Summary
Major depression is a prevalent and long-lasting psychiatric illness with severe functional impairment and high suicide rate. We have previously shown that the ventrolateral orbital cortex (VLO) plays a key role in the stress responses in mice, but the underlying mechanisms remains unclear. Here, we used proteomic method to identify differentially expressed proteins in VLO of chronic unpredictable mild stress (CUMS) mice. Of 4,953 quantified proteins, 45 proteins were differentially expressed following CUMS. The integrated pathway analyses identified 14-3-3ε and TrkB signaling as differentially downregulated in association with stress-induced depressive-like behaviors. 14-3-3ε overexpression in VLO relieved the depressive-like behaviors by rescue of Bad-mediated apoptosis. Moreover, treatment with the 14-3-3ε stabilizer FC-A precluded neuronal apoptotic signaling in VLO of depressed mice. Because 14-3-3ε provides significant protection against chronic stress, boosting 14-3-3ε expression, pharmacological stabilization of 14-3-3s (e.g. with FC-A) is identified as an exciting therapeutic target for major depression.
Subject areas: Neuroscience, Cell Biology, Proteomics
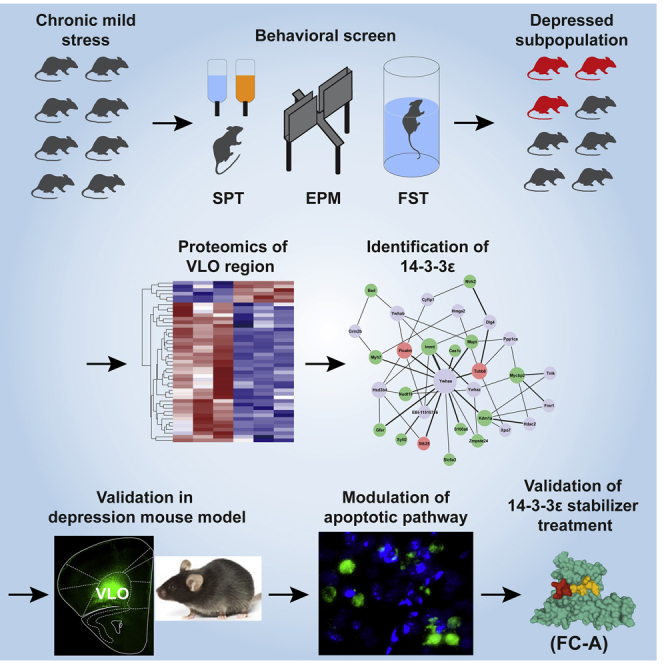
Graphical abstract
Highlights
-
•
Novel screening of chronic mild stress-induced depression phenotypes in mice
-
•
Proteomics identify 14-3-3ε as a key modulator of depressive behaviors in VLO
-
•
14-3-3ε partially reversed depressive behaviors through neural apoptotic pathway
-
•
14-3-3ε stabilizer FC-A ameliorates depression phenotypes after chronic mild stress
Neuroscience; Cell Biology; Proteomics
Introduction
Major depression (MD) is a common and debilitating psychiatric illness with nearly 350 million sufferers worldwide (Kessler et al., 2005). Symptoms of the illness include apathy, anhedonia, anxiety, sleep disturbance, changes in appetite or weight, and impaired attention (Pinheiro et al., 2015). MD can be long-lasting or recurrent, substantially impairing quality of life and functioning, which significantly contribute to financial burden, increased hospitalization, functional impairment, and suicide (Saarni et al., 2007). However, substantial molecular and signaling pathway dysregulation are involved in MD, making the pathophysiology of this multifactorial affective disorder complicated and largely unknown. External stress is widely acknowledged as a predisposing and precipitating factor of MD especially in genetically predisposed individuals. In animal models, the chronic unpredictable mild stress (CUMS) rodent model mimics the variable and unpredictable physical and mental irritations encountered in human daily life and generates a variety of behavioral changes, which are thought to reflect some of the core symptoms seen in depressed humans (e.g., anhedonia, anxiety and despair) (Willner, 2005). Although MD patients show a highly diverse set of combination of symptoms, CUMS-exposed animals are often considered as a homogeneous population. Therefore, in order to study the symptom-specific neural correlates, it is urgent to establish a novel strategy that recognize the heterogeneity of CUMS-induced behavioral phenotypes.
The orbitofrontal cortex has been widely implicated in reward processing, hedonic behavior, and stress responses in both animals and human (Elliott et al., 2010; Rolls, 2004), suggesting a role for orbitofrontal cortex in mood disorders. Indeed, neuroimaging, neuropathological, and lesion studies have provided converging evidence for structural and functional abnormalities of the orbitofrontal cortex in MD individuals (Drevets, 2007). Reductions of gray matter volume and cortex thickness, indicated by magnetic resonance imaging, have been demonstrated more specifically in the orbitofrontal cortex (Nugent et al., 2006; Taylor et al., 2007). In addition, depressed patients showed abnormalities of regional cerebral blood flow and metabolism in the orbitofrontal cortex compared with non-depressed patients (Drevets et al., 2002; Nugent et al., 2006; Ring et al., 1994). Postmortem studies have characterized the histopathologic correlates of the reductions in neuronal and glial size, decreased synaptic density, decreased expression of synaptic markers, increased neuronal apoptosis, and abnormalities in glial-to-neuron ratios in MD and bipolar disorder in the orbitofrontal cortex (Drevets, 2007; Johnston-Wilson et al., 2000; Ongur et al., 1998). A series of research from our team have showed that the ventrolateral orbital cortex (VLO), a major subdivision of orbitofrontal cortex, was critically involved in the stress-related memory formation, depressive-like behaviors, and antidepressant effects in mice (Xing et al., 2011; Zhao et al., 2013, 2017). Based on this evidence, VLO dysfunction may partially explain the pathophysiology of MD, which requires further investigation. However, most of the previous studies of this area have focused on changes in a single or several signaling pathways in MD; very little is known about global proteomic profiles in VLO in animal models of depression.
The isobaric tags for relative and absolute quantification (iTRAQ) is a classic quantitative proteomic technique. It can simultaneously quantify proteins in 8-plex samples, making this method a powerful tool for the identification of disease biomarkers (Evans et al., 2012). A combined iTRAQ-liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics approach is one of the most sensitive proteomics technologies and can detect and quantitatively analyze low-abundance proteins in complex biological samples (Liu et al., 2009).
Over the last few years, the iTRAQ combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis has become a powerful quantitative proteomic approach due to its remarkable advantages over traditional proteomic methods, such as its high-throughput setup and outstanding accuracy (Jiang et al., 2020). Moreover, iTRAQ is sensitive to subtle fold changes in proteins, especially in quantifying medium to high abundance proteins. Recently, iTRAQ-based quantitative proteomics are rapidly being applied to neurobiological investigations of various neuropsychiatric disorders, including autism-spectrum disorders (Shen et al., 2018), schizophrenia (Velasquez et al., 2019), and drug addiction (Zhu et al., 2017). In the present study, we first used a method based on the multiple behavioral assays to identify CUMS-induced depressed mice with multifaceted long-term depression-like symptoms, which is a typical and obligatory feature for clinical depression. Then, we used iTRAQ-LC-MS/MS to screen for the expression of various proteins in VLO of depressed mice. The differentially expressed proteins were identified and analyzed for gene ontology annotation, signaling pathway analysis, and protein-protein interaction. Furthermore, based on the proteomic data, we identified that the differentially expressed 14-3-3 epsilon is a key regulator of the TrkB/PI3K/Akt pathway and neural apoptosis in VLO of depressed mice. Our findings revealed that boosting 14-3-3ε expression, pharmacological stabilization of 14-3-3s (e.g. with FC-A) or its protein partners is an exciting potential therapeutic target for the treatment of MD.
Results
Experiment 1: identification of the CUMS-induced depressed mouse
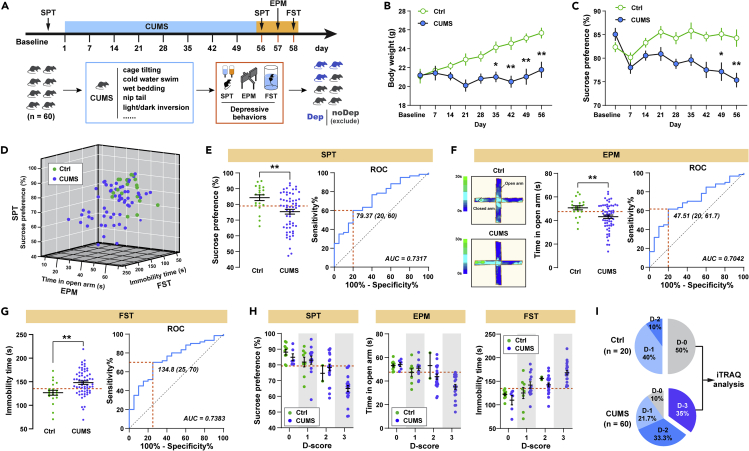
The timeline of the CUMS regime and behavioral assessments is showed in Figure 1A. The body weight and sucrose preference test (SPT) were recorded weekly. The timeline figure only shows SPT before and after CUMS. Mice were exposed to 8 weeks of CUMS and subsequently analyzed them using three different behavioral tests. We observed significantly decreased body weight in CUMS-exposed mice (n = 60) after 35 days of CUMS compared with non-stressed control mice (n = 20) (Figure 1B). At the end of the CUMS, we used SPT to determine whether the mice showed anhedonia, which is a core symptom of depression (Duman, 2007). As showed in Figure 1C, CUMS mice exhibited decreased sucrose preference at both day 49 and day 56, indicating that this CUMS regime is sufficient to induce anhedonia in mice. We also assessed anxiety-related behaviors and pathological motivational impairments, which are commonly seen in MD patients, by the elevated plus maze (EPM) and forced swim test (FST), respectively. Despite a great variability in the depressive-like behaviors for individual mice (Figure 1D), the CUMS-exposed mice showed decreased sucrose preference in SPT (Figure 1E), spent significantly less time in the open arms in the EPM (Figure 1F), and struggled significantly less in the FST (Figure 1G), indicating that, on average, these animals develop anhedonia, anxiety, and despair in response to CUMS exposure. Moreover, significant correlations were found between sucrose preference, open-arm time, and immobile time within the same animals (Table 1), suggesting that these measures are suitable to assess a similar behavioral phenotype. To establish cut-off criteria based on the three depressive phenotype measures, we used the receiver operating characteristic (ROC) algorithm, which is an objective method that has been used extensively in clinical epidemiology for the evaluation of binary classifiers (Berrar and Flach, 2012). As shown in Figures 1E–1G, the area under curve (AUC) for all measures are higher than 0.7. Specifically, cut-off criteria were defined as the Youden's index (Youden, 1950), which allowed us to make an unbiased decision as to whether an individual animal is positive for a given behavioral phenotype. The cut-off values that have the combination of highest true-positive and lowest false-positive rates are 79.37% for SPT sucrose preference, 47.51s for EPM open-arm time, and 134.8s for FST immobile time.
