Abstract
Circular RNA (circRNA) is a widely expressed non-coding RNA element characterized by a covalently closed continuous loop. Emerging evidence suggests important roles of circRNAs in the pathogenesis of human cancers. However, the functions and underlying mechanisms of circRNAs in glioma remain largely unclear. Previously, our studies uncovered a batch of abnormally expressed circRNAs in glioma tissue, among which circPARP4 was significantly upregulated with the top fold change. Here, we focused on the functional investigation toward circPARP4 in glioblastoma progression and looked for insight into its underlying mechanisms. The results confirmed the elevated expression of circPARP4 in glioma and found its association with glioma pathological grade. Gain- and loss-of-function strategies showed that circPARP4 could obviously promote glioma cell proliferation, migration, invasion, and epithelial-mesenchymal transition. Mechanistically, in vivo and in vitro studies demonstrated that circPARP4, as a miRNA sponge, directly interacted with miR-125a-5p, which then regulated FUT4 to exert the oncogenic effect on glioma behavior. Our findings illustrate functions of circPARP4 in modulating glioma progression through miR-125a-5p/FUT4 pathway, which provides a novel and potential target for glioma therapy.
Keywords: Glioma, circular RNA, circPARP4, miRNA-125a-5p, FUT4
Introduction
Gliomas represent the most widespread primary form of central nervous system tumors and account for approximately 80% of intracranial malignancies, more than half of which is the most malignant histologic subtype glioblastoma (GBM) [1-3]. Despite the aggressive treatments, including maximum surgical resection followed by radiotherapy and temozolomide chemotherapy, patients with GBM have a median survival of only 14-16 months [4,5]. However, additional therapies, such as personalized therapies against molecular targets, have so far also been unsuccessful in clinical trials. Therefore, identifying critical treatment targets underlying tumor progression for glioma is of paramount importance.
Circular RNA (circRNA) is a widely expressed non-coding RNA that is generated by back-splicing and characterized by a covalently closed loop structure, which enables its stable existing [6,7]. Previous studies have demonstrated that circRNA can act as a competing endogenous RNA (ceRNA) that mainly sponges miRNA or binds proteins to regulate cellular biological processes [8]. Emerging evidence indicates the functional roles of circRNA in tumorigenesis and disease progression. For instance, circSLC8A1 suppressed bladder cancer progression through sponging miR-130b/miR-494 via regulating PTEN [9], circTADA2A could readily sponge miR-203a-3p to upregulate the expression of CREB3 that facilitated osteosarcoma progression and metastasis [10], and circAKT3 upregulated PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression [11]. Moreover, emerging studies have also been focusing the functions of circRNAs and their contributing pathways in glioma. Our previous work revealed a total of 417 abnormally expressed circRNAs in GBM tissue samples and hsa_circ_0005198 (circPARP4) was upregulated with the top fold change [12]. Then, the role of circPARP4 in the development and progression of glioma still needs to be determined.
In this study, we confirmed elevated expression of circPARP4 in glioma, which was positively associated with tumor pathological grade. FISH analysis showed circPARP4 was expressed in cytoplasm, and gain- and loss-function studies found circPARP4 affected the proliferation, migration, and invasion abilities of glioma cells in vitro and in vivo. Then, miRNA microarray combined with RNA immunoprecipitation (RIP) assay were performed and discovered circPARP4 as a sponge to negatively control miR-125a-5p. Moreover, it is revealed that FUT4 mediated the oncogenic function of circPARP4/miR-125a-5p axis by using a bioinformatics method. Our findings provide an insight into the complicated regulatory network of non-coding RNAs in glioma progression and a potential target for developing therapeutic strategies.
Materials and methods
Clinic samples
The present study recruited patients who joined the organization between 2017 and 2019. In total, 36 high-grade gliomas (WHO grade III-IV) and 40 low-grade gliomas (WHO grade I-II) were collected and pathologically confirmed. Meanwhile, 10 non-tumor tissue specimens were obtained from severe brain trauma patients who required decompression treatment. The gliomas were graded according to the WHO pathological diagnostic standard [13]. No patients had received radiotherapy or chemotherapy prior to tumor resection. The clinicopathological features of the included patients are detailed in Table 1. Samples were divided and either frozen in liquid nitrogen and stored at -80°C or kept in RNAlater (Ambion, Austin, TX, USA) at -20°C. Ethical approval for the study was provided by the Independent Ethics Committee of Wuxi Clinical College of Anhui Medical University. Informed and written consents were supplied by all patients or their advisors according to Ethics Committee guidelines.
Table 1.
Relationships between circPARP4 expression in human glioma tissues and clinicopathological features
| Clinicopathological features | No. of cases | circPARP4 (n, %) | p Value | |
|---|---|---|---|---|
|
| ||||
| High | Low | |||
| Gender | ||||
| Male | 35 | 18 | 17 | >0.05 |
| Female | 41 | 17 | 24 | |
| Age | ||||
| <45 | 31 | 17 | 14 | >0.05 |
| ≥45 | 45 | 21 | 23 | |
| WHO Grade | ||||
| I | 17 | 2 | 15 | <0.05 |
| II | 23 | 4 | 19 | |
| III | 21 | 15 | 6 | |
| IV | 15 | 11 | 4 | |
WHO = World Health Organization.
Cell culture
Human malignant glioma cell lines U87, U138, U251 and SHG-44 were purchased from the Cell Bank at the Chinese Academy of Sciences (Shanghai, China), and HEB cells (human astrocyte) were stored in our laboratory. They were all cultured in DMEM containing penicillin G (100 U/mL), 8% FBS, as well as streptomycin (100 mg/mL), and maintained at 37°C in monolayer culture with 5% carbon dioxide humidified air. Subsequently, cell viability was determined by trypan blue staining.
Cell transfection
Glioma cells were transfected with appropriate amount of vector by using Lipofectamine 2000 (Invitrogen, MA, U.S.) and then cultured for 48 hours according to the manufacturer’s protocol.
Vector construction
The full-length cDNA of circPARP4 (Supplementary Table 1) was synthesised by GeneChem (Shanghai, China) and then cloned into the circRNA vector (GV535, purchased from GeneChem). Three siRNAs against circPARP4, shown in Supplementary Table 1, were also synthesized by GeneChem (Shanghai, China). The expression efficiency was examined using qPCR in glioma cells transfected with vector or siRNA. As to potently knock down circPARP4, si-circ-1 was selected for further research. MiR-125a-5p mimc, inhibitor and control (miR-NC) were all created by Hanbio (Shanghai, China), which are presented in Supplementary Table 1. And the FUT4 overexpression vector and control vector were synthesized and purchased from GeneChem (Shanghai, China).
