Abstract
Ovarian cancer is one of the leading causes of gynecological cancer-related deaths worldwide. We investigated the role of a newly discovered long noncoding RNA, NR_026689, in cell proliferation, metastasis, and apoptosis in ovarian cancer cells. Our results showed that NR_026689 was overexpressed in both clinical ovarian cancer patients and cultured ovarian cancer cells. Knockdown of NR_026689 in HO-8910PM cells significantly decreased the cell proliferative rate and the ability to form colonies. Transwell assays revealed that depletion of NR_026689 suppressed cell migration ability by 68% and cell invasive capacity by 71% in HO-8910PM cells. Moreover, specific shRNAs against NR_026689 notably promoted cell apoptosis in HO-8910PM cells by upregulating the expression of proapoptotic proteins, including caspase 3, caspase 9, cytochrome C, and PARP. Our study suggests an oncogenic potential of NR_026689 in ovarian cancer and might provide novel clues for the diagnosis and treatment of ovarian cancer in the clinic.
Key words: NR_026689, Ovarian cancer, Proliferation, Metastasis, Apoptosis
INTRODUCTION
Ovarian cancer is among the most common causes of cancer-related deaths for females around the world. More than 230,000 new ovarian cancer cases and approximately 152,000 deaths were reported in 20121. Although great efforts have been made to improve the diagnosis and treatment of ovarian cancer, more than 75% of ovarian cancer patients are diagnosed at a very late stage2. Clinical treatment for these patients at an advanced stage is cytoreductive surgery combined with chemotherapy3, which is only effective in the initial period. A majority of these patients will relapse with a median progression-free survival (PFS) of 18 months and eventually die from ovarian cancer4. Thus, accumulative studies have been focusing on targeted therapies and proposing the application of biomarkers in the diagnosis and treatment of ovarian cancer at a relatively early stage5,6. However, no reliable molecular biomarkers have been discovered that are capable of identifying ovarian cancer before there is radiographic or biochemical evidence of progression.
Long noncoding RNAs (lncRNAs) are a category of RNAs that have a length of more than 200 nucleotides and are incapable of encoding proteins because of a lack of open reading frames7. According to their genomic locations, lncRNAs are classified into four categories, namely, intergenic, intronic, antisense, and enhancer lncRNAs8. Recently, the role of lncRNAs has been increasingly recognized in tumorigenesis. For instance, it was found that lncRNA MALAT-1 was highly expressed in patients with ovarian cancer, and a high expression of MALAT-1 promoted cell proliferation and cell metastasis9. lncRNA ANRIL also showed its potential for promoting cell proliferation and cell cycle progression and suppressing cell apoptosis and senescence in ovarian cancer10. However, the detailed mechanism of how these lncRNAs regulated tumorigenesis in ovarian cancer is still an enigma for researchers. Furthermore, the effects of other lncRNAs on ovarian cancer also need to be uncovered.
lncRNA NR_026689 is a newly discovered lncRNA that could potentially serve as a biomarker for lung carcinogenesis11. Researchers identified this lncRNA in 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK)-injected rats and analyzed this with the Arraystar rat lncRNA array. Upon NNK injection, NR_026689 was significantly upregulated in lung cancer tissues and rat whole blood. However, a more detailed knowledge of the involvement of NR_026689 in the carcinogenesis of the lung and other organs remains largely unclear.
In our study, the expression of NR_026689 in ovarian cancer was explored in vivo and in vitro. Detailed roles of NR_026689 in cell proliferation and metastasis were also assessed in ovarian cancer HO-8910PM cells. Furthermore, the effects of NR_026689 on cell apoptosis and its preliminary mechanism were examined by knockdown of NR_026689 with specific shRNAs in HO-8910PM cells. Our study indicated the oncogenic potential of NR_026689 in ovarian cancer and might provide novel clues for the diagnosis and treatment of ovarian cancer in the clinic.
MATERIALS AND METHODS
Human Tissues
This study was approved by the ethics committee of Xi’an Jiaotong University. A total of 70 ovarian cancer patients gave their full intention to participate in our study, and written consent from each patient was obtained. These patients underwent clinical surgeries in The Second Affiliated Hospital of Xi’an Jiaotong University. No chemotherapy or radiotherapy preceded these surgeries. The tumor tissues and their adjacent noncancerous counterparts were dissected and frozen in liquid nitrogen immediately and subjected to RNA extraction.
