Abstract
The Atlantic sea nettle (Chrysaora quinquecirrha) has an important evolutionary position due to its high ecological value. However, due to limited sequencing technologies and complex jellyfish genomic sequences, the current C. quinquecirrha genome assembly is highly fragmented. Here, we used the most advanced high-throughput chromosome conformation capture (Hi-C) technology to obtain high-coverage sequencing data of the C. quinquecirrha genome. We then anchored these data to the previously published contig-level assembly to improve the genome. Finally, a high-continuity genome sequence of C. quinquecirrha was successfully assembled, which contained 1 882 scaffolds with a N50 length of 3.83 Mb. The N50 length of the genome assembly was 5.23 times longer than the previously released one, and additional analysis revealed that it had a high degree of genomic continuity and accuracy. Acquisition of the high-continuity genome sequence of C. quinquecirrha not only provides a basis for the study of jellyfish evolution through comparative genomics but also provides an important resource for studies on jellyfish growth and development.
Keywords: Jellyfish;, Genome, Assembly, Hi-C, High continuity, Evolution
DEAR EDITOR,
The Atlantic sea nettle (Chrysaora quinquecirrha) has an important evolutionary position due to its high ecological value. However, due to limited sequencing technologies and complex jellyfish genomic sequences, the current C. quinquecirrha genome assembly is highly fragmented. Here, we used the most advanced high-throughput chromosome conformation capture (Hi-C) technology to obtain high-coverage sequencing data of the C. quinquecirrha genome. We then anchored these data to the previously published contig-level assembly to improve the genome. Finally, a high-continuity genome sequence of C. quinquecirrha was successfully assembled, which contained 1 882 scaffolds with a N50 length of 3.83 Mb. The N50 length of the genome assembly was 5.23 times longer than the previously released one, and additional analysis revealed that it had a high degree of genomic continuity and accuracy. Acquisition of the high-continuity genome sequence of C. quinquecirrha not only provides a basis for the study of jellyfish evolution through comparative genomics but also provides an important resource for studies on jellyfish growth and development.
Jellyfishes belong to the phylum Cnidaria, which are lower invertebrate umbrella-shaped gelatinous zooplankton. Jellyfish, especially C. quinquecirrha, have substantial ecological impact due to their wide distribution, ranging from the southern coast of New England to tropical areas of the eastern coast of North America (Decker et al., 2007). Atlantic sea nettles are fertile in late spring and early summer, and large populations can have a significant impact on fisheries (Olesen et al., 1996). Additionally, continuous blooms of gelatinous zooplankton can permanently disrupt natural food webs (Oguz et al., 2012). This disruption is because jellyfish consume eggs, larvae, and juveniles, and thus can have long-term effects on commercially important fishery species (Finenko et al., 2013).
Acquisition of the genome sequence could help in C. quinquecirrha research, including on their developmental processes. Fortunately, the first reference genome of C. quinquecirrha was assembled and released recently (Xia et al., 2020). However, due to its complexity and high heterozygosity, the assembled genome is very fragmented, thereby hindering further study of this species. Several sequencing technologies, such as Bionano optical mapping (http://www.bionanogenomics.com), 10x genomics (https://www.10xgenomics.com), and Hi-C (Pal et al., 2019), have been developed to help the assembly of high-continuity genomes (Chen et al., 2020; Dudchenko et al., 2017; Ghurye et al., 2019). The Hi-C technique has been widely used for the assembly of high-quality genomes (Chen et al., 2020; Dudchenko et al., 2017; Ghurye et al., 2019). With second-generation sequencing, Hi-C can obtain high-throughput data of the genomic loci and measure physical interactions. Moreover, Hi-C can measure the frequency of interactions within and between different chromosomes, including the number of interactions between chromosome fragments (Pal et al., 2019). Additionally, different chromosomes can be distinguished by identifying differences in the frequency of direct interactions between different regions, thereby constructing a genome at the chromosome level (Pal et al., 2019).
