Abstract
The advent of digital, electronic, and molecular technologies has allowed the study of complete genomes. Integrating this information into drug development has opened the door for pharmacogenomic (PGx) interventions in direct patient care. PGx allows clinicians to better identify drug of choice and optimize dosing regimens based on an individual’s genetic characteristics. Integrating PGx into pediatric care is a priority for the Sanford Children’s Genomic Medicine Consortium, a partnership of ten children’s hospitals across the US committed to the innovation and advancement of genomics in pediatric care. In this white paper, we review the current state of PGx research and its clinical utility in pediatrics, a largely understudied population, and make recommendations for advancing cutting-edge practice in pediatrics.
Subject terms: Drug screening, Drug development
Introduction
As early as 1959, Friedrich Vogel, a German human geneticist, proposed that our genes play an important role in determining drug response and coined the term pharmacogenetics [1]. With current advances in technology, pharmacogenomics (PGx) is often touted as among the most actionable areas of clinical innovation within the broadly defined precision medicine initiative [2]. Efforts to optimize the efficacy of drug therapy and minimize adverse drug reactions are consistently at the forefront of healthcare quality improvement programs. Adverse drug reactions remain a major cause of morbidity and death in the USA, especially in the pediatric population [3]. A single institution study reported that 3% of unplanned hospital admissions were attributed to adverse drug reactions [4]. While it contained limited pediatric data, a large meta-analysis identified that 6.7% of hospitalized patients experienced a serious adverse drug reaction, with a fatality rate of 0.32% [5], and significant impacts on care quality. PGx testing represents a new medication safety tool that may be used to decrease adverse drug reactions, optimize efficacy of drug therapy and reduce healthcare costs. In the last decade there has been a steady increase in PGx-guided dosing recommendations provided by different research groups, drug manufacturers, and in-house pharmacy/pharmacology committees for both older generic drugs as well as recent FDA-approved drugs. Most of this information, however, focuses on drugs used to treat adults. Selecting the right dose can be challenging in the case of pediatric populations where the effects of maturation (ontogeny) and evidence-based practice has been considerably more limited [6]. While more outcomes and pharmacoeconomic studies need to be completed, it is anticipated that the implementation of PGx in clinical practice for both adults and children will lead to the optimization of pharmacotherapy to decrease trial and error cycles, promote medication adherence, improve the safety of medications and elevate overall health outcomes.
It has long been recognized that drug metabolizing enzyme activities demonstrate large inter-individual variability. Polymorphisms in the genes coding these enzymes partially explain this variability [7]. The earliest pharmacogenetic research collected DNA from individuals who experienced treatment failure or unexpected side effects/toxicities identifying genetic variation in enzymes that metabolize a particular drug [8]. Depending on the nature of the polymorphism, enzyme activity may be characterized into five phenotypes: poor metabolizers, intermediate metabolizers, normal (formerly extensive) metabolizers, rapid metabolizers, and ultra-rapid metabolizers. Given the rapid advances in sequencing technologies, PGx research can expand beyond evaluation of single pharmacogenes to the study of molecular mechanisms of drug response as determined by both pharmacokinetic (PK) and pharmacodynamic (PD) considerations [8]. While still in early stages, there is growing evidence that evaluating inherited variation in drug transporter function (e.g., SLCO1B1/simvastatin), drug targets (e.g., SLC6A4/selective serotonin reuptake inhibitors), and unintended targets (e.g. HLA-B/abacavir) can help define critical elements of variability in drug response and toxicity.
One of the barriers to the implementation of PGx testing is the complexity of translating genetic laboratory results into medication decisions. The U.S. Food and Drug Administration (FDA) drug label, a primary resource of drug information used by prescribers, contains PGx information to guide medication use. Recently the FDA has also published a “Table of Pharmacogenetic Associations” (https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations#ft1) identifying medications with sufficient scientific evidence that a gene/drug interaction may impact outcome. On this website, the FDA identifies 48 medications for which the scientific data support therapeutic management recommendations, 17 medications where gene/drug interactions may impact safety or response and 36 medications where gene/drug interactions affect PK properties. There is limited information regarding children and adolescents presented on this website. Other than identifying these gene/drug interactions as having sufficient scientific evidence that may impact outcome, the FDA is clear that they are not advocating for preemptive PGx testing unless the test is considered a companion diagnostic.
Two National Institutes of Health (NIH) funded organizations are also tackling the challenge of translating PGx test results into medication management decisions. The Clinical Pharmacogenetics Implementation Consortium (CPIC®) was established in 2009 with the purpose of curating and posting freely available, peer-reviewed, evidence-based, and updatable pharmacogenomics-related clinical practice guidelines [9]. In a recent publication, CPIC authors report the publication of 23 guidelines covering 19 genes and 46 drugs across several therapeutic areas [10]. The application of CPIC guidelines in the pediatric setting is discussed in detail below. The Pharmacogenomics Knowledgebase (PharmGKB), another NIH funded entity, is a knowledge resource that curates, annotates and disseminates PGx clinical information. PharmGKB annotates FDA label information on its website and categorizes PGx information in the FDA label as “Testing Required”, “Testing Recommended”, “Actionable PGx” and “Informative PGx.” (https://www.pharmgkb.org/page/drugLabelLegend, accessed 10 May 2020).
Current implementations in pediatrics
Despite a growing volume of research supporting the implementation of PGx in clinical practice, uptake has been slow, occurring predominantly in academic health centers. More recently, some health systems, including nonacademic centers, have begun implementation of PGx testing in support of precision medicine programs. There are two approaches or methods for most PGx implementation projects. The first is to provide preemptive PGx testing for future medication considerations or reactive screening when a high-risk medication is indicated for a patient’s care. These are not mutually exclusive and programs may be a mixture of both approaches. The goals of all of these implementation programs are to provide optimal, safe, and economical use of medications for patients receiving these high-risk medications.
