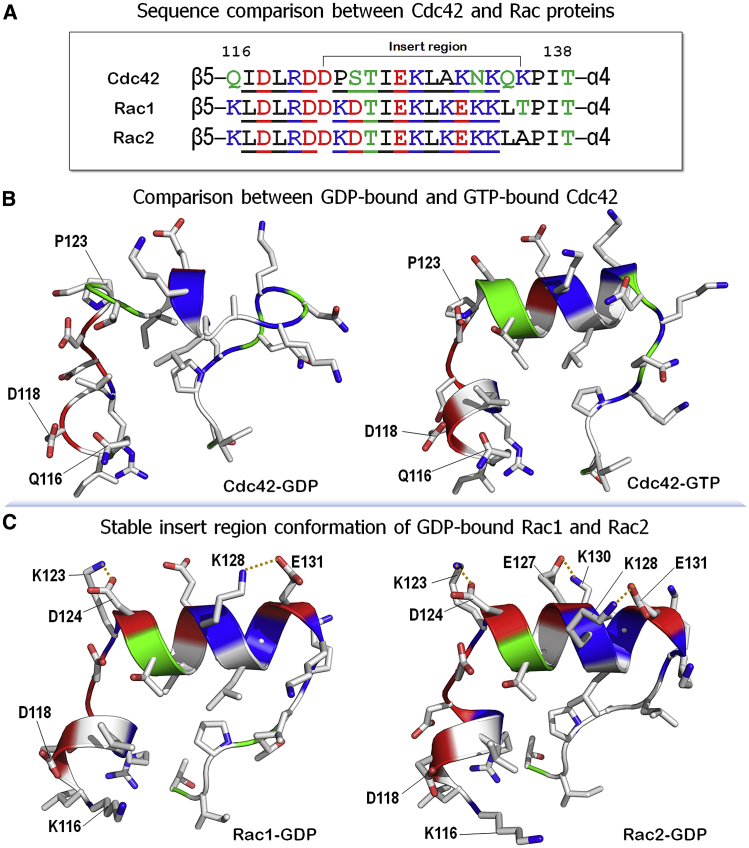
To corroborate the conformational instability of the insert region in the GDP-bound Cdc42, we compared the sequences of the insert region and its adjacent loop regions between Cdc42 and Rac proteins (Fig. 8 A). Although these proteins share a similar sequence for the insert region, some unmatched residues significantly contribute to the stability of insert region. To compare the region between β5 and the insert region for GTP-bound and GDP-bound Cdc42 (Fig. 8 B), the 117xDLRx121 motif in the GTP-bound state folds into an α-helix, whereas the motif is disordered in the GDP-bound state. It appears that the α-helical moiety in the motif is critical to preserve the α-helical structure of the insert region. For Cdc42-GDP, the large fluctuations of switch I and switch II lead to GDP slipping from the catalytic site, resulting in the interactions of Asp118 and Gln116 with the guanine group of GDP, which becomes unstable (Fig. S7). This induces large fluctuations in the 117xDLRx121 motif and, hence, the unstable insert region. Pro123 plays a role in reducing the stability of the insert region, disrupting the α-helical secondary structure. We found that in the Cdc42-GDP, Lys131 in the insert region forms a salt bridge with Glu91 in helix α3. The salt bridge, appearing in ∼10% of the conformations, may help in maintaining the disordered helical conformation.
Figure 8.
Comparison between Cdc42 and Rac1/Rac2 proteins. (A) Sequences of the insert region and its adjacent regions for Cdc42, Rac1, and Rac2. In the sequence, hydrophobic, polar/glycine, positively charged, and negatively charged residues are colored black, green, blue, and red, respectively. Underlining denotes the residues presenting the α-helical structure. The former is the helix-forming 117xDLRx121 motif, and the latter is the insert region. Snapshots representing the final structures of insert region for (B) GDP-bound (left panel) and GTP-bound (right panel) Cdc42 and (C) Rac1-GDP (left panel) and Rac2-GDP (right panel) are shown. Rac proteins preserved a highly stable insert region during a 1-μs simulation. In the structures, colors are the same as in the sequence, except for the hydrophobic resides with white. Dotted lines denote salt bridges. To see this figure in color, go online.