Cell Culture and Transfection
Human normal ovarian cell line HUM-CELL-0088 and ovarian cancer cell line OVCAR-3 were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Three other ovarian cancer cell lines—HO-8910, HO-8910PM, and SK-OV-3—were commercially purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, P.R. China). Cells were cultured within the recommended medium, supplied with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), and kept in a 5% CO2 atmosphere at 37°C. Cell transfection was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s protocols.
Real-Time Polymerase Chain Reaction (RT-PCR)
Total RNAs form both clinical tissues and cultured cells were extracted by TRIzol reagents (TaKaRa, Dalian, P.R. China) at a concentration of 1 ml/well in six-well plates. NanoDrop 2000 was used to quantify the extracted RNAs by collecting absorbance of 280 and 260 nm. cDNAs were reverse transcribed by PrimeScript RT Master Mix Reagent Kit (TaKaRa), and 10 ng from each sample was subjected to PCR analysis. The following primers were used: NR_026689, 5′-TCATCCATCACCTTCCAACA-3′ (forward) and 5′-ACCGCTCGCTTCTTAGCAAT-3′ (reverse); GAPDH, 5′-ACAGCAACAGGGTGGTGGAC-3′ (forward) and 5′-TTTGAGGGTGCAGCGAACTT-3′ (reverse). Briefly, the protocol for the PCR assay was 95°C for 30 s followed by 40 cycles at 95°C for 5 s and 60°C for 1 min. GAPDH was included here as an internal control. The cycle threshold values were calculated with the ABI7900 software (ABI; Applied Biosystems, Foster City, CA, USA).
Cell Proliferation
HO-8910PM cells were transfected with scramble or specific shRNAs against NR_026689 for 48 h and then seeded into 96-well plates (3,000 cells/well) and allowed to grow overnight. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide methyl thiazolyl tetrazolium (MTT) solution (2 mg/ml) was mixed into each experimental well. After incubation for 4 h at 37°C, culture medium was removed, and 200 μl of DMSO was added into cells in each group. The plate was shaken for 5 min, and the optical density was obtained at 570 nm. Cell proliferation assay was performed for 5 consecutive days with each group repeated in triplicate.
Colony Formation Assay
Human ovarian cancer HO-8910PM cells were cultured in 10-cm plates and treated with shRNAs targeting scramble or NR_026689 sequences. After 48 h of incubation, infected cells were washed with PBS, reseeded in new plates (800 cells/well), and allowed to grow naturally. After 2 weeks of incubation, treated HO-8910PM cells were washed twice with PBS, fixed with paraformaldehyde, and then stained with crystal violet (1%) for 10 min. Afterward, colonies were washed with ddH2O and photographed under an inverted microscope with five random fields in each experimental group. Colonies that contained more than 50 cells were manually counted and calculated.
Transwell Assay
HO-8910PM cells were cultured in 24-well plates and infected with specific shRNAs. After 48 h, cells were harvested with serum-free medium, and 200 μl of cell suspension (approximately 3 × 104 cells) was added into the upper chamber (Corning, NY, USA). The lower chamber was filled with 600 μl of culture medium supplied with 10% FBS. For the invasion assay, the chamber was precoated with Matrigel (Corning) for 6 h at 37°C before seeding cells. After incubation in a 37°C incubator for an additional 12 h, cells were fixed with precold methanol for 5 min and then stained with crystal violet (1%) for 5 min. The images were then captured under a Nikon microscope with five random fields in each experimental group.
Cell Apoptosis Assay
Morphological analysis was performed to evaluate the apoptotic cells using Hoechst 33258 staining. HO-8910PM cells were treated with distinct shRNAs for 48 h and then washed with cold PBS and fixed in methanol/acetic acid (3:1) for 15 min at 37°C. Fixed cells were then stained with 5 μg/ml Hoechst 33258 for 10 min. Morphological changes in the nuclei of cells were imaged using a fluorescence microscope (Leica, Germany), and cell apoptotic rate was calculated.
Determination of Caspase Activities
The relative activities of caspase 3, caspase 8, and caspase 9 were determined by the Caspase 3 Activity Kit, Caspase 8 Activity Kit, and Caspase 9 Activity Kit, respectively, following the instructions from Beyotime (Nantong, P.R. China). Briefly, cell lysates were collected by centrifuging after shRNA treatment. Aliquots of 10-μl proteins from cell lysates were added into 96-well plates and coincubated with 80 μl of reaction buffer containing caspase substrate (caspase 3, caspase 8, and caspase 9; 2 mM for each). After incubation for an additional 4 h at 37°C, caspase activities were obtained with a TECAN reader at an absorbance of 405 nm.