Quantitative RT-PCR
Total RNA was isolated from glioma cells using TRIzol reagent (Invitrogen). Subsequently, cDNA was prepared with M-MLV kit (Promega, Madison, USA). Quantitative RT-PCR was performed in triplicates using SYBR Green Master Mix (TaKaRa, Tokyo, Japan) on 7500 Fast Real-Time PCR system (Applied Biosystems, Life Technology, Foster City, USA). β-actin and small nuclear RNA U6 was used as internal controls of mRNA and miRNA, respectively. The expression levels were calculated using the 2ΔΔCT method. The primers used as follows: circPARP4 forward primer, CTTGGAGAAAGTGGGAATGG and reverse primer, TTCTTCTGCTGCTGAGGTAAG; miR-125a-5p forward primer, ACACTCCAGCTGGGTCCCTGAGACCCTTTAAC and reverse primer, GTGCAGGGTCCGAGGT; FUT4 forward primer, TCCTACGGAGAGGCTCAG and reverse primer, TCCTCGTAGTCCAACACG; β-actin forward primer, GGCGGCACCACCATGTACCCT and reverse primer, AGGGGCCGGACTCGTCATACT; U6 forward primer, CTCGCTTCGGCAGCACA and reverse primer, AACGCTTCACGAATTTGCGT.
Fluorescence in situ hybridization (FISH)
In situ hybridization was carried out using cy5 labeled probes complementary to the circPARP4 sequence as previously described [14]. Cell nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI). All procedures were conducted according to the manufacturer’s protocol (Genepharma, Shanghai, China).
CircRNA target prediction
Prediction of the circPARP4-miRNA-target gene was performed using the STARBASE website (http://starbase.sysu.edu.cn/) [15].
Luciferase reporter assay
Transfection was accomplished by using Lipofectamine 2000 (Invitrogen). Glioma cells were co-transfected with plasmids harboring the 3’-UTR of wildtype or mutant fragments from FUT4, or predicted binding sequence from circPARP4 and miRNA mimics. Firefly and Renilla luciferase activities were measured consecutively 48 hours after transfection by using a dual-luciferase reporter assay system (Promega, Fitchburg, WI, USA). Each assay was performed independently in triplicate.
Library preparation for small RNA sequencing
A total amount of 3 μg total RNA from each sample was used as input material for the small RNA library. The NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, USA) was utilized to process the sequencing libraries as per the instructions provided by the manufacturer. And index codes were added for sample identification. In brief, 3’ end of the NEB SR Adaptor was precisely ligated to the 3’ end of siRNA, miRNA, as well as piRNA directly. Next, the SR RT Primer hybridized to the excess 3’ SR Adaptor were used to generate a double-stranded DNA from a single-stranded (ss) DNA adaptor. This step prevented adaptor-dimer formation and ligation of ssDNA to 5’ SR Adaptor in the next step of ligation. Then, 5’ ends adapter was ligated to 5’ ends of miRNAs, siRNA and piRNA. To generate the first strand cDNA, the M-MuLV Reverse Transcriptase (RNase H) was employed to perform reverse transcription. Thereafter, PCR amplification was performed using LongAmp Taq 2X Master Mix, SR Primer for illumina and index (X) primer. Next, the PCR products were purified by running them on a polyacrylamide gel (8%, 100 V) for 1 hour 20 minutes. The DNA fragments that were 140~160 bp long (the size of small ncRNA with the 3’ and 5’ adaptors) were recovered and then dissolved in 8 μL elution buffer. Finally, library quality was assessed on the Agilent Bioanalyzer 2100 system using DNA High Sensitivity Chips.
Clustering and sequencing
Clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq SR Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s protocol. Subsequently, the library preparations were sequenced on an Illumina Hiseq 2500/2000 platform and 50 bp single-end reads were generated.
Known miRNA alignment
Mapped small RNA tags were used to identifying known miRNAs. miRBase20.0 served as a reference, and modified software mirdeep2 and srna-tools-cli were used to obtain the potential miRNA and draw the secondary structures. Custom scripts were used to determine the miRNA counts as well as base bias on the first position of identified miRNA with certain length and on each position of all the uncovered miRNAs respectively.
Western blot
Total proteins were isolated from lysed cells (50 μg) and separated by 10% SDS-PAGE, then transferred to nitrocellulose membranes. Blocking was done for 2 hours, subsequently, overnight incubation of the membranes with primary antibodies was performed, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies. Imaging of bands was carried out by the ECL Plus Detection Reagent (Applygen, Beijing, China).
Cell proliferation assay
Cell counting kit-8 (CCK8) (Solarbio, Beijing, China) was employed to evaluate the proliferative ability of the cells following the instructions of the manufacturer. In brief, glioma cells (1 × 104) were seeded into 96-well plates and incubated overnight. Subsequently, cells were washed three times with PBS after removing the medium. DMEM (90 µl) and CCK8 (10 µl) were added to each well. Incubation of the mixture was done at 37°C for 1 hour and 30 minutes. A microplate reader (EL × 800, BIO-TEK, Winooski, USA) was used to measure optical density at 450 nm.
Cell migration and invasion assays
The migration and invasion abilities of glioma cells were analyzed using transwell plates (Millipore, Billerica, MA, USA). Briefly, glioma cells were seeded in uncoated (migration assays) or Matrigel-coated (invasion assays) plates with a diameter of 8 μm (BD Bioscience, Bedford, MA, USA). The top chamber contained 2 × 104 cells/well in serum-free medium, while FBS with 10% serum was loaded into the lower chamber. For invasion assays, Matrigel-coated chambers were used. The plates were incubated for 24 hours, after which the cells that remained on the top filter surface were removed using a cotton swab, whereas those that migrated to the lower chamber were counted using an optical inverted microscope (Nikon, Tokyo, Japan).
RNA immunoprecipitation (RIP)
According to the manufacturer’s protocol, RNA immunoprecipitation was performed in glioma cells 48 hours after transfection with either the miR-125a-5p overexpression or miR-NC vectors using the Magna RIPTM RNA Binding Protein Immunoprecipitation Kit (Millipore). A total of 1 × 107 cells were lysed in RNA lysis buffer. The cell lysate was introduced to a magnetic beads solution conjugated to either human anti-Argonaute 2 (AGO2) antibodies (Millipore) or control mouse IgG molecules (Millipore). The RIP immunoprecipitation buffer was incubated together, while the samples were incubated with proteinase K (Gibco, Grand Island, NY, USA). Following incubation, IP RNA was extracted from the samples and analyzed using RT-PCR to assess circPARP4 enrichment.