Unlike Cdc42-GDP, both Rac1-GDP and Rac2-GDP yield stable α-helical structures for both the 117xDLRx121 motif and the insert region (Fig. 8 C). During the simulations, we observed that stable salt bridge interactions between the side chains of the residues enhance the stability of the insert region for both Rac proteins, preserving the α-helical secondary structure (Fig. S7). Based on the sequence, we note that Rac proteins intrinsically possess a more stable insert region than Cdc42. For Rac proteins, in addition to Asp118, Lys116 plays a critical role in enhancing the interaction with the guanine group of GDP, preventing GDP’s slip from the catalytic site due to large fluctuations of switch I and switch II. Lys116 of Rac proteins forms a salt bridge with GDP, but Gln116 of Cdc42 does not (Fig. 8). Unlike Pro123 of Cdc42, Lys123 of Rac proteins contributes the stable insert region α-helix through the salt bridge formation with Asp12, which is absent from Cdc42.
We also simulated and analyzed two mutants, G12V and Q61L, that play a role in GTPase activation (44,71). Our results show that the differences in dynamics and structure between the wild type and the mutants are smaller than the differences between the wild-type GTP and GDP. The main differences involve the conformational dynamics of the switch I and switch II regions, which are associated with IQGAP2 binding. Collectively, our results can shed light on the dynamic differences between the active and inactive conformation caused by the differences in the conformational flexibility of the insert region, which may explain their different levels of activity.
Author Contributions
N.H., H.J, and R.N. conceived and designed the study. N.H. performed MD simulations and prepared the first draft. N.H. and H.J. analyzed the data, generated figures, and wrote the manuscript. All authors edited and approved the manuscript.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This research was supported (in part) by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research. The simulations were run in part on the UMass Boston research cluster and the Massachusetts Green High Performance Computing Center (MGHPCC) cluster and in part on the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (https://hpc.nih.gov/).
Editor: Alexandr Kornev.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.12.007.
Supporting Material
References
- 1.Zong H., Kaibuchi K., Quilliam L.A. The insert region of RhoA is essential for Rho kinase activation and cellular transformation. Mol. Cell. Biol. 2001;21:5287–5298. doi: 10.1128/MCB.21.16.5287-5298.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ridley A.J. Rho GTPases and cell migration. J. Cell Sci. 2001;114:2713–2722. doi: 10.1242/jcs.114.15.2713. [DOI] [PubMed] [Google Scholar]
- 3.Owen D., Campbell L.J., Mott H.R. The IQGAP1-Rac1 and IQGAP1-Cdc42 interactions: interfaces differ between the complexes. J. Biol. Chem. 2008;283:1692–1704. doi: 10.1074/jbc.M707257200. [DOI] [PubMed] [Google Scholar]
- 4.Jang H., Muratcioglu S., Nussinov R. Membrane-associated Ras dimers are isoform-specific: K-Ras dimers differ from H-Ras dimers. Biochem. J. 2016;473:1719–1732. doi: 10.1042/BCJ20160031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L., Zhang L., Gu L. Non-canonical notch signaling regulates actin remodeling in cell migration by activating PI3K/AKT/Cdc42 pathway. Front. Pharmacol. 2019;10:370. doi: 10.3389/fphar.2019.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narumiya S., Thumkeo D. Rho signaling research: history, current status and future directions. FEBS Lett. 2018;592:1763–1776. doi: 10.1002/1873-3468.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pichaud F., Walther R.F., Nunes de Almeida F. Regulation of Cdc42 and its effectors in epithelial morphogenesis. J. Cell Sci. 2019;132:jcs217869. doi: 10.1242/jcs.217869. [DOI] [PubMed] [Google Scholar]