Western Blot Analysis
Proteins from HO-8910PM cells, which were transfected with shRNAs in the presence or absence of NR_026689 depletion, were extracted for subsequent immunoblot assays when cell growth confluence was approximately 90%. Briefly, an equal amount of 50 μg of protein from each group was loaded into each well in a 10% SDS-PAGE gel. Immunoreactivity was developed with enhanced chemoluminescence autoradiography (Thermo Scientific, Pittsburgh, PA, USA) by the Las3000 machine. Primary antibodies against caspase 3, caspase 9, and cytochrome C (cyt C) were purchased from Abcam (Cambridge, MA, USA). Primary antibody against PARP and GAPDH as well as the secondary antibodies were ordered from Santa Cruz Biotechnology (Santa Cruz, CA, USA). GAPDH was included as an internal control.
Statistical Analysis
The results were presented as means ± standard deviation (SD). Statistical analysis was carried out with the Student’s t-test. Any difference with a value of p < 0.05 was considered statistically significant. All experiments were repeated in triplicate.
RESULTS
Long Noncoding RNA NR_026689 Was Overexpressed in Human Ovarian Cancers
Cancer tissue and adjacent noncancerous tissue from a total of 70 ovarian cancer patients were collected and dissected for RT-PCR analysis. As shown in Figure 1A, the relative transcript level of NR_026689 was notably increased by approximately 3.8-fold in tumor tissues compared with their control counterparts. Next, the transcript level of NR_026689 was assessed in ovarian cancer cell lines. HUM-CELL-0088 is a normal ovarian cell line and was also included, whereas HO-8910, HO-8910PM, OVCAR-3, and SK-OV-3 cell lines are derived from ovarian cancer patients, of which the HO-8910PM cells have the strongest migration potential. As shown in Figure 1B, compared with HUM-CELL-0088 cells, all of the ovarian cancer cell lines showed a higher expression of NR_026689, which were increased by 1.5-fold (HO-8910 cells), 4.5-fold (HO-8910PM), 2.2-fold (OVCAR-3), and 2.5-fold (SK-OV-3), respectively. Of note, the relative transcript level of NR_026689 was the highest in HO-8910PM cells, indicating a potential association between the expression of NR_026689 and cell metastasis. Our data suggested a higher expression of NR_026689 in ovarian cancer in vivo and in vitro.
Figure 1.
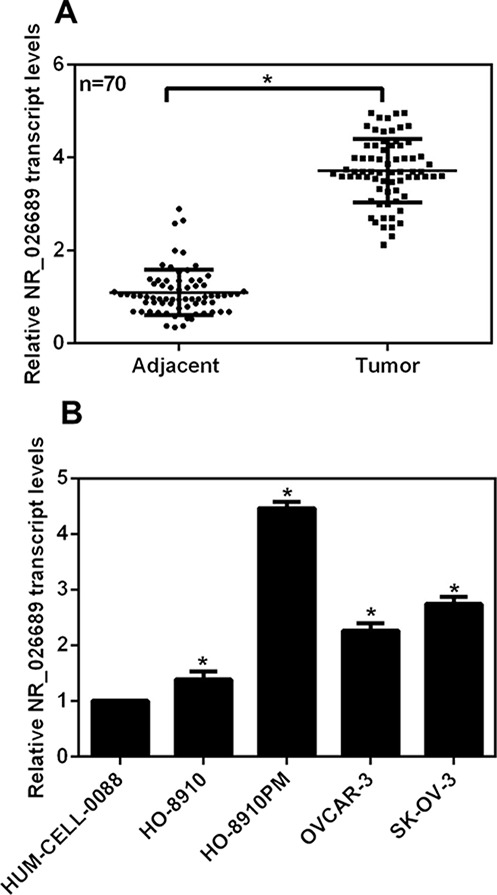
Long noncoding RNA NR_026689 was overexpressed in human ovarian cancers. (A) A total of 70 ovarian cancer patients participated in our study, and their tumor tissues as well as their adjacent noncancerous tissues were obtained and subjected to RT-PCR analysis to detect the expression of long noncoding RNA NR_026689. *p < 0.05, Tumor versus Adjacent. (B) Human normal ovarian cells (HUM-CELL-0088) and four ovarian cancer cell lines (HO-8910, HO-8910PM, OVCAR-3, and SK-OV-3) were included in the RT-PCR assays. The relative transcript level of NR_026689 in each cell line was detected. *p < 0.05 versus HUM-CELL-0088.