Immunohistochemistry
Immunohistochemical staining was performed using a method described previously [20]. Briefly, thawed samples were fixed in 4% formalin and embedded in paraffin for histopathological analysis. Samples were deparaffinized with xylol and then sliced into 4 µm sections. Sections were rehydrated using a graded ethanol series. A heat-induced epitope protocol was used for antigen-retrieval (95°C for 40 min). Samples were incubated in methanol containing 0.3% hydrogen peroxide to block endogenous peroxidase. Samples were blocked with protein serum (Vectastain Elite ABC kit; Vector Laboratories, Inc., Burlingame, CA, USA) and then incubated overnight at 4°C with polyclonal rabbit anti-human FUT4 or Ki67 antibody at 1:1,000 (MBL International Corporation, Nagoya, Japan). After washing three times in TBST (150 mM NaCl, 10 mM Tris-HCl, pH 7.6), sections were incubated with a secondary antibody for 20 min at room temperature. Peroxidase-conjugated biotin-streptavidin complex (Dako, Glostrup, Denmark) was then applied to the sections for 20 min. Sections were visualized with 3, 3’-diaminobenzidine and counterstained with hematoxylin. Nonimmune serum instead of a primary antibody was employed as the negative control.
Tumor xenograft model
BALB/c nude mice (n = 5) were acquired from the Beijing Vital River Animal Company (Beijing, China). U87 and U251 cells transfected with circPARP4 siRNA or miR-125a-5p inhibitor were collected and inoculated subcutaneously into the right flank regions of the mice. A Vernier caliper was used to quantify the width of the tumors on a weekly basis. To determine the tumor volume, the following equation was used: tumor volume = (length × width2)/2. Mice were euthanized 5 weeks post-inoculation, and then xenograft tumors were excised. All experiments involving mice were conducted in accordance with the Institutional Guidelines for the Care and Use of Laboratory Animals of Wuxi Clinical College at the Anhui Medical University.
TUNEL assay
For quantificational analysis of apoptosis, U87 and U251 cells in xenograft were examined in situ using a TUNEL assay via the Apoptosis Detection Kit (POD, Roche, Switzerland) according to the manufacturer’s instructions. Samples of 3 μm thickness were deparaffinized, rehydrated with xylene and ethanol, and permeabilized with 20 μg/ml proteinase K (Gibco). Covering the sample with 3% H2O2 inactivated endogenous peroxidase. The sections were rinsed with PBS, then immersed in TdT buffer for 60 minutes at 37°C, incubated with anti-digoxigenin peroxidase conjugate for 30 minutes, followed by treatment of the peroxidase substrate. Finally, counterstaining the slices with 0.5% (wt/vol) methyl green was performed.
Statistical analysis
The SPSS 22.0 software (IBM, SPSS, Chicago, IL, USA) was conducted for statistical analyses. The differences of measurement data between groups were analyzed using one-way ANOVA. Chi-square test was applied to analyze the correlations between circPARP4 expression and clinicopathological characterization of GBM patients. Linear correlation analysis was used to evaluate mathematical relationship of two variables. P<0.05 was considered statistically significant in all tests.
Results
Expression of circPARP4 is upregulated in glioma and associated with pathological grade
Our previous research using circRNA microarray identified a total of 417 abnormally expressed circRNAs in GBM tissues compared to matched adjacent normal brain samples [12]. CircPARP4 remains the top fold-change upregulated circRNA, which is located in chr13:25072253-25077915 (Figure 1A) and derived from two PARP4 gene exons. To further validate the expression of circPARP4 in glioma, we collected 36 high-grade gliomas, 40 low-grade gliomas and 10 non-tumor tissue samples to detect its level by RT-PCR. Results revealed a significant elevation of circPARP4 in glioma as comparing to the control (Figure 1B). Moreover, the level of circPARP4 expression positively correlated with the pathological grade of glioma (Table 1), indicating a contributing role of circPARP4 in tumorigenesis and glioma progression. We then detected circPARP4 in four glioma cell lines and found its higher expression in SHG-44, U87, U251 and U138 compared to that in HEB cells (Figure 1C). U87 and U251 were selected to study the functions and mechanisms of circPARP4 in further experiments.
Figure 1.
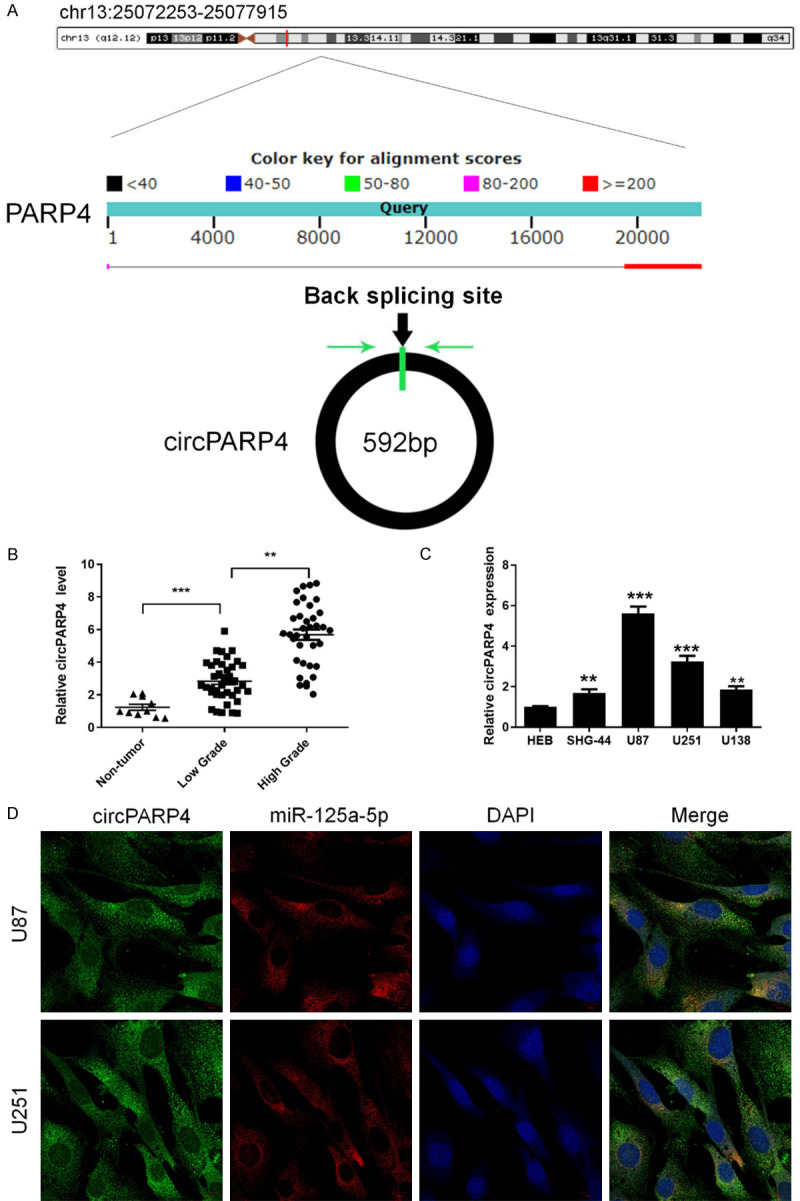
circPARP4 is upregulated in glioma and located in the cytoplasm. A. The loci of the PARP4 gene and circPARP4 element are shown. Green arrows indicate the back-splicing site. B. The circPARP4 levels were detected by RT-PCR in low- and high-grade glioma samples and control brain tissue. C. Expression of circPARP4 in glioma cell lines and HEB cells (n = 3) was analyzed by RT-PCR. D. FISH assay determined the subcellular localization of circPARP4 and miR-125a-5p. Scale bar, 10 μm. Data indicate the mean ± SD, n = 3. *P<0.05, **P<0.01, ***P<0.001 vs. control.