A review of published work by several of the early implementers of pediatric focused clinical PGx programs as identified on the CPIC website indicates wide variability in genotyping methodologies ranging from single gene assays to panels covering greater than 200 genes (https://cpicpgx.org/implementation/) (See Table 1 for additional details). Pharmacogenomic services may be provided in both inpatient and ambulatory care settings. A description of specific clinical activities is beyond the scope of this review and has been described elsewhere [11–14].
Table 1.
Pediatric healthcare institutions with PGx implementation projects.
| Healthcare institution | Genes implemented | Associated drugs |
|---|---|---|
| St. Jude Children’s Research Hospitala | TPMT/NUDT15 | Thiopurines |
| CYP2C9 | Celecoxib | |
| CYP2C19 | Clopidogrel, amitriptyline, clomipramine, imipramine, trimipramine, doxepin, voriconazole, PPIs | |
| CYP2D6 | Codeine, oxycodone, tramadol, amitriptyline, clomipramine, imipramine, trimipramine, doxepin, fluoxetine, paroxetine, ondansetron, | |
| CYP3A5 | Tacrolimus | |
| DPYD | Fluoropyrimidines | |
| SLCO1B1 | Simvastatin | |
| UGT1A1 | Atazanavir | |
| mt-RNR1 | Aminoglycoside antibiotics | |
| RYR1/CACNA1S | Succinylcholine and volatile fluorinated anesthetics | |
| Children’s Minnesotab | TPMT/NUDT15 | Thiopurines |
| CYP2C9 | Phenytoin, fosphenytoin | |
| CYP2C19 | Citalopram, escitalopram, amitriptyline, clomipramine, imipramine, trimipramine, doxepin, voriconazole | |
| CYP2D6 | Atomoxetine, aripiprazole, amitriptyline, clomipramine, imipramine, trimipramine, doxepin, desipramine, nortriptyline, paroxetine, fluvoxamine, ondansetron, tramadol | |
| CYP3A5 | Tacrolimus | |
| DPYD | 5-Fluorouracil | |
| RYR1/CACNA1S | Succinylcholine and volatile fluorinated anesthetics | |
| University of Florida in partnership with Nemours Children’s Specialty Care, Jacksonville, FL and Nemours Children’s Hospital, Orlando, FLc | TPMT/NUDT15 | Thiopurines |
| CYP2C19 | PPIs | |
| Boston Children’s Hospitald | TPMT/NUDT15 | Thiopurines |
| CYP2C9 | Phenytoin, fosphenytoin, glimepiride, glipizide, glipizide-metformin, glyburide, glyburide-metformin, glimepiride-pioglitazone, glimepiride-rosiglitazone, irbesartan, hydrochlorothiazide-irbesartan, losartan, hydrochlorothiazide-losartan, tolbutamide, warfarin | |
| CYP2C19 | Amitriptyline, amitriptyline/chlordiazepoxide, amitriptyline/perphenazine, citalopram, clobazam, clomipramine, clopidogrel, doxepin, escitalopram, esomeprazole, esomeprazole/naproxen, imipramine, lansoprazole, lansoprazole/amoxicillin/clarithromycin, lansoprazole/naproxen, omeprazole, omeprazole/sodium bicarbonate, omeprazole/clarithromycin/amoxicillin, pantoprazole, rabeprazole, sertraline, voriconazole | |
| VKORC1 | Warfarin | |
| HLA-A | Carbamazepine | |
| HLA-B | Abacavir, allopurinol, carbamazepine, fosphenytoin, oxcarbazepine, phenytoin | |
| Cincinnati Children’s Hospital Medical Centere | TPMT/NUDT15 | Thiopurines |
| CYP2C19 | Amitriptyline, clomipramine, imipramine, trimipramine, doxepin, clopidogrel, voriconazole, PPIs | |
| CYP2D6 | Codeine, hydrocodone, oxycodone, tramadol, aripiprazole, haloperidol, olanzapine, perphenazine, risperidone, thioridazine, amitriptyline, clomipramine, imipramine, trimipramine, doxepin, desipramine, maprotiline, nortriptyline, fluoxetine, paroxetine, fluvoxamine, venlafaxine |
aHaidar et al. [15].
bUnpublished data, courtesy of D Gregornik, PharmD, Children’s Minnesota.
cWeitzel et al. [19] and Cicali et al. [20].
dManzi et al. [13] and Kim et al. [46].
eRamsey et al. [21], https://www.cincinnatichildrens.org/service/g/genetic-pharmacology/education, accessed 20 Nov 2019.
One of the earliest pediatric implementation programs, St. Jude Children’s Research Hospital opened a clinical research protocol called PG4KDS in 2011 that established preemptive PGx testing as a standard of care (www.stjude.org/pg4kds). Genotyping on the PG4KDS protocol is completed using the PharmacoScan array (ThermoFisher Scientific, Waltham, MA) which interrogates 4627 variants in 1191 genes. As of June 2019, St. Jude has integrated information on 11 genes and 35 drugs into their electronic health record (EHR). These results are used to deliver point of care electronic clinical decision support (eCDS) to clinicians when high-risk medications are ordered for an affected patient [15]. Investigators at St. Jude have demonstrated the clinical utility of preemptive PGx testing for TPMT in patients with acute lymphoblastic leukemia receiving 6-mercaptopurine and for CYP2D6 in their sickle cell population where codeine is used in the management of pain crises [16, 17].