Knockdown of NR_026689 Inhibited Cell Proliferation and Colony Formation in Ovarian Cancer Cells
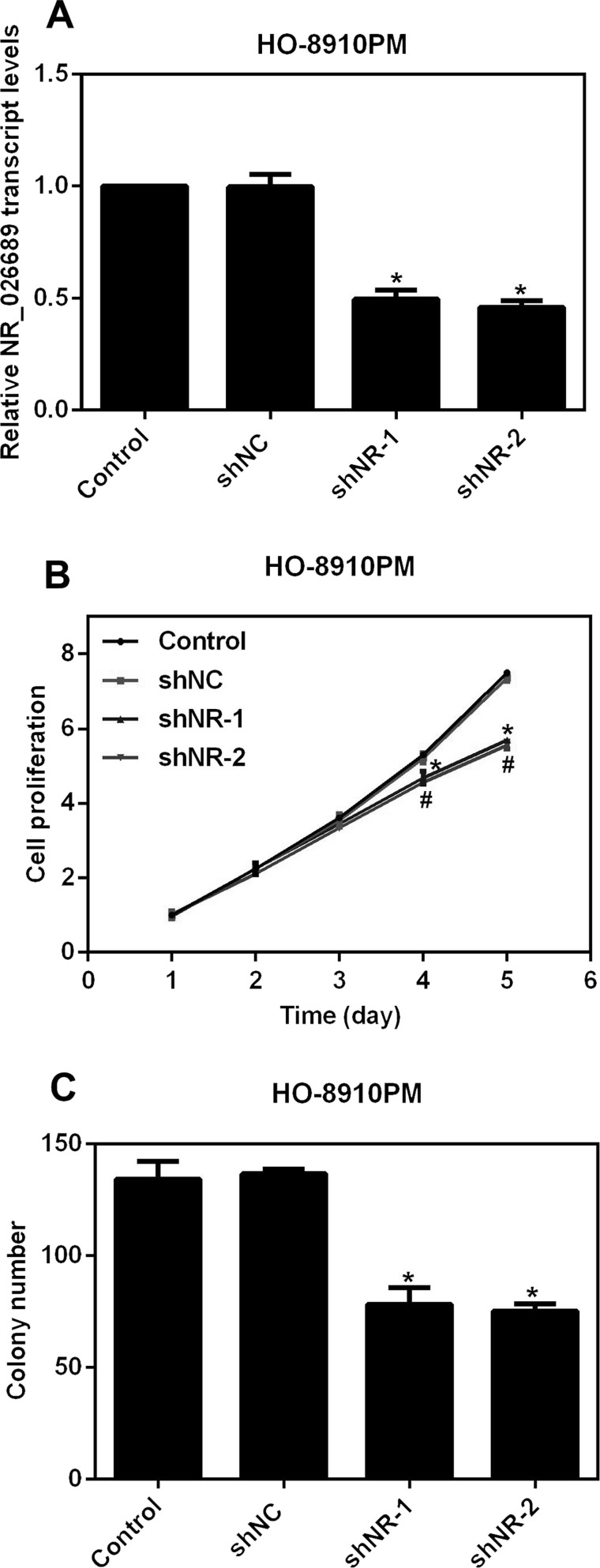
To elucidate the detailed role of NR_026689 in ovarian cancer, two specific shRNAs against NR_026689 were designed and synthesized, shortly named shNR-1 and shNR-2. When HO-8910PM cells were transfected with shRNAs, the relative transcript level of NR_026689 was significantly decreased by 50% and 45%, respectively, in shNR-1-treated cells and shNR-2-treated cells (Fig. 2A), demonstrating the high efficiency of designed shRNAs and transfection processes. Afterward, we explored the role of NR_026689 in cell proliferation. As shown in Figure 2B, control shRNA (shNC) caused no notable difference in cell proliferation in HO-8910PM cells throughout the time period. However, transfection of shNR-1 in HO-8910PM cells inhibited the cell proliferative rate by 12% on the fourth day and 24% on the fifth day, while the inhibitory effects caused by NR-2 were somewhat stronger on the fourth and fifth days, accounting for 14% and 26%, respectively (Fig. 2B). Furthermore, more than 130 colonies were formed in control HO-8910PM cells after a 2-week incubation, whereas only 78 colonies were observed in shNR-1-treated cells, and 75 colonies were counted in shNR-2-infected cells (Fig. 2C). These data revealed that knockdown of NR_026689 in HO-8910PM cells suppressed cell proliferation and colony formation.