circPARP4 is located in cytoplasm and promotes glioma cell proliferation, migration, invasion and EMT
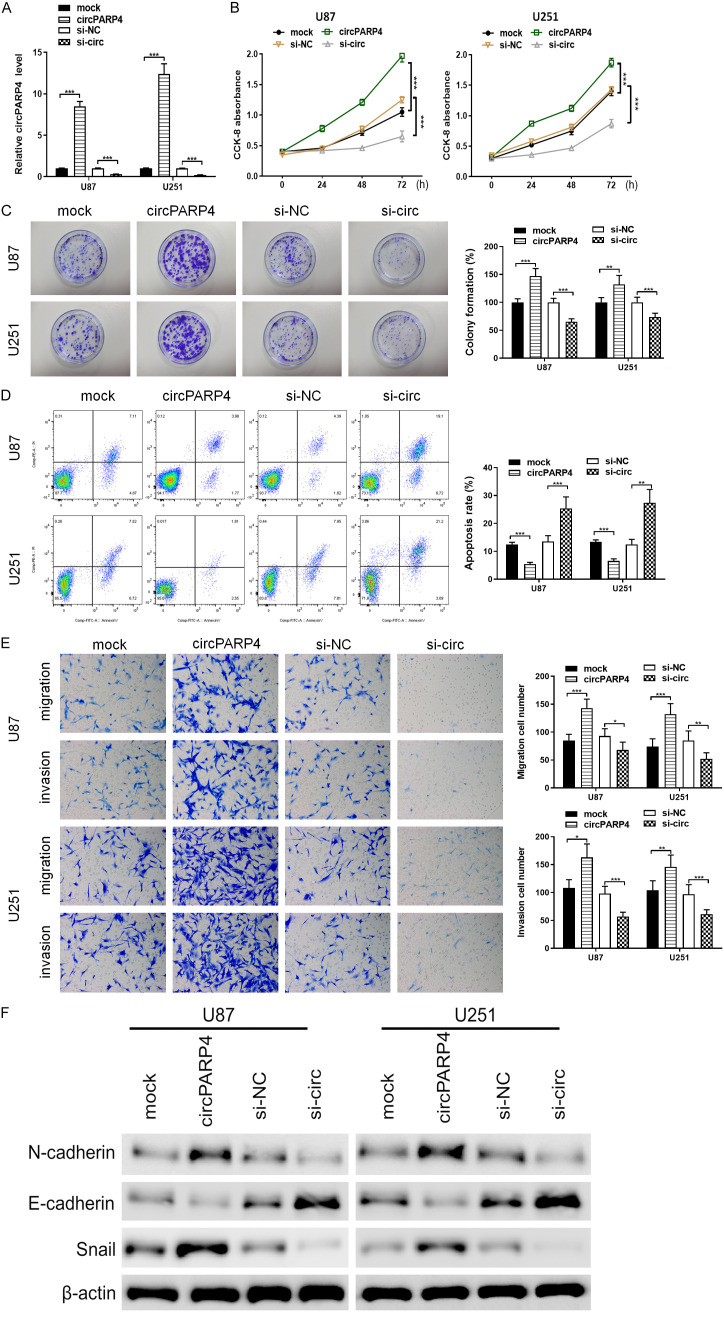
To investigate the functions of circPARP4 in glioma cells, we constructed siRNA against circPARP4 (si-circ) and circPARP4-overexpressing vectors, and stable transfection was established. qRT-PCR assays revealed that si-circ vector significantly downregulated circPARP4 expression, while circPARP4 was increased in the presence of the circPARP4-overexpressing vector (Figure 2A). CCK8 assay found that circPARP4 could promote proliferation of U87 and U251 cells, which would be inhibited by the silencing of circPARP4 (Figure 2B). Cell apoptotic and colony formation abilities were then examined. The results indicated a delay in cell colony formation but increased apoptosis after knocking down circPARP4 compared with the controls, while opposite results occurred when overexpressing circPARP4 (Figure 2C and 2D). Transwell assay was performed to analyze whether circPARP4 affect glioma cells phenotype by altering the cell migration and invasion. As shown in Figure 2E, the number of migrated or invasive U87 and U251 cells in circPARP4 overexpression group was more than that in control group, while downregulating circPARP4 decreased their migration and invasion capacity (Figure 2E). To explore the relationship between circPARP4 and epithelial-mesenchymal transition (EMT), the expression of EMT-related proteins (N-cadherin, E-cadherin and Snail) were further examined by western blot. Consequently, upregulated N-cadherin and Snail expression, but downregulated E-cadherin were seen in circPARP4-overexpressing glioma cells, and silencing circPARP4 inhibited N-cadherin and Snail and increased E-cadherin expression (Figure 2F). These findings revealed that circPARP4 has the ability to promote cell proliferation, colony formation, migration, invasion and EMT in vitro.
Figure 2.
The phenotypes of glioma cell are regulated by circPARP4 in vitro. A. RT-PCR was used to examine the expression efficiency of circPARP4 in U87 and U251 cells with circPARP4 overexpression or knockdown. beta-actin was used as the control. B. The proliferation of glioma cells was assessed by CCK-8 at different time intervals. C. Colony number was assessed by colony forming assays and quantified at 72 h. D. Flow cytometry detected the changes of apoptosis in glioma cells under different conditions. E. Glioma cell migration and invasion were assessed and quantified by transwell assays. F. The expression of EMT markers was determined by western blot. Data indicate the mean ± SD, n = 3. **P<0.01, ***P<0.001 vs. control.
