Abstract
Depression is a prevalent mental disorder that is associated with aging and contributes to increased mortality and morbidity. The overall prevalence of geriatric depression with clinically significant symptoms is currently on the rise. Recent studies have demonstrated that altered expressions of long non-coding RNAs (lncRNAs) in the brain affect neurodevelopment and manifest modulating functions during the depression. However, most lncRNAs have not yet been studied. Herein, we analyzed the transcriptome of dysregulated lncRNAs to reveal their expressions in a mouse model exhibiting depressive-like behaviors, as well as their corresponding response following antidepressant fluoxetine treatment. A chronic unpredictable mild stress (CUMS) mouse model was applied. A six-week fluoxetine intervention in CUMS-induced mice attenuated depressive-like behaviors. In addition, differential expression analysis of lncRNAs was performed following RNA-sequencing. A total of 282 lncRNAs (134 up-regulated and 148 down-regulated) were differentially expressed in CUMS-induced mice relative to non-stressed counterparts (P<0.05). Moreover, 370 differentially expressed lncRNAs were identified in CUMS-induced mice after fluoxetine intervention. Gene Ontology (GO) analyses showed an association between significantly dysregulated lncRNAs and protein binding, oxygen binding, and transport activity, while the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicated that these dysregulated lncRNAs might be involved in inflammatory response pathways. Fluoxetine effectively ameliorated the symptoms of depression in CUMS-induced mice by regulating the expression of lncRNAs in the hippocampus. The findings herein provide valuable insights into the potential mechanism underlying depression in elderly people.
Keywords: Depression, Elderly, Fluoxetine, LncRNA, Chronic unpredictable mild stress
INTRODUCTION
Depression is a prevalent psychiatric disorder associated with chronic and stressful life events. It results in increased mortality and morbidity (Fiske et al., 2009). Despite efforts to reduce the prevalence of depression, variation in its onset and difficulty in diagnosis of elderly patients have resulted in limited success in controlling the disease. The overall prevalence of geriatric depression, with clinically significant symptoms, is estimated to be 8%–16% (Beyer, 2007). Elderly people with depression often experience diminished function and decreased quality of life, leading to increased mortality from suicide and other medical illnesses (Blazer, 2003; Casey, 1994; Fiske et al., 2009). Several symptoms in this patient group may resemble normal age-related conditions or other illnesses. Thus, the lack of proper treatment for elderly depressed patients may be partly attributed to misdiagnosis. Therefore, effective interventions for depression are critically dependent on early diagnosis. Further, the development of effective antidepressants relies on the identification of biological substrates underpinning depression.
The diagnosis of late-life depression is performed using both clinical and physical examinations, supplemented with instructive laboratory evaluation. Recently, there have been substantial efforts aimed at improving the diagnosis and treatment of depression (Gururajan et al., 2016; Strawbridge et al., 2017). However, accurate diagnosis of elderly patients has been challenged by inconsistencies in findings (Jani et al., 2015). For example, increased levels of various proinflammatory markers in patients with depression were identified as potential biological markers for depression (Miller & Raison, 2016; Raison et al., 2006). Although these biomarkers have not been used in practical settings to diagnose depression, they have provided a basis for understanding the molecular mechanisms of depression.
Numerous antidepressants are currently available for treating depression. Among them, selective serotonin reuptake inhibitors (SSRIs) are commonly prescribed (Beyer, 2007; Sampson, 2001). Fluoxetine, a second-generation antidepressant categorized as an SSRI, blocks serotonin reuptake in the presynaptic terminal, ultimately resulting in increased concentration of 5-hydroxytryptamine (5-HT) in widespread brain areas without affecting other neurotransmitter systems (Perez-Caballero et al., 2014). Due to various side effects and safety issues, previously-developed classes of antidepressants, such as tricyclic antidepressants, are less commonly used in clinics (Sampson, 2001). Given its minimal adverse effects, fluoxetine is widely employed in the treatment of depression (Bremner, 1984). However, the precise mechanism of action of this drug is not completely understood.
Depression is a multifactorial disease caused by psychological, social, and biological factors. Genetic effects, including dysregulated expressions of microRNAs (miRNAs) and lncRNAs, as well as epigenetic modifications by DNA methylation, are associated with depression (Chen et al., 2017; Leung & Sharp, 2010; Liu et al., 2014). LncRNAs are transcripts of over 200 nucleotides in the genome without protein-coding functions. They play important roles in various biological processes, such as epigenetic regulation, chromosome imprinting, cell-cycle control and cell apoptosis, by regulating gene expression (Wapinski & Chang, 2011). Additionally, lncRNAs have been implicated in the regulation of gene expression in various psychiatric disorders, including major depressive disorder (MDD), schizophrenia, autism spectrum disorder (ASD), and neurodegenerative diseases via both cis- and trans-acting pathways (Gibbons et al., 2018; Huang et al., 2017; Riva et al., 2016; Tang et al., 2017). It is hypothesized that identifying dysregulated lncRNAs in depression, alongside deciphering their roles and corresponding pathophysiological mechanisms, could guide the development of therapeutic strategies. A model of chronic unpredictable mild stress (CUMS) has been developed to investigate depressive symptoms in rodents and elucidate relevant molecular substrates and effects of pharmacological interventions(Nollet et al., 2013; Surget & Belzung, 2008; Willner et al., 1987). Herein, we aimed to investigate the antidepressant-like effects of fluoxetine on CUMS-induced mice and identify differentially expressed lncRNAs (DElnRNAs) following fluoxetine treatment to explore their role in depression.
MATERIALS AND METHODS
Experimental animals and chemicals
Thirty-six BALB/c male mice purchased and maintained under the standard conditions with 12 h light/dark cycle and received ad libitum food and water routinely from the animal center of Inner Mongolia Medical University (Huhhot, China) were used in behavioral testing. We used the mice at 13-month-old at the beginning of our study. Generally, mice over 15 months are elderly correlated with humans (Flurkey et al., 2007). All the mice were divided into three groups (n=12 per group) and cage-housed separately: Group I: a non-stressed control group was administered vehicle (NOR); Group II: a CUMS induced stressed group was administered vehicle as a control (CUMS); and Group III: a CUMS induced group was chronically treated with fluoxetine (FLU). All experimental procedures were approved by the Academic Ethics Committee of Inner Mongolia Medical University (approval No.: YKD2016024) and performed under the Guide for Care and Use of Laboratory Animals. Fluoxetine hydrochloride (catalog No.: 4525A) was purchased from Patheon France distributed by Lilly Suzhou Pharmaceutical Co., Ltd. Fluoxetine, dissolved in 0.3% carboxymethyl cellulose (CMC), was administered to the CUMS-induced mice at the dose of 20 mg/kg of body weight at the fixed time every day. The same volume of 0.3% CMC was administered to the control group. Fluoxetine administration lasted continuously for six weeks. The enzyme-linked immunosorbent assay (ELISA) kits for concentration measurements of 5-HT (catalog No.: E-EL-0033c) and brain-derived neurotrophic factor (BDNF, catalog No.: E-EL-M0203c) were purchased from Elabscience Biotechnology Co. Ltd, China.
CUMS procedures and fluoxetine intervention
The CUMS protocol is a classical method to study the behavioral and neurobiological mechanisms of depression. The depressive-like behaviors in mice were induced by mild daily stressors. Briefly, the non-stressed control animals were housed six per cage without contacting with the stressed groups. For the other two groups, mice were housed separately and exposed to the CUMS procedures. The weekly stress protocol consists of 24 h light/dark cycle disruption, 12 h wet cage housing, 12 h food or water deprivation, 30 min intermittent white noise, 4 h physical restraint, 5 min unpredictable shocks (15 mA, one shock/5 s, 10 s duration), 12 h housing cage tilted (45 degrees), 5 min cold water swimming, 1 h shaking stress with 160 r/min, 12 h bedding materials deprivation, 1 min tail clipping, and 30 min intermittent flash. One of these stressors was applied daily at a different time to minimize its adaptability. All stressors were randomized from the second week of induction for unpredictability. The CUMS procedure lasted for eight weeks. Fluoxetine was administered from the third week after the beginning of CUMS. All the behavioral tests were performed at the end of the protocol.
Behaviors assessments
The behaviors of all the mice were assessed by sucrose preference test (SPT), forced swim test (FST), and tail suspension test (TST). The SPT was performed on day 1 after the CUMS procedure. Subsequently, the FST and TST were performed.
SPT is widely used to measure an anhedonic response with less sucrose consumption (Liu et al., 2018). Initially, the mice were exposed to two bottles containing 1% (w/v) sucrose for 72 h; then the bottles were replaced by tap water. After 48 h, the mice were under food and water deprivation for 24 h. All the mice received one bottle of 1% sucrose or one bottle of tap water for 24 h. Two bottles changed positions once during the test to avoid the predictability by mice. Then, the sucrose preference was evaluated according to the formula: sucrose preference=(sucrose intake/(sucrose intake+water intake))×100, as described previously (Liu et al., 2018).