Figure 2.
Knockdown of NR_026689 inhibited cell proliferation and colony formation in ovarian cancer cells. (A) Two specific shRNAs against NR_026689 were designed and synthesized and transfected into HO-8910PM cells to knock down the expression of NR_026689 for the subsequent analysis. *p < 0.05 versus Control. (B) Cell proliferative rate was explored in 5 consecutive days upon the transfection of shRNAs in HO-8910PM cells. Knockdown of NR_026689 significantly suppressed cell proliferation on the fourth day and fifth day. *p < 0.05 versus Control by shNR-1. #p < 0.05 versus Control by shNR-2. (C) Colony formation assays were performed upon NR_026689 depletion in HO-8910PM cells. *p < 0.05 versus Control.
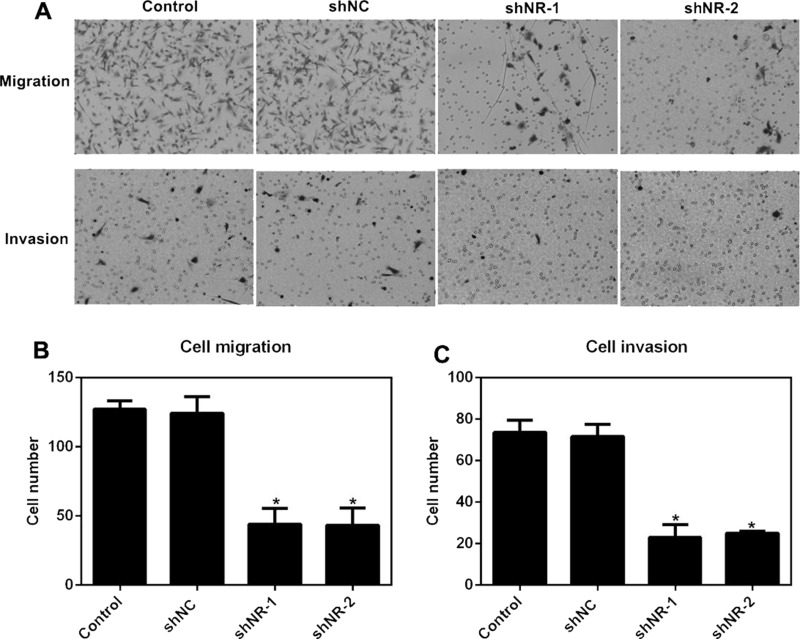
Knockdown of NR_026689 Suppressed Cell Migration and Invasion in HO-8910PM Cells
We hypothesized the association between the high expression of NR_026689 and cell metastasis after the observations in Figure 1B. To this end, we performed Transwell assays. Cells were transfected with shRNAs in the presence or absence of NR_026689 knockdown. It was found that cells that migrated through the membrane and invaded to the lower surface of the chamber were remarkably decreased by either shNR-1 or shNR-2 transfection (Fig. 3A). Quantification of Transwell analysis revealed that the ability of cells to migrate was retarded by 58% by shNR-1 and 59% by shNR-2 in HO-8910PM cells (Fig. 3B). Moreover, the potential of cells to invade was suppressed by more than 70% by both shRNAs against NR_026689 in HO-8910PM cells (Fig. 3C). These results verified our prior assumption that upregulated NR_026689 in ovarian cancer was positively related to cell metastasis.
Figure 3.
Knockdown of NR_026689 suppressed cell migration and invasion in HO-8910PM cells. (A) Representative images of cell migration and cell invasion assays are shown. Five random sights were photographed and quantified after cells were stained with crystal violet. (B) Quantification of cell migration assay revealed that knockdown of NR_026689 in HO-8910PM cells inhibited cell migration abilities. (C) Quantification of cell invasion assay revealed that NR_026689 depletion in HO-8910PM cells suppressed cell-invasive potentials. *p < 0.05 versus Control.