Based upon the St. Jude PG4KDS model, the Children’s Minnesota health-system began a clinical PGx program in 2016 and to date has leveraged technology within their EHR to develop custom rules for 10 genes and 26 medications with point-of-care alerts to guide medication selection and dosing in the Children’s Minnesota hospitals and hospital based clinics. The process at Children’s Minnesota is agnostic to PGx testing platform, allowing patients who have been tested elsewhere to have their PGx results entered into the EHR to benefit from the eCDS (Gregornik, D., personal communication with Boston Children’s Hospital). The University of Florida implemented a custom array with 256 clinically actionable genetic polymorphisms [18]. Pediatric focused gene implementations by the University of Florida include TPMT/thiopurines [19] and CYP2C19/proton pump inhibitors (in partnership with Nemours Children’s Specialty Care, Jacksonville, FL and Nemours Children’s Hospital, Orlando, FL [20]. Boston Children’s Hospital has focused on children with significant adverse drug reactions or failed treatment using either single gene sequencing tests for TPMT and CYP2C9/VKORC1 or microarray panels testing up to 225 SNPs [13]. Boston Children’s Hospital reports that they have eCDS available for TPMT, CYP2C9, CYP2C19, and VKORC1 that is linked to more than 30 medications based upon the CPIC guidelines [13]. In a recently published paper, Cincinnati Children’s Hospital Medical Center describes the evolution of their Genetic-Pharmacology Service and the development of single gene tests as well as specialty genotyping panels for psychiatry, opioids, and warfarin [21].
The Sanford Children’s Genomic Medicine Consortium
Founded in September 2016, the Sanford Children’s Genomic Medicine Consortium (SCGMC) was established as a partnership of ten pediatric hospitals across the United States dedicated to transforming care for children through genomic medicine. The charter membership included six institutions (https://www.sanfordhealth.org/initiatives/childrens-genomic-medicine). Inspired by the vision of philanthropist Denny Sanford, the Sanford Consortium Member Institutions was expanded and includes: Banner Children’s at Diamond Children’s Medical Center, Tucson, AZ; Children’s Hospital Colorado, Denver, CO; Children’s Hospital Los Angeles, Los Angeles, CA; Children’s Minnesota, Minneapolis & St Paul, MN; Lucile Packard Children’s Hospital Stanford, Palo Alto, CA; Nicklaus Children’s Hospital, Miami, FL; Rady Children’s, San Diego, CA; Sanford Children’s, Fargo, ND & Sioux Falls, SD and Seattle Children’s Hospital, Seattle, WA. The Consortium is committed to improving children’s health by integrating genomic medicine into pediatric care through collaborations that:
Improve diagnosis and treatment of a host of common and rare childhood diseases.
Improve pediatric care through innovative joint research programs and genomic health-care workforce development.
Improve genetic and genomic discovery through access to a large, racially, and ethnically diverse population.
Develop relationships with industry and governmental entities.
Provide training for physicians, pharmacists, genetic counselors, and other health professionals.
Research projects funded by SCGMC to date have focused on rapid whole genome sequencing of newborns and next generation sequencing (NGS) of pediatric tumors. SCGMC has recently provided a total of $500,000 in seed funding for projects to be completed by consortium members in calendar year 2020. With respect to PGx, SCGMC member institutions are in various stages of implementation from reactive testing of single gene/drug pairs in specialty care services to preemptive testing of pharmacogenes for general primary and specialty care. The SCGMC is now considering options for a consortium-wide implementation project for PGx.
Genetic testing technology
Genotyping assays are the typical methods for evaluating genetic variants in drug metabolizing enzymes and transporters. These tests range from single gene assays to multigene panels evaluating hundreds to thousands of variants, with costs ranging from $100 to more than $1000. Despite the current focus on genotyping, it is important to note that NGS technologies such as whole exome or whole genome sequencing are increasingly utilized in clinical care. The use of NGS in PGx is still in the initial stages of research development. The primary limitation to wider uptake is the need to confirm NGS with Sanger sequencing. The current cost and turn-around time to incorporate Sanger sequencing is prohibitive to wide implementation across patient populations in hospitals and health systems [22]. Nevertheless, the trend in utilizing NGS can be expected to continue as the accuracy of this technology improves while costs and turn-around time quickly decrease. A review of genetic testing technologies is described elsewhere and beyond the scope of this report. The reader is referred to these reviews for details [23, 24].
A major outcome of the decreased costs of genetic testing is the rise of direct-to consumer marketing of genetic tests. One of the most popular tests on the market today is 23andMe (https://www.23andme.com/), which offers both ancestry and health information based on a genotyping array. To date, 23andMe is the only direct-to-consumer genetic test with FDA approval to provide health predisposition reports to their customers. The FDA has also approved 23andMe to provide PGx reports to individuals, but has not yet started offering this service on their website. While direct-to-consumer testing may identify some genetic variants associated with increased health risks or drug response, the platforms employed are not comprehensive and subject to considerable variability. The FDA recommends that direct-to-consumer testing should not be used to make medical decisions and 23andMe includes this disclaimer on their website. A recent evaluation of direct-to-consumer testing has shown that up to 40% of the variants reported for a variety of genes may be false positives [25]. As such, confirmatory testing from a Clinical Laboratory Improvement Amendments certified laboratory is required to validate direct-to-consumer test findings. Likewise, PGx testing should only be completed in a certified clinical laboratory before medication selections or dosing decisions are made based on such testing.
Challenges to clinical implementation of PGx in pediatrics
Many of the barriers to implementing PGx in clinical practice are similar in both children and adults and include: (1) the lack of clinician understanding of how to use pharmacogenomic test results in clinical practice; (2) the absence of infrastructure in EHRs to store structured genetic data; (3) the lack of portability of data when patients are seen in multiple health systems and; (4) concerns about cost and insurance reimbursement. These challenges are well documented and will not be addressed further here [11, 14, 26, 27]. The remainder of this section will address challenges and barriers specific to children and adolescents such as the effect of maturation on pharmacogenomic phenotypes, paucity of pharmacogenomic data obtained in pediatric populations (requiring extrapolation of adult data), and differences in disease states between children and adults where medications with pharmacogenomic implications may be utilized.