The mouse FST was conducted as described previously (Yankelevitch-Yahav et al., 2015), with some modifications. Briefly, mice were individually forced to swim in an open glass container (20 cm in diameter), containing warm water at 25 ℃. The height of the water was adjusted to fit the mice’s size. In the test, the time of immobility was measured during a 6 min period. Immobility was assigned when no additional activity was observed other than that required to keep the mice head above the water.
The modified TST was performed by measuring the total duration of immobility induced by the tail suspension (Cryan et al., 2005). Briefly, each mouse was suspended 50 cm above the floor by a hand grasping approximately 1 cm from the tip of the tail. Mice were considered immobile only when they hung passively and completely motionless. Immobility time was recorded every 6 min using Smart3 Small Animal tracking system (Panlab, Spain).
The enzyme-linked immunosorbent assay (ELISA)
All the tissue samples were collected after the intervention for further experiments. The hippocampi were homogenized on ice after cold PBS washing and subjected to centrifuge, then the supernatants were collected for ELISA. The ELISA procedures were followed as the manufacturer’s instructions (Elabscience Biotechnology, China). Briefly, 50 µL of prepared standards and samples were added to wells followed by adding 50 µL of diluted biotinylated detection antibody and incubate at room temperature for 90 min, then washing the wells thoroughly four times after decanting solutions. 100 µL of diluted HRP conjugate was added and the plate was incubated for 45 min. After incubation, 5 times of washing were performed and 90 µL of the chromogenic substrate (TMB) was added to each well with 30 mins incubation at room temperature in dark. Finally, 50 µL of stop solution was added to each well to stop the reaction. The absorbance of each well at 450 nm was read in an ELx808TM absorbance microplate reader (BioTek Instruments, USA). A four-parameter logistic curve fit to the standards using KC junior software (BioTek Instruments, USA) was used to calculate concentrations of HT or BDNF for all the samples.
RNA isolation and measurements
Total RNA was extracted from the hippocampi of 12 mice of three groups according to a previously reported method (Xiao et al., 2005). Briefly, RNAs were isolated using TRIzol reagent (Invitrogen, USA) and RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer's protocol. The purity, concentration, and integrity of total RNA samples were determined using the NanoPhotometer® spectrophotometer (IMPLEN, USA), Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, USA) and RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, USA).
Library preparation, clustering, and sequencing
A total amount of 20 ng RNA per sample was used for the library preparation and subsequent sequencing was done by the Experimental Department of Novogene, China according to the previously reported protocol (Huang et al., 2019). Briefly, ribosomal RNA was removed and cleaned by Epicentre Ribo-zeroTM rRNA Removal Kit (Epicentre, USA) and ethanol precipitation, respectively. Subsequently, libraries for sequencing were generated using the rRNA-depleted RNA by NEBNext® UltraTM Directional RNA Library Prep Kit for Illumina® (New England Biolabs, USA) following the manufacturer's recommendations. The library quality was measured on the Agilent Bioanalyzer 2100 system. The index-coded samples were clustered on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina, USA) according to the manuline-height-add:-0.35ptfacturer’s instructions. Subsequently, the pair-end sequencing on an Illumina Hiseq 2500 platform was performed, and finally, 125 bp paired-end reads were generated.
Sequence tag preprocessing and mapping
The clean data were obtained by removing reads containing adapter, ploy-N, and low-quality reads from raw data. At the same time, the Phred score (Q20, Q30) and GC content of the clean data were calculated. All the downstream analyses were based on clean data with high quality. The paired-end clean reads were then mapped to the GRCm38/mm10 reference genome from the Mouse Genomics Database (http://genome-asia.ucsc.edu/cgi-bin/hgGateway) using HISAT2 v2.0.4 (Kim et al., 2019). The mapped reads of each sample were assembled by StringTie (v1.3.3) (Pertea et al., 2015) in a reference-based approach.
Coding potential and conservation analysis
The transcripts predicted with coding potential by CNCI (Coding-Non-Coding-Index) (v2) (Sun et al., 2013), CPC2 (v0.1) (Kong et al., 2007), Pfam Scan (v1.3) (Punta et al., 2012), and PhyloCSF (phylogenetic codon substitution frequency) (v20121028) (Lin et al., 2011) tools were filtered out, and those without coding potential were our candidate set of lncRNAs. The phyloFit (a program of Phast software) (Siepel et al., 2005) was applied to compute phylogenetic models for conserved and non-conserved regions among species. Then, the phyloP was used to compute a set of conservation scores of lncRNA and coding transcripts based on the computed phylogenetic model and HMM transition parameters.
Gene expression calculation
Gene expression was calculated using the RPKM (reads per kilobase transcriptome per million mapped reads) method by StringTie (v2.1.1). Transcripts with a P-adjust (Q-value) <0.05 or P-value<0.05 were assigned as differentially expressed. The differential expression of the biological replicated count data was conducted by edgeR software package (Robinson et al., 2010).
LncRNAs-mRNAs colocalization and co−expression analysis
The cis role of target genes was predicted by searching the 100kb upstream and downstream regions of lncRNAs and followed by their function analysis (Guil & Esteller, 2012). Besides, the expressed Pearson's correlation coefficient between lncRNAs and coding genes with R function "cor.test" was calculated.
Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
The GO enrichment analysis of differentially expressed genes or lncRNA target genes was implemented by the GOseq R package. GO terms with corrected P-value less than 0.05 were considered significantly enriched by differential expressed genes. KEGG pathway analysis was performed using KOBAS 2.0 (http://kobas.cbi.pku.edu.cn/), and false discovery rate (FDR) corrected P-value (Q-value) cut-off was set at 0.05.
Quantitative RT−PCR (qRT-PCR)
The qRT-PCR was performed to verify the expressions of lncRNAs in CUMS mice and fluoxetine-treated mice. Purified total RNAs isolated from 14 samples (except the samples for RNA-seq and ELISA) were reverse-transcribed into cDNA according to the manufacturer’s instructions. Data were analyzed using the 2−△△CT relative quantification method and normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to calculate relative lncRNA concentrations. All primers are shown in Table 1.
Table 1. qRT-PCR primers used for validation of lncRNA expression identified from RNA-seq.
| lncRNA transcript ID | Transcript symbol name | Forward primer sequence (5'–3') | Reverse primer sequence (5'–3') | PCR product size (bp) |
| –: Not available. | ||||
| LNC_007274 | – | CTTGGTCAGAAGCATCTGGA | AAGAACAGGCTTCGAGAACG | 273 |
| ENSMUST00000148687.1 | Gm16638 | ATCCAGCAGACAGCACTATG | GTTGCCTCTGTGTTCAGAAG | 278 |
| LNC_001203 | – | CACAGTTGCTTCTAAGCCAG | ATACAGAGAGCGCAGCATTC | 259 |
| LNC_004971 | – | TGGTGACATTCTTCAGCTCC | ATTGCAGAAGAGGCCGATAG | 145 |
| ENSMUST00000139987.8 | Tmem134 | GATCTTCATCTACTGCGCTG | CAGGTGTAGGTTGGAGGAAT | 159 |
| ENSMUST00000182520.1 | Gm26917 | AATGGTGCTACCGGTCATTC | ACACCTCTCTTATCCGCTCT | 194 |
| ENSMUST00000152663.7 | 4933431E20Rik | GACTCAGATCCATCCGTTCA | CAGTGTCTGGACCTGTTCAT | 280 |
| ENSMUST00000174808.1 | Malat1 | CCTTCCTGTGTGGCAAGAAT | CTGCAAGCACAACTTGAGGT | 223 |
Statistical analysis
The results are expressed as mean±standard error mean (SEM). A statistical significance P-value of less than 0.05 was set as the thresholds for filtering the differentially expressed lncRNAs. GO and pathway enrichment analysis was done using Fisher's test. P-value<0.05 was considered statistically significant.