In this study, we generated high-coverage Hi-C sequencing data of C. quinquecirrha, which we then anchored to the previously published genome to generate a high-continuity assembly. Fresh muscle samples of C. quinquecirrha were dissected and used for high-quality DNA extraction with a Qiagen Blood & Cell Culture DNA Mini Kit (Germany). The Hi-C library was then prepared via digestion of the DpnII restriction enzyme and polymerase chain reaction (PCR) enrichment by Novogene (China) on the NovaSeq 6000 sequencing platform (Illumina, USA) with a read length of 150 bp. Raw Hi-C sequencing reads with more than 30% low-quality bases or 10% unknown bases were filtered. Duplicate reads, which may be produced during PCR, and adaptor sequences were also removed as described in previous study (Chen et al., 2020). All remaining sequencing reads were used for further analysis.
The clean Hi-C sequencing reads were then used for high-continuity genome assembly. We used Juicer (v1.5.6) (Durand et al., 2016b) to align all clean Hi-C reads to the previously published contig-level genome (Xia et al., 2020), with obviously duplicated mapping regions removed. The default parameters of Juicer were used, except –S was set to “early”. We then anchored the contigs into long sequences using 3D de novo assembly software (v170123) (Dudchenko et al., 2017) with parameters “-m haploid -i 15000 -r 0”. We used Juicebox (v1.9.8) (Durand et al., 2016a) to visualize the chromosome assembly after raw genome construction. According to the sequence interactions, we modified the fragments with obvious assembly errors. We obtained the final high-continuity genome after adjusting minor errors in connection order.
The genome annotation workflow used was the same as in the previous study (Xia et al., 2020), except additional de novo prediction software were used. Specifically, we used SNAP (v2006-07-28) (Korf, 2004) with the HMM library (mam54.hmm) and default parameters. We next predicted the coding-region using Genscan (v1.0) (Burge & Karlin, 1997) with the HumanIso library. In addition, GlimmerHMM (v3.0.1) (Majoros et al., 2004) was used in the prediction of the coding regions. We analyzed genome synteny between C. quinquecirrha and Aurelia aurita (GCA_004194415.1_ABSv1) using LAST (v802) (Kiełbasa et al., 2011) with parameters “-m 100 –E 0.05”. The one-to-one comparison areas in the obtained maf file were selected for plotting, and the syntenic blocks between genomes were plotted using Circos (v0.69-6) (Krzywinski et al., 2009).
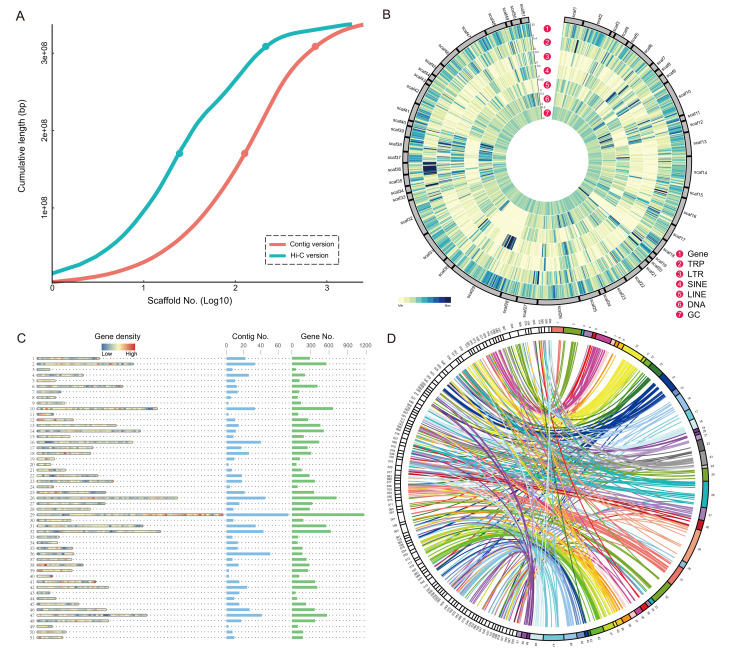
Although several jellyfish genome assemblies have been published in recent years, most are highly fragmented (Jiang et al., 2019; Leclère et al., 2019). For C. quinquecirrha, the published genome contig N50 is 733 647 kb. To acquire a high-continuity assembly for the C. quinquecirrha genome, we sequenced high-coverage (~272 Gb) Hi-C data, which were mapped to the previously published genome (Xia et al., 2020). To obtain a more accurate genome assembly, we constructed long sequences using 3D de novo assembly software, allowing broken contig sequences. Finally, we identified 51 scaffolds with obvious edges, and ~67.18% of the total contig length was assembled into super-scaffolds (Supplementary Table 1; Supplementary Figures 1, 2). Results showed that this assembly, with a N50 of 3.83 Mb (Table 1, >Supplementary Table 2), was 5.23 times longer than the earlier version (Xia et al., 2020). In addition, the cumulative assembly length showed by the L50 (smallest number of sequences that make up at least 50% of the total assembly) statistics between the two genome versions (contig and Hi-C) indicated substantial improvement in connectivity degree of C. quinquecirrha (Figure 1A). The BUSCO scores were also significantly improved compared to the contig version (Supplementary Table 3).