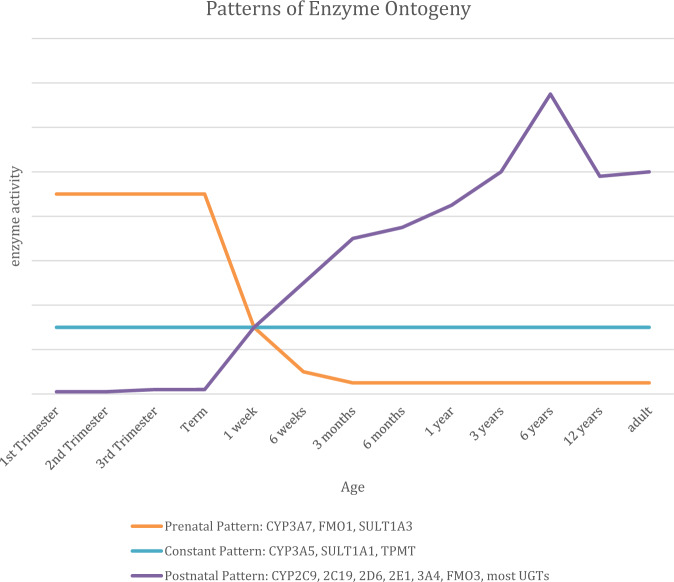
Healthcare providers working with children and adolescents have long recognized that there are developmental changes that impact response to medications. Grey-baby syndrome associated with chloramphenicol is one of the first observations of immature drug metabolism reported. Chloramphenicol is metabolized by UGT2B7, which is known to be immature at birth [28]. Ontogeny, the impact of maturation on drug metabolizing enzymes and transporters, is an intrinsic factor of drug response that complicates the implementation of PGx in pediatric populations [29]. Figure 1 demonstrates three typical patterns of drug metabolizing enzyme ontogeny. Prior to birth, the fetus lacks many drug metabolism activities. For example, CYP2D6 has negligible activity at birth and reaches adult activity by 2 weeks of age [30]. CYP2C9, CYP2C19 as well as most of the UDP glucuronosyltranferases (UGTs) follow a similar pattern [28, 31]. The apparent outcome of genetic polymorphisms on enzymes and transporters will not be measurable until the gene product is fully expressed as the individual matures. The impact of development and maturation on the utility of PGx-guided drug selection and dosing information in FDA labeling and CPIC guidelines can create significant challenges when applying PGx testing to neonates and infants [32].
Fig. 1. Patterns of drug metabolizing enzyme ontogeny.
Modified from de Wildt et al. [28].
Historically, finding evidence-based information to guide the use of medications in children and adolescents is an ongoing challenge for pediatric healthcare workers. Adult data are frequently extrapolated to the use of drugs in children and adolescents [33]. Clinical utilization of PGx in the pediatric population is no different. The majority of CPIC guidelines have a pediatric section with a general statement addressing the use of the guidelines in children: “… there is no reason to suspect that gene name genetic variation will affect this drug’s metabolism differently in children compared with adults.” (https://cpicpgx.org/guidelines/). Several of the guidelines suggest “caution” when applying PGx-guided dosing to children. When sufficient pediatric evidence exists, CPIC guidelines include specific recommendations for PGx-guided dosing in children (e.g., atomoxetine, voriconazole).
Prescribers are familiar with the FDA drug label as a primary medication reference that includes PGx information when available. Green et al. published a systematic review assessing the suitability of applying FDA label information to children and adolescents [34]. The review identified 65 drugs with pediatric labeling that included PGx information (See Table 2). Based on the PharmGKB annotations of the FDA drug label, 16 out of the 65 labels (25%) list “Testing required” for the related PGx biomarker. Additional two drug labels are annotated as “Testing recommended.” Only nine (14%) of the drug labels contained information derived from pediatric studies. In 56 of the 65 (86%) drug labels, PGx data were extrapolated from adult studies. Green et al. classified 16 of the studies (25%) where application of PGx information to the pediatric population was “unclear.” It is evident that more studies on the use of PGx testing in children and adolescents need to be completed to further expand precision medicine to this population.
Table 2.
Medications used in pediatric patients with PGx information in FDA-approved drug label.
| Medication | PGx biomarker | PGx on FDA labela | PGx data from pediatric studies (Y/N) | CPIC levelb | CPIC guideline (Y/N) |
|---|---|---|---|---|---|
| Abacavir | HLA-B | Testing required | Nc | A | Y |
| Anastrazole | Estrogen receptor | Testing required | Nd | N/A | N |
| Aripiprazole | CYP2D6 | Actionable PGx | Nc | B | N |
| Arsenic trioxide | PML/RYRα | Testing required | Nc | N/A | N |
| Atomoxetine | CYP2D6 | Actionable PGx | Y | A | Y |
| Atorvastatin | LDL receptor | Informative PGx | Nc | D | N |
| Busulfan | Ph chromosome | Actionable PGx | Nc | N/A | N |
| Capecitabine | DPYD | Actionable PGx | Nc | A | Y |
| Carbamazepine | HLA-B | Actionable PGx | Nc | A | Y |
| Carglumic Acid | NAGS | Testing required | Y | B | N |
| Carisoprodol | CYP2C19 | Actionable PGx | Nc | B/C | N |
| Carvedilol | CYP2D6 | Actionable PGx | Nd | C | N |
| Celecoxib | CYP2C9 | Actionable PGx | Nc | B | Ne |
| Cetuximab | K-Ras, EGFR | Testing required | Nd | D | N |
| Chloroquine | G6PD | Actionable PGx | Nc | B | N |
| Citalopram | CYP2C19 | Actionable PGx | Nd | A | Y |
| Clobazam | CYP2C19 | Actionable PGx | Nc | C | N |
| Clomipramine | CYP2C19/CYP2D6 | Actionable PGx | Nc | B | Y |
| Clopidogrel | CYP2C19 | Actionable PGx | Nd | A | Y |
| Codeine | CYP2D6 | Actionable PGx | Nd | A | Y |
| Dapsone | G6PD | Actionable PGx | Nc | B | N |
| Diazepam | CYP2C19 | Actionable PGx | Nc | C | N |
| Drospirenone-ethinyl estradiol | CYP2C19 | Informative PGx | Nc | N/A | N |
| Elosulfase alfa | GALNS | Testing required | Y | N/A | N |