- 8.Huang Q.-Y., Lai X.-N., Xiong L.-X. Cdc42: a novel regulator of insulin secretion and diabetes-associated diseases. Int. J. Mol. Sci. 2019;20:179. doi: 10.3390/ijms20010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustelo X.R. RHO GTPases in cancer: known facts, open questions, and therapeutic challenges. Biochem. Soc. Trans. 2018;46:741–760. doi: 10.1042/BST20170531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maldonado M.D.M., Dharmawardhane S. Targeting Rac and Cdc42 GTPases in cancer. Cancer Res. 2018;78:3101–3111. doi: 10.1158/0008-5472.CAN-18-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris L.E., Bloom G.S., Powell S.M. Nucleotide variants within the IQGAP1 gene in diffuse-type gastric cancers. Genes Chromosomes Cancer. 2005;42:280–286. doi: 10.1002/gcc.20150. [DOI] [PubMed] [Google Scholar]
- 12.Cardama G.A., Gonzalez N., Gomez D.E. Rho GTPases as therapeutic targets in cancer (Review) Int. J. Oncol. 2017;51:1025–1034. doi: 10.3892/ijo.2017.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamas I., Merlini L., Martin S.G. Optogenetics reveals Cdc42 local activation by scaffold-mediated positive feedback and Ras GTPase. PLoS Biol. 2020;18:e3000600. doi: 10.1371/journal.pbio.3000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maldonado M.D.M., Medina J.I., Dharmawardhane S. Targeting Rac and Cdc42 GEFs in metastatic cancer. Front. Cell Dev. Biol. 2020;8:201. doi: 10.3389/fcell.2020.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pálfy G., Vida I., Perczel A. 1H, 15N backbone assignment and comparative analysis of the wild type and G12C, G12D, G12V mutants of K-Ras bound to GDP at physiological pH. Biomol. NMR Assign. 2020;14:1–7. doi: 10.1007/s12104-019-09909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 17.Stengel K., Zheng Y. Cdc42 in oncogenic transformation, invasion, and tumorigenesis. Cell. Signal. 2011;23:1415–1423. doi: 10.1016/j.cellsig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aspenström P. Activated Rho GTPases in cancer-the beginning of a new paradigm. Int. J. Mol. Sci. 2018;19:3949. doi: 10.3390/ijms19123949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wennerberg K., Rossman K.L., Der C.J. The Ras superfamily at a glance. J. Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 20.Castellano E., Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer. 2011;2:216–231. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vetter I.R., Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 22.Liao T.-J., Jang H., Nussinov R. Allosteric KRas4B can modulate SOS1 fast and slow Ras activation cycles. Biophys. J. 2018;115:629–641. doi: 10.1016/j.bpj.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris K.M., Henderson R., Adams P.D. Intrinsic GTP hydrolysis is observed for a switch 1 variant of Cdc42 in the presence of a specific GTPase inhibitor. Small GTPases. 2016;7:1–11. doi: 10.1080/21541248.2015.1123797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Zhang Y., Gerwert K. Specific substates of Ras to interact with GAPs and effectors: revealed by theoretical simulations and FTIR experiments. J. Phys. Chem. Lett. 2018;9:1312–1317. doi: 10.1021/acs.jpclett.8b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudas B., Merzel F., Balog E. Nucleotide-specific autoinhibition of full-length K-Ras4B identified by extensive conformational sampling. Front. Mol. Biosci. 2020;7:145. doi: 10.3389/fmolb.2020.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar A.P., Verma C.S., Lukman S. Structural dynamics and allostery of Rab proteins: strategies for drug discovery and design. Brief. Bioinform. 2020 doi: 10.1093/bib/bbz161. Published online January 17, 2020. [DOI] [PubMed] [Google Scholar]
- 27.Wu W.J., Leonard D.A., Manor D. Interaction between Cdc42Hs and RhoGDI is mediated through the Rho insert region. J. Biol. Chem. 1997;272:26153–26158. doi: 10.1074/jbc.272.42.26153. [DOI] [PubMed] [Google Scholar]
- 28.Walker S.J., Wu W.J., Brown H.A. Activation of phospholipase D1 by Cdc42 requires the Rho insert region. J. Biol. Chem. 2000;275:15665–15668. doi: 10.1074/jbc.M000076200. [DOI] [PubMed] [Google Scholar]
- 29.Miyano K., Koga H., Sumimoto H. The insert region of the Rac GTPases is dispensable for activation of superoxide-producing NADPH oxidases. Biochem. J. 2009;422:373–382. doi: 10.1042/BJ20082182. [DOI] [PubMed] [Google Scholar]
- 30.Kukimoto-Niino M., Tsuda K., Shirouzu M. Structural basis for the dual substrate specificity of DOCK7 guanine nucleotide exchange factor. Structure. 2019;27:741–748.e3. doi: 10.1016/j.str.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Kang N., Liu J., Zhao Y. Dissociation mechanism of GDP from Cdc42 via DOCK9 revealed by molecular dynamics simulations. Proteins. 2019;87:433–442. doi: 10.1002/prot.25665. [DOI] [PubMed] [Google Scholar]