Table 1. Statistics of two genome versions.
| Statistical item | Length (bp) | Number | Length (bp) | Number |
| Version | Contig-version (Xia et al., 2020) | Hi-C version (this study) | ||
| N90 | 66 354 | 666 | 227 000 | 195 |
| N80 | 205 342 | 365 | 582 500 | 109 |
| N70 | 395 469 | 249 | 873 000 | 62 |
| N60 | 555 468 | 178 | 2 312 428 | 36 |
| N50 | 733 647 | 125 | 3 825 607 | 24 |
| Average length (bp) | 134 943 | 179 287 | ||
| Max length (bp) | 4 015 784 | 15 257 941 | ||
| Total length (bp) | 336 819 409 | 337 419 359 | ||
| Total number | 2 496 | 1 882 | ||
| Number ≥1 000 (bp) | 2 496 | 1 880 | ||
Figure 1.
Statistics and evaluation of Hi-C-based genome assembly
A: Cumulative assembly length of sequences from two genome assemblies of C. quinquecirrha. Two versions, including previously published contig version (Xia et al., 2020) and Hi-C version from this study, were used in statistical analysis. Dots above and below each line indicate L50 and L90 values, respectively. B: Scaffold-level genome assembly of C. quinquecirrha. Assembly results are shown in Circos diagram, with outer to inner rings showing distribution of protein-coding genes, tandem repeats (TRP), long tandem repeats (LTR), short interspersed repetitive elements (SINE), long interspersed repetitive elements (LINE), DNA elements, and GC content, respectively. C: Distribution of contigs and coding genes in each scaffold. Plot shows gene density distribution, contig number, and coding gene number in each scaffold, from left to right. D: Synteny of genomes between scaffold-level C. quinquecirrha and A. aurita. Syntenic blocks are linked between two genomes with a Circos plot.
To obtain more accurate information about the distribution of repetitive sequences (including interspersed nuclear elements, tandem repeats (TRP), and DNA elements), coding genes, and GC content of each assembled sequence, the genome was cut with a 200 kb slide-window and plotted the results with Circos (v0.69-6) (Figure 1B). Results revealed that the GC content of each scaffold (scaf) exhibited little difference, ranging from 37.37% (scaf35) to 41.48% (scaf54). However, the ratio of repetitive sequences on different scaffolds was highly variable, ranging from 17.40% (scaf51, repeat length: 842 697 bp) to 61.94% (scaf44, repeat length: 1 888 601 bp). According to the Circos plot, the distribution of GC content was the most uniform, followed by the distribution of coding genes (Figure 1B). In some scaffolds, the repetitive sequence distribution was quite uneven, such as scaf29 (Figure 1B). The distribution ranges of short interspersed repetitive elements (SINE, length: 138 bp, 0.0009% in length of scaf29) and TRP (length: 1.32 Mb, 8.69% in length of scaf29) are shown in different colors in the two circles in Figure 1B. The distribution of TRP was relatively more concentrated in scaf36 (SINE, length: 1 944 bp, 0.064% in length of scaf36; TRP, length: 1.36 Mb, 44.65% in length of scaf36), while the distribution of SINE was more concentrated in scaf29 (Figure 1B).