| Esomeprazole | CYP2C19 | Actionable PGx | Nd | B | Ne |
| Everolimus | HER2/Neu | Testing required | Nd | N/A | N |
| Fluoxetine | CYP2D6 | Informative PGx | Nc | C | N |
| Fluvoxamine | CYP2D6 | Actionable PGx | Nc | A | Y |
| Fulvestrant | Estrogen receptor | Testing required | Nd | N/A | N |
| Glimepiride | G6PD | Actionable PGx | Nc | B | N |
| Imatinib | C-kit, Ph Chromosome, PDGFR, FIP1L1-PDGFRα | Testing required | Nc | N/A | N |
| Irinotecan | UGT1A1 | Actionable PGx | Nd | A | N |
| Isoniazid-pyrazinamide-rifampin | NAT1-2 | Informative PGx | Nd | N/A | N |
| Ivacaftor | CFTR | Testing required | Y | A | Y |
| Lansoprazole | CYP2C19 | Informative PGx | Y | B | Ne |
| Mercaptopurine | TPMT, NUDT15 | Testing recommended | Nc | A | Y |
| Metoclopramide | CYB5R1-4 | Actionable PGx | Nd | D | N |
| Metoprolol | CYP2D6 | Actionable PGx | Nc | C | N |
| Modafinil | CYP2D6 | Actionable PGx | Nc | C | N |
| Mycophenolic acid | HPRT1 | Actionable PGx | Nc | B | N |
| Omeprazole | CYP2C19 | Actionable PGx | Y | B | Ne |
| Paroxetine | CYP2D6 | Informative PGx | Nc | A | Y |
| Perphenazine | CYP2D6 | Actionable PGx | Nc | B | N |
| Phenytoin | HLA-B | Actionable PGx | Nc | A | Y |
| Phenytoin | CYP2C19 | Actionable PGx | Nd | N/A | N |
| Phenytoin | CYP2C9 | Actionable PGx | Nd | A | Y |
| Pimozide | CYP2D6 | Testing required | Nc | B | N |
| Pantoprazole | CYP2C19 | Actionable PGx | Y | B | Ne |
| Pravastatin | LDL receptor | Informative PGx | Nc | N/A | N |
| Quinine | G6PD | Actionable PGx | Nc | B | N |
| Rabeprazole | CYP2C19 | Actionable PGx | Y | B | Ne |
| Rasburicase | G6PD | Testing required | Nc | A | Y |
| Risperidone | CYP2D6 | Informative PGx | Nc | B | N |
| Rosuvastatin | SLCO1B1 | Actionable PGx | Nd | C | N |
| Sevoflurane | RYR1 | Actionable PGx | Nd | A | Y |
| Sodium phenylacetate-Sodium benzoate | NAGS; CPS1; ASS1; OTC; ASL; ARG1 | Testing required | Y | N/A | N |
| Sodium phenylbutyrate | NAGS; CPS1; ASS1; OTC; ASL; ARG1 | Testing required | Y | N/A | N |
| Succimer | G6PD | Informative PGx | Nc | N/A | N |
| Sulfamethoxazole-trimethoprim | G6PD | Actionable PGx | Nc | B | N |
| Tamoxifen | CYP2D6 | Actionable PGx | Nd | A | Y |
| Thioguanine | TPMT, NUDT15 | Testing recommended | Nc | A | Y |
| Thioridazine | CYP2D6 | Actionable PGx | Nc | C | N |
| Tretinoin | PML/RARα | Testing required | Nc | N/A | N |
| Valproic acid | POLG | Testing required | Nc | B | N |
| Valproic acid | OTC | Actionable PGx | Nc | B | N |
| Venlafaxine | CYP2D6 | Actionable PGx | Nc | B | N |
| Voriconazole | CYP2C19 | Actionable PGx | Nc | A | Y |
| Warfarin | CYP2C9-VKORC1 | Actionable PGx | Nd | A | Y |
Note that only CPIC level A and B gene/drug pairs have sufficient evidence for at least one prescribing action to be recommended. CPIC level C and D gene/drug pairs are not considered to have adequate evidence or actionability to have prescribing recommendations. (https://cpicpgx.org/prioritization/#leveldef, accessed 13 May 2020).
aPharmGKB (https://pharmgkb.org, accessed 13 May 2020).
bThe CPIC levels (A, B, C, and D) assigned are subject to change; only those gene/drug pairs that have been the subject of guidelines have had sufficient in-depth review of evidence to provide definitive CPIC level assignments.
cSuitable to extrapolate adult data to pediatric patients [34].
dUnclear applicability of adult data to pediatric patients [34].
eCPIC proton pump inhibitor guideline under development (https://cpicpgx.org, accessed 20 Nov 2019).
When evaluating medication use, it is important to consider the effects of disease state on drug PDs and PKs, with changes in drug transport and compartmentation most prominently featured in conjunction with polymorphisms in drug metabolizing enzymes. The PD effect of a drug may vary depending upon the condition being treated and can be expected to be different between children, adolescents, and adults. For instance, Kawasaki disease affects children but does not have an equivalent adult disease state. Clopidogrel is frequently used to treat children who have Kawasaki disease with moderate-to-large aneurysms. Similar to adults, children have demonstrated significant variability in response to clopidogrel [35, 36]. CPIC guidelines recommend alternative agents to clopidogrel for CYP2C19 intermediate and poor metabolizers in adults undergoing percutaneous transluminal coronary angioplasty, but impact of CYP2C19 polymorphisms on the activity of clopidogrel in Kawasaki disease has not been studied. Similarly, tamoxifen, an established medication for the prevention and treatment of breast cancer, is frequently used in the management of pubertal gynecomastia. The role of endoxifen (an active metabolite of tamoxifen produced by CYP2D6) and the impact of CYP2D6 polymorphisms on the outcome of pubertal gynecomastia has likewise not been evaluated in this population. These examples raise the need for more research of the PD response to medications in children and adolescents while also studying the application of PGx in these conditions.