RESULTS
Antidepressant effects of fluoxetine on CUMS-induced depression in mice
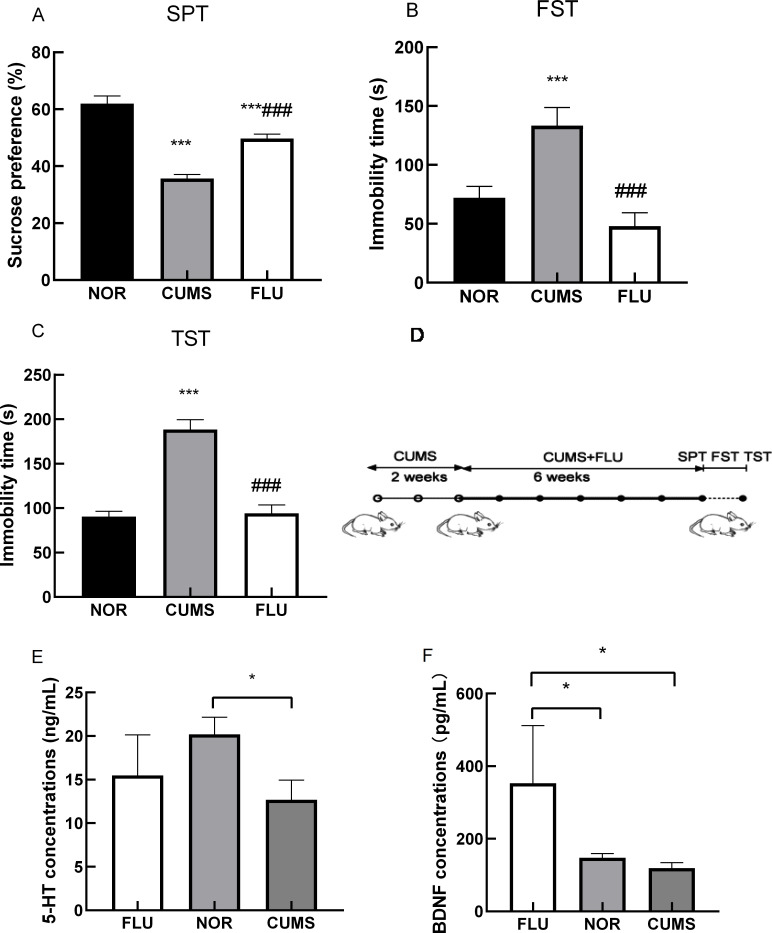
A total of 36 BALB/c mice were exposed to 8 weeks of CUMS. Fluoxetine was administered from the 3rd week following the CUMS procedure. From the 9th week, all mice were assessed for depressive behaviors using SPT, FST, and TST. In SPT, mice exposed to CUMS displayed decreased sucrose preference when compared to NOR counterparts. These effects were ameliorated after exposure to fluoxetine (Figure 1A). In FST and TST, immobility time was longer in the CUMS group. These effects were reversed in both FLU and NOR groups (Figure 1B, C). Stressed BALB/c mice manifested depression-like behaviors. The experimental scheme is depicted in Figure 1D. Depressive-like behavior in CUMS mice was ameliorated after fluoxetine administration, suggesting that fluoxetine effectively attenuated the adverse effects of stress on depression-like behavior.
Figure 1.
Behavioral assessments and the schematic diagram of the indcution process of depressive-like hehaviors in mice
In the SPT assessment (A), mice exposed to CUMS displayed decreased sucrose preference when compared to NOR mice; these effects were ameliorated after fluoxetine treatment. In the FST (B) and TST (C) assessments, immobility time was longer in the CUMS group than in the FLU and the NOR groups. * P-value<0.001. D: Schematic diagram showing the induction process of depressive-like behaviors in mice. Changes in the expression of 5-HT (E) and BDNF (F) in samples from CUMS, FLU, and NOR groups (n=4 per group). A significant decrease in 5-HT concentration was observed in the CUMS group compared to the NOR group. 5-HT concentration was increased after fluoxetine treatment (E). BDNF concentration increased after fluoxetine treatment in the FLU group compared to the CUMS group, in which it was decreased (F). Data indicate mean±SEM. *P-value<0.05. NOR: Normal control group; CUMS: CUMS induced depression group; FLU: Fluoxetine-treated group; SPT: Sucrose preference test; FST: Forced swim test; TST: Tail suspension test.
5-HT and BDNF expression increased in CUMS-induced mice after fluoxetine treatment
The concentrations of 5-HT and total BDNF (mature and its precursor proBDNF) in mouse hippocampi across the three groups were examined using ELISA following the examination of the phenotype of CUMS-induced mice after fluoxetine treatment. There was a significant decrease in 5-HT concentrations in the hippocampi of CUMS-induced mice compared to those in NOR mice (P<0.05). 5-HT concentration was increased after fluoxetine treatment in fluoxetine-treated mice compared to CUMS-induced mice (Figure 1E). There was an increase in the total BDNF concentration after fluoxetine treatment in fluoxetine-treated mice compared to CUMS-induced mice, in which it was decreased (Figure 1F). These findings implied that fluoxetine alleviated depression-like symptoms of CUMS-induced mice by increasing hippocampal 5-HT and BDNF concentrations.
Identification of lncRNAs and genomic context analysis
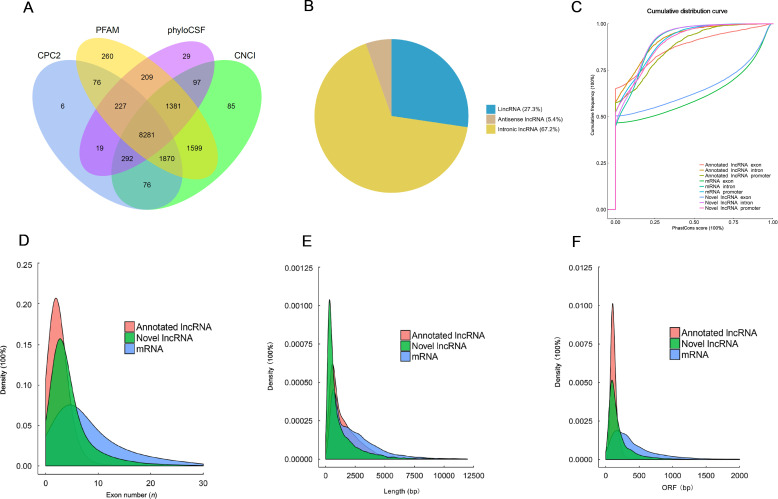
Hippocampal tissues from the three groups of mice (n=3 per group) were isolated after behavioral tests and used for the identification of lncRNAs. A computational approach and stepwise filtering procedures were applied to identify high-confidence lncRNAs from the RNA-seq cohort. The transcripts predicted with coding potential by CNCI, CPC2, Pfam Scan, and PhyloCSF tools were filtered out. A final total of 8 281 lncRNAs were identified and used for further analysis (Figure 2A). The identified lncRNAs were classified into long intergenic non-coding RNA (lincRNA), anti-sense lncRNA, and intronic lncRNA (Figure 2B). Among them, intronic lncRNA occurred frequently at 67.2%. Generally, lncRNAs are characterized by shorter lengths, are less conserved, and have lower expression levels than those of protein-coding transcripts. However, we observed that the identified lncRNAs were more conserved than messenger RNAs (mRNAs) and the predicted novel lncRNAs (Figure 2C). Further, the number of exons and lengths of identified lncRNAs were comparatively shorter than those of mRNA transcripts (Figure 2D, E). The length of open reading frames (ORFs) in most identified lncRNAs was also less than 300 bp, consistent with the features of lncRNA (Figure 2F).
Figure 2.
Transcriptome-wide identification and characterization of lncRNAs
A: A total of 8 281 lncRNAs were identified using the CNCI, CPC2, Pfam Scan, and PhyloCSF tools. B: Identified lncRNAs were mainly classified as lincRNA, anti-sense lncRNA, and intronic lncRNA. C: Conservation comparison of the identified lncRNAs, mRNAs, and predicted novel lncRNAs. D, E: The exon number (D) and length (E) of lncRNAs identified were comparatively shorter than mRNA transcripts. F: Length of ORF in most identified lncRNAs was less than 300 bp.
Expression profiling of lncRNA in the hippocampus of CUMS-induced mice after fluoxetine treatment
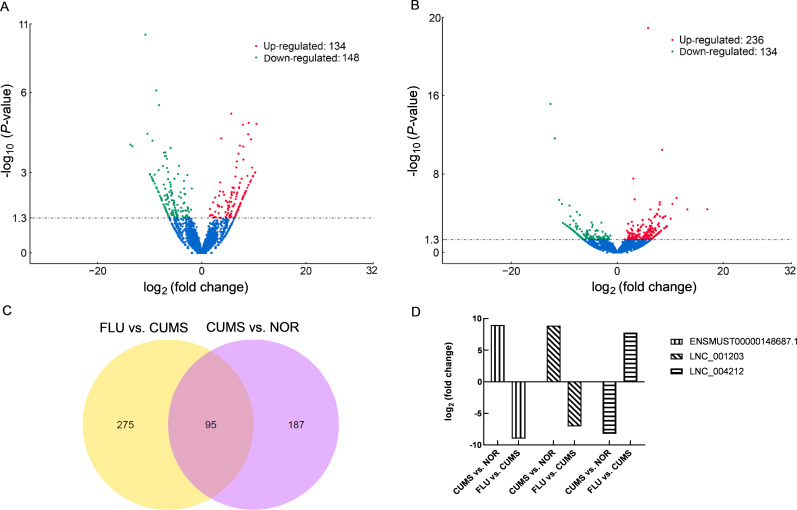
Genome-wide RNA sequencing was performed to identify DElnRNAs in the hippocampi of CUMS-induced and NOR mice. We compared the expressions of lncRNAs from the hippocampus of CUMS-induced and NOR mice based on read counts and RPKM values. In total, 282 lncRNAs differentially expressed between CUMS-induced and NOR mice were identified (P<0.05, Supplementary Table S1). Of these, 134 were up-regulated while 148 were down-regulated in CUMS-induced mice compared to the NOR group. A volcano map of all differentially regulated lncRNAs, based on a group-wise comparison between CUMS and NOR groups, is shown inFigure 3A. To avoid false positives and further narrow down on DElnRNAs, Q-value was used as the statistical parameter, with Q<0.05 considered statistically significant. Consequently, 20 lncRNAs (10 up-regulated and 10 down-regulated) exhibited altered expression during comparison (Table 2), suggesting that DElnRNAs could be potential biomarkers for the diagnosis of depression.