circPARP4 functions as a miR-125a-5p sponge in glioma
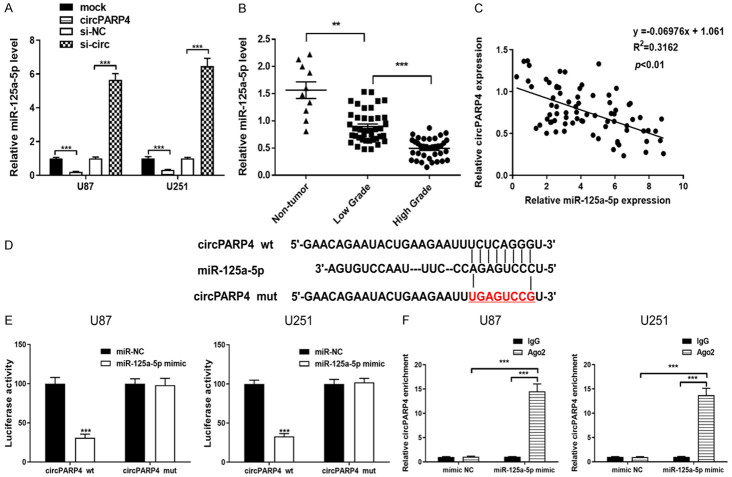
FISH analysis revealed that circPARP4 predominately localized to the cytoplasm (Figure 1D), which suggested that circPARP4 might function as a miRNA sponge to regulate gene expression. To study the downstream pathways of circPARP4, small RNA sequencing was performed comparing si-circ to si-NC U251 cells, as the most upregulated miRNAs were shown in Table 2. Among these miRNAs, miR-125a-5p was selected as a candidate due to it has a binding site for circPARP4 (Figure 3D) through bioinformatics prediction (STARBASE, version 2.0). Moreover, co-localization of circPARP4 and miR-125a-5p in the cytoplasm further supported the interaction between the two molecules (Figure 1D). As determined by RT-PCR, the expression of miR-125a-5p was significant increased after knocking down circPARP4, but decreased in circPARP4-overexpressing U87 and U251 cells (Figure 3A). Then, an inverse correlation between circPARP4 and miR-125a-5p expression was also found in glioma tissue (Figure 3C). Further analysis revealed that miR-125a-5p expression was associated with the pathological grade as the highest level in non-tumor tissues and the lowest one in high-grade glioma samples (Figure 3B). To validate the miRNA binding to circPARP4, we constructed a wild-type or mutant circPARP4 expression vector containing a luciferase gene. After co-transfection of the miR-125a-5p mimics with the wild-type reporter vector, decreased luciferase activity was observed, but not in the miR-125a-5p mimics and mutated vector co-transfection group (Figure 3E). Given that mRNA translation is suppressed by miRNAs in an AGO2-dependent manner, we performed an anti-AGO2 immunoprecipitation (RIP) assay, in which miR-125a-5p overexpression was used to pull down circRNAs by anti-AGO2 antibody or control IgG. RT-PCR revealed an enrichment of circPARP4 in glioma cells overexpressing miR-125a-5p compared to the mimic NC control (Figure 3F). These results suggest that circPARP4 functions as a sponge for miR-125a-5p.
Table 2.
Upregulated miRNA in the si-circ U251 cells compared to si-NC U251 cells
| miRNAs | Fold-change | p value |
|---|---|---|
| hsa-miR-516b-5p | 23.67840504 | 0.00087853 |
| hsa-miR-146b-5p | 8.476683963 | 1.28E-89 |
| hsa-miR-125a-5p | 5.020739942 | 1.36E-124 |
| hsa-miR-335-5p | 4.835952252 | 0.00043182 |
| hsa-miR-328-3p | 4.667661693 | 0.00000101 |
| hsa-miR-99a-5p | 4.518362437 | 0 |
| hsa-miR-132-5p | 4.311829107 | 2.19E-14 |
| hsa-miR-125b-5p | 4.227779105 | 0 |
| hsa-miR-26a-5p | 4.156300602 | 0 |
| hsa-miR-181a-5p | 3.974297855 | 1.3E-196 |
| hsa-let-7c-5p | 3.731873283 | 2.55E-190 |
| hsa-miR-3681-5p | 3.710465004 | 2.37E-13 |
| hsa-miR-3615 | 3.624042816 | 0.0000226 |
| hsa-miR-145-5p | 3.597263897 | 0.00013819 |
| hsa-miR-342-3p | 3.595518917 | 0.00000657 |
Figure 3.
circPARP4 can function as a miRNA sponge to negatively regulate miR-125a-5p in glioma cells. A. The expression of miR-125a-5p was determined by RT-PCR in circPARP4 overexpressing or inhibiting glioma cells. B. The level of miR-125a-5p level was detected in low- and high-grade glioma samples and compared to that in control. C. The correlation between miR-125a-5p and circPARP4 expression in glioma tissue was determined by Spearman’s correlation analysis. D, E. Wild-type (wt) or mutant (mut) circPARP4 vector was transfected into the glioma cell lines with or without synthetic miR-125a-5p mimics, and the corresponding relative luciferase activity was detected by luciferase assays. F. Anti-AGO2 RIP was performed in U87 and U251 cells transfected with miR-125a-5p mimics or miR-NC to detect circPARP4 expression. Data indicate the mean ± SD, n = 3. **P<0.01, ***P<0.001 vs. control.
circPARP4 exerts oncogenic functions via downregulating miR-125a-5p in glioma cells
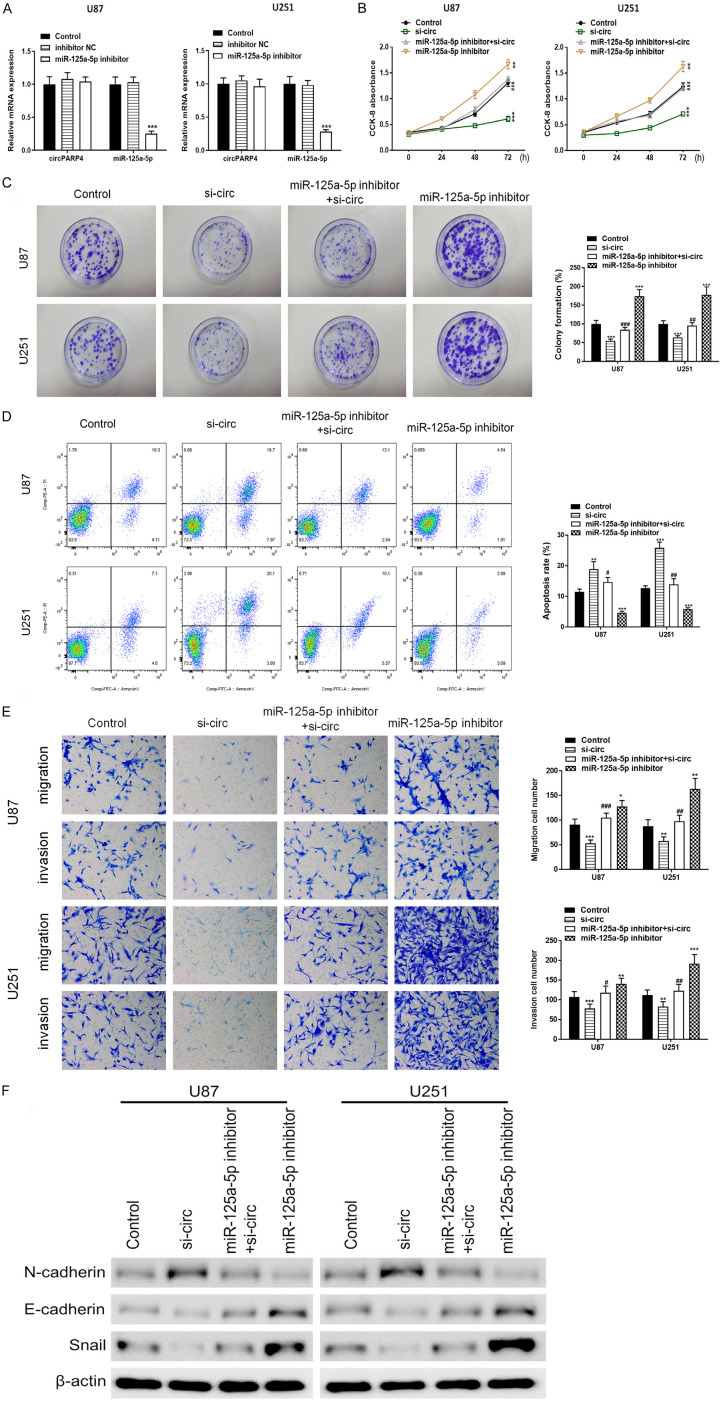
To further explore the functional role of miR-125a-5p underlying circPARP4 in glioma cells, effects of miR-125a-5p on the abilities of cell proliferation, colony formation, apoptosis, migration, invasion and EMT were also examined. The results indicated that miR-125a-5p downregulation didn’t change circPARP4 expression (Figure 4A), but significantly promoted glioma cell proliferation, colony formation, migration, invasion, N-cadherin and Snail expression, and inhibited apoptosis and E-cadherin (Figure 4B-F). While introducing miR-125a-5p inhibitors into U87 and U251 cells stably transfected by si-circ, these phenotypes of glioma cells conferred by knockdown of circPARP4 could be reverted by the miR-125a-5p inhibitor. As shown in Figure 4B-F, cell proliferation, colony formation, migration, invasion and EMT abilities were improved and apoptosis rate was decreased in dual-transfection of miR-125a-5p inhibitor and si-circ group comparing to these in si-circ alone group. These results indicate that miR-125a-5p functions as a suppressor in glioma and circPARP4 promotes tumor progression partly via downregulating miR-125a-5p.