Opportunities
Screening for genetic disorders at birth can prevent lifelong consequences by identifying at risk newborns allowing early intervention [37]. While more evidence is needed to fully endorse the clinical utility of preemptive PGx testing, it is anticipated that using a broad panel covering important pharmacogenes may guide selection and dosing of medications across multiple conditions including behavioral health, pain management, and cancer for both chemotherapy and supportive care. Beyond the current focus on metabolism, future drug development will benefit from the ability to produce medications that better target specific biochemical pathways, RNA and protein molecules directly associated with the disease of interest. As such, this represents an area of tremendous opportunity for the scientific and medical communities.
In the next decade, the degree to which different healthcare systems adopt PGx testing will likely vary greatly throughout the USA. We believe it is safe to predict that such practice will become more the norm than the exception. How quickly the practice is incorporated into the pediatric sphere remains to be fully defined, especially in primary care medical practice. However, such practice will ultimately help reduce the cost of healthcare when patients begin treatment with the medication most likely to work best for them.
Recommendations
The above challenges can be viewed as a “gap analysis” that identifies opportunities for the SCGMC to consider in promoting the implementation of PGx across its member institutions. Below, we describe strategies for advancing clinical pediatric PGx.
Standardization of Practice. With established consortia like CPIC, PharmGKB, IGNITE, and eMERGE currently leading standardized implementation efforts, partnership with these groups should be pursued to advance the interests of children and adolescents in the standardization of clinical PGx practice. For example, this would include prioritization of gene/drug pairs for implementation, evaluation and modification of CDS to specifically support pediatric practice (point of care alert language and dose recommendations) as well as increasing pediatric presence in ongoing implementation studies being conducted by these groups.
Multi-site demonstration project. To date, most published work describes PGx implementation projects at single institutions. Consortia wide demonstration projects may identify new challenges and implementation models for larger health systems. They will also expand implementation knowledge specific to the pediatric and adolescent populations. Outcomes to be studied are expected to demonstrate best practices for education of providers in pediatric primary and specialty care, assessment of parent/caregiver understanding and acceptance of PGx testing, and evaluation of a CDS system that is agnostic to type of EHR.
Pharmacogenomic databases. As noted above, a large part of the evidence justifying the use of PGx results in children and adolescents is extrapolated from adult data. Due to the effects of maturation or different disease states, this extrapolation may not always be appropriate. To this end, the creation of pediatric PGx registry/database should be considered. These resources will not only store de-identified PGx test results and outcomes related to the impact of ontogeny on PGx but also large amounts of phenotype data (e.g. PK results, adverse drug reactions, etc.) extracted from EHRs via natural language processing and artificial intelligence, similar to that used by consortium member Rady Children’s Institute for Genomic Medicine.
Workforce Training and Development. While not restricted to pediatrics, one of the largest existing barriers is the lack of standard practice models and experiential training available for medical professionals [38, 39]. The SCGMC is poised to consider projects to evaluate practice models perhaps including collaborations between pharmacists and genetic counselors where each discipline may bring their expertise to the clinic/bedside to advance prescriber and patient understanding of clinical PGx [40]. The consortium may consider partnering with early implementers of clinical PGx to sponsor a larger number of post-graduate year 2 pharmacy specialty residencies or other experiential training in clinical PGx. Similar strategies can be used for other health professionals.
Payer Engagement. Another benefit of creating a consortium PGx registry will be to facilitate pediatric focused PGx research further expanding the evidence available to provide evidence-based data for use by insurance payors in assessing the benefits of clinical PGx testing. Using evidence generated from the registry, the SCGMC and other groups can more effectively advocate to insurance payors on behalf of our patients who are unable to pay for this testing out-of-pocket.
Concluding remarks
Clearly, there are limitations to clinical PGx testing. While test results may provide a lifelong benefit, at the present time we are limited by the numbers of genes and variants that can be reliably tested. New evidence continues to emerge that may require reinterpretation of results (e.g., recent harmonization of CYP2D6 [41]), additional variants may be identified that may become important to test, necessitating repeat testing. Lastly, PGx data may account for only a small proportion of the variability observed in drug response [42]. Despite these limitations, clinical PGx testing presents a significant opportunity to implement genetic testing to optimize pharmacotherapy. As cost, availability, and efficiency of PGx testing improves, the opportunities in PGx are increasingly tangible. Clinician education, research, and institutional support and infrastructure are needed to make PGx programs successful. Implementation of the above recommendations can be a foundation from which to expand PGx services for pediatric patients to optimize pharmacotherapy, reduce adverse effects, and improve healthcare outcomes for this underserved population.
Executive summary
Despite documented evidence-based benefits of PGx in pediatric oncology, pain management, organ transplantation, and immunosuppression [43, 44], significant challenges to its implementation remain unresolved.
The advent of digital, electronic, and molecular technologies has facilitated the study of complete genomes and allowed the integration of molecular insight into drug development and has opened the door for actionable PGx interventions that can transform healthcare.
PGx allows clinicians to more precisely optimize drug treatment, thus enabling identification of drug of choice, as well as optimal dosing regimens based on the individual’s genetic characteristics [45].
Acknowledgements
The authors of the white paper are grateful to the Sanford Children’s Genomic Medicine Consortium member hospitals for their support of our efforts: Sanford Children’s (Sioux Falls and Fargo), Children’s Minnesota, Nicklaus Children’s (Miami), American Family Children’s/UW Health (Madison), Seattle Children’s, Lucile Packard Children’s at Stanford, Children’s Los Angeles, Rady Children’s (San Diego), Colorado Children’s and Banner Children’s at Diamond Children’s Medical Center (Tucson).