Figure 3.
Differentially expressed lncRNAs (DElnRNAs) comparison between CUMS vs. NOR groups and FLU vs. CUMS groups
A, B: Volcano map of 282 and 370 differentially regulated lncRNAs based on a group-wise comparison of CUMS vs. NOR groups (A), and FLU vs. CUMS groups (B), respectively (P-value<0.05). C: Venn map showing 95 commonly expressed lncRNAs identified in a comparison between FLU vs. CUMS groups and CUMS vs. NOR groups (P-value<0.05). D: Three lncRNAs (LNC_004212, LNC_001203, and ENSMUST00000148687.1) with contrasting expression patterns between FLU and CUMS groups vs. CUMS and NOR groups based on RNA-seq analysis (Q-value<0.05).
Table 2. DElnRNAs between CUMS-induced mice and normal controls (adjusted P-value <0.05) .
| LncRNA transcript_id | CUMS/NOR fold change (log2 ratio) | P-value | Q-value |
| LNC_001649 | –13.699 2 | 8.93E–05 | 0.029 085 |
| LNC_002578 | –13.348 5 | 0.000102 | 0.031 665 |
| LNC_007352 | –10.429 3 | 3.50E–05 | 0.014 861 |
| LNC_001657 | –9.480 77 | 6.31E–05 | 0.022 996 |
| LNC_007186 | –8.750 24 | 8.35E–07 | 0.000 832 |
| LNC_004212 | –8.231 64 | 3.01E–06 | 0.002 228 |
| LNC_003004 | –7.265 23 | 0.00018 | 0.045 952 |
| LNC_002630 | –7.001 84 | 0.000174 | 0.044 864 |
| LNC_003171 | –5.972 24 | 0.000118 | 0.034 934 |
| LNC_001871 | 3.741 192 | 5.21E–05 | 0.020 057 |
| LNC_002576 | 5.643 55 | 6.22E–06 | 0.004 041 |
| LNC_005505 | 7.062 69 | 0.000196 | 0.048 071 |
| LNC_000648 | 7.296 817 | 9.84E–05 | 0.030 782 |
| LNC_002056 | 7.889 765 | 1.61E–05 | 0.008 33 |
| ENSMUST00000139987.8 | 7.965 873 | 0.000105 | 0.032 184 |
| LNC_001203 | 8.904 771 | 3.63E–05 | 0.015 174 |
| ENSMUST00000148687.1 | 8.997 071 | 1.36E–05 | 0.007 63 |
| LNC_003699 | 9.458 857 | 5.65E–05 | 0.021 329 |
| LNC_007164 | 10.498 72 | 1.48E–05 | 0.007 939 |
Because fluoxetine administration attenuated the depressive phenotype in CUMS-induced mice, therefore we investigated the differences in lncRNA expression across the FLU groups. We observed that 370 lncRNAs (236 up-regulated and 134 down-regulated) exhibited differential expression (P<0.05,Figure 3B; Supplementary Table S2). A more stringent analysis resulted in 28 lncRNAs that were differentially expressed between the FLU and the CUMS groups (Table 3). A comparison of altered lncRNAs between the FLU and the CUMS groups, as well as between the CUMS and the NOR groups indicated that 95 lncRNAs were commonly expressed and exhibited contrasting expression patterns (Figure 3C; Supplementary Table S3). Based on Q<0.05, three lncRNAs (LNC_004212, LNC_001203, and ENSMUST00000148687.1) exhibited a contrasting fold change expression between the FLU and the CUMS groups, as well as between the CUMS and the NOR groups (Figure 3D). These results suggested that CUMS-induced dysregulation of hippocampal lncRNA levels contribute to depressive behaviors. Also, fluoxetine may have played an antidepressant-like role by reversing the altered expression of novel LNC_004212 and LNC_001203 and annotated ENSMUST00000148687.1 lncRNA transcripts in CUMS-induced mice.
Table 3. DELnRNAs between fluoxetine-treated CUMS-induced mice and CUMS-induced mice (adjusted P-value <0.05) .
| LncRNA transcript_id | FLU/CUMS fold change (log2 ratio) | P-value | Q-value |
| LNC_000804 | –12.589 2 | 7.49E–16 | 2.95E–12 |
| LNC_004168 | –11.758 1 | 2.41E–12 | 5.29E–09 |
| LNC_007274 | –10.935 3 | 4.54E–06 | 0.001 971 |
| LNC_005854 | –10.426 8 | 1.12E–05 | 0.003 987 |
| ENSMUST00000148687.1 | –9.016 46 | 1.63E–05 | 0.005 382 |
| LNC_004971 | –8.843 16 | 0.000 204 | 0.036 612 |
| LNC_004518 | –7.637 87 | 7.27E–05 | 0.017 453 |
| LNC_001203 | –7.069 03 | 0.000 151 | 0.029 595 |
| LNC_001138 | 3.050 837 | 3.00E–08 | 3.39E–05 |
| LNC_004347 | 3.307 226 | 3.81E–06 | 0.001 737 |
| ENSMUST00000182520.1 | 5.833 658 | 1.36E–23 | 2.54E–19 |
| LNC_007241 | 6.265 049 | 4.08E–05 | 0.011 235 |
| LNC_003126 | 7.047 628 | 0.000 264 | 0.043 714 |
| LNC_002186 | 7.188 645 | 0.000 182 | 0.034 032 |
| LNC_003822 | 7.450 247 | 0.000 115 | 0.02 4392 |
| LNC_007358 | 7.570 922 | 0.000 133 | 0.027 026 |
| LNC_004212 | 7.790 258 | 4.21E–05 | 0.011 393 |
| LNC_003821 | 8.000 836 | 1.37E–05 | 0.004 665 |
| LNC_000872 | 8.030 34 | 7.94E–06 | 0.00 313 |
| LNC_001392 | 8.045 196 | 0.000 21 | 0.037 484 |
| LNC_001105 | 8.478 254 | 3.56E–11 | 7.19E–08 |
| LNC_000238 | 8.580 248 | 0.000 309 | 0.048 759 |
| LNC_002941 | 8.760 41 | 0.000 251 | 0.042 275 |
| LNC_007536 | 9.109 974 | 0.000 196 | 0.035 421 |
| ENSMUST00000123998.1 | 10.423 95 | 1.09E–05 | 0.003 939 |
| LNC_003023 | 11.197 15 | 2.82E–06 | 0.001 406 |
| LNC_000806 | 13.241 01 | 4.17E–05 | 0.011 379 |
| ENSMUST00000174808.1 | 16.987 89 | 3.93E–05 | 0.010 935 |
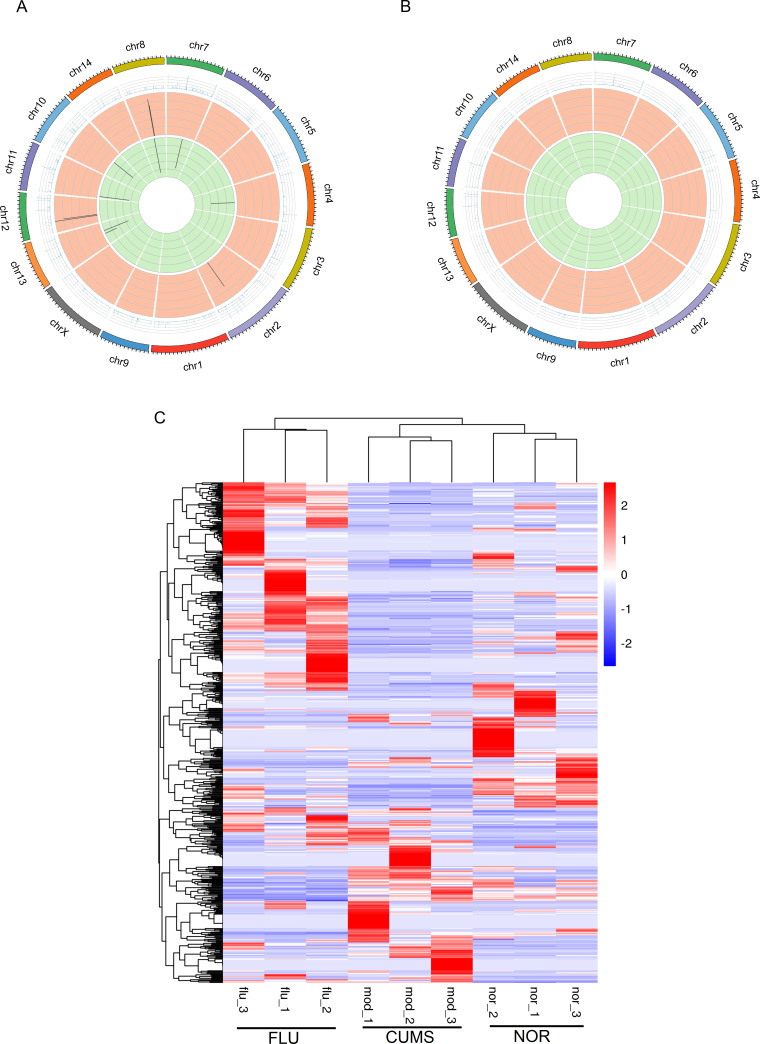
Chromosomal localization and hierarchical clustering analysis of DElnRNAs in CUMS, FLU, and NOR mice
Transcripts located closer together on a chromosome may perform similar biological functions or participate in the same pathway. we thus dissected the chromosomal localization of DElnRNAs. The distribution patterns of differential transcripts on chromosomes and the differential expression profiles of associated transcripts may have important biological implications, which could be vital for understanding the molecular mechanisms underlying depression. The 282 DElnRNAs (P<0.05) were distributed on all chromosomes in CUMS-induced mice compared to the NOR group (Figure 4A). Among them, the 134 up-regulated and 148 down-regulated lncRNAs were enriched on chromosomes 1, 2, 7, 9, 10, 11 and 2, 4, 5, 8, 10, 12, 15, respectively. After fluoxetine treatment, the 236 up-regulated and 134 down-regulated lncRNAs mapped to chromosomes 4, 6, 10, 11, 17 and 2, 7, 9, 11, 17, respectively (Figure 4B). These results indicated that genes close to the DElnRNAs on the same chromosomes might perform similar biological functions in the pathophysiology of depression. LncRNA transcripts with contrasting expression changes in CUMS-induced mice and fluoxetine-treated mice were enriched on chromosomes 10, 11, 15 and 19. We believe that DElnRNAs with predicted mRNA targets on chromosomes 10 and 11 might be involved in depression. Moreover, hierarchical clustering analysis revealed four main groups of DElnRNAs when comparing FLU and CUMS, as well as CUMS and NOR mice (P<0.05) (Figure 4C). Samples from the same group were clustered together, indicating minimal effects of variation among samples from the same group. LncRNAs clustered together and exhibited differential expression patterns across CUMS, FLU, and NOR mice. These results were consistent with earlier experiments on lncRNA identification whereby the expression of certain lncRNAs with reversed regulatory pattern in CUMS-induced mice treated with fluoxetine was observed. Hierarchical clustering analysis also indicated that DElnRNAs with similar expression patterns may be involved in the same processes or pathways resulting in depressive-like behaviors.