Figure 4.
Inhibition of miR-125a-5p reverses the silencing effect of circPARP4 in glioma cells. A. The expression of circPARP4 and miR-125a-5p was determined in U87 and U251 cells transfected with miR-125a-5p inhibitor or control. B. The proliferation ability of glioma cells in different conditions of miR-125a-5p and circPARP4 expression pattern was assessed by CCK-8. C. The colony number of glioma cells with different treatments was measured via colony formation assays and quantified at 72 h. D. The effects of miR-125a-5p inhibition and knocking down circPARP4 on glioma cell apoptosis were analyzed by flow cytometry and quantified at 72 h. E. Cell migration and invasion of U87 and U251 in different conditions were assessed by transwell assays. F. The expression of EMT markers affected by miR-125a-5p and circPARP4 was determined by western blot. Data indicate the mean ± SD, n = 3. **P<0.01, ***P<0.001 vs. control and ##P<0.01, ###P<0.001 vs. si-circ.
The activity of circPARP4/miR-125a-5p pathway in glioma cells is mediated by FUT4
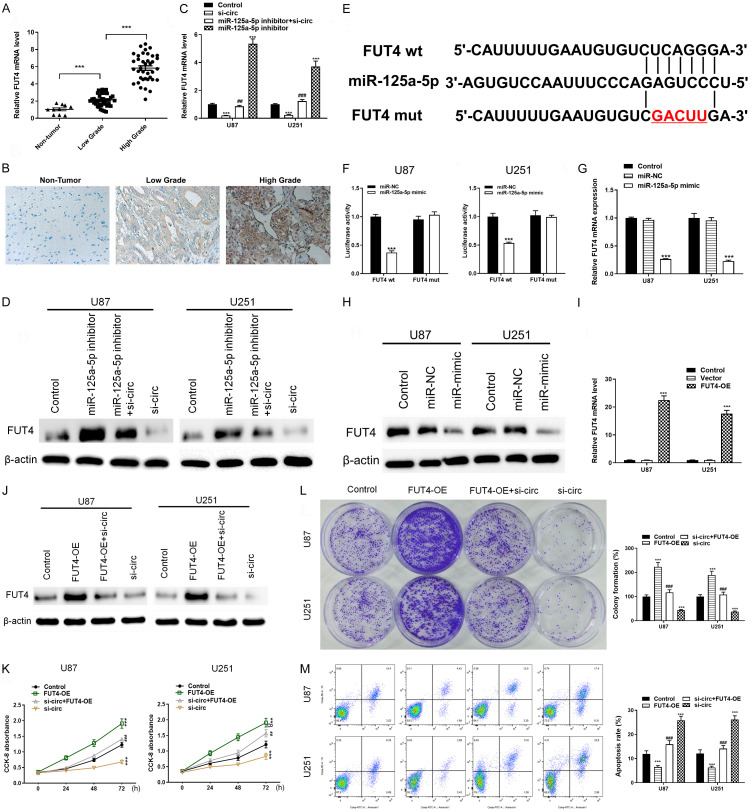
Bioinformatics analysis using several databases showed several genes potentially regulated by miR-125a-5p, of which FUT4 has demonstrated as a target of this miRNA and exerted oncogenic functions in a previous study [16]. Here, we further detected the role of FUT4 in glioma and found that the expression level of FUT4 was higher in glioma tissue than that in normal control and positively correlated with the pathological grade of glioma (Figure 5A, 5B). Moreover, high expression of FUT4 was also associated with poor prognosis in GBM patients (Supplementary Figure 1), implying cancer-promoting activity of FUT4. At the cellular level, it was revealed that si-circ significantly decreased the expression of FUT4 while inhibition of miR-125a-5p reversed the suppression of both FUT4 mRNA and protein (Figure 5C and 5D). Moreover, overexpression of miR-125a-5p significantly suppressed the expression of FUT4 (Figure 5G and 5H). As determined by luciferase assay, FUT4 acted as a direct target of miR-125a-5p that co-transfection of miR-125a-5p mimics and reporter plasmids visibly reduced luciferase activity, while co-transfection of miR-125a-5p mimics and FUT4 mutated vectors displayed no significant effect (Figure 5E, 5F). Furthermore, overexpression vector of FUT4 was transfected into glioma cells (Figure 5I, 5J), which significantly enhanced the proliferation and colony formation abilities and suppressed apoptosis of tumor cells, and reversed the inhibitory effect of si-circ (Figure 5L, 5M). These results reveal FUT4 possibly mediate the function of circPARP4/miR-125a-5p pathway in glioma progression.
Figure 5.