- 32.Sahai E., Alberts A.S., Treisman R. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 1998;17:1350–1361. doi: 10.1093/emboj/17.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansson T., Castillo-Castrejon M., Rosario F.J. Down-regulation of placental Cdc42 and Rac1 links mTORC2 inhibition to decreased trophoblast amino acid transport in human intrauterine growth restriction. Clin. Sci. (Lond.) 2020;134:53–70. doi: 10.1042/CS20190794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozdemir E.S., Jang H., Nussinov R. Unraveling the molecular mechanism of interactions of the Rho GTPases Cdc42 and Rac1 with the scaffolding protein IQGAP2. J. Biol. Chem. 2018;293:3685–3699. doi: 10.1074/jbc.RA117.001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heo W.D., Meyer T. Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell. 2003;113:315–328. doi: 10.1016/s0092-8674(03)00315-5. [DOI] [PubMed] [Google Scholar]
- 36.Vatansever S., Erman B., Gümüş Z.H. Comparative effects of oncogenic mutations G12C, G12V, G13D, and Q61H on local conformations and dynamics of K-Ras. Comput. Struct. Biotechnol. J. 2020;18:1000–1011. doi: 10.1016/j.csbj.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melendez J., Grogg M., Zheng Y. Signaling role of Cdc42 in regulating mammalian physiology. J. Biol. Chem. 2011;286:2375–2381. doi: 10.1074/jbc.R110.200329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinelli S., Krumbach O.H.F., Mirzaa G.M., University of Washington Center for Mendelian Genomics Functional dysregulation of CDC42 causes diverse developmental phenotypes. Am. J. Hum. Genet. 2018;102:309–320. doi: 10.1016/j.ajhg.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandrashekar R., Salem O., Adams P.D. A switch I mutant of Cdc42 exhibits less conformational freedom. Biochemistry. 2011;50:6196–6207. doi: 10.1021/bi2004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu S., Jang H., Zhang J. Ras conformational ensembles, allostery, and signaling. Chem. Rev. 2016;116:6607–6665. doi: 10.1021/acs.chemrev.5b00542. [DOI] [PubMed] [Google Scholar]
- 41.Bera A.K., Lu J., Westover K.D. GTP hydrolysis is modulated by Arg34 in the RASopathy-associated KRASP34R. Birth Defects Res. 2020;112:708–717. doi: 10.1002/bdr2.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zanuy D., Kotla R., Haspel N. Sequence dependence of C-end rule peptides in binding and activation of neuropilin-1 receptor. J. Struct. Biol. 2013;182:78–86. doi: 10.1016/j.jsb.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tichauer R.H., Favre G., Brut M. Water distribution within wild-type NRas protein and Q61 mutants during unrestrained QM/MM dynamics. Biophys. J. 2018;115:1417–1430. doi: 10.1016/j.bpj.2018.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arrington M.E., Temple B., Campbell S.L. The molecular basis for immune dysregulation by the hyperactivated E62K mutant of the GTPase RAC2. J. Biol. Chem. 2020;295:12130–12142. doi: 10.1074/jbc.RA120.012915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khrenova M.G., Mironov V.A., Nemukhin A.V. Modeling the role of G12V and G13V Ras mutations in the Ras-GAP-catalyzed hydrolysis reaction of guanosine triphosphate. Biochemistry. 2014;53:7093–7099. doi: 10.1021/bi5011333. [DOI] [PubMed] [Google Scholar]
- 46.Grigorenko B.L., Kots E.D., Nemukhin A.V. Diversity of mechanisms in Ras-GAP catalysis of guanosine triphosphate hydrolysis revealed by molecular modeling. Org. Biomol. Chem. 2019;17:4879–4891. doi: 10.1039/c9ob00463g. [DOI] [PubMed] [Google Scholar]
- 47.Hunter J.C., Manandhar A., Westover K.D. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res. 2015;13:1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 48.Lu S., Jang H., Zhang J. The structural basis of oncogenic mutations G12, G13 and Q61 in small GTPase K-Ras4B. Sci. Rep. 2016;6:21949. doi: 10.1038/srep21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nussinov R., Jang H., Cheng F. Precision medicine review: rare driver mutations and their biophysical classification. Biophys. Rev. 2019;11:5–19. doi: 10.1007/s12551-018-0496-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chakrabarti M., Jang H., Nussinov R. Comparison of the conformations of KRAS isoforms, K-Ras4A and K-Ras4B, points to similarities and significant differences. J. Phys. Chem. B. 2016;120:667–679. doi: 10.1021/acs.jpcb.5b11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.LeCour L., Jr., Boyapati V.K., Worthylake D.K. The structural basis for cdc42-induced dimerization of IQGAPs. Structure. 