Figure 1.
Classification of the behavioral phenotypes in the chronic unpredictable mild stress (CUMS) mice model of depression
(A) Schematic showing the CUMS regime used to induce depressive-like behaviors in mice. Mice in the CUMS group (n = 60) was exposed to 8 weeks of consecutive CUMS, whereas mice in the control group (n = 20) received only regular handling. At the end of CUMS regime, the behavioral tests were carried out to assess the depressive phenotype.
(B) CUMS-induced significant body weight loss compared with control group (two-way RM ANOVA, CUMS: F 1, 702 = 40.44, p < 0.0001, followed by Sidak's post hoc test).
(C) CUMS-induced decreased sucrose preference compared with control group (two-way RM ANOVA, CUMS: F 1, 702 = 29.49, p < 0.0001, Sidak's post hoc test).
(D) 3D plot showing variability in the depressive-like behaviors from SPT, EPM, and FST for individual Ctrl (green) and CUMS (blue) mice.
(E) Left: sucrose preference for Ctrl and CUMS mice in SPT (unpaired t test, t 78 = 3.119, p < 0.0001; dashed line: cutoff value = 79.37%); right: receiver operating characteristic (ROC) curve (blue line). Orange line, maximum Youden index.
(F) Left: open arm time for Ctrl and CUMS mice in EPM (unpaired t test, t 78 = 2.814, p < 0.01; dashed line: cutoff value = 47.51s); right: ROC curve (blue line). Orange line, maximum Youden index.
(G) Left: immobile time for Ctrl and CUMS mice in FST (unpaired t test, t 78 = 3.101, p < 0.01; dashed line: cutoff value = 134.8s); right: ROC curve (blue line). Orange line, maximum Youden index.
(H) Ctrl and CUMS mice positive for 0, 1, 2, or 3 behavioral criteria. An animal was considered positive if it scored below the corresponding cutoff value (orange line).
(I) Percentage of the total population of Ctrl and CUMS mice positive for D-score 0–3. Only the mice that met all three positive criteria were defined as the depressed (Dep) mice and were selected for further proteomic assessment (D-3, n = 21). The control group (Ctrl) only included the mice showing no depressive phenotype (D-0, n = 10). Data represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, compared with the Ctrl group.
Table 1.
Correlation coefficients across markers of depressive-like behaviors following CUMS regime
| EPM open-arm time | FST immobility time | |
|---|---|---|
| SPT sucrose preference | r = 0.4063∗∗ | r = −0.2717∗ |
| EPM open-arm time | NA | r = −0.3513∗∗ |
Pearson's correlation: ∗p < 0.05, ∗∗p < 0.01.
In order to identify the most vulnerable individuals, we separated both CUMS-exposed mice and non-stressed control mice into four groups according to the number of positive criteria met and assigned them a depression score (D-score) between 0 and 3 (Cerniauskas et al., 2019). Strikingly, both CUMS and control groups contained animals that were positive for one to three criteria or showed no depressive behavior (Figure 1H). Mice that met all three positive criteria (i.e. D-3) represented 35% of the CUMS group, and none of control mice fell into this category (Figure 1I). Only a small fraction of CUMS animals (10%) did not test positive in any depressive behaviors at all (i.e. D-0), whereas the proportion of control mice in this category was much higher (50%). Therefore, we identified the depressed mice (Dep), as those met all three positive criteria (D-3, n = 21). The mice showing only two, one, or no phenotype (D-2 to D-0, total n = 39) were identified as the undepressed mice and were excluded from subsequent experiments. The control group (Ctrl) only included the mice showing no depressive phenotype (D-0, n = 10). This method was used for subsequent experiments to screen out the depressed mice in CUMS model.
Experiment 2: iTRAQ-based quantitative proteomic analysis of VLO
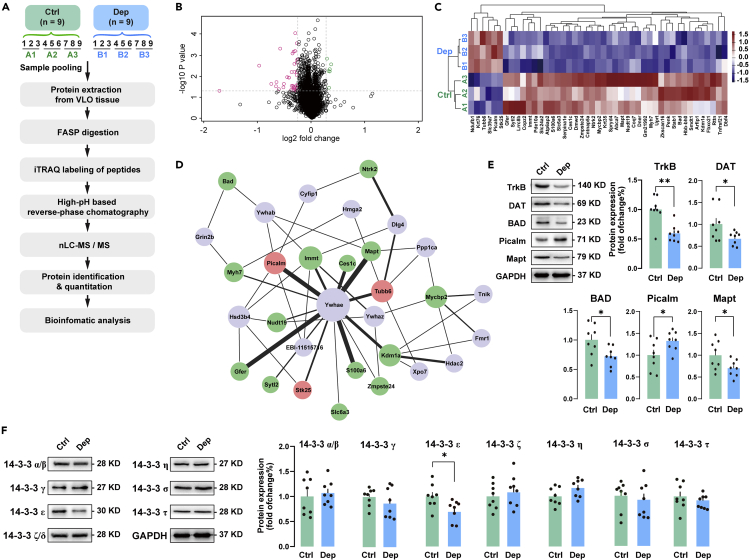
The sample pooling and workflow of the triple biological replicates of iTRAQ-based quantitative proteomic analysis is showed in Figure 2A. Within the VLO tissue, a total of 4,953 proteins were identified and quantitated from both the Dep and Ctrl groups. Differentially expressed proteins were identified by the criteria of 1.2-fold change (>1.20 or <0.84) and false discovery rate (FDR) < 12% between Dep and Ctrl groups (Tables 2 and S1). Of these proteins, 6 were upregulated and 39 were downregulated. These differentially expressed proteins were displayed using a volcano plot map (Figure 2B). For a better systematic view of the expressional alterations between the Dep and Ctrl groups, we performed a hierarchical cluster analysis of the differentially expressed proteins from the iTRAQ experiments. The resulting heatmap clearly indicates that several protein clusters were uniquely changed in Ctrl, whereas same proteins in the Dep group altered in the opposite direction, which emphasizes the dysregulations of protein expressions in the VLO of Dep mice (Figure 2C).
Figure 2.
iTRAQ-based proteomic analysis of VLO proteins from the depressed mice
(A) iTRAQ workflow diagram. The samples included for iTRAQ proteomic analysis were chosen randomly. The VLO samples from nine mice of each group were pooled into three different biological replicates and subjected to iTRAQ labeling followed by mass-spectrometric analysis.
(B) Volcano plot of 4,953 proteins quantified by proteomic analysis from the Dep and Ctrl groups, respectively. Forty-five differentially expressed proteins are identified in the two groups. Black, non-significantly regulated proteins; Red, significantly downregulated proteins; Green, significantly upregulated proteins.
(C) Heatmap illustration of the differential proteins showing expression trends and hierarchical clusters, with the blue to red gradient depicting downregulation to upregulation for each of the Dep and Ctrl groups.
(D) The PPI network of the differentially expressed proteins. Red and green nodes represent differentially expressed upregulated and downregulated proteins, respectively. Gray nodes represent second shell of interactors. Node size represents PPI degrees. Line thickness indicates the strength of data support.
(E) The expression level of the differentially expressed proteins was validated by immunoblots. Unpaired t test was used to determine the difference between Dep and Ctrl groups.
(F) Expression of the seven distinct isoforms of the 14-3-3 protein family. 14-3-3ε expression was significantly decreased in Dep mice (unpaired t test, t 14 = 2.505, p < 0.05), whereas other 14-3-3 isoforms were unchanged. n = 8 per group. Data represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, compared between the two indicated groups.
Table 2.