Author contributions
All authors made substantial contributions to the conception and design of the work and participated in drafting and revising it critically for important intellectual content. All authors approved of the final version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
David Gregornik, Email: David.Gregornik@childrensmn.org.
Kenneth Ramos, Email: kramos@ibt.tamhsc.edu.
References
- 1.Roden DM, Mcleod HL, Relling MV, Williams MS, Mensah GA, Peterson JS, et al. Pharmacogenomics. The Lancet. 2019;394:521–32. doi: 10.1016/S0140-6736(19)31276-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva PJ, Schaibley VM, Ramos KS. Academic medical centers as innovation ecosystems to address population -omics challenges in precision medicine. J Transl Med. 2018;16:28. doi: 10.1186/s12967-018-1401-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elzagallaai AA, Greff M, Rieder MJ. Adverse drug reactions in children: the double-edged sword of therapeutics. Clin Pharmacol Ther. 2017;101:725–35. doi: 10.1002/cpt.677. [DOI] [PubMed] [Google Scholar]
- 4.Gallagher RM, Mason JR, Bird KA, Kirkham JJ, Peak M, Williamson PR, et al. Adverse drug reactions causing admission to a paediatric hospital. PLoS ONE. 2012;7:e50127. doi: 10.1371/journal.pone.0050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. Jama. 1998;279:1200–5. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 6.Maagdenberg H, Vijverberg SJ, Bierings MB, Carleton BC, Arets HGM, de Boer A, et al. Pharmacogenomics in Pediatric Patients: Towards Personalized Medicine. Paediatr Drugs. 2016;18:251–60. doi: 10.1007/s40272-016-0176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tracy TS, Chaudhry AS, Prasad B, Thummel KE, Schuetz EG, Zhong XB, et al. Interindividual variability in cytochrome P450-mediated drug metabolism. Drug Metab Dispos. 2016;44:343–51. doi: 10.1124/dmd.115.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther. 2007;81:328–45. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–7. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Caudle KE. The Clinical Pharmacogenetics Implementation Consortium: 10 years later. Clin Pharmacol Ther. 2019. 10.1002/cpt.1651. [DOI] [PMC free article] [PubMed]
- 11.Dunnenberger HM, Crews KR, Hoffman JM, Caudle KE, Broeckel U, Howard SC, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Ann Rev Pharmacol. Toxicol. 2015;55:89–106. doi: 10.1146/annurev-pharmtox-010814-124835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arwood MJ, Chumnumwat S, Cavallari LH, Nutescu EA, Duarte JD. Implementing pharmacogenomics at your institution: establishment and overcoming implementation challenges. Clin Transl Sci. 2016;9:233–45. doi: 10.1111/cts.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manzi SF, Fusaro VA, Chadwick L, Brownstein C, Clinton C, Mandl KD, et al. Creating a scalable clinical pharmacogenomics service with automated interpretation and medical record result integration - experience from a pediatric tertiary care facility. J Am Med Inform Assoc. 2017;24:74–80. doi: 10.1093/jamia/ocw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weitzel KW, Elsey AR, Langaee TY, Burkley B, Nessl DR, Obeng AO, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166C:56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haidar CE, Relling MV, Hoffman JM. Preemptively precise: returning and updating pharmacogenetic test results to realize the benefits of preemptive testing. Clin Pharmacol Ther. 2019;106:942–4. doi: 10.1002/cpt.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gammal RS, Crews KR, Haidar CE, Hoffman JM, Baker DK, Barker PJ, et al. Pharmacogenetics for safe codeine use in sickle cell disease. Pediatrics. 2016;138:e20153479. [DOI] [PMC free article] [PubMed]
- 17.Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, Kornegay NM, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;166C:45–55. doi: 10.1002/ajmg.c.31391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson JA, Elsey AR, Clare-Salzler MJ, Nessl D, Conlon M, Nelson DR. Institutional profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics. 2013;14:723–6. doi: 10.2217/pgs.13.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weitzel KW, Smith DM, Elsey AR, Duong BQ, Burkley B, Clare-Salzler M, et al. Implementation of standardized clinical processes for TPMT testing in a diverse multidisciplinary population: challenges and lessons learned. Clin Transl Sci. 2018;11:175–81. doi: 10.1111/cts.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cicali EJ, Blake K, Gong Y, Mougey EB, Al-Atrash H, Chambers N, et al. Novel Implementation of genotype-guided proton pump inhibitor medication therapy in children: a pilot, randomized, multisite pragmatic trial. Clin Transl Sci. 2019;12:172–9. doi: 10.1111/cts.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey LB, Prows CA, Zhang K, Saldana SN, Sorter MT, Pestian JP, et al. Implementation of pharmacogenetics at Cincinnati Children’s Hospital Medical Center: lessons learned Over 14 years of personalizing medicine. Clin Pharmacol Ther. 2019;105:49–52. doi: 10.1002/cpt.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mu W, Lu HM, Chen J, Li S, Elliott AM. Sanger confirmation is required to achieve optimal sensitivity and specificity in next-generation sequencing panel testing. J Mol Diagn. 2016;18:923–32. doi: 10.1016/j.jmoldx.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Alekseyev YO, Fazeli R, Yang S, Basran R, Maher T, Miller NS, et al. A next-generation sequencing primer-how does it work and what can it do? Acad Pathol. 