Figure 4.
Chromosomal distribution and hierarchical clustering analysis of DElnRNAs in CUMS, FLU and NOR mice
A: Distribution of DElnRNAs on all chromosomes in CUMS vs. NOR groups. Among the 282 DElnRNAs, the 134 up-regulated and 148 down-regulated lncRNAs were enriched on chromosome 1, 2, 7, 9, 10, 11 and 2, 4, 5, 8, 10, 12, 15, respectively. B: Distribution of DElnRNAs on all chromosomes in FLU vs. CUMS groups. After fluoxetine treatment, the 236 up-regulated and 134 down-regulated lncRNAs mapped to chromosomes 4, 6, 10, 11, 17 and 2, 7, 9, 11, 17, respectively. P-value<0.05. C: Hierarchical clustering showing that lncRNAs differentially expressed (P-value<0.05) in FLU vs. CUMS and CUMS vs. NOR groups were clustered into four main categories. DElnRNAs in the same category may participate in similar processes resulting in depressive-like behaviors. Samples from the same group were clustered together, indicating minimal variation in samples from the same group.
Prediction of target lncRNA genes
LncRNAs regulate neighboring genes in the cis or trans patterns, although the mechanisms by which they interact with mRNA remain unclear. We predicted the biological functions of lncRNAs based on their co-location and co-expression with protein-coding genes. We first selected mRNA transcripts co-localized 100 kb upstream and downstream of DElnRNAs. We then performed functional enrichment analysis of lncRNA co-localized mRNA transcripts to predict their functions. We observed that 154 mRNA transcripts were co-localized with 28 dysregulated lncRNAs in FLU and CUMS comparison groups (Q<0.05) (Supplementary Table S4), suggesting that they were target genes for dysregulated lncRNAs. Conversely, 86 mRNAs were identified as possible target genes for DElnRNAs between CUMS and NOR groups (Supplementary Table S4). We predicted lncRNA target genes using a correlation analysis among samples. We used Pearson’s coefficient to analyze the correlation between lncRNAs and mRNAs. mRNAs with a correlation value greater than 0.95 were used for functional enrichment analysis to predict lncRNA function. The accuracy of this co-expression-based prediction method largely depends on the sample number. A total of 679 mRNAs were predicted targets for the 28 dysregulated lncRNAs between FLU and CUMS-induced mice (Supplementary Table S5). In addition, 436 mRNA transcripts were identified as target genes for 20 dysregulated lncRNAs between the CUMS and the NOR groups (Supplementary Table S5). Of note, the identified target genes differed according to the prediction methods even when comparing the same groups.
GO and KEGG pathway analyses of putative target genes
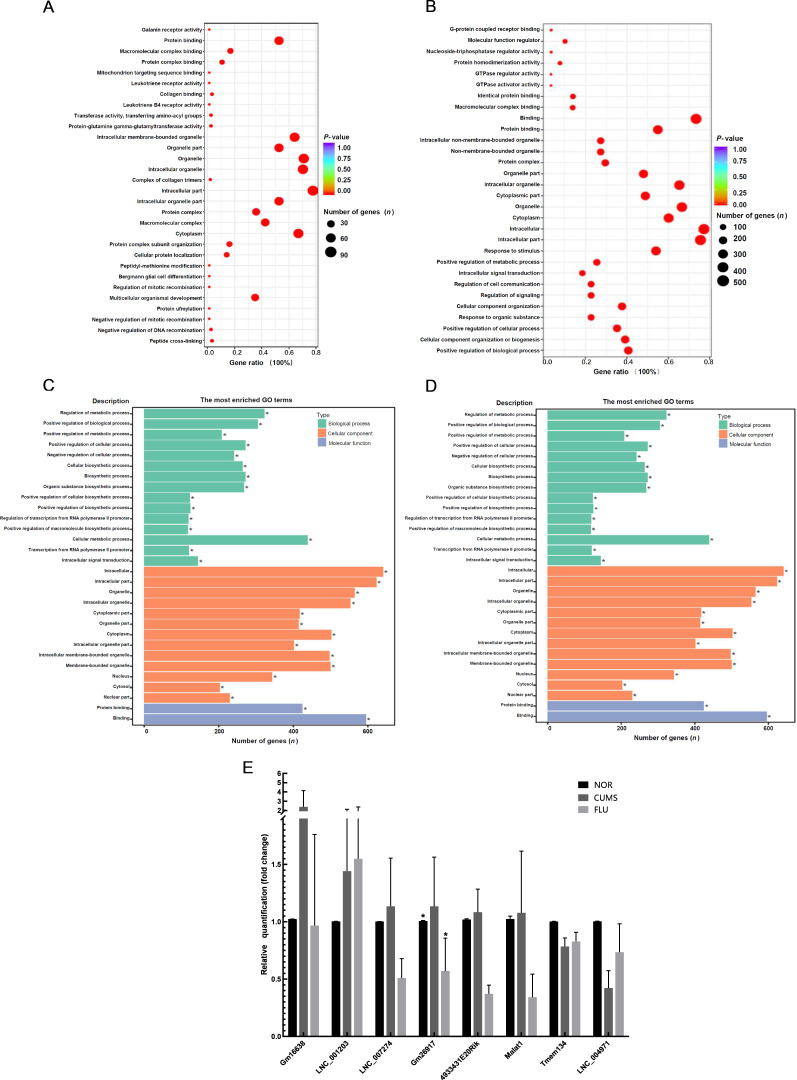
To decipher the functions of DElnRNAs in CUMS-induced depressive mice following fluoxetine treatment, a GO analysis targeting cellular component (CC), molecular function (MF) and biological process (BP) was performed using the predicted targets of altered lncRNAs. A total of 154 co-localized differentially expressed transcripts were identified to be enriched with GO terms (Figure 5A). These genes were related to multicellular organismal development (GO:0007275), intracellular part (GO:0044424), organelles (GO:0043226), and macromolecular complex binding (GO:0044877) in the FLU vs. CUMS comparison. The GO analysis of 679 co-expressed target genes of DElnRNAs indicated an association with the BPs of responses to stimuli (GO:0050896) and MF of protein binding (GO:0005515) in the FLU vs. the CUMS group (Figure 5B). The predicted co-expressed target genes were possibly enriched in the intracellular part or cytoplasm of the cell (GO:0044424 and GO:0005737) and organelles (GO:0043226) (Figure 5B). Detailed GO analyses using the Protein ANalysis THrough Evolutionary Relationships (PANTHER) classification system (http://www.pantherdb.org/) revealed that lncRNAs differentially expressed with co-localized mRNAs in fluoxetine-treated mice compared to CUMS-induced mice may function in cellular processes, including cellular component organization (GO:0016043), cellular responses to stimuli (GO:0051716), cellular metabolic process (GO:0044237), and signal transduction (GO:0007165) (data not shown). Pathway analysis using PANTHER based on the same predicted target genes identified gonadotropin-releasing hormone receptor (P06664) and endothelin signaling pathways (P00019) (Supplementary Figure S1). We further identified integrin signaling (P00034), angiogenesis (P00005), GABA-B receptor II signaling (P05731), and other signaling pathways involved in the pathogenesis of fluoxetine-treated CUMS-induced mice (Supplementary Figure S1).