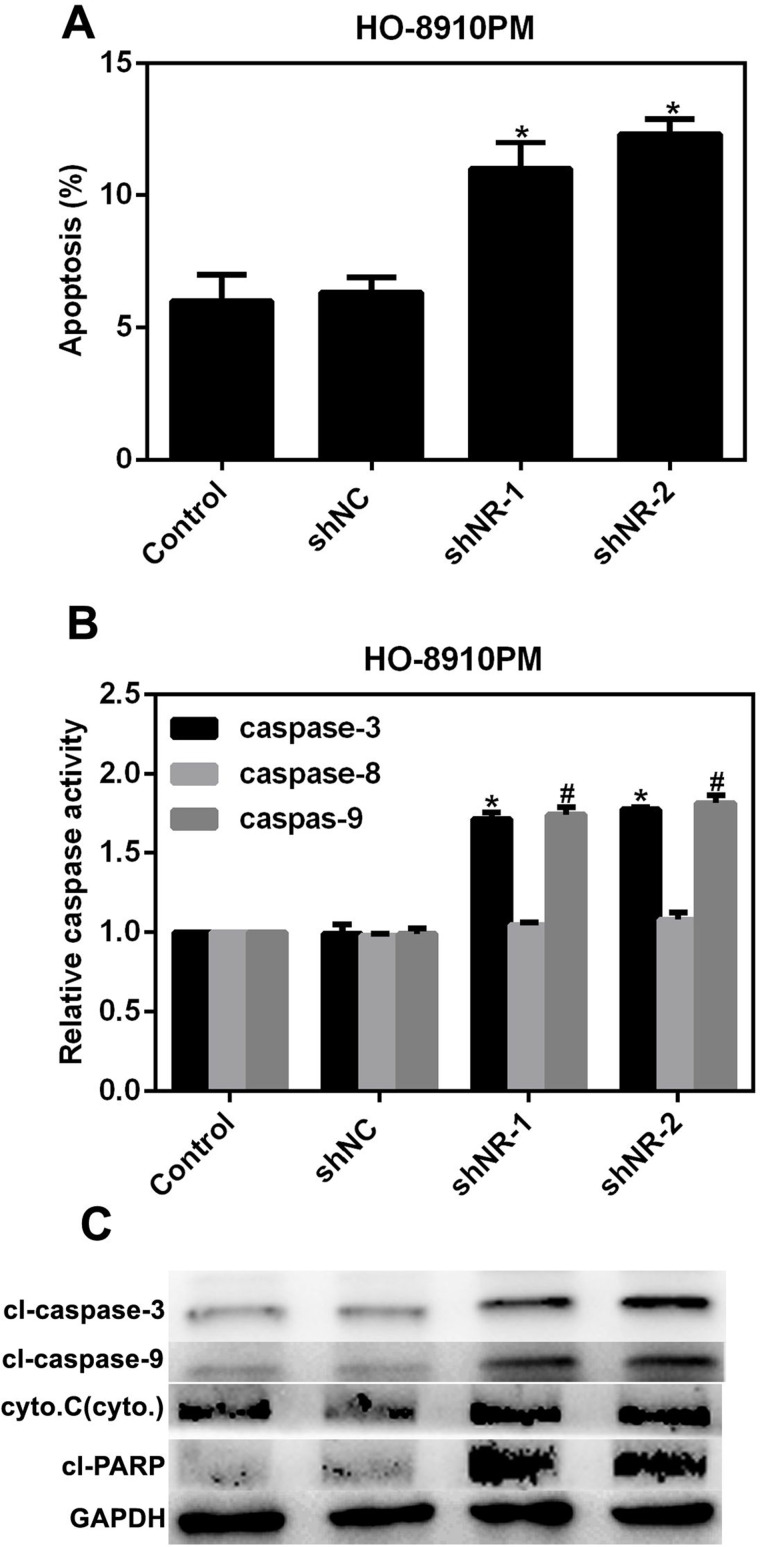
NR_026689 Depletion in HO-8910PM Cells Promoted Cell Apoptosis and the Activities of Related Caspases
Inhibition of cell apoptosis is a good basis for the aggressive activities of cancer cells. Thus, we further explored the effects of depletion of NR_026689 on cell apoptosis in ovarian cancer HO-8910PM cells. As shown in Figure 4A, the apoptotic rate of cells was prominently increased by 5% in shNR-1-treated HO-8910PM cells and by 6% in shNR-2-transfected cells, respectively. Next, we examined the relative activities of the key molecules in cell apoptotic signaling pathways. It was revealed that upon depletion of NR_026689 in HO-8910PM cells, the relative activities of caspase 3 and caspase 9 were remarkably increased by 1.5-hold compared to the control. However, the activity of caspase 8 remained unchanged. These data indicated that NR_026689 might affect cell apoptosis through the intrinsic signaling pathway (Fig. 4B). Furthermore, the protein levels of caspase 3, caspase 9, cyt C, and PARP were significantly increased in shNR-treated HO-8910PM cells (Fig. 4C). These data suggested that depletion of NR_026689 increased cell apoptosis by promoting the activities of key molecules in the intrinsic apoptotic signaling pathway.