2016;24:1499–1508. doi: 10.1016/j.str.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lam B.D., Hordijk P.L. The Rac1 hypervariable region in targeting and signaling: a tail of many stories. Small GTPases. 2013;4:78–89. doi: 10.4161/sgtp.23310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson D.I. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson J.L., Erickson J.W., Cerione R.A. C-terminal di-arginine motif of Cdc42 protein is essential for binding to phosphatidylinositol 4,5-bisphosphate-containing membranes and inducing cellular transformation. J. Biol. Chem. 2012;287:5764–5774. doi: 10.1074/jbc.M111.336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beaumont V.A., Reiss K., Loria J.P. Allosteric impact of the variable insert loop in vaccinia H1-related (VHR) phosphatase. Biochemistry. 2020;59:1896–1908. doi: 10.1021/acs.biochem.0c00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruan Z., Kannan N. Altered conformational landscape and dimerization dependency underpins the activation of EGFR by αC-β4 loop insertion mutations. Proc. Natl. Acad. Sci. USA. 2018;115:E8162–E8171. doi: 10.1073/pnas.1803152115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeung W., Ruan Z., Kannan N. Emerging roles of the αC-β4 loop in protein kinase structure, function, evolution, and disease. IUBMB Life. 2020;72:1189–1202. doi: 10.1002/iub.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pantsar T. KRAS(G12C)-AMG 510 interaction dynamics revealed by all-atom molecular dynamics simulations. Sci. Rep. 2020;10:11992. doi: 10.1038/s41598-020-68950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khaled M., Gorfe A., Sayyed-Ahmad A. Conformational and dynamical effects of Tyr32 phosphorylation in K-Ras: molecular dynamics simulation and Markov state models analysis. J. Phys. Chem. B. 2019;123:7667–7675. doi: 10.1021/acs.jpcb.9b05768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pantsar T. The current understanding of KRAS protein structure and dynamics. Comput. Struct. Biotechnol. J. 2019;18:189–198. doi: 10.1016/j.csbj.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knihtila R., Volmar A.Y., Mattos C. Titration of ionizable groups in proteins using multiple neutron data sets from a single crystal: application to the small GTPase Ras. Acta Crystallogr. F Struct. Biol. Commun. 2019;75:111–115. doi: 10.1107/S2053230X18018125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brooks B., Bruccoleri R.E., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 63.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jang H., Banerjee A., Nussinov R. The higher level of complexity of K-Ras4B activation at the membrane. FASEB J. 2016;30:1643–1655. doi: 10.1096/fj.15-279091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 66.Bakan A., Meireles L.M., Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nouri K., Timson D.J., Ahmadian M.R. New model for the interaction of IQGAP1 with CDC42 and RAC1. Small GTPases. 2020;11:16–22. doi: 10.1080/21541248.2017.1321169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tee W.-V., Guarnera E., Berezovsky I.N. Disorder driven allosteric control of protein activity. Curr. Res. Struct. Biol. 2020;2:191–203. doi: 10.1016/j.crstbi.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guarnera E., Berezovsky I.N. Allosteric drugs and mutations: chances, challenges, and necessity. Curr. Opin. Struct. Biol. 2020;62:149–157. doi: 10.1016/j.sbi.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 70.Tee W.-V., Guarnera E., Berezovsky I.N. On the allosteric effect of nsSNPs and the emerging importance of allosteric polymorphism. J. Mol. Biol. 2019;431:3933–3942. doi: 10.1016/j.jmb.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 71.Hodge R.G., Schaefer A., Der C.J. RAS and RHO family GTPase mutations in cancer: twin sons of different mothers? Crit. Rev. Biochem. Mol. Biol. 2020;55:386–407. doi: 10.1080/10409238.2020.1810622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.