We counted the coding gene number and contig number in each scaffold to better understand the distribution of protein-coding genes and contigs in the scaffolds of C. quinquecirrha. We identified the longest 51 scaffolds (≥1 Mb), which showed a maximum of nearly 15 Mb. The longest scaffold (scaf29) was also comprised of the largest number (72) of contigs (Figure 1C), implying that complex scaffold composition may be the cause of the fragmented genome assembly in previous research. The longest scaffold (scaf29) also contained the most genes (1 170 genes), suggesting a positive relationship between gene number and scaffold length (Figure 1C). This was verified by the positive Pearson correlation coefficient (r=0.982) between chromosome length and gene number.
To clarify the syntenic block relationship of C. quinquecirrha with other jellyfish species, we performed a genome-wide collinearity comparison between C. quinquecirrha and Aurelia aurita using the whole genome sequences. The two regions that showed similar sequences between the scaffolds of the two species were connected in the resulting plot (Figure 1D). As shown in Figure 1D (only sequences longer than 1 Mb are shown), we found that the collinearity between these two jellyfishes was good but was poor when we compared other species groups with short divergence time (such as among mammals). Among the C. quinquecirrha scaffolds, scaf26 (Figure 1D) had the longest collinearity with A. aurita, and the length of the collinear region was 401.33 kb. After removing several matched fragments with overlapping regions, the total lengths of the collinear regions were 8.23 Mb (3.65% of the whole genome) and 10.23 Mb (5.22% of the whole genome) in C. quinquecirrha and A. aurita, respectively. Based on comparison of their scaffold-level genomes, the two jellyfish showed few collinear regions (Figure 1D), which may result from differences in chromosome number and many genetic variation sites in each evolutionary process with the long divergence time of ~475 million years (Xia et al., 2020).
Through systematic investigation of the Animal Genome Size Database (http://www.genomesize.com), we found that the C-value of the groups, including hydrozoan and scyphozoan, ranged from 0.26 to 1.49, indicating differences in the size of their genomes. In addition, though there are many extant species of jellyfish (likely more than 250), only a few species’ genomes have been published and most are very fragmented, suggesting complex genome composition or karyotypes in jellyfish. The previously published genome of C. quinquecirrha is very fragmented. Genome assembly can be difficult, especially regarding differences in the assembly of the genomes of Anura and Urodela species in Amphibia. The C-values in Anura range from 0.95 to 12.40 (Olmo & Morescalchi, 2005), but range from 10.02 to 120.6 in Urodela (Goin et al., 1968). In addition, the differences in published genome papers between these two groups also reflect the impact of genome size on assembly. To date, only one Urodela genome has been published (Nowoshilow et al., 2018) in comparison to the many genomes of Anura, e.g., Nanorana parkeri (Sun et al., 2015), Xenopus laevis (Session et al., 2016), and Xenopus tropicalis (Hellsten et al., 2010). Therefore, it may be that difficulty in genome assembly is closely related to the content of repetitive sequences and genome size. It remains a huge technological challenge to analyze high-quality genomes of these complex species.