Figure 4.
NR_026689 depletion in HO-8910PM cells promoted cell apoptosis and the activities of related caspases. (A) Knockdown of NR_026689 by each shRNA against NR_026689 consistently promoted cell apoptosis in HO-8910 PM cells. *p < 0.05 versus Control. (B) The relative activities of caspase 3, caspase 8, and caspase 9 were determined in HO-8910PM cells upon the transfection of NR_026689 shRNAs. *p < 0.05 versus Control by shNR-1. #p < 0.05 versus Control by shNR-2. (C) Western blot analysis also revealed that the protein levels of caspase 3, caspase 9, cytochrome C, and PARP were upregulated in ovarian cancer HO-8910PM cells treated with NR_026689 shRNAs.
DISCUSSION
Since 1996, the standard regimen after surgery for ovarian cancer has been a systemic platinum/taxane-based method12. Unfortunately, most ovarian cancer patients experience recurrence and require further therapy despite high initial response rates. Thus, it is a high priority to find new clues for the diagnosis and treatment of ovarian cancer. lncRNAs have been reported to play a significant role in the tumorigenesis of many kinds of cancers, including ovarian cancer. As a result, we sought to explore the detailed role of lncRNA NR_026689 in ovarian cancer progression.
NR_026689 is a new lncRNA identified by Wu et al. in 201611. However, in their article, they only elucidated that NR_026689 was overexpressed in rat lung cancer induced by NNK, and the detailed role of NR_026689 in tumor progression was not shown. Cell proliferation and metastasis are two main manifestations of malignant tumors and are studied widely13–15. Therefore, we explored the effects of NR_026689 on ovarian cancer cells by cell proliferation assays, colony formation assays, and Transwell assays. Afterward, we probed the role of NR_026689 in cell apoptosis. We found that NR_026689 was significantly upregulated in ovarian cancer tissues and highly invasive ovarian cancer cells. Depletion of NR_026689 inhibited cell proliferation and metastasis, and promoted cell apoptosis in HO-8910PM cells. These observations show conclusively that NR_026689 exerts an oncogenic potential in ovarian cancer.
Cell apoptosis is a process of programmed cell death that occurs in multicellular organisms and various organs. There are two signaling pathways involved in the apoptotic process: intrinsic pathway and extrinsic pathway, depending on the apoptotic signal origin. The intrinsic pathway, namely, the mitochondrial pathway, is activated by intracellular signals generated when cells are stressed and depends on the release of proteins from the intermembrane space of the mitochondria16,17. cyt C is also released from the mitochondria in the intrinsic pathway and serves a regulatory function as it precedes morphological change associated with apoptosis18. Once cyt C is released, it can form a complex with apoptotic protease-activating factor 1 (Apaf-1) and ATP19,20, which binds to procaspase 9 to create a new protein complex, named apoptosome. The apoptosome cleaves the procaspase to its active form of caspase 9, which in turn activates the effector caspase 321. However, caspase 8 is a typical indicator of extrinsic pathway22. In our study, we demonstrated the upregulated expression of intrinsic pathway proteins including caspase 3, caspase 9, and cyt C and that the relative activity of caspase 8 remained stable upon NR_026689 depletion in HO-8910PM cells, indicating that NR_026689 might promote cell apoptosis via the intrinsic pathway.
Our study is the first that elucidated the detailed role of the newly identified lncRNA NR_026689 in ovarian cancer. Knockdown of NR_026689 in ovarian cancer cells inhibited cell proliferation and metastasis and also promoted cell apoptosis via the intrinsic pathway. This oncogenic potential of NR_026689 might pave the way for novel clues to the diagnosis and treatment of ovarian cancer in the clinic.