The involvement of circPARP4/miR-125a-5p in glioma cells is mediated by FUT4 modulation. A. The level of FUT4 mRNA was measured in low- and high-grade glioma samples and control tissues. B. Immunohistochemistry staining of FUT4 in glioma and brain tissues. C and D. The changes of FUT4 were analyzed by RT-PCR and western blot in glioma cells with different circPARP4 and miR-125a-5p expression patterns. E. The binding sites of miR-125a-5p were predicted in the 3’UTR of FUT4. F. The relative luciferase activity was measured in U87 and U251 cells 48 h after transfection with the miR-125a-5p mimic/control or the 3’UTR of FUT4 wt/mut constructs. G and H. The expression of FUT4 in glioma cells treated with miR-125a-5p mimics or control was determined by RT-PCR and western blot. I. FUT4 expression in glioma cells treated with FUT4 overexpression vector (FUT4-OE) or control was measured by RT-PCR. J. The expression of FUT4 in glioma cells with different transfection patterns of circPARP4 and FUT4 was determined by western blot. K. The proliferation ability of glioma cells in different conditions of FUT4 and circPARP4 expression pattern was assessed by CCK-8. L. The colony number of glioma cells with different treatments was measured via colony formation assays and quantified at 72 h. M. The effects of FUT4 overexpression and knocking down circPARP4 on glioma cell apoptosis were explored by flow cytometry and quantified at 72 h. Data indicate the mean ± SD, n = 3. *P<0.05, **P<0.01, ***P<0.001 vs. control and ##P<0.01, ###P<0.001 vs. si-circ.
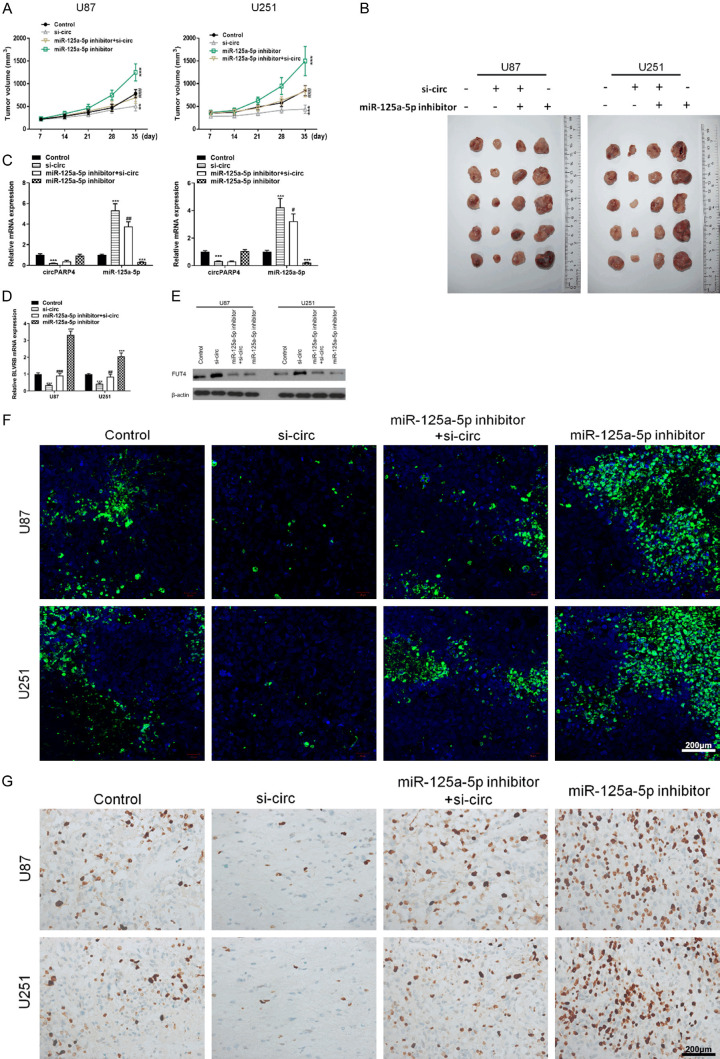
Targeting circPARP4 inhibits tumor formation in vivo
To confirm the oncogenic functions of circPARP4 in vivo, we established a xenograft mouse model in which si-circ U87 or U251 cells were injected subcutaneously (n = 6 for each group). The results showed that silencing circPARP4 significantly reduced tumor volume compared to the control, whereas inhibition of miR-12a-5p reversed the effect of si-circ and restored tumor size to the level comparable with the control (Figure 6A and 6B). Moreover, knocking down circPARP4 could increase miR-125a-5p level and downregulate FUT4 expression in xenograft tumors (Figure 6C-E). Furthermore, TUNEL assays showed that samples from si-circ tumors had a higher rate of apoptosis, while inhibition of miR-125a-5p reduced the number of apoptotic cells (Figure 6F). In addition, immunohistochemical staining found a lower level of Ki67 in the si-circ group comparing to the control (Figure 6G). These results imply that miR-125a-5p can function underlying circPARP4 pathway in vivo and targeting circPARP4 has the potential to treat glioma.
Figure 6.
Silencing circPARP4 inhibits in vivo tumor formation of xenografts. A. Tumor sizes were measured in the different groups at scheduled time interval. B. Representative images showed xenograft tumors isolated from nude mice in the different groups. C. The expressions of circPARP4 and miR-125a-5p in xenograft tumors were determined by RT-PCR. D and E. The level of FUT4 expression in xenograft tumors was analyzed by RT-PCR and western blot. F. The TUNEL assay (400 ×) was performed to determine the apoptotic index in the four groups. Scale bar, 200 μm. G. Immunohistochemical staining of Ki67 (400 ×) indicated the proliferation index of glioma in different xenograft tumors. Scale bar, 200 μm. Data indicate the mean ± SD, n = 5. *P<0.05, **P<0.01, ***P<0.001 vs. control and ##P<0.01, ###P<0.001 vs. si-circ.
Discussion
Although circRNAs have been found several decades ago, their novel functions have remained unclear until recently. Emerging evidence indicated that circRNAs were expressed ubiquitously and involved in diverse biological processes [17]. Dysregulated circRNAs have been demonstrated in a variety of cancers and contribute to tumor development and progression [7]. In this study, we demonstrated that circPARP4 was overexpressed in glioma, functioned with pro-tumor activity and acted as a sponge of miR-125a-5p, whose expression was downregulated. Moreover, inhibiting miR-125a-5p could revert the phenotypes induced by knockdown of circPARP4. Further, FUT4 was found as a direct target of miR-125a-5p and potentially mediated the oncogenic function of circPARP4/miR-125a-5p pathway. Investigating the expression, function and mechanism of the novel regulatory axis of circPARP4/miR-125a-5p/FUT4 extends our knowledge of circNRAs in glioma progression and provides more clinical implications.