Differential expressed proteins in VLO identified by iTRAQ proteomics
| Accession number | Gene symbol | Protein description | Unique peptides | Coverage% | Fold change | FDR% | |
|---|---|---|---|---|---|---|---|
| 1 | P23953 | Ces1c | Carboxylesterase 1C | 3 | 9.9278 | 0.8064↓ | 0.8935 |
| 2 | Q00898 | Serpina1e | Alpha-1-antitrypsin 1-5 | 1 | 20.5811 | 0.8237↓ | 1.3709 |
| 3 | Q3UHE3 | Ntrk2 | BDNF/NT-3 growth factors receptor (predicted), isoform 2 | 1 | 19.7479 | 0.6644↓ | 1.3827 |
| 4 | P14069 | S100a6 | S100 calcium binding protein A6 | 1 | 8.9888 | 0.8053↓ | 1.3827 |
| 5 | E9PVS5 | Immt | MICOS complex subunit MIC60 | 2 | 46.8085 | 0.7894↓ | 1.3827 |
| 6 | Q3UVW5 | Slc6a3 | Sodium-dependent dopamine transporter | 1 | 1.2924 | 0.7791↓ | 1.4085 |
| 7 | Q3U6Y0 | Stk25 | Serine/threonine-protein kinase 25 | 1 | 17.8404 | 1.2128↑ | 1.8732 |
| 8 | Q9CYN9 | Atp6ap2 | Renin receptor | 3 | 13.4286 | 0.8321↓ | 1.8732 |
| 9 | Q61337 | Bad | Bcl2-associated agonist of cell death | 1 | 14.2157 | 0.8273↓ | 1.8732 |
| 10 | Q0V8T9 | Cntnap5a | Contactin-associated protein like 5-1 | 1 | 1.1503 | 0.8209↓ | 2.3267 |
| 11 | Q80W54 | Zmpste24 | CAAX prenyl protease 1 homolog | 1 | 3.3684 | 0.7983↓ | 2.3502 |
| 12 | Q14C53 | Slc39a7 | Solute carrier family 39 (Zinc transporter), member 7 | 1 | 2.7311 | 1.2805↑ | 2.3502 |
| 13 | Q497I4 | Krt35 | Keratin, type I cuticular Ha5 | 1 | 3.0769 | 0.5067↓ | 2.5003 |
| 14 | P0DN34 | Ndufb1 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1 | 2 | 31.5789 | 1.2573↑ | 3.4552 |
| 15 | Q7M6Y3 | Picalm | Phosphatidylinositol-binding clathrin assembly protein | 1 | 36.5152 | 1.2392↑ | 3.5233 |
| 16 | Q922F4 | Tubb6 | Tubulin beta-6 chain | 2 | 50.1119 | 1.2565↑ | 3.5233 |
| 17 | Q08274 | Dmwd | Dystrophia myotonica WD repeat-containing protein | 3 | 8.8722 | 0.7986↓ | 3.5233 |
| 18 | E9PUJ6 | Mycbp2 | E3 ubiquitin-protein ligase MYCBP2 | 1 | 0.2528 | 0.7699↓ | 3.5233 |
| 19 | P22005 | Penk | Proenkephalin-A | 1 | 8.9552 | 0.8220↓ | 3.5233 |
| 20 | A2ALW2 | Zkscan16 | Zinc finger with KRAB and SCAN domains 16 | 1 | 1.9231 | 0.8220↓ | 4.6454 |
| 21 | Q7TPG2 | Pde10a | cAMP and cAMP-inhibited cGMP 3′,5′-cyclic phosphodiesterase 10A | 1 | 2.0075 | 0.7269↓ | 5.3662 |
| 22 | Q9DB10 | Smdt1 | Essential MCU regulator, mitochondrial | 1 | 9.3458 | 0.7435↓ | 5.3662 |
| 23 | E5RKB6 | Stab1 | Stabilin-1 | 1 | 0.2723 | 0.7440↓ | 5.3662 |
| 24 | P04443 | Hbb-bh0 | Hemoglobin subunit beta-H0 | 1 | 6.8027 | 0.6334↓ | 8.3718 |
| 25 | F7AV51 | Dbf4 | Protein DBF4 homolog A | 1 | 15.7895 | 0.5926↓ | 9.4245 |
| 26 | B2RY26 | Myh7 | Myosin-7 protein | 1 | 1.1370 | 0.6966↓ | 9.4245 |
| 27 | Q9EPU5 | Tnfrsf21 | Tumor necrosis factor receptor superfamily member 21 | 1 | 1.9847 | 0.8265↓ | 9.9930 |
| 28 | Q8BUN9 | Slc24a2 | Sodium/potassium/calcium exchanger 2 (predicted), isoform 1 | 1 | 16.6667 | 0.7833↓ | 10.2352 |
| 29 | Q91WK1 | Spryd4 | SPRY domain-containing protein 4 | 2 | 12.0773 | 0.8290↓ | 10.2352 |
| 30 | G5E8V9 | Arfip1 | ADP-ribosylation factor-interacting protein 1 | 2 | 5.8981 | 0.7707↓ | 10.2352 |
| 31 | A3KG93 | Kdm1a | Lysine-specific histone demethylase 1A | 1 | 1.1455 | 0.6293↓ | 10.4007 |
| 32 | Q3TYT1 | Coq7 | 5-Demethoxyubiquinone hydroxylase | 1 | 3.2641 | 0.7900↓ | 10.4782 |
| 33 | Q8VDH1 | Fbxo21 | F box only protein 21 | 2 | 5.4226 | 0.6925↓ | 10.7308 |
| 34 | Q547J4 | Mapt | Microtubule-associated protein tau | 1 | 60.4839 | 0.7937↓ | 10.7308 |
| 35 | Q3UV17 | Krt76 | Keratin, type II cytoskeletal 2 oral | 1 | 5.3872 | 1.2468↑ | 10.7495 |
| 36 | Q99N50 | Sytl2 | Synaptotagmin-like protein 2 | 1 | 1.1579 | 0.8270↓ | 10.9079 |
| 37 | F7BGR7 | Gm21992 | Rbm14-Rbm4 readthrough (predicted) isoform 1 | 1 | 11.3208 | 0.7999↓ | 11.0185 |
| 38 | P11930 | Nudt19 | Nucleoside diphosphate-linked moiety X motif 19 | 1 | 3.9216 | 0.6532↓ | 11.5098 |
| 39 | Q5DU41 | Lrrc8b | Volume-regulated anion channel subunit LRRC8B | 2 | 2.7397 | 0.7913↓ | 11.5098 |
| 40 | B1AVZ0 | Uprt | Uracil phosphoribosyltransferase homolog | 1 | 3.2258 | 0.7773↓ | 11.5098 |
| 41 | Q8JZM4 | Dner | Delta and Notch-like epidermal growth factor-related receptor | 1 | 1.8996 | 0.7642↓ | 11.5098 |
| 42 | P56213 | Gfer | FAD-linked sulfhydryl oxidase ALR | 2 | 17.1717 | 0.8226↓ | 11.5098 |
| 43 | B2RWU5 | Abca7 | ATP-binding cassette sub-family A member 7 | 2 | 1.5690 | 0.8127↓ | 11.5098 |
| 44 | F6WZN3 | Copz2 | Coatomer subunit zeta-2 | 1 | 50.9804 | 0.8243↓ | 11.5098 |
| 45 | Q8R4Y8 | Rttn | Rotatin | 1 | 0.4492 | 0.3038↓ | 11.7727 |
Differentially expressed proteins were also mapped to KEGG pathways, which were enriched for 9 specific pathways with p < 0.05 (Table S2). The overrepresented pathways, which were also relevant to the subject of depression, included cytokine-cytokine receptor interaction (ko04060), Alzheimer disease (ko05010), neurotrophin signaling pathway (ko04722), alcoholism (ko05034), and cocaine addiction (ko05030). These findings further point to abnormalities in neuronal functions in the VLO of Dep mice. Next, we searched for known and predicted interactions for the differentially expressed proteins in the IntAct PPI database and constructed a PPI network (confidence >0.4; Figure 2D). The network predicted a central role of Ywhae gene (encodes the tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon polypeptide or the 14-3-3 epsilon protein), which was not identified among the differentially expressed proteins (Ywhae: 81.17% peptides coverage, 22 unique peptides, 0.8691-fold change). In our hands, the network predicted interactions between 14-3-3ε and 14 of identified differentially expressed proteins (Picalm, Tubb6, Stk25, Myh7, Immt, Ces1c, Mapt, Kdm1a, Zmpste24, S100a6, Slc6a3, Sytl2, Gfer, Nudt19). Some of the interactions have strong supporting experimental evidences, such as the interaction between Kdm1a and Ywhae, Picalm and Ywhae, and Myh7 and Ywhae. Moreover, some predicted interactions are implicated in nervous system diseases such as Alzheimer disease (Mapt and Ywhae), drug addiction (Slc6a3 and Ywhae), and neurobiological processes such as neurotrophin receptor activity (Dlg4 and Ntrk2) and neuronal apoptosis (Bad and Grin2b). Overall, the interactions between and pathways linking differentially expressed proteins may have important roles in CUMS-induced depression.
According to the above bioinformatics analysis, the following proteins were selected for further study: TrkB (Ntrk2), DAT (Slc6a3), BAD, Picalm, Mapt, and 14-3-3ε (Ywhae). These proteins were selected due to (1) intracellular region location; (2) high expression fold-change in differentially expressed proteins; (3) the role they play in neurobiological processes and pathways; and (4) involved or a hub node in protein-protein interaction. Figure 2E showed that the expression level of TrkB, DAT, BAD, and MAPT protein was significantly decreased in the Dep group compared with the Ctrl group. In contrast, PICALM showed increased expression in the VLO after CUMS. Next, we analyzed the expression of the seven distinct isoforms (α/β, γ, ε, ζ/δ, η, σ, τ) of the 14-3-3 protein family (Figure 2F). Interestingly, 14-3-3ε expression was significantly decreased in Dep mice, whereas other 14-3-3 isoforms were unchanged. These results were consistent with the protein expression level obtained in iTRAQ approach, validating the proteomics analysis.
Experiment 3: 14-3-3ε prevents stress-induced depressive phenotype via rescue of Bad-mediated apoptosis in VLO
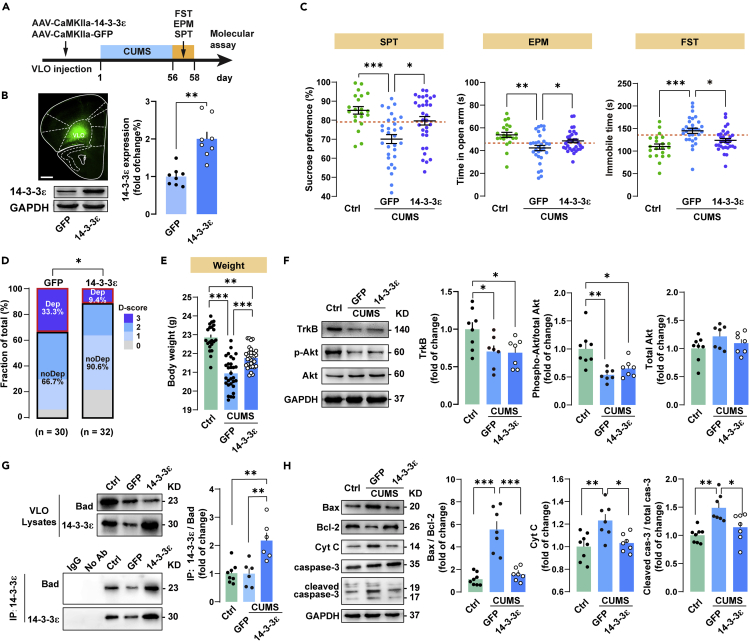
To find out whether 14-3-3ε plays a role in CUMS-induced depressive-like behaviors, we first upregulated 14-3-3ε in VLO by a bilateral stereotaxic injection of AAV that expressed 14-3-3ε (AAV9-CaMKIIa-bGlobin-14-3-3ε-eGFP-3FLAG) (Figure 3A). Mice were allowed to recover for 2 weeks to allow for adequate gene expression before CUMS. As showed in Figure 3B, compared with the GFP control, the level of 14-3-3ε protein was significantly increased by injecting the 14-3-3ε overexpressing virus. Behavioral assays indicated that CUMS-exposed mice with VLO 14-3-3ε overexpression (n = 32) exhibited significant increased sucrose preference in SPT and open-arm time in EPM and significantly decreased immobility time in FST, compared with the GFP mice (n = 30) (Figure 3C). Moreover, 14-3-3ε overexpression within VLO significantly changed the constituent ratio of depressive phenotypes and effectively reduced the number of Dep mice (D-3: 9.4%) compared with the GFP mice (D-3: 33.3%) (Figure 3D). In addition, 14-3-3ε overexpressed mice have higher body weights than GFP mice (Figure 3E), whereas no significant change in locomotion was found in the 14-3-3ε overexpressing mice (Figure S1). These results suggest that elevation of 14-3-3ε in the VLO prevents chronic stress-induced depressive phenotype in mice.
Figure 3.