ACKNOWLEDGMENTS
This study is sponsored by the Natural Science Foundation of Shanxi Province. Xin Zhang designed the study and conducted most of the experiments; Shaowen Li and Chunli Dong contributed to the clinical tissue collection; and Xiuying Xie and Yunping Zhang helped with the cell proliferation assays.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA CancerJ Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2. Salani R, Backes FJ, Fung MF, Holschneider CH, Parker LP, Bristow RE, Goff BA. Posttreatment surveillance and diagnosis of recurrence in women with gynecologic malignancies: Society of Gynecologic Oncologists recommendations. Am J Obstet Gynecol. 2011;204(6):466–78. [DOI] [PubMed] [Google Scholar]
- 3. McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med. 1996;334(1):1–6. [DOI] [PubMed] [Google Scholar]
- 4. Meirelles K, Benedict LA, Dombkowski D, Pepin D, Preffer FI, Teixeira J, Tanwar PS, Young RH, MacLaughlin DT, Donahoe PK, Wei X. Human ovarian cancer stem/progenitor cells are stimulated by doxorubicin but inhibited by Mullerian inhibiting substance. Proc Natl Acad Sci USA 2012;109(7):2358–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor DD, Gercel-Taylor C, Parker LP. Patient-derived tumor-reactive antibodies as diagnostic markers for ovarian cancer. Gynecol Oncol. 2009;115(1):112–20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 6. Buckanovich RJ, Sasaroli D, O’Brien-Jenkins A, Botbyl J, Hammond R, Katsaros D, Sandaltzopoulos R, Liotta LA, Gimotty PA, Coukos G. Tumor vascular proteins as biomarkers in ovarian cancer. J Clin Oncol. 2007;25(7):852–61. [DOI] [PubMed] [Google Scholar]
- 7. Huang T, Alvarez A, Hu B, Cheng SY. Noncoding RNAs in cancer and cancer stem cells. Chin J Cancer 2013;32(11):582–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Y, Xu X, Lv H, Wen Q, Li J, Tan L, Li J, Sheng X. The long noncoding RNA MALAT-1 is highly expressed in ovarian cancer and induces cell growth and migration. PLoS One 2016;11(5):e155250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qiu JJ, Wang Y, Liu YL, Zhang Y, Ding JX, Hua KQ. The long non-coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer. Oncotarget 2016;7(22):32487–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu J, Li X, Xu Y, Yang T, Yang Q, Yang C, Jiang Y. Identification of a long non-coding RNA NR_026689 associated with lung carcinogenesis induced by NNK. Oncotarget 2016;7(12):14486–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med. 1996;334(1):1–6. [DOI] [PubMed] [Google Scholar]
- 13. Huang W, Li J, Guo X, Zhao Y, Yuan X. miR-663a inhibits hepatocellular carcinoma cell proliferation and invasion by targeting HMGA2. Biomed Pharmacother. 2016;81:431–8. [DOI] [PubMed] [Google Scholar]
- 14. Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget 2016;7(27):42183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lei X, Xu JF, Chang RM, Fang F, Zuo CH, Yang LY. JARID2 promotes invasion and metastasis of hepatocellular carcinoma by facilitating epithelial-mesenchymal transition through PTEN/AKT signaling. Oncotarget 2016;7(26):40266–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science 1995;267(5203):1456–62. [DOI] [PubMed] [Google Scholar]
- 17. Kumar S. Regulation of caspase activation in apoptosis: Implications in pathogenesis and treatment of disease. Clin Exp Pharmacol Physiol. 1999;26(4):295–303. [DOI] [PubMed] [Google Scholar]
- 18. Dejean LM, Martinez-Caballero S, Kinnally KW. Is MAC the knife that cuts cytochrome c from mitochondria during apoptosis? Cell Death Differ. 2006;13(8):1387–95. [DOI] [PubMed] [Google Scholar]
- 19. Kinnally KW, Martinez-Caballero S, Dejean LM. Detection of the mitochondrial apoptosis-induced channel (MAC) and its regulation by Bcl-2 family proteins. Curr Protoc Toxicol. 2006;Chapter 2:Unit2.12. [DOI] [PubMed] [Google Scholar]
- 20. Dejean LM, Martinez-Caballero S, Manon S, Kinnally KW. Regulation of the mitochondrial apoptosis-induced channel, MAC, by BCL-2 family proteins. Biochim Biophys Acta 2006;1762(2):191–201. [DOI] [PubMed] [Google Scholar]
- 21. Murphy KM, Ranganathan V, Farnsworth ML, Kavallaris M, Lock RB. Bcl-2 inhibits Bax translocation from cytosol to mitochondria during drug-induced apoptosis of human tumor cells. Cell Death Differ. 2000;7(1):102–11. [DOI] [PubMed] [Google Scholar]
- 22. Chen W, Li N, Chen T, Han Y, Li C, Wang Y, He W, Zhang L, Wan T, Cao X. The lysosome-associated apoptosis-inducing protein containing the pleckstrin homology (PH) and FYVE domains (LAPF), representative of a novel family of PH and FYVE domain-containing proteins, induces caspase-independent apoptosis via the lysosomal-mitochondrial pathway. J Biol Chem. 2005;280(49):40985–95. [DOI] [PubMed] [Google Scholar]