CircPARP4 is located in chromosome 13 and translated from the spanning junction ORF formed by the covalent connection of exon 2 and exon 6 of the PARP4 gene, which codes for the largest member of the poly (ADP-ribose) polymerases family [18]. PARP4 exhibits poly-ADP ribosyltransferase activity at the post-translational level and has been predicted to be involved in the DNA base excision repair [19]. Previous study showed that inhibition of PARP enzyme activity led to increased genetic instability and recombination and reduced tumor formation of Hela cells in nude mice [20]. However, colon and lung cancer models with PARP4 (-/-) mice have no obvious changes in phenotype. Following exposure to the carcinogen dimethylhydrazine, there was an increase in colon cancer incidence, which was not observed in a lung cancer mice model with PARP4 (-/-) after urethane injection [21]. Moreover, as analyzed in breast cancer datasets, patients with low expression of PARP4 showed shorter survival time, suggesting the possible role of PARP4 as a tumor suppressor [22]. Taken together, these results reveled that PARP4 might function dually in a tissue-dependent manner. Utilizing TCGA GBM dataset in Betastasis (www.betastasis.com), we could see that the expression of PARP4 was upregulated in GBM and poor overall survival was seen in high-expression PARP4 group, indicating that PARP4 might play an oncogenic role in glioma. Given the elevated expression and function of circPARP4 and its host PARP4 in glioma, we hypothesized that the transcription of circPARP4 and PARP4 was possibly in a concerted way and they might serve as “double insurances” to promote tumor progression, as there could exist post-transcriptional regulation of PARP4 [23].
Our study also revealed the subcellular localization of circPARP4 in cytoplasm, where circRNAs usually act as sponges of miRNAs to regulate their activity. We screened several predicted miRNAs by bioinformatics analysis, and finally confirmed that miR-125a-5p was able to bind with circPARP4 in an AGO2-dependent manner and its expressed was suppressed. Aberrant expression of miR-125a-5p has been found in various cancers and associated with tumor progression. In non-small cell lung cancer, miR-125a-5p has been reported to be downregulated and decrease migration and invasion of lung cancer cell [24]. Moreover, decreased expression of miR-125a-5p also appeared in hepatocellular carcinoma and gastric cancer, and this miRNA worked as a suppressor to induce hepatocellular carcinoma cell cycle arrest, inhibit gastric cell proliferation more potently in combination with trastuzumab, and impedes colorectal cancer cell epithelial-mesenchymal transition, invasion and migration [25-27]. However, higher expression level of miR-125a-5p was observed in nasal pharyngeal cancer, multiple myeloma and prostate cancer [28-30]. MiR-125a-5p promoted proliferation, migration and invasion of nasal pharyngeal cancer cells, and inhibition of miR-125a-5p dampened cell growth and cell migration in multiple myeloma [28,29]. In addition, overexpression of miR-125a-5p was associated with imatinib resistance in gastrointestinal stromal tumors [31]. These studies reveal that miR-125a-5p biology can assume varied roles. As for glioma, it provides an enabling environment for tumor suppressor role of miR-125a-5p. MiR-125a-5p has been reported to inhibit glioma cell proliferation and promote cell differentiation by targeting TAZ, suppression of miR-125a-5p can restore malignant phenotypes after inhibiting the oncogene BCYRN1 in glioma [32,33]. Furthermore, down-regulated miR-125a-3p has been seen in CD133+ stem-like GBM cells compared with the CD133+ cells, and it is able to induce the differentiation of stem-like GBM cells, suggesting its involvement in the regulation of glioma stem cells [32,34]. Consistently, our study confirmed miR-125a-3p as a tumor suppressor underlying the regulation of oncogenic circPARP4.
As determined by our experiments, FUT4 applying as a direct target of miR-125a-3p, at least in part, mediated the oncogenic functions of circPARP4. FUT4 showed upregulated expression and a tumor-promoting activity in this study, which is consistent with previous findings that this oncogenic glycogene could promote tumor progression through multiple signaling pathways. As reported, FUT4 mediated the multidrug resistance in hepatocellular carcinoma associated with the activation of the PI3K/Akt pathway and the expression of MRP1 [35], induced activation of PI3K/Akt, and inactivation of GSK3b and nuclear translocation of NF-κB, resulting in the induction of epithelial-mesenchymal transition in breast cancer [36], and FUT4 promoted the malignant behaviors of leukemia stem cells by regulating fucosylated CD44 via Wnt/β-catenin pathway in acute myeloid leukemia [37]. Interestingly, miR-125a-3p also has been found involved in the PI3K/Akt pathway that miR-125a-3p inhibited cell proliferation, migration, invasion and pathological angiogenesis via suppression of the PI3K/Akt pathway in colorectal cancer, liver cancer and ovarian carcinoma [38-40]. It is hypothesized that circPARP4 may regulate the PI3K/Akt signaling pathway via miR-125a-3p/FUT4 to participate in glioma progression, which needs further investigations. Moreover, FUT4 is also proposed to be a marker of stem-like cells derived from brain tumors that FUT4+ cells recapitulated the original disease in medulloblastoma and the FUT4+ cells isolated from GBM had an increased expression of stem cell genes and were capable of self-renewal and multilineage differentiation [41-43]. Given the potential function of miR-125a-3p in the induction of differentiation of stem-like GBM cells, the circPARP4/miR-125a-3p/FUT4 axis possibly has a role in modulating the activity of stem-like GBM cells during tumor development and progression.
Conclusions
CircPARP4 plays an oncogenic function in glioma progression via sponging miR-125a-3p and modulating FUT4, targeting the regulatory circPARP4/miR-125a-3p/FUT4 pathway offers a potential therapeutic strategy for glioma patients.
Acknowledgements
This study was supported by grants from Young Top Medical Professionals of Jiangsu Province (QNRC2016884), Shanghai Sailing Program (19YF1448200) and the National Natural Science Foundation of China (81902538 and 81872072).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weller M, Wick W, Aldape K, Brada M, Berger M, Pfister SM, Nishikawa R, Rosenthal M, Wen PY, Stupp R, Reifenberger G. Glioma. Nat Rev Dis Primers. 2015;1:15017. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- 3.de Robles P, Fiest KM, Frolkis AD, Pringsheim T, Atta C, St Germaine-Smith C, Day L, Lam D, Jette N. The worldwide incidence and prevalence of primary brain tumors: a systematic review and meta-analysis. Neuro Oncol. 2015;17:776–783. doi: 10.1093/neuonc/nou283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, Jeraj R, Brown PD, Jaeckle KA, Schiff D, Stieber VW, Brachman DG, Werner-Wasik M, Tremont-Lukats IW, Sulman EP, Aldape KD, Curran WJ Jr, Mehta MP. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weller M, van den Bent M, Tonn JC, Stupp R, Preusser M, Cohen-Jonathan-Moyal E, Henriksson R, Rhun EL, Balana C, Chinot O, Bendszus M, Reijneveld JC, Dhermain F, French P, Marosi C, Watts C, Oberg I, Pilkington G, Baumert BG, Taphoorn MJB, Hegi M, Westphal M, Reifenberger G, Soffietti R, Wick W European Association for Neuro-Oncology (EANO) Task Force on Gliomas. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18:