14-3-3ε prevents stress-induced depressive phenotype via rescue of Bad-mediated apoptosis in VLO
(A) Schematic of experiment. Mice were injected with virus that expressed 14-3-3ε (AAV9-CaMKIIa-bGlobin-14-3-3ε-eGFP-3FLAG; n = 32) or the GFP control virus (AAV9-CaMKIIa-bGlobin-eGFP-3FLAG; n = 30) into bilateral VLO. Then, these mice were allowed to recover for 2 weeks to allow for adequate gene expression before 8 weeks of CUMS exposure. At the end of CUMS regime, the behavioral tests were carried out to assess the depressive phenotype and molecular changes in VLO.
(B) Left: eGFP-labeled cells was observed in the VLO 80 days after injection of AAV-CaMKIIa-14-3-3ε-eGFP. Scale bar = 1 mm. Right: immunoblot showing the expression level of 14-3-3ε in VLO was significantly increased by the 14-3-3ε overexpressing virus (n = 8 per group; unpaired t test, t 14 = 4.871, p < 0.01).
(C) CUMS mice with 14-3-3ε overexpression in VLO exhibited increased sucrose preference in SPT (one-way ANOVA, followed by Tukey's post hoc, 14-3-3ε versus GFP, t 80 = 3.893, p < 0.05) and open arm time in EPM (one-way ANOVA, Tukey's post hoc, 14-3-3ε versus GFP, t 80 = 3.698, p < 0.05), and significantly decreased immobility time in the FST (one-way ANOVA, Tukey's post hoc, 14-3-3ε versus GFP, t 80 = 3.942, p < 0.05).
(D) 14-3-3ε overexpression within VLO significantly changed the constituent ratio of depressive phenotypes (Fisher's exact test, p < 0.05) and effectively reduced the number of Dep (D-3) mice. No-depressive mice (noDep) were defined as those with D-score 0, 1 and 2.
(E) 14-3-3ε overexpression mice have obvious body weight gains than GFP mice (one-way ANOVA, Tukey's post hoc, 14-3-3ε versus GFP, t 80 = 8.836, p < 0.0001).
(F) TrkB and phospho-AktThr308 were decreased in both GFP and 14-3-3ε overexpressing mice compared with the non-stressed Ctrl mice. Phosphorylated Akt was expressed as phosphorylated Akt to total Akt. Protein expression levels were analyzed with one-way ANOVA, followed by Tukey's post hoc test.
(G) Co-immunoprecipitation revealed that 14-3-3ε specifically bound to Bad. The GFP group has similar ratio of 14-3-3ε to Bad with that in the non-stressed Ctrl mice. The ratio of 14-3-3ε to Bad is significantly higher in the 14-3-3ε overexpression group than that in the GFP group (n = 6–8 per group; one-way ANOVA, Tukey's post hoc, 14-3-3ε versus GFP, t 17 = 5.786, p < 0.01).
(H) The expression of the apoptotic markers such as Bax, Cyt c, and cleaved caspase-3 were increased in the GFP mice and decreased by 14-3-3ε overexpression in VLO of CUMS mice. Protein levels were analyzed with one-way ANOVA, followed by Tukey's post hoc test. Data represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.0001 compared between the two indicated groups.
To understand how overexpression of 14-3-3ε in VLO leads to the preventing effect on depressive phenotype, we turned our attention to the neurotrophin signaling pathway, especially the TrkB-Akt-Bad-mediated apoptosis pathway, which was implicated by the iTRAQ-integrated pathway analysis. As showed in Figure 3F, the level of TrkB and phosphorylated AktThr308 (expressed as phosphorylated Akt to total Akt) was decreased in CUMS-exposed GFP mice compared with the non-stressed Ctrl mice. However, these proteins were also reduced to the same extent in the CUMS-exposed 14-3-3ε overexpressing mice, indicating that the 14-3-3ε-induced rescue of depressive behavior was not mediated by 14-3-3ε-enhanced TrkB-Akt signaling. A fundamental cellular function of 14-3-3 proteins is suppression of apoptosis via sequestration of proapoptotic factors (Masters and Fu, 2001). Moreover, significant reductions in gray matter volume and increased neuronal apoptosis have been found in MD in the orbital cortex (Drevets, 2007). Therefore, we turned our attention to Bad, which is a proapoptotic factor and is identified as one of the differentially expressed proteins by iTRAQ analysis.
Firstly, we determined the protein levels of Bad and 14-3-3ε in the full lysates (Figure 3G). Consistent with the result of experiment 2, protein expression of Bad was decreased in CUMS-treated GFP mice as compared with Ctrl mice. Moreover, Bad levels in the 14-3-3ε mice were similar as in the GFP mice, suggesting that expression of Bad was not changed by 14-3-3ε overexpression. Next, we investigated whether Bad binds with 14-3-3ε by co-immunoprecipitation (co-IP) test (Figure 3G). In the Ctrl mice, 14-3-3ε pull-down assay confirmed the interaction between 14-3-3ε and Bad. In the GFP mice, the amount of immunoprecipitated Bad was reduced, consistent with the level of 14-3-3ε being decreased in the VLO. In contrast, 14-3-3ε overexpression significantly increased the pull-down of Bad. We further quantified the ratio of 14-3-3ε to the Bad pull-down. The GFP group has similar ratio of 14-3-3ε to Bad with that in the Ctrl group. Interestingly, the ratio of 14-3-3ε to Bad is about 2 times higher in the 14-3-3ε overexpression group. In summary, our co-immunoprecipitation experiments clearly revealed that 14-3-3ε specifically bind to Bad in the VLO lysates.
Bad is a member of the Bcl-2 family that promotes cell death by displacing Bax from binding to Bcl-2 and Bcl-xL, allowing Bax to initiate mitochondrial cell death (Bui et al., 2018). Therefore, we next examined the expression of the apoptotic markers. Increased levels of Bax, Cyt c, and cleaved caspase-3 were found in the GFP mouse VLO lysates compared with the levels in lysates from Ctrl mice (Figure 3H). In contrast, Bax, Cyt c, and cleaved caspase-3 were significantly decreased by 14-3-3ε overexpression in VLO of mice, similar to the levels observed in Ctrl mice. These results suggest that CUMS activates apoptotic signaling in VLO, and 14-3-3ε suppressed this apoptotic signaling by binding to Bad to restrain its proapoptotic activity.
Experiment 4: treatment with the fusicoccin-A (FC-A), a small-molecule 14-3-3s stabilizer, relieves depressive-like behaviors and restores VLO neuronal apoptosis in CUMS mice
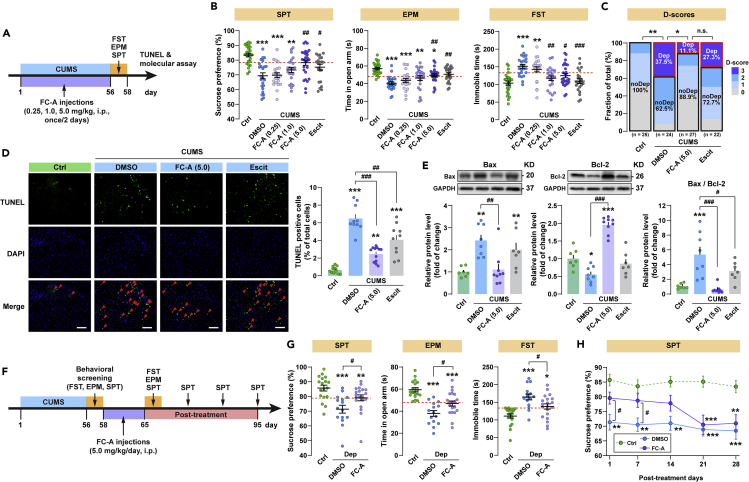
The fusicoccane family of natural small molecules is well characterized for their ability to stabilize binding between 14-3-3s and their client proteins. Intriguingly, FC-A have been shown to stabilize a variety of 14-3-3:client protein interactions by binding to a pocket within the target-binding groove located at the interface of the two proteins in which the client motif turns out from, or terminates within, the groove (Kaplan et al., 2017b; Wurtele et al., 2003). Recent work reports that 14-3-3s stabilizers have strong effects on axonal growth and neuronal survival (Kaplan et al., 2017b), suggesting that they may serve as a potential therapy for the treatment of CNS diseases. Therefore, we next investigated whether treatment with the 14-3-3s stabilizer FC-A relieves depressive-like behaviors in CUMS mice. The timeline of the experiment is showed in Figure 4A. FC-A (#20108-30-9, Sigma-Aldrich, St. Louis, MO, USA) were dissolved in dimethyl sulfoxide (DMSO) and diluted with PBS to <1% DMSO. FC-A (0.25, 1.0 and 5.0 mg/kg body weight in 10 mL/kg volume) was given intraperitoneally (i.p.) every two days during the whole CUMS period. A classic anti-depressant escitalopram (Escit, 20 mg/kg, i.p., once daily (Aguiar et al., 2019); #219861-08-2, Sigma-Aldrich, St. Louis, MO, USA) was given during the whole CUMS period as the positive control. An equal volume of 1% DMSO was given i.p. as vehicle control. As displayed in Figure 4B, CUMS mice treated with 0.25 and 1.0 mg/kg of FC-A (n = 22/group) displayed similar depressive-like behaviors as the CUMS + DMSO mice (n = 24). In contrast, 5.0 mg/kg FC-A treatment (n = 27) significantly alleviated the depressive-like behaviors in CUMS mice, which was similar to CUMS mice treated with Escit (n = 22). In addition, weight loss was significantly rescued by 5.0 mg/kg FC-A treatment (Figure S2A). No difference was found between these groups in total distance traveled (Figure S2B), indicating intact locomotion in these mice. Based on these results, we selected treatment with FC-A at 5.0 mg/kg body weight for the following experiments. We first examined the impact of FC-A on CUMS-induced depressive phenotypes in mice. As showed in Figure 4C, Fisher's exact test revealed significant effect of FC-A treatment on the constituent ratio of depressive phenotypes (D-3 vs. D-0–2). FC-A treatment effectively reduced the number of mice in D-3 (11.1%) compared with the DMSO mice (37.5%). Next, we examined the effect of 5.0 mg/kg FC-A on CUMS-induced neuronal apoptosis in VLO by TUNEL staining. As showed in Figure 4D, CUMS significantly increased TUNEL positive cells in the VLO compared with the Ctrl group. However, FC-A treatment effectively rescued the CUMS-induced neuronal apoptosis and to a greater degree than escitalopram treatment. Consistent with the increased apoptosis, increased levels of Bax protein and Bax/Bcl-2 ratio were found in the CUMS + DMSO mouse VLO lysates compared with Ctrl mice (Figure 4E). In contrast, the abnormalities in these apoptosis markers were significantly reversed by FC-A treatment, similar to the levels observed in Ctrl mice.