Figure 5.
GO analysis of putative target genes
A: GO analysis of mRNAs co-localized with DElnRNAs between FLU and CUMS mice. A total of 154 co-localized differentially expressed transcripts were found to be enriched with the GO terms of organismal development, intracellular part, organelle, and macromolecular complex binding when comparing FLU and CUMS groups. B: GO analysis of mRNAs co-expressed with DElnRNAs between FLU and CUMS mice. A total of 679 co-expressed transcripts indicated an association with the GO terms of response to stimulus, intracellular part, organelle or cytoplasm of the cell, and protein binding. C, D: GO annotation of mRNA targets co-localized with 48 DElnRNAs which were up-regulated in CUMS-induced mice compared to NOR mice, and down-regulated in FLU treated compared to CUMS mice. These 48 lncRNA transcripts were predicted to be associated with oxygen binding and transport activity. E: Validation of lncRNA expressions in the hippocampus of CUMS, FLU, and NOR mice by qRT-PCR. Six of eight randomly selected lncRNAs were up-regulated, whereas Tmem134 and LNC_004971 were down-regulated in CUMS mice compared to NOR controls. Up-regulated lncRNAs in CUMS mice compared to NOR controls were down-regulated after fluoxetine treatment, thereby validating the RNA-seq results.
The GO analysis revealed that most genes were related to binding and catalytic activity (Supplementary Figure S2A), including those involved in cellular and metabolic processes (Supplementary Figure S2B) and located within the intracellular part (GO:0005622) and cellular anatomical entity (GO:0110165), based on the co-expressed mRNA transcripts (Supplementary Figure S2C). The encoded proteins were grouped as metabolite interconversion enzyme (PC00262), protein modifying enzyme (PC00260), transporter (PC00227), and other protein families (Supplementary Figure S2D).
In the CUMS vs. the NOR group, predicted co-localized target mRNAs for DElnRNAs were located in the cytosol (GO:0005829) and had the ciliary neurotrophic factor receptor binding function (GO:0005127) (data not shown). In addition, multiple signaling pathways, including those involved in CCKR signaling, were identified to be involved in the pathophysiology of CUMS-induced mice (data not shown). These results suggested that fluoxetine intervention could reverse the depressive effects in CUMS-induced mice by regulating the expression of lncRNAs through multiple signaling pathways.
To further elucidate the specific mechanism of CUMS induced-depression, we used GO annotation to analyze the functions of 95 commonly expressed lncRNAs that had contrasting expression patterns between FLU vs. CUMS and CUMS vs. NOR groups based on the predicted mRNA genes regulated in cis or trans. We observed that 48 transcripts up-regulated in CUMS-induced mice but down-regulated after fluoxetine intervention were associated with oxygen binding and transport activity (Figure 5C). This suggested that immune response activation caused oxidative stress in CUMS-induced mice. After fluoxetine treatment, a reduction in the expression of initially over-expressed transcripts following oxidative stress was observed. This may have attenuated the symptoms of depression in CUMS-induced mice. We then analyzed the functions of possible co-expressed and regulated target genes using GO and found enrichment in the regulation of metabolic processes (BP category), with a majority of genes associated with protein binding (MF category) and located in the cellular portion and organelles (CC category) (Figure 5D).
qRT-PCR-based validation of hippocampal lncRNA expression
To validate the expression of lncRNAs identified by RNA-Seq, we randomly selected seven significantly altered lncRNAs (LNC_007274, Gm16638, LNC_001203, LNC_004971, Tmem134, Gm26917-201, and 4933431E20Rik) (P<0.05) and one metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in the hippocampi of CUMS, FLU, and NOR mice (n=3 per group) and analyzed their expressions using qRT-PCR. No significant changes in the expression of MALAT1 in the CUMS vs. the NOR group were observed. Fluoxetine treatment decreased MALAT1 expression (Figure 5E). The seven altered lncRNAs showed a similar expression pattern in CUMS-induced mice compared to NOR mice, consistent with the RNA-seq data, except for Tmem134 (ENSMUST00000139987.8) and Gm26917 (ENSMUST00000182520.1) (Figure 5E). Six lncRNAs were up-regulated, whereas Tmem134 and LNC_004971 were down-regulated in CUMS-induced mice compared to NOR controls (Figure 5E). LncRNAs were up-regulated in CUMS mice compared to normal controls but down-regulated after fluoxetine treatment, which validated the results from RNA-seq and further confirmed that fluoxetine regulated the expression of lncRNAs in the hippocampus of CUMS-induced mice to alleviate depressive-like behaviors.
DISCUSSION
This is the first study to explore the expression of lncRNAs in the hippocampus of old CUMS-induced mice coupled with a pharmacological intervention using the antidepressant drug fluoxetine. In particular, we applied the CUMS procedure in BALB/c mice to identify changes in lncRNA expression on a genome-wide scale with and without fluoxetine intervention concerning depressive-like behaviors. Because it is not possible to obtain brain samples from living patients to study the molecular mechanisms of depression, the CUMS model has been applied as a mild daily stressor to induce depression in mice (Nollet et al., 2013; Surget & Belzung, 2008).
Diagnosis of depression in older adults is complicated because symptoms are different from those in younger populations (Fiske et al., 2009). In this study, we used the CUMS procedure to induce depression in 13-month-old BALB/c mice, representing late-life according to the mouse lifespan. Fluoxetine treatment attenuated depressive-like behaviors in the SPT, FBT, and TST, consistent with previous reports in which depressive-like behaviors were reversed by fluoxetine in CUMS-induced mice (Yalcin et al., 2008). Although fluoxetine exhibits an anti-depressant effect on depression in CUMS-induce mice, its specific underlying molecular mechanism remains unclear.
Based on the classical monoamine deficiency hypothesis of depression (López-Muñoz & Alamo, 2009), we examined the hippocampal expression of 5-HT across three groups (CUMS, FLU, and NOR mice). Fluoxetine increased the expression levels of 5-HT in the hippocampus of CUMS-induced mice compared to NOR mice, suggesting that fluoxetine-mediated inhibition of 5-HT reuptake reversed depressive-like behaviors in old CUMS mice. This observation highlights the efficacy of fluoxetine in the geriatric population. The neurotrophic factor BDNF is considered a molecular mediator of neuroplasticity (Bramham & Messaoudi, 2005). Based on the neurotrophic hypothesis of depression, a reduction in BDNF levels is expected in the brain of individuals diagnosed with depression, although antidepressants can induce an increase in BDNF (Neto et al., 2011). Based on this, we profiled the expression levels of BDNF in CUMS-induced mice and observed decreased BDNF levels in old CUMS-induced depressive mice. However, these levels increased after fluoxetine treatment. Previous studies have reported that BDNF levels are decreased in MDD (Sen et al., 2008) and increased after electroconvulsive stimuli or Alzheimer’s disease (AD) medication in both animal models (Angelucci et al., 2002) and MDD patients (Brunoni et al., 2008; Sen et al., 2008). Conversely, BDNF levels have been reported to increase in antidepressant drug-naïve MDD patients during the early stages of the disorder (Stelzhammer et al., 2014). Overall, these findings demonstrated that viable neuroplasticity was affected at the onset of depression. Our results were consistent with previous findings of decreased BDNF levels in MDD patients and increased BDNF expression after the medical intervention (Sen et al., 2008). This suggests a common regulatory mechanism underpinning both geriatric and late-onset depression.
Numerous studies have reported that the regulation of depression is multigenic (Huang et al., 2017). However, the role of genetics in geriatric depression is poorly understood. LncRNAs are promising candidates for managing psychiatric disorders. They are highly expressed in the central nervous system, and affect neural stem cell maintenance, neurogenesis and gliogenesis, brain patterning, synaptic and stress responses, neural plasticity, and cognitive function. Abnormal expression of lncRNAs has been observed in multiple human diseases, including MDD (Liu et al., 2014; Wapinski & Chang, 2011). For instance, a microarray-based analysis of lncRNA from peripheral blood of MDD patients revealed that 2007 lncRNAs were dysregulated (Liu et al., 2014). In addition, a recent study reported a significant down-regulation of six lncRNAs (TCONS_00019174, ENST00000566208, NONHSAG045500, ENST00000517573, NONHSAT034045, and NONHSAT142707) in patients with MDD compared to healthy controls, highlighting the potential diagnostic and therapeutic value of lncRNAs for MDD (Cui et al., 2016). Of these, TCONS_00019174 has been observed to be down-regulated in depressive mice, whereas the over-expression of TCONS_00019174 improved depressive symptoms in mice (Ni et al., 2018). TCONS_00019174 over-expression increased the expression of phosphorylated-GSK3β and β-catenin in the hippocampus. This study suggested that the Wnt/β-catenin pathway regulated by lncRNA TCONS_00019174 might be involved in the pathogenesis of depression (Ni et al., 2018). Roy et al. (2018) reported that altered expression of lncRNAs associated with zinc finger binding proteins, such as Zbtb20 and Zfp385b, in Holtzman rats exhibited learned or non-learned helplessness in a microarray-based study. Despite the identification of numerous lncRNAs and their association with MDD and other depressive behaviors, the specific roles they play in these symptoms are vague.