2018;5:2374289518766521. doi: 10.1177/2374289518766521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slatko BE, Gardner AF, Ausubel FM. Overview of next-generation sequencing technologies. Curr Protoc Mol Biol. 2018;122:e59. doi: 10.1002/cpmb.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tandy-Connor S, Guiltinan J, Krempely K, LaDuca H, Reineke P, Gutierrez S, et al. False-positive results released by direct-to-consumer genetic tests highlight the importance of clinical confirmation testing for appropriate patient care. Genet Med. 2018;20:1515–21. doi: 10.1038/gim.2018.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman JM, Dunnenberger HM, Kevin Hicks J, Caudle KE, Whirl-Carrillo M, Freimuth RR, et al. Developing knowledge resources to support precision medicine: principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC) J Am Med Inform Assoc. 2016;23:796–801. doi: 10.1093/jamia/ocw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borden BA, Galecki P, Wellmann R, Danahey K, Lee SM, Patrick-Miller L, et al. Assessment of provider-perceived barriers to clinical use of pharmacogenomics during participation in an institutional implementation study. Pharmacogenet Genom. 2019;29:31–8. doi: 10.1097/FPC.0000000000000362. [DOI] [PubMed] [Google Scholar]
- 28.De Wildt SN, Tibboel D, Leeder JS. Drug metabolism for the paediatrician. Arch Dis Child. 2014;99:1137–42. doi: 10.1136/archdischild-2013-305212. [DOI] [PubMed] [Google Scholar]
- 29.Hines RN. Ontogeny of human hepatic cytochromes P450. J Biochem Mol Toxicol. 2007;21:169–75. doi: 10.1002/jbt.20179. [DOI] [PubMed] [Google Scholar]
- 30.Blake MJ, Gaedigk A, Pearce RE, Bomgaars LR, Christensen ML, Stowe C, et al. Ontogeny of dextromethorphan O- and N-demethylation in the first year of life. Clin Pharmacol Ther. 2007;81:510–6. doi: 10.1038/sj.clpt.6100101. [DOI] [PubMed] [Google Scholar]
- 31.Koukouritaki SB, Manro JR, Marsh SA, Stevens JC, Rettie AE, McCarver DG, et al. Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Experimental therapeutics. 2004;308:965–74. doi: 10.1124/jpet.103.060137. [DOI] [PubMed] [Google Scholar]
- 32.Leeder JS, Kearns GL. Interpreting pharmacogenetic data in the developing neonate: the challenge of hitting a moving target. Clin Pharmacol Ther. 2012;92:434–6. doi: 10.1038/clpt.2012.130. [DOI] [PubMed] [Google Scholar]
- 33.Leeder JS, Kearns GL, Spielberg SP, Van Den, Anker J. Understanding the relative roles of pharmacogenetics and ontogeny in pediatric drug development and regulatory science. J Clin Pharmacol. 2010;50:1377–87. doi: 10.1177/0091270009360533. [DOI] [PubMed] [Google Scholar]
- 34.Green DJ, Mummaneni P, Kim IW, Oh JM, Pacanowski M, Burckart GJ. Pharmacogenomic information in FDA-approved drug labels: application to pediatric patients. Clin Pharmacol Ther. 2016;99:622–32. doi: 10.1002/cpt.330. [DOI] [PubMed] [Google Scholar]
- 35.Li JS, Yow E, Berezny KY, Bokesch PM, Takahashi M, Graham TP, et al. Dosing of clopidogrel for platelet inhibition in infants and young children: primary results of the Platelet Inhibition in Children On cLOpidogrel (PICOLO) trial. Circulation. 2008;117:553–9. doi: 10.1161/CIRCULATIONAHA.107.715821. [DOI] [PubMed] [Google Scholar]
- 36.Maltz LA, Gauvreau K, Connor JA, Jenkins KJ. Clopidogrel in a pediatric population: prescribing practice and outcomes from a single center. Pediatric Cardiol. 2009;30:99–105. doi: 10.1007/s00246-008-9289-x. [DOI] [PubMed] [Google Scholar]
- 37.Boyle CA, Bocchini JA, Jr., Kelly J. Reflections on 50 years of newborn screening. Pediatrics. 2014;133:961–3. doi: 10.1542/peds.2013-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown JT, Gregornik D, Kennedy MJ, Advocacy, Research C. The role of the pediatric pharmacist in precision medicine and clinical pharmacogenomics for children. J Pediatr Pharmacol Ther. 2018;23:499–501. doi: 10.5863/1551-6776-23.6.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avello K, Bell M, Landsverk M, Hoyme E. Perspectives of pediatric providers regarding clinical use of pharmacogenetics. National Society of Genetic Counselors; 2018. [PubMed]
- 40.Zierhut HA, Campbell CA, Mitchell AG, Lemke AA, Mills R, Bishop JR. Collaborative counseling considerations for pharmacogenomic tests. Pharmacotherapy. 2017;37:990–9. doi: 10.1002/phar.1980. [DOI] [PubMed] [Google Scholar]
- 41.Caudle KE, Sangkuhl K, Whirl-Carrillo M, Swen JJ, Haidar CE, Klein TE et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci. 2019. 10.1111/cts.12692. [DOI] [PMC free article] [PubMed]
- 42.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526:343–50. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abaji R, Krajinovic M. Current perspective on pediatric pharmacogenomics. Expert Opin Drug Metab Toxicol. 2016;12:363–5. doi: 10.1517/17425255.2016.1145656. [DOI] [PubMed] [Google Scholar]
- 44.Najafzadeh M, Garces JA, Maciel A. Economic evaluation of implementing a novel pharmacogenomic test (IDgenetix((R))) to guide treatment of patients with depression and/or anxiety. PharmacoEconomics. 2017;35:1297–310. doi: 10.1007/s40273-017-0587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valdes R, Jr., Yin DT. Fundamentals of pharmacogenetics in personalized, precision medicine. Clin Lab Med. 2016;36:447–59. doi: 10.1016/j.cll.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 46.Kim H, Chadwick L, Alzaidi Y, Picker J, Poduri A, Manzi S. HLA-A*31:01 and Oxcarbazepine-Induced DRESS in a patient with seizures and complete DCX deletion. Pediatrics. 2018;141:S434–8. doi: 10.1542/peds.2017-1361. [DOI] [PubMed] [Google Scholar]