Figure 4.
Treatment with the fusicoccin-A (FC-A) relieves depressive-like behaviors and restores VLO neuronal apoptosis in CUMS mice
(A) Schematic of experiment for panel B–E. FC-A (0.25, 1.0 and 5.0 mg/kg body weight) was given i.p. every two days during the whole CUMS period. Escitalopram (Escit) was given once daily (20 mg/kg, i.p.) as the positive control. An equal volume of 1% DMSO was given i.p. as vehicle control. At the end of CUMS regime, the behavioral tests were carried out to assess the depressive phenotype and molecular changes in VLO.
(B) CUMS mice with 5.0 mg/kg FC-A treatment showed increased sucrose preference in SPT (post hoc, DMSO versus FC-A 5.0 mg/kg, t 136 = 3.716, p < 0.01), increased open arm time in EPM (post hoc, DMSO versus FC-A 5.0 mg/kg, t 136 = 4.982, p < 0.01), and decreased immobile time in FST (post hoc, DMSO versus FC-A 5.0 mg/kg, t 136 = 4.354, p < 0.05) compared with DMSO group. Data were analyzed with one-way ANOVA, followed by Sidak's post hoc test.
(C) 5.0 mg/kg FC-A treatment significantly changed the constituent ratio of depressive phenotypes (Fisher's exact test, DMSO versus FC-A 5.0 mg/kg, p < 0.05) and effectively reduced the number of Dep (D-3) mice as compared with DMSO group (n = 11/27 versus 2/24). No-depressive mice (noDep) were defined as those with D-score 0, 1, and 2.
(D) Left: confocal images showing the TUNEL-positive cells in the VLO of the mice in the 4 groups: non-stressed Ctrl, DMSO + CUMS, FC-A (5.0 mg/kg) + CUMS, and Escit + CUMS. n = 10–12 per group. Red arrow indicates the TUNEL-positive apoptotic cell. Scale bar: 200 μm. Right: quantification of TUNEL-positive cells demonstrated that treatment with FC-A rescued the CUMS-induced neural apoptosis in VLO (one-way ANOVA, Tukey's post hoc test, FC-A versus DMSO, t 40 = 11.11, p < 0.01).
(E) The CUMS-induced abnormal expression of Bax (left), Bcl-2 (middle), and ratio of Bax/Bcl-2 (right) were effectively rescued by systemic FC-A treatment. Protein levels were analyzed with one-way ANOVA, followed by Tukey's post hoc test.
(F) Schematic of experiment for panel G and H. After CUMS and behavioral screening, Dep mice were treated with FC-A (5.0 mg/kg, i.p., once per day) for 7 days. After the FC-A treatment, depressive-like behaviors were assessed by SPT, EPM, and FST.
(G) Dep mice with FC-A treatment showed increased sucrose preference in SPT (post hoc, FC-A versus DMSO, t 50 = 3.499, p < 0.05), increased open arm time in EPM (post hoc, FC-A versus DMSO, t 50 = 3.774, p < 0.05), and decreased immobile time in FST (post hoc, FC-A versus DMSO, t 50 = 3.670, p < 0.05). Depressive behaviors were analyzed with one-way ANOVA, followed by Tukey's post hoc test.
(H) Sucrose preference in the FC-A + CUMS group was significantly higher than the DMSO + CUMS group 7 days after the FC-A treatment (two-way RM ANOVA, followed by Sidak's post hoc test, day 7: FC-A vs. DMSO, t 250 = 2.346, p < 0.05). Data represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.0001 compared with the Ctrl group; #p < 0.05, ##p < 0.05, ###p < 0.0001 compared with the DMSO group. n.s., not significant. LO and relieves depressive-like behaviors following CUMS.
We next determined whether FC-A treatment could rescue depressive-like behaviors in CUMS-induced depressed mice. After CUMS and behavioral screening, Dep mice were treated with FC-A (5.0 mg/kg, i.p., once per day) for 7 days. After the FC-A treatment, depressive-like behaviors were assessed by SPT, EPM, and FST (Figure 4F). As showed in Figure 4G, FC-A-treated Dep mice (n = 20) exhibited significant increased sucrose preference in SPT, increased open-arm time in EPM, and decreased immobility time in FST, compared with the DMSO mice (n = 15). These results suggested that FC-A treatment ameliorates depressive phenotype and protected against VLO neuronal apoptosis in CUMS mice. We also determined whether FC-A treatment could result in a long-lasting rescue of depressive-like behavior. Compared with DMSO mice, which had consistently reduced sucrose preference for 28 days, FC-A-treated mice showed significantly elevated sucrose preference at least for 7 days after the treatment (Figure 4H). In addition, there was a strong trend (near-significant, p = 0.055) effect of FC-A on reducing the time spent immobile in the forced swim test 7 days after treatment of the Dep mice (Figure S3). These results indicated that FC-A treatment leads to an alleviation of the depressive-like behaviors, such as anhedonia and motivational impairment, in depressed mice and that the therapeutic effect could last for at least 7 days.
Discussion
In the present study, we used a model of chronic unpredicted mild stress (CUMS) in mice to investigate the pathophysiological mechanisms underlying MD. Focusing on the VLO, due to its dysfunction in MD, proteomics identified 45 proteins that were differentially regulated between chronic stress and control mice. In silico network analysis predicted a central role of YWHAE gene encoding for 14-3-3ε in mediating the CUMS-induced behavioral changes. This prediction was confirmed experimentally whereby CUMS-induced depressive behaviors were reversed by overexpression of 14-3-3ε in the VLO or its stabilization with FC-A treatment, as well as preventing the indication of apoptotic markers in this brain regions. These data strongly implicate the expression of 14-3-3ε and its regulation of Bad and the mitochondrial death signaling pathway in controlling stress-induced anhedonia and depressive-like behaviors and identify a possible candidate treatment (the 14-3-3ε stabiliser FA-C) that can reverse these molecular pathways.
Depression is a complex mood disorder with high individual heterogeneity. Despite distinct pathophysiologies underlying the clinical manifestations of depression, MD patients with symptomatic differences are often categorized into the same diagnostic group (Beijers et al., 2019; Musil et al., 2018). Like humans, rodents display great heterogeneity in their response to stress and adversity. However, insufficient attention has been paid to individual variability of behavioral phenotypes in responding to CUMS in rodents (Febbraro et al., 2017). To address this issue, we used an approach to screen out depressed animals before studying underlying neurobiological mechanisms, similar to what had been reported by Cerniauskas et al. (Cerniauskas et al., 2019). The behavioral screening of the depressed animals is based on paradigms that are widely used to measure depression-related behaviors in rodents, such as anhedonia (Liu et al., 2018), anxiety (Ducottet and Belzung, 2004, 2005), and behavioral despair (Castagne et al., 2011). Moreover, additional measures of depression (e.g., weight gain or loss, and locomotion) were also included to further support classification of subgroups. In the present study, after CUMS exposure, we found that only a subpopulation of mice displayed most salient depressive-like behaviors. We retrieved three metrics, sucrose preference in SPT (anhedonia), open arm time in EPM (anxiety), and immobility in FST (behavioral despair), directly from three well-established paradigms for depressive-like behaviors of mice with CUMS exposure. We used ROC curves and Youden's index based on these metrics to define cutoff criteria for these mice. We found that 35% (n = 21) of CUMS mice met all three behavioral criteria, and 65% (n = 39) of mice showed only one, two, or no depression phenotype. Thus, we defined the mice that met all three positive criteria (i.e. D-score = 3) as the depressed mice, which were selected for subsequent analyses. Moreover, the control group only included the mice with no positive depressive behaviors (i.e. D-score = 0). This approach helped reduce the heterogeneity of mice and enhance the specificity of identified differentially expressed proteins in VLO.
Using an iTRAQ-based quantitative proteomics approach, we identified 45 proteins from the VLO of depressed mice that were differentially expressed relative to controls. Among these proteins, 6 were upregulated and 39 were downregulated under chronic stress conditions. We performed a complete bioinformatical analysis including clustering and integrated pathway analysis. As a result, 9 specific biological pathways including ubiquinone biosynthesis, cytokine-cytokine receptor interaction, neurotrophin signaling pathway, and MAPK signaling pathway were identified as significantly altered in depressed mice. This result shed light on a number of altered biological/biochemical functions in VLO in anhedonia and responses to chronic stress. Based on these changes, the PPI network analysis predicted a central role of chaperone protein 14-3-3ε (YWHAE gene product), which was significantly compromised in VLO of depressed mice.
The 14-3-3 protein was initially described as an acidic, abundant brain protein by Moore & Perez in 1967 (Moore BW, 1967). The name is derived from the combination of its characteristic elution on chromatography and its migration pattern in gel electrophoresis. 14-3-3 is now recognized as a family of relatively small (25–30 kDa), acidic, ubiquitous, highly conserved homologous proteins encoded by separate genes. To date, seven different 14-3-3 isoforms have been isolated in eukaryotic cells (α/β, γ, ε, ζ/δ, η, σ, and τ; α and δ are actually the phosphorylated forms of β and ζ) (Aitken et al., 1995). These proteins are highly abundant in CNS and comprise about 1% of total brain soluble protein (Dougherty and Morrison, 2004). The 14-3-3ε is widely expressed in the central nervous system and is especially abundant in frontal lobe and cerebellum (Yue et al., 2014). 14-3-3ε is found to control multiple developmental processes and regulate neuronal morphogenesis and differentiation (Gittenberger-de Groot et al., 2016; McConnell et al., 1995). Recent studied found that complete ablation of the YWHAE gene resulted in multiple defects in neuropsychiatric behaviors in mice (Wachi et al., 2017). However, in the depressed mice, only 14-3-3ε isoform levels were significantly downregulated in VLO, as were a number of its trophic signaling partners. In contrast, overexpression of 14-3-3ε in VLO of CUMS-exposed mice reversed their weight loss, increased their preference of sucrose, and promoted swimming in the forced swim test. Importantly, treatment with the 14-3-3ε stabilizer FC-A (Figure 5A) led to an alleviation of the depression phenotype in depressed mice, and the therapeutic effect lasted for at least 7 days. Together, these findings indicate that 14-3-3ε plays a critical role in VLO neuronal function in response to chronic stress, which results in depressive-like behaviors in mice.
Figure 5.