In the current study, 282 lncRNAs including 134 up-regulated and 148 down-regulated lncRNAs were identified to be differentially expressed between the CUMS-induced and the NOR mice. In contrast, 370 lncRNAs (236 up-regulated and 134 down-regulated) were differentially expressed between the FLU and the CUMS groups. Of note, 95 lncRNAs were commonly expressed and exhibited contrasting expression patterns between the two comparative groups. Three lncRNAs (LNC_004212, LNC_001203, and ENSMUST00000148687.1) were identified to have contrasting expression changes (Q<0.05). These results demonstrated that CUMS-induced depressive behaviors could be ameliorated by fluoxetine intervention through regulation of hippocampal lncRNA levels. LNC_004212 and LNC_001203 were novel lncRNAs. In contrast, ENSMUST00000148687.1, located on Chromosome 7, has previously been annotated and encodes RP24-252B21.2 transcript. Prediction of target genes revealed that RP24-252B21.2 regulates interleukin-16 (IL-16) expressionin cis. Previous studies have reported that treatments using antidepressants are associated with a decrease in inflammatory markers (Hiles et al., 2012). Alterations in inflammatory cytokines (IL-6, C-reactive protein, and IL-10) and other inflammation-related proteins (increased levels of pro-inflammatory proteins EN-RAGE, IL-16, MIF, and tenascin-C) have previously been identified in blood-cerebrospinal fluid and post-mortem studies of MDD patients (Maes et al., 1995; Raedler, 2011). Furthermore, the side effects of cytokine treatment could induce depression symptoms in patients diagnosed with certain cancers and hepatitis C. There was an increase in inflammatory-associated factors regulated by lncRNAs in CUMS-induced depressive mice and a decrease in corresponding lncRNA regulators after fluoxetine treatment. This highlights RP24-252B21.2 as a promising candidate in the neuro-immune interplay that may link depressive symptoms and altered immune state. Our findings suggested that stress alters hippocampal expression of lncRNAs and mRNAs, and activates the inflammatory pathways, leading to the release of cytokine factors such as IL-16, ultimately contributing to the development of depression. This hypothesis was consistent with inflammatory responses during the depression. In addition, the predicted functions of the 95 lncRNAs showing contrasting expression patterns in FLU vs. CUMS and CUMS vs. NOR groups were associated with oxygen-binding, transport activity, protein binding, and metabolic regulation processes. Reactive oxygen species (ROS) are involved in the pathology of MDD, which is also associated with metabolic disorders. LncRNAs identified in the present study may regulate target genes involved in inflammatory responses and metabolic processes.
Interestingly, our RNA-seq result showed that lncRNA Gm26917 (ENSMUST00000182520.1) was up-regulated in fluoxetine-treated CUMS mice compared to the CUMS depressive counterpart. Generally, lncRNAs function as “miRNA sponges” and regulate the expression of miRNAs on their target mRNA transcripts (Cesana et al., 2011). Gm26917, a novel competing endogenous RNA (ceRNA) for miRNA-29b was regulated by transcription factor FoxM1 involved in muscle satellite cells (SCs) survival (Chen et al., 2018). This was the first report of Gm26917 function related to cell survival. Chen et al. also predicted that seven miRNAs, including miR-425, miR-486, miR-28-5p, miR-29b, miR-100, miR-125b, and miR-320, showed potential matching parts with Gm26917 (Chen et al., 2018). The miR-29b-3p expression was increased in the prefrontal cortex of the depressive-like rats and primary neurons after ketamine treatment, resulting in a decrease of metabotropic glutamate receptor 4 (GRM4) expression (Wan et al., 2018). Hence, lncRNA Gm26917/miR-29b-3p/GRM4 pathway could be the key regulators of both fluoxetine and ketamine interventions in CUMS-induced depressive mice and could be the potential target for depression treatment. In addition, miR-425-3p expression was changed in MDD patients after 8 weeks of treatment by duloxetine and was involved in MAPK/Wnt signaling pathways (Lopez et al., 2017). MiR-320 was found significantly down-regulated in MDD patients (Camkurt et al., 2015). Taken together, several predicted miRNA targets with Gm29617 were associated with the pathophysiology of depression. It indicated that Gm26917 could bind to specific miRNAs and regulate the expression of their target mRNAs in depression and become the promising therapeutic candidate for antidepressant treatment.
In summary, 370 DElnRNAs were identified in CUMS-induced mice after fluoxetine intervention. These dysregulated lncRNAs were associated with protein-binding, oxygen binding and transport, and inflammatory response pathways. The specific roles played by novel lncRNAs require further elucidation. Overall, our findings suggest that inflammation regulates lncRNAs and their targets in the hippocampus in stress-induced depression. Our study provides insight into the roles of lncRNAs in the pathogenesis of depression and may facilitate the diagnosis, treatment, and prevention of depression in the elderly. Further studies on the specific roles of these DElnRNAs in attenuating depression are required.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare no competing interests.
AUTHORS’ CONTRIBUTIONS
C.L.Z. secured the funds, conceived and designed the experiments, collected the data, performed analysis, and prepared the original draft. Y.J.L., S.L. and T.Z. collected the data and performed preliminary analysis. R.X. and H.R.L. supervised the experiments, reviewed and edited the draft. All the authors discussed and revised the draft. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We thank Elsevier Ltd., UK and FreeScience, China for their assistance in English editing of the manuscript.
Funding Statement
This work was supported by the Scientific Research Projects of Universities in Inner Mongolia Autonomous Region (NJZY111) and Natural Scientific Research Projects of Inner Mongolia Autonomous Region (2020MS03060)
Contributor Information
Rui Xiao, Email: xiaorui79@hotmail.com.
Huai-Rong Luo, Email: lhr@swmu.edu.cn.
References
- 1.Angelucci F, Aloe L, Jiménez-Vasquez P, Mathé AA Electroconvulsive stimuli alter the regional concentrations of nerve growth factor, brain-derived neurotrophic factor, and glial cell line-derived neurotrophic factor in adult rat brain. The Journal of ECT. 2002;18(3):138–143. doi: 10.1097/00124509-200209000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Beyer JL Managing depression in geriatric populations. Annals of Clinical Psychiatry. 2007;19(4):221–238. doi: 10.1080/10401230701653245. [DOI] [PubMed] [Google Scholar]
- 3.Blazer DG Depression in late life: review and commentary. The Journals of Gerontology: Series A. 2003;58(3):M249–M265. doi: 10.1093/gerona/58.3.M249. [DOI] [PubMed] [Google Scholar]
- 4.Bramham CR, Messaoudi E BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Progress in Neurobiology. 2005;76(2):99–125. doi: 10.1016/j.pneurobio.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Bremner JD Fluoxetine in depressed patients: a comparison with imipramine. The Journal of Clinical Psychiatry. 1984;45(10):414–419. [PubMed] [Google Scholar]
- 6.Brunoni AR, Lopes M, Fregni F A systematic review and meta-analysis of clinical studies on major depression and BDNF levels: implications for the role of neuroplasticity in depression. International Journal of Neuropsychopharmacology. 2008;11(8):1169–1180. doi: 10.1017/S1461145708009309. [DOI] [PubMed] [Google Scholar]
- 7.Camkurt MA, Acar Ş, Coşkun S, Güneş M, Güneş S, Yılmaz MF, et al Comparison of plasma MicroRNA levels in drug naive, first episode depressed patients and healthy controls. Journal of Psychiatric Research. 2015;69:67–71. doi: 10.1016/j.jpsychires.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 8.Casey DA Depression in the elderly. South Medcal Jounal. 1994;87(5):559–563. [PubMed] [Google Scholar]
- 9.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen DM, Meng L, Pei F, Zheng Y, Leng JY A review of DNA methylation in depression. Journal of Clinical Neuroscience. 2017;43:39–46. doi: 10.1016/j.jocn.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Bu NP, Qiao XH, Zuo ZX, Shu YH, Liu ZL, et al Forkhead Box M1 transcriptionally regulates the expression of long noncoding RNAs Snhg8 and Gm26917 to promote proliferation and survival of muscle satellite cells. Stem Cells. 2018;36(7):1097–1108. doi: 10.1002/stem.2824. [DOI] [PubMed] [Google Scholar]
- 12.Cryan JF, Mombereau C, Vassout A The tail suspension test as a model for assessing antidepressant activity: review of pharmacological and genetic studies in mice. Neuroscience & Biobehavioral Reviews. 2005;29(4–5):571–625. doi: 10.1016/j.neubiorev.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Cui XL, Sun XY, Niu W, Kong LM, He MJ, Zhong AF., et al Long non-coding RNA: potential diagnostic and therapeutic biomarker for major depressive disorder. Medical Science Monitor. 2016;22:5240–5248. doi: 10.12659/MSM.899372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiske A, Wetherell JL, Gatz M Depression in older adults. Annual Review of Clinical Psychology. 2009;5:363–389. doi: 10.1146/annurev.clinpsy.032408.153621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flurkey K, Currer JM, Harrison DE. 2007. Mouse models in aging research. In: Fox JG, Davisson MT, Quimby FW, Barthold SW, Newcomer CE, Smith AL. The Mouse in Biomedical Research. 2nd ed. Amsterdam: Elsevier, 637−672.