Sequencing technology has developed rapidly in recent years. In particular, the emergence of Hi-C techniques has been of considerable benefit for comparative genomics (Cali et al., 2019) and provided unprecedented accuracy and convenience for obtaining high-quality chromosome-level genomes (Pal et al., 2019). For example, high-continuity genomes have been obtained for many species using Hi-C technology (Dudchenko et al., 2017). With Hi-C, more high-continuity genomes of jellyfish species can be assembled in the future. In this study, we successfully assembled a high-continuity genome of C. quinquecirrha by generating high-coverage Hi-C sequencing data. Compared to the previously published version (Xia et al., 2020), the N50 length was substantially improved (Figure 1A). Genome synteny analysis showed a collinear relationship between C. quinquecirrha and A. aurita. The assembled high-continuity C. quinquecirrha genome could help improve our knowledge on the evolution of genomes and have practical application in studies on conservation biology and population genetics. It could also improve our understanding of the genomes of jellyfish, which should help in studies on the growth, development, and reproduction of C. quinquecirrha.
DATA AVAILABILITY
The raw Hi-C sequencing data and genome assembly of Chrysaora quinquecirrha were deposited in the National Center for Biotechnology Information (NCBI) database under accession No. PRJNA658826. The annotation file was uploaded to the DRYAD database (https://datadryad.org/stash/share/RY8FQAETuLZR_O0_hEdQsvROvJnMcsPZ9UtJBZsnqlQ).
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
X.C.G. and Y.W.L. conceived and supervised the project and revised the manuscript. W.X.X. and Y.W.L. collected samples. W.X.X. and H.R.L. performed bioinformatics analyses. W.X.X. and J.H.G. wrote the manuscript. Y.W.L., H.H.L., Y.H.S., H.Z.W., H.F.G., and Y.X.D. revised the manuscript. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the Province of China (202011840014) Shaanxi College Students’ Innovation and Entrepreneurship Training Program (S202011840014), Xi’an Medical University College Students’ Innovation and Entrepreneurship Training Program (121520014), National Natural Science Foundation of China (31760671), Joint Special Project of Agricultural Basic Research in Yunnan Province (2018FG001-041), Yunnan Provincial Department of Education Research Fund (2020J0251), Scientific Research Fund of Shaanxi Provincial Education Department (20JS143), and Natural Science Basic Research Plan in Shaanxi Province of China (2020JQ-876)
Contributor Information
Yao-Wen Liu, Email: yaowenliu@foxmail.com.
Xing-Chun Gou, Email: gouxingchun@189.cn.
References
- 1.Burge C, Karlin S Prediction of complete gene structures in human genomic DNA. Journal of Molecular Biology. 1997;268(1):78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 2.Cali DS, Kim JS, Ghose S, Alkan C, Mutlu O Nanopore sequencing technology and tools for genome assembly: computational analysis of the current state, bottlenecks and future directions. Briefings in Bioinformatics. 2019;20(4):1542–1559. doi: 10.1093/bib/bby017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen MS, Niu LJ, Zhao ML, Xu CJ, Pan BZ, Fu QT, et al De novo genome assembly and Hi-C analysis reveal an association between chromatin architecture alterations and sex differentiation in the woody plant Jatropha curcas . GigaScience. 2020;9(2):giaa009. doi: 10.1093/gigascience/giaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decker MB, Brown CW, Hood RR, Purcell JE, Gross TF, Matanoski JC, et al Predicting the distribution of the scyphomedusa Chrysaora quinquecirrha in Chesapeake Bay . Marine Ecology Progress Series. 2007;329:99–113. doi: 10.3354/meps329099. [DOI] [Google Scholar]