Proposed signaling pathway for 14-3-3ε as a regulator in chronic stress-induced depression
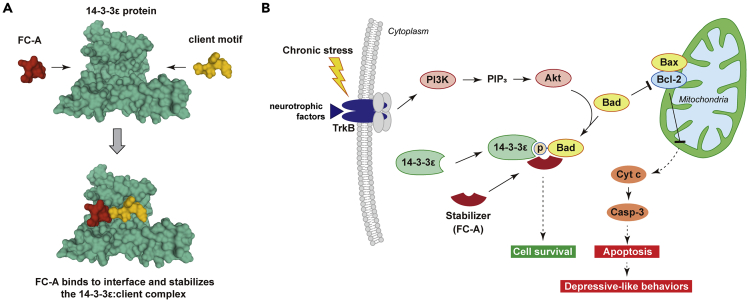
(A) FC-A (red) binds to a solvent-exposed hydrophobic pocket created by the docking of a client motif (yellow); for example the phosphorylated S112 and S136 sites of Bad, within the 14-3-3 binding groove (green), simultaneously stabilizes both proteins (generated with PDB: 1O9F).
(B) The hypothesized molecular mechanism underlying the antidepressant-like effect of FC-A in the VLO of Dep mice. Chronic stress induces downregulation of TrkB/PI3K/Akt signaling pathway and alters the expression of 14-3-3ε, leading to the activation of Bad-mediated mitochondrial apoptotic signaling. Treatment with FC-A promotes the stabilization of 14-3-3ε:Bad complex and therefore prevents neuronal apoptotic signaling in the in the VLO and relieves depressive-like behaviors following CUMS.
Regulation of 14-3-3β and 14-3-3ζ isoforms has been reported in cells and animal models after chronic antidepressant treatment (Carboni et al., 2006; Cecconi et al., 2007; Malki et al., 2012), and a YWHAE gene polymorphism was reported to be associated with schizophrenia (Ikeda et al., 2008); however, in another study the association was not replicated (Liu et al., 2011). As 14-3-3 proteins can bind to an extensive number of partners, small changes in 14-3-3s abundance could therefore have major impacts on cell signaling. 14-3-3 proteins bind to serine- and threonine-phosphorylated client proteins via a highly conserved binding groove. Binding of 14-3-3 can influence client protein activity, localization, stability, or PPIs (Bridges and Moorhead, 2005) and has been implicated in a variety of biological processes that could account for the multiple cellular or signaling alterations. 14-3-3 functions include exocytosis (Morgan and Burgoyne, 1992), regulation of signal transduction (Abramow-Newerly et al., 2006), neurodevelopmental regulation (Skoulakis and Davis, 1998), programmed cell death (Zha et al., 1996), and transcriptional control of gene expression in response to environmental changes (Yaffe, 2002). 14-3-3ε has been reported to be a critical regulators of axon guidance, neurite formation, and cell signaling transduction (Cornell et al., 2016; Kaplan et al., 2017a; Wachi et al., 2017). In addition to 14-3-3ε downregulation, TrkB expression and Akt phosphorylation were significantly decreased in VLO of depressed mice, in line with previous reports that chronic stress can induce decrease in BDNF-PI3K/Akt signaling and promote neuronal apoptosis (Liu et al., 2017; Wu et al., 2018; Xia et al., 2016). Indeed, it has been suggested that reduced BDNF expression and downstream signaling in the VLO is the mechanism underlying CUMS-induced depressive-like behaviors (Zhao et al., 2017). However, TrkB and phospho-Akt protein levels were unchanged by the rAAV-mediated 14-3-3ε overexpression, suggesting that 14-3-3ε-induced behavioral effect on depression was not caused by restoring TrkB-Akt signaling.
Although numerous imaging studies document significant reductions of frontal cortex, amygdala, and hippocampus volume in MD patients (Belleau et al., 2019; Campbell and Macqueen, 2004; Kim et al., 2016; Treadway et al., 2015), the exact underlying cellular mechanisms are unclear. Clinical and experimental studies indicate that multiple factors, such as dendritic retraction, suppressed neurogenesis, putative glial changes, and neuronal death, may contribute to the structural changes in the volume of the frontal cortex and hippocampus (Czeh and Lucassen, 2007; Drzyzga et al., 2009; Lucassen et al., 2001). Although postmortem studies show little cellular death in the cortical area of MD patients, emerging animal studies suggest that elevated apoptotic signaling in MD may associate with pathogenesis of depression (Fan et al., 2018; Salehpour et al., 2019; Xiao et al., 2020). In the present study, the Bcl-2 antagonist of death (Bad), a downstream proapoptotic factor, was also identified as one of the differentially expressed proteins by iTRAQ analysis. Bad, similar to other Bcl-2 homologs, is capable of dimerizing with various family members, and its levels are dynamically regulated by apoptotic stimuli (Bui et al., 2018). Bad is thought to induce apoptosis by binding and inactivating Bcl-2/Bcl-xl on the mitochondria to displace Bax and promotes cell death (Yang et al., 1995; Zhang et al., 2017). Recent studies have reported that Akt phosphorylates Bad at the S112 or S136 sites and directly facilities binding of Bad to 14-3-3ζ (Datta et al., 2000; Pozuelo-Rubio, 2011). Our co-immunoprecipitation experiments revealed a direct interaction between Bad and 14-3-3ε, which was reduced following CUMS correlating with the reduction in Akt phosphorylation. Although increased expression of 14-3-3ε did not increase trophic signaling, it did restore the proportion of Bad bound and presumably inhibited by 14-3-3ε. Furthermore, increased 14-3-3ε levels reversed the high levels of the mitochondrial apoptotic signaling molecules Bax, Cyt c, and cleaved caspase-3 in the VLO of depressed mice. Based on these results, we suggest that one mechanism by which CUMS induces depressive-like behaviors is by upregulation neuronal apoptotic signaling in the VLO, which concurs with the reports that MD patients had reduced orbital cortex volume and thickness (Drevets, 2007; Taylor et al., 2007).
We propose that in the default state, the neurotrophic factor BDNF binds to TrkB and activates the PI3K/Akt signaling pathway (Figure 5B). In turn, Akt phosphorylates Bad, which promotes its binding with 14-3-3ε, inhibiting Bad's proapoptotic activity. Chronic stress not only induces downregulation of TrkB/PI3K/Akt signaling pathway but also alters the expression of 14-3-3ε, which leads to the increased Bad unbound by 14-3-3ε. Released Bad binds to Bcl-2/Bcl-xl, provoking dissociation of Bax-Bcl-2 allowing Bax to penetrate the mitochondrial outer membrane and triggering cytochrome c release from the intermembrane space to the cytosol, where it activates the caspase cascade and leads to neuronal degeneration. Increased 14-3-3ε levels, induced by overexpression, shifts the balance of Bad/Bcl-2/Bax proteins toward survival signaling and therefore rescues VLO neurons from Bad-mediated apoptosis.
Given that 14-3-3ε exerts a vital role in suppression of stress-induced apoptosis in the VLO via sequestration of proapoptotic factors, its upregulation is one promising strategy to promote neuronal survival in the context of chronic stress. The small molecule FC-A is a chemical tool used to stabilize 14-3-3ε:client complexes (Figure 5A). We discovered that daily treatment with FC-A at 5 mg/kg body weight is sufficient to relieve depressive-like behaviors and CUMS-induced neuronal apoptosis for at least 7 days in depressed mice. Previous evidence of trophic effects of FC-A is scarce, but a recent study reported that that FC-A stimulates neurite outgrowth in vitro, reduces corticospinal axon dieback after spinal cord injury, and stimulates optic nerve regeneration (Kaplan et al., 2017a). Our results demonstrate that manipulation of 14-3-3ε binding to Bad, among other partners, with FC-A can prevent neuronal apoptotic signaling in the VLO and relieve depressive-like behaviors following CUMS without noticeable side effects. In addition, the level of Bcl2 in FC-A group is increased compared with the levels in controls. We also observed changes in Bax and Bcl2 levels either following CUMS regime or FC-A treatment. We have experimentally confirmed that FC-A promotes neuronal survival against CUMS-induced apoptosis by stabilizing the 14-3-3ε:Bad complexes. Therefore, systemic treatment with FC-A as an effective approach for prophylactic treatment of MD in the context of chronic unpredictable stress is worth exploring further.
In conclusion, quantitative proteomics profiling to investigate changes in the expression of VLO proteins after CUMS treatment of mice identified 14-3-3ε and TrkB signaling as differentially downregulated in association with stress-induced depressive-like behaviors. We demonstrated that the promotion of 14-3-3ε expression in the VLO prevented stress-induced depression phenotypes via rescue of Bad-mediated apoptosis. Treatment with the 14-3-3ε stabilizer FC-A precluded neuronal apoptotic signaling in the VLO of depressed mice and relieved the depressive-like behaviors. Because 14-3-3ε provides significant protection after exposure to chronic unpredictable stress, boosting 14-3-3ε expression, pharmacological stabilization of 14-3-3s (e.g. with FC-A) or its protein partners is identified as an exciting potential therapeutic target for the treatment of MD.
Limitations of the study
Our iTRAQ results are based on the triple biological replicates, which is a relative low number experiment with plenty of biological follow-up. Thus, we have to use a relative loose criterion to screen the differentially expressed proteins, and we might have missed some important proteins during the quantitative proteomic analysis, as iTRAQ is a fold-change sensitive technique, and a 10% decrease in a key scaffold protein could well be significant. In this case, p value correction such as FDR is mandatory to control the error rate in multiple tests. Moreover, the proteomic data shed light on many other pathways implicated in chronic-stress-induced depression, such as mitochondrial apoptotic pathway, MAPK cascade, complement and coagulation cascades, and several proteins involved in Alzheimer disease. These points were beyond the scope of our study but will be an essential part for understanding the functional features of the stress-induced depression and are definitely worth future study.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yunpeng Wang (wyp033@xjtu.edu.cn).
Materials availability
All resources and reagents are available on request without restriction.
Data and code availability
This study did not generate datasets or analyze codes.
Methods
All methods can be found in the accompanying Transparent methods supplemental file.
Acknowledgments
The authors would like to thank Dr. Zijun Xiong for his help and valuable advices on bioinformation analysis. This study was supported by the National Natural Science Foundation of China (81501175 and 81501636), the Science and Technology Innovation Base-Open and Sharing Platform of Science and Technology Resources Project of Shaanxi Province (2019PT-26), and the Foundation of Xi'an Medical University (2018XNRC08, 2018XNRC06, 2018XNRC07, 05041901, 2017GJFY31).
Author contributions
Conceptualization, Y.W. and Y.Z.; Data Acquisition and Investigation, S.W., X.S., J.Z., B.S., H.X., Y.D., Y.C., and J.C.; Writing and Supervision, Y.W.; Reviewing & Editing, E.C.
Declaration of interests
The authors declare no competing interests.
Published: February 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102043.
Contributor Information
Yunpeng Wang, Email: wyp033@xjtu.edu.cn.