- 16.Gibbons A, Udawela M, Dean B Non-coding RNA as novel players in the pathophysiology of schizophrenia. Non-Coding RNA. 2018;4(2):11. doi: 10.3390/ncrna4020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guil S, Esteller M Cis-acting noncoding RNAs: friends and foes. Nature Structural & Molecular Biology. 2012;19(11):1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 18.Gururajan A, Clarke G, Dinan TG, Cryan JF Molecular biomarkers of depression. Neuroscience & Biobehavioral Reviews. 2016;64:101–133. doi: 10.1016/j.neubiorev.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Hiles SA, Baker AL, de Malmanche T, Attia J Interleukin-6, C-reactive protein and interleukin-10 after antidepressant treatment in people with depression: a meta-analysis. Psychological Medicine. 2012;42(10):2015–2026. doi: 10.1017/S0033291712000128. [DOI] [PubMed] [Google Scholar]
- 20.Huang X, Luo YL, Mao YS, Ji JL The link between long noncoding RNAs and depression. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2017;73:73–78. doi: 10.1016/j.pnpbp.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Huang XY, Sun WY, Yan ZQ, Shi HR, Yang QL, Wang PF, et al Integrative analyses of long non-coding RNA and mRNA involved in piglet ileum immune response to Clostridium perfringens Type C infection . Frontiers in Cellular and Infection Microbiology. 2019;9:130. doi: 10.3389/fcimb.2019.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jani BD, McLean G, Nicholl BI, Barry SJE, Sattar N, Mair FS, et al Risk assessment and predicting outcomes in patients with depressive symptoms: a review of potential role of peripheral blood based biomarkers. Frontiers in Human Neuroscience. 2015;9:18. doi: 10.3389/fnhum.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei LP, et al CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Research. 2007;35(S2):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung AKL, Sharp PA MicroRNA functions in stress responses. Molecular Cell. 2010;40(2):205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin MF, Jungreis I, Kellis M PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27(13):i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu MY, Yin CY, Zhu LJ, Zhu XH, Xu C, Luo CX, et al Sucrose preference test for measurement of stress-induced anhedonia in mice. Nature Protocols. 2018;13(7):1686–1698. doi: 10.1038/s41596-018-0011-z. [DOI] [PubMed] [Google Scholar]
- 28.Liu ZF, Li XR, Sun N, Xu Y, Meng YQ, Yang CX, et al Microarray profiling and co-expression network analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. PLoS One. 2014;9(3):e93388. doi: 10.1371/journal.pone.0093388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez JP, Fiori LM, Cruceanu C, Lin RX, Labonte B, Cates HM, et al MicroRNAs 146a/b-5 and 425–3p and 24–3p are markers of antidepressant response and regulate MAPK/Wnt-system genes. Nature Communications. 2017;8:15497. doi: 10.1038/ncomms15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-Muñoz F, Alamo C Monoaminergic neurotransmission: the history of the discovery of antidepressants from 1950s until today. Current Pharmaceutical Design. 2009;15(14):1563–1586. doi: 10.2174/138161209788168001. [DOI] [PubMed] [Google Scholar]
- 31.Maes M, Smith R, Scharpe S The monocyte-T-lymphocyte hypothesis of major depression. Psychoneuroendocrinology. 1995;20(2):111–116. doi: 10.1016/0306-4530(94)00066-J. [DOI] [PubMed] [Google Scholar]
- 32.Miller AH, Raison CL The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nature Reviews Immunology. 2016;16(1):22–34. doi: 10.1038/nri.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neto FL, Borges G, Torres-Sanchez S, Mico JA, Berrocoso E Neurotrophins role in depression neurobiology: a review of basic and clinical evidence. Current Neuropharmacology. 2011;9(4):530–552. doi: 10.2174/157015911798376262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni XQ, Liao YZ, Li LM, Zhang XL, Wu ZZ Therapeutic role of long non-coding RNA TCONS_00019174 in depressive disorders is dependent on Wnt/β-catenin signaling pathway . Journal of Integrative Neuroscience. 2018;17(2):125–132. doi: 10.31083/JIN-170052. [DOI] [PubMed] [Google Scholar]
- 35.Nollet M, Le Guisquet AM, Belzung C Models of depression: unpredictable chronic mild stress in mice. Current Protocols in Pharmacology. 2013;61(1):5.65. doi: 10.1002/0471141755.ph0565s61. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Caballero L, Torres-Sanchez S, Bravo L, Mico JA, Berrocoso E Fluoxetine: a case history of its discovery and preclinical development. Expert Opinion on Drug Discovery. 2014;9(5):567–578. doi: 10.1517/17460441.2014.907790. [DOI] [PubMed] [Google Scholar]
- 37.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al The Pfam protein families database. Nucleic Acids Research. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raedler TJ Inflammatory mechanisms in major depressive disorder. Current Opinion in Psychiatry. 2011;24(6):519–525. doi: 10.1097/YCO.0b013e32834b9db6. [DOI] [PubMed] [Google Scholar]
- 40.Raison CL, Capuron L, Miller AH Cytokines sing the blues: inflammation and the pathogenesis of depression. Trends in Immunology. 2006;27(1):24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riva P, Ratti A, Venturin M The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Current Alzheimer Research. 2016;13(11):1219–1231. doi: 10.2174/1567205013666160622112234. [DOI] [PubMed] [Google Scholar]
- 42.Robinson MD, McCarthy DJ, Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy B, Wang QZ, Dwivedi Y Long noncoding RNA-Associated transcriptomic changes in resiliency or susceptibility to depression and response to antidepressant treatment. The International Journal of Neuropsychopharmacology. 2018;21(5):461–472. doi: 10.1093/ijnp/pyy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampson SM Treating depression with selective serotonin reuptake inhibitors: a practical approach. Mayo Clinic Proceedings. 2001;76(7):739–744. doi: 10.4065/76.7.739. [DOI] [PubMed] [Google Scholar]
- 45.Sen S, Duman R, Sanacora G Serum brain-derived neurotrophic factor, depression, and antidepressant medications: meta-analyses and implications. Biological Psychiatry. 2008;64(6):527–532. doi: 10.1016/j.biopsych.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou MM, Rosenbloom K, et al Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Research. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stelzhammer V, Haenisch F, Chan MK, Cooper JD, Steiner J, Steeb H, et al Proteomic changes in serum of first onset, antidepressant drug-naïve major depression patients. International Journal of Neuropsychopharmacology. 2014;17(10):1599–1608. doi: 10.1017/S1461145714000819. [DOI] [PubMed] [Google Scholar]
- 48.Strawbridge R, Young AH, Cleare AJ Biomarkers for depression: recent insights, current challenges and future prospects. Neuropsychiatric Disease and Treatment. 2017;13:1245–1262. doi: 10.2147/NDT.S114542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun L, Luo HT, Bu DC, Zhao GG, Yu KT, Zhang CH, et al Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Research. 2013;41(17):e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surget A, Belzung C. 2008. Unpredictable chronic mild stress in mice. In: Kalueff AV, LaPorte JL. Experimental Animal Models in Neurobehavioral Research. New York, NY: Nova Science, 79−112.
- 51.Tang J, Yu YZ, Yang W Long noncoding RNA and its contribution to autism spectrum disorders. CNS Neuroscience & Therapeutics. 2017;23(8):645–656. doi: 10.1111/cns.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan YQ, Feng JG, Li M, Wang MZ, Liu L, Liu XR, et al Prefrontal cortex miR-29b-3p plays a key role in the antidepressant-like effect of ketamine in rats. Experimental & Molecular Medicine. 2018;50(10):1–14. doi: 10.1038/s12276-018-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wapinski O, Chang HY Long noncoding RNAs and human disease. Trends in Cell Biology. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Willner P, Towell A, Sampson D, Sophokleous S, Muscat R Reduction of sucrose preference by chronic unpredictable mild stress, and its restoration by a tricyclic antidepressant. Psychopharmacology. 1987;93(3):358–364. doi: 10.1007/BF00187257. [DOI] [PubMed] [Google Scholar]
- 55.Xiao R, Park Y, Dirisala VR, Zhang YP, Um SJ, Lee HT, et al Identification of genes differentially expressed in wild type and Purkinje cell degeneration mice. Molecules and Cells. 2005;20(2):219–227. [PubMed] [Google Scholar]
- 56.Yalcin I, Belzung C, Surget A Mouse strain differences in the unpredictable chronic mild stress: a four-antidepressant survey. Behavioural Brain Research. 2008;193(1):140–143. doi: 10.1016/j.bbr.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 57.Yankelevitch-Yahav R, Franko M, Huly A, Doron R The forced swim test as a model of depressive-like behavior. Journal of Visualized Experiments. 2015;(97):e52587. doi: 10.3791/52587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.