- 5.Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, et al De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds . Science. 2017;356(6333):92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, et al Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Systems. 2016a;3(1):99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, et al Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Systems. 2016b;3(1):95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finenko GA, Abolmasova GI, Romanova ZA, Datsyk NA, Anninskii BE Population dynamics of the ctenophore Mnemiopsis leidyi and its impact on the zooplankton in the coastal regions of the black sea of the Crimean coast in 2004-2008 . Oceanology. 2013;53(1):80–88. doi: 10.1134/S0001437012050074. [DOI] [Google Scholar]
- 9.Ghurye J, Rhie A, Walenz BP, Schmitt A, Selvaraj S, Pop M, et al Integrating Hi-C links with assembly graphs for chromosome-scale assembly. PLoS Computational Biology. 2019;15(8):e1007273. doi: 10.1371/journal.pcbi.1007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goin OB, Goin CJ, Bachmann K DNA and amphibian life history. Copeia. 1968;1968(3):532–540. doi: 10.2307/1442021. [DOI] [Google Scholar]
- 11.Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, et al The genome of the western clawed frog Xenopus tropicalis . Science. 2010;328(5978):633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang JB, Quattrini AM, Francis WR, Ryan JF, Rodríguez E, McFadden CS A hybrid de novo assembly of the sea pansy (Renilla muelleri) genome . GigaScience. 2019;8(4):giz026. doi: 10.1093/gigascience/giz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC Adaptive seeds tame genomic sequence comparison. Genome Research. 2011;21(3):487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korf I Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al Circos: an information aesthetic for comparative genomics. Genome Research. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leclère L, Horin C, Chevalier S, Lapébie P, Dru P, Peron S, et al The genome of the jellyfish Clytia hemisphaerica and the evolution of the cnidarian life-cycle . Nature Ecology & Evolution. 2019;3(5):801–810. doi: 10.1038/s41559-019-0833-2. [DOI] [PubMed] [Google Scholar]
- 17.Majoros WH, Pertea M, Salzberg SL TigrScan and glimmerHMM: two open source ab initio eukaryotic gene-finders . Bioinformatics. 2004;20(16):2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 18.Nowoshilow S, Schloissnig S, Fei JF, Dahl A, Pang AWC, Pippel M, et al The axolotl genome and the evolution of key tissue formation regulators. Nature. 2018;554(7690):50–55. doi: 10.1038/nature25458. [DOI] [PubMed] [Google Scholar]
- 19.Oguz T, Salihoglu B, Moncheva S, Abaza V Regional peculiarities of community-wide trophic cascades in strongly degraded black sea food web. Journal of Plankton Research. 2012;34(4):338–343. doi: 10.1093/plankt/fbs002. [DOI] [Google Scholar]
- 20.Olesen NJ, Purcell JE, Stoecker DK Feeding and growth by ephyrae of scyphomedusae Chrysaora quinquecirrha . Marine Ecology Progress Series. 1996;137(1-3):149–159. [Google Scholar]
- 21.Olmo E, Morescalchi A Genome and cell sizes in frogs: a comparison with salamanders. Experientia. 2005;34:44–46. [Google Scholar]
- 22.Pal K, Forcato M, Ferrari F Hi-C analysis: from data generation to integration. Biophysical Reviews. 2019;11(1):67–68. doi: 10.1007/s12551-018-0489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, et al Genome evolution in the allotetraploid frog Xenopus laevis . Nature. 2016;538(7625):336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun YB, Xiong ZJ, Xiang XY, Liu SP, Zhou WW, Tu XL, et al Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes . Proceedings of the National Academy of Sciences of the United States of America. 2015;112(11):E1257–E1262. doi: 10.1073/pnas.1501764112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia WX, Li HR, Cheng WM, Li HH, Mi YJ, Gou XC, et al High-quality genome assembly of Chrysaora quinquecirrha provides insights into the adaptive evolution of jellyfish . Frontiers in Genetics. 2020;11:535. doi: 10.3389/fgene.2020.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
The raw Hi-C sequencing data and genome assembly of Chrysaora quinquecirrha were deposited in the National Center for Biotechnology Information (NCBI) database under accession No. PRJNA658826. The annotation file was uploaded to the DRYAD database (https://datadryad.org/stash/share/RY8FQAETuLZR_O0_hEdQsvROvJnMcsPZ9UtJBZsnqlQ).