Shuang Wang, Email: wangshuang78213@163.com.
Supplemental information
References
- Abramow-Newerly M., Ming H., Chidiac P. Modulation of subfamily B/R4 RGS protein function by 14-3-3 proteins. Cell Signal. 2006;18:2209–2222. doi: 10.1016/j.cellsig.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Aguiar R.P., Soares L.M., Meyer E., da Silveira F.C., Milani H., Newman-Tancredi A., Varney M., Prickaerts J., Oliveira R.M.W. Activation of 5-HT1A postsynaptic receptors by NLX-101 results in functional recovery and an increase in neuroplasticity in mice with brain ischemia. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2019;99:109832. doi: 10.1016/j.pnpbp.2019.109832. [DOI] [PubMed] [Google Scholar]
- Aitken A., Howell S., Jones D., Madrazo J., Patel Y. 14-3-3 alpha and delta are the phosphorylated forms of raf-activating 14-3-3 beta and zeta. In vivo stoichiometric phosphorylation in brain at a Ser-Pro-Glu-Lys MOTIF. J. Biol. Chem. 1995;270:5706–5709. doi: 10.1074/jbc.270.11.5706. [DOI] [PubMed] [Google Scholar]
- Beijers L., Wardenaar K.J., van Loo H.M., Schoevers R.A. Data-driven biological subtypes of depression: systematic review of biological approaches to depression subtyping. Mol. Psychiatry. 2019;24:888–900. doi: 10.1038/s41380-019-0385-5. [DOI] [PubMed] [Google Scholar]
- Belleau E.L., Treadway M.T., Pizzagalli D.A. The impact of stress and major depressive disorder on hippocampal and medial prefrontal cortex morphology. Biol. Psychiatry. 2019;85:443–453. doi: 10.1016/j.biopsych.2018.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrar D., Flach P. Caveats and pitfalls of ROC analysis in clinical microarray research (and how to avoid them) Brief Bioinformatics. 2012;13:83–97. doi: 10.1093/bib/bbr008. [DOI] [PubMed] [Google Scholar]
- Bridges D., Moorhead G.B. 14-3-3 proteins: a number of functions for a numbered protein. Sci. STKE. 2005;2005:re10. doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- Bui N.L., Pandey V., Zhu T., Ma L., Basappa, Lobie P.E. Bad phosphorylation as a target of inhibition in oncology. Cancer Lett. 2018;415:177–186. doi: 10.1016/j.canlet.2017.11.017. [DOI] [PubMed] [Google Scholar]
- Campbell S., Macqueen G. The role of the hippocampus in the pathophysiology of major depression. J. Psychiatry Neurosci. 2004;29:417–426. [PMC free article] [PubMed] [Google Scholar]
- Carboni L., Vighini M., Piubelli C., Castelletti L., Milli A., Domenici E. Proteomic analysis of rat hippocampus and frontal cortex after chronic treatment with fluoxetine or putative novel antidepressants: CRF1 and NK1 receptor antagonists. Eur. Neuropsychopharmacol. 2006;16:521–537. doi: 10.1016/j.euroneuro.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Castagne V., Moser P., Roux S., Porsolt R.D. Rodent models of depression: forced swim and tail suspension behavioral despair tests in rats and mice. Curr. Protoc. Neurosci. 2011;8:Unit 8 10A. doi: 10.1002/0471142301.ns0810as55. [DOI] [PubMed] [Google Scholar]
- Cecconi D., Mion S., Astner H., Domenici E., Righetti P.G., Carboni L. Proteomic analysis of rat cortical neurons after fluoxetine treatment. Brain Res. 2007;1135:41–51. doi: 10.1016/j.brainres.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Cerniauskas I., Winterer J., de Jong J.W., Lukacsovich D., Yang H., Khan F., Peck J.R., Obayashi S.K., Lilascharoen V., Lim B.K. Chronic stress induces activity, synaptic, and transcriptional remodeling of the lateral habenula associated with deficits in motivated behaviors. Neuron. 2019;104:899–915 e898. doi: 10.1016/j.neuron.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell B., Wachi T., Zhukarev V., Toyo-Oka K. Regulation of neuronal morphogenesis by 14-3-3epsilon (Ywhae) via the microtubule binding protein, doublecortin. Hum. Mol. Genet. 2016;25:4405–4418. doi: 10.1093/hmg/ddw270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeh B., Lucassen P.J. What causes the hippocampal volume decrease in depression? Are neurogenesis, glial changes and apoptosis implicated? Eur. Arch. Psychiatry Clin. Neurosci. 2007;257:250–260. doi: 10.1007/s00406-007-0728-0. [DOI] [PubMed] [Google Scholar]
- Datta S.R., Katsov A., Hu L., Petros A., Fesik S.W., Yaffe M.B., Greenberg M.E. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol. Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- Dougherty M.K., Morrison D.K. Unlocking the code of 14-3-3. J. Cell Sci. 2004;117:1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- Drevets W.C. Orbitofrontal cortex function and structure in depression. Ann. N. Y Acad. Sci. 2007;1121:499–527. doi: 10.1196/annals.1401.029. [DOI] [PubMed] [Google Scholar]
- Drevets W.C., Bogers W., Raichle M.E. Functional anatomical correlates of antidepressant drug treatment assessed using PET measures of regional glucose metabolism. Eur. Neuropsychopharmacol. 2002;12:527–544. doi: 10.1016/s0924-977x(02)00102-5. [DOI] [PubMed] [Google Scholar]
- Drzyzga L.R., Marcinowska A., Obuchowicz E. Antiapoptotic and neurotrophic effects of antidepressants: a review of clinical and experimental studies. Brain Res. Bull. 2009;79:248–257. doi: 10.1016/j.brainresbull.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Ducottet C., Belzung C. Behaviour in the elevated plus-maze predicts coping after subchronic mild stress in mice. Physiol. Behav. 2004;81:417–426. doi: 10.1016/j.physbeh.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Ducottet C., Belzung C. Correlations between behaviours in the elevated plus-maze and sensitivity to unpredictable subchronic mild stress: evidence from inbred strains of mice. Behav. Brain Res. 2005;156:153–162. doi: 10.1016/j.bbr.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Duman R.S. A silver bullet for the treatment of depression? Neuron. 2007;55:679–681. doi: 10.1016/j.neuron.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Elliott R., Agnew Z., Deakin J.F. Hedonic and informational functions of the human orbitofrontal cortex. Cereb. Cortex. 2010;20:198–204. doi: 10.1093/cercor/bhp092. [DOI] [PubMed] [Google Scholar]
- Evans C., Noirel J., Ow S.Y., Salim M., Pereira-Medrano A.G., Couto N., Pandhal J., Smith D., Pham T.K., Karunakaran E. An insight into iTRAQ: where do we stand now? Anal Bioanal. Chem. 2012;404:1011–1027. doi: 10.1007/s00216-012-5918-6. [DOI] [PubMed] [Google Scholar]
- Fan C., Song Q., Wang P., Li Y., Yang M., Yu S.Y. Neuroprotective effects of ginsenoside-rg1 against depression-like behaviors via suppressing glial activation, synaptic deficits, and neuronal apoptosis in rats. Front. Immunol. 2018;9:2889. doi: 10.3389/fimmu.2018.02889. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Febbraro F., Svenningsen K., Tran T.P., Wiborg O. Neuronal substrates underlying stress resilience and susceptibility in rats. PLoS One. 2017;12:e0179434. doi: 10.1371/journal.pone.0179434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittenberger-de Groot A.C., Hoppenbrouwers T., Miquerol L., Kosaka Y., Poelmann R.E., Wisse L.J., Yost H.J., Jongbloed M.R., Deruiter M.C., Brunelli L. 14-3-3epsilon controls multiple developmental processes in the mouse heart. Dev. Dyn. 2016;245:1107–1123. doi: 10.1002/dvdy.24440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Hikita T., Taya S., Uraguchi-Asaki J., Toyo-oka K., Wynshaw-Boris A., Ujike H., Inada T., Takao K., Miyakawa T. Identification of YWHAE, a gene encoding 14-3-3epsilon, as a possible susceptibility gene for schizophrenia. Hum. Mol. Genet. 2008;17:3212–3222. doi: 10.1093/hmg/ddn217. [DOI] [PubMed] [Google Scholar]
- Jiang H., Wei L., Wang D., Wang J., Zhu S., She R., Liu T., Tian J., Quan R., Hou L. ITRAQ-based quantitative proteomics reveals the first proteome profiles of piglets infected with porcine circovirus type 3. J. Proteomics. 2020;212:103598. doi: 10.1016/j.jprot.2019.103598. [DOI] [PubMed] [Google Scholar]
- Johnston-Wilson N.L., Sims C.D., Hofmann J.P., Anderson L., Shore A.D., Torrey E.F., Yolken R.H. Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. The Stanley Neuropathology Consortium. Mol. Psychiatry. 2000;5:142–149. doi: 10.1038/sj.mp.4000696. [DOI] [PubMed] [Google Scholar]
- Kaplan A., Morquette B., Kroner A., Leong S., Madwar C., Sanz R., Banerjee S.L., Antel J., Bisson N., David S. Small-molecule stabilization of 14-3-3 protein-protein interactions stimulates axon regeneration. Neuron. 2017;93:1082–1093 e1085. doi: 10.1016/j.neuron.2017.02.018. [DOI] [PubMed] [Google Scholar]
- Kaplan A., Ottmann C., Fournier A.E. 14-3-3 adaptor protein-protein interactions as therapeutic targets for CNS diseases. Pharmacol. Res. 2017;125:114–121. doi: 10.1016/j.phrs.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Kessler R.C., Chiu W.T., Demler O., Merikangas K.R., Walters E.E. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the national comorbidity survey replication. Arch. Gen. Psychiatry. 2005;62:617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.K., Nunes P.V., Oliveira K.C., Young L.T., Lafer B. Neuropathological relationship between major depression and dementia: a hypothetical model and review. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2016;67:51–57. doi: 10.1016/j.pnpbp.2016.01.008. [DOI] [PubMed] [Google Scholar]
- Liu J., Zhou G., Ji W., Li J., Li T., Wang T., Li Y., Zeng Z., Hu Z., Zheng L. No association of the YWHAE gene with schizophrenia, major depressive disorder or bipolar disorder in the Han Chinese population. Behav. Genet. 2011;41:557–564. doi: 10.1007/s10519-010-9426-1. [DOI] [PubMed] [Google Scholar]
- Liu M.Y., Yin C.Y., Zhu L.J., Zhu X.H., Xu C., Luo C.X., Chen H., Zhu D.Y., Zhou Q.G. Sucrose preference test for measurement of stress-induced anhedonia in mice. Nat. Protoc. 2018;13:1686–1698. doi: 10.1038/s41596-018-0011-z. [DOI] [PubMed] [Google Scholar]
- Liu T., Hu J., Li H. iTRAQ-based shotgun neuroproteomics. Methods Mol. Biol. 2009;566:201–216. doi: 10.1007/978-1-59745-562-6_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu Y., Jin H., Cong P., Zhang Y., Tong C., Shi X., Liu X., Tong Z., Shi L. Cold stress-induced brain injury regulates TRPV1 channels and the PI3K/AKT signaling pathway. Brain Res. 2017;1670:201–207. doi: 10.1016/j.brainres.2017.06.025. [DOI] [PubMed] [Google Scholar]
- Lucassen P.J., Muller M.B., Holsboer F., Bauer J., Holtrop A., Wouda J., Hoogendijk W.J., De Kloet E.R., Swaab D.F. Hippocampal apoptosis in major depression is a minor event and absent from subareas at risk for glucocorticoid overexposure. Am. J. Pathol. 2001;158:453–468. doi: 10.1016/S0002-9440(10)63988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki K., Campbell J., Davies M., Keers R., Uher R., Ward M., Paya-Cano J., Aitchinson K.J., Binder E., Sluyter F. Pharmacoproteomic investigation into antidepressant response in two mouse inbred strains. Proteomics. 2012;12:2355–2365. doi: 10.1002/pmic.201100306. [DOI] [PubMed] [Google Scholar]
- Masters S.C., Fu H. 14-3-3 proteins mediate an essential anti-apoptotic signal. J. Biol. Chem. 2001;276:45193–45200. doi: 10.1074/jbc.M105971200. [DOI] [PubMed] [Google Scholar]
- McConnell J.E., Armstrong J.F., Hodges P.E., Bard J.B. The mouse 14-3-3 epsilon isoform, a kinase regulator whose expression pattern is modulated in mesenchyme and neuronal differentiation. Dev. Biol. 1995;169:218–228. doi: 10.1006/dbio.1995.1139. [DOI] [PubMed] [Google Scholar]
- Moore BW P.V. Specific acidic proteins of the nervous system. In: FD C., editor. In Physiological and Biochemical Aspects of Nervous Integration. Prentice-Hall; 1967. pp. 343–359. [Google Scholar]
- Morgan A., Burgoyne R.D. Interaction between protein kinase C and Exo1 (14-3-3 protein) and its relevance to exocytosis in permeabilized adrenal chromaffin cells. Biochem. J. 1992;286(Pt 3):807–811. doi: 10.1042/bj2860807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musil R., Seemuller F., Meyer S., Spellmann I., Adli M., Bauer M., Kronmuller K.T., Brieger P., Laux G., Bender W. Subtypes of depression and their overlap in a naturalistic inpatient sample of major depressive disorder. Int. J. Methods Psychiatr. Res. 2018;27:e1569. doi: 10.1002/mpr.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent A.C., Milham M.P., Bain E.E., Mah L., Cannon D.M., Marrett S., Zarate C.A., Pine D.S., Price J.L., Drevets W.C. Cortical abnormalities in bipolar disorder investigated with MRI and voxel-based morphometry. Neuroimage. 2006;30:485–497. doi: 10.1016/j.neuroimage.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Ongur D., Drevets W.C., Price J.L. Glial reduction in the subgenual prefrontal cortex in mood disorders. Proc. Natl. Acad. Sci. U S A. 1998;95:13290–13295. doi: 10.1073/pnas.95.22.13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro M.B., Ferreira M.L., Refshauge K., Ordonana J.R., Machado G.C., Prado L.R., Maher C.G., Ferreira P.H. Symptoms of depression and risk of new episodes of low back pain: a systematic review and meta-analysis. Arthritis Care Res. (Hoboken) 2015;67:1591–1603. doi: 10.1002/acr.22619. [DOI] [PubMed] [Google Scholar]
- Pozuelo-Rubio M. Regulation of autophagic activity by 14-3-3zeta proteins associated with class III phosphatidylinositol-3-kinase. Cell Death Differ. 2011;18:479–492. doi: 10.1038/cdd.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring H.A., Bench C.J., Trimble M.R., Brooks D.J., Frackowiak R.S., Dolan R.J. Depression in Parkinson's disease. A positron emission study. Br. J. Psychiatry. 1994;165:333–339. doi: 10.1192/bjp.165.3.333. [DOI] [PubMed] [Google Scholar]
- Rolls E.T. The functions of the orbitofrontal cortex. Brain Cogn. 2004;55:11–29. doi: 10.1016/S0278-2626(03)00277-X. [DOI] [PubMed] [Google Scholar]
- Saarni S.I., Suvisaari J., Sintonen H., Pirkola S., Koskinen S., Aromaa A., Lonnqvist J. Impact of psychiatric disorders on health-related quality of life: general population survey. Br. J. Psychiatry. 2007;190:326–332. doi: 10.1192/bjp.bp.106.025106. [DOI] [PubMed] [Google Scholar]
- Salehpour F., Farajdokht F., Cassano P., Sadigh-Eteghad S., Erfani M., Hamblin M.R., Salimi M.M., Karimi P., Rasta S.H., Mahmoudi J. Near-infrared photobiomodulation combined with coenzyme Q10 for depression in a mouse model of restraint stress: reduction in oxidative stress, neuroinflammation, and apoptosis. Brain Res. Bull. 2019;144:213–222. doi: 10.1016/j.brainresbull.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Zhang K., Feng C., Chen Y., Li S., Iqbal J., Liao L., Zhao Y., Zhai J. iTRAQ-based proteomic analysis reveals protein profile in plasma from children with autism. Proteomics Clin. Appl. 2018;12:e1700085. doi: 10.1002/prca.201700085. [DOI] [PubMed] [Google Scholar]
- Skoulakis E.M., Davis R.L. 14-3-3 proteins in neuronal development and function. Mol. Neurobiol. 1998;16:269–284. doi: 10.1007/BF02741386. [DOI] [PubMed] [Google Scholar]
- Taylor W.D., Macfall J.R., Payne M.E., McQuoid D.R., Steffens D.C., Provenzale J.M., Krishnan K.R. Orbitofrontal cortex volume in late life depression: influence of hyperintense lesions and genetic polymorphisms. Psychol. Med. 2007;37:1763–1773. doi: 10.1017/S0033291707000128. [DOI] [PubMed] [Google Scholar]
- Treadway M.T., Waskom M.L., Dillon D.G., Holmes A.J., Park M.T.M., Chakravarty M.M., Dutra S.J., Polli F.E., Iosifescu D.V., Fava M. Illness progression, recent stress, and morphometry of hippocampal subfields and medial prefrontal cortex in major depression. Biol. Psychiatry. 2015;77:285–294. doi: 10.1016/j.biopsych.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez E., Martins-de-Souza D., Velasquez I., Carneiro G.R.A., Schmitt A., Falkai P., Domont G.B., Nogueira F.C.S. Quantitative subcellular proteomics of the orbitofrontal cortex of schizophrenia patients. J. Proteome Res. 2019;18:4240–4253. doi: 10.1021/acs.jproteome.9b00398. [DOI] [PubMed] [Google Scholar]
- Wachi T., Cornell B., Toyo-Oka K. Complete ablation of the 14-3-3epsilon protein results in multiple defects in neuropsychiatric behaviors. Behav. Brain Res. 2017;319:31–36. doi: 10.1016/j.bbr.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner P. Chronic mild stress (CMS) revisited: consistency and behavioural-neurobiological concordance in the effects of CMS. Neuropsychobiology. 2005;52:90–110. doi: 10.1159/000087097. [DOI] [PubMed] [Google Scholar]
- Wu Z., Wang G., Wei Y., Xiao L., Wang H. PI3K/AKT/GSK3beta/CRMP-2-mediated neuroplasticity in depression induced by stress. Neuroreport. 2018;29:1256–1263. doi: 10.1097/WNR.0000000000001096. [DOI] [PubMed] [Google Scholar]
- Wurtele M., Jelich-Ottmann C., Wittinghofer A., Oecking C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 2003;22:987–994. doi: 10.1093/emboj/cdg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B., Chen C., Zhang H., Xue W., Tang J., Tao W., Wu R., Ren L., Wang W., Chen G. Chronic stress prior to pregnancy potentiated long-lasting postpartum depressive-like behavior, regulated by Akt-mTOR signaling in the hippocampus. Sci. Rep. 2016;6:35042. doi: 10.1038/srep35042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao P., Zhang X., Li Y., Ma Z., Si S., Gao X. miR-9 inhibition of neuronal apoptosis and expression levels of apoptosis genes Bcl-2 and Bax in depression model rats through Notch pathway. Exp. Ther. Med. 2020;19:551–556. doi: 10.3892/etm.2019.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing B., Zhao Y., Zhang H., Dang Y., Chen T., Huang J., Luo Q. Microinjection of valproic acid into the ventrolateral orbital cortex exerts an antidepressant-like effect in the rat forced swim test. Brain Res. Bull. 2011;85:153–157. doi: 10.1016/j.brainresbull.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B. How do 14-3-3 proteins work?-- Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- Yang E., Zha J., Jockel J., Boise L.H., Thompson C.B., Korsmeyer S.J. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- Youden W.J. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha J., Harada H., Yang E., Jockel J., Korsmeyer S.J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang H., Chen S., Xu Y., Yao A., Liao Q., Han L., Zou Z., Zhang X. Dihydromyricetin induces mitochondria-mediated apoptosis in HepG2 cells through down-regulation of the Akt/Bad pathway. Nutr. Res. 2017;38:27–33. doi: 10.1016/j.nutres.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wang S., Chu Z., Dang Y., Zhu J., Su X. MicroRNA-101 in the ventrolateral orbital cortex (VLO) modulates depressive-like behaviors in rats and targets dual-specificity phosphatase 1 (DUSP1) Brain Res. 2017;1669:55–62. doi: 10.1016/j.brainres.2017.05.020. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Xing B., Dang Y.H., Qu C.L., Zhu F., Yan C.X. Microinjection of valproic acid into the ventrolateral orbital cortex enhances stress-related memory formation. PLoS One. 2013;8:e52698. doi: 10.1371/journal.pone.0052698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Li X., Chen H., Wang H., Zhu X., Hou H., Hu Q. iTRAQ proteomic analysis of the hippocampus in a rat model of nicotine-induced conditioned place preference. Biochem. Biophys. Res. Commun. 2017;486:971–977. doi: 10.1016/j.bbrc.2017.03.141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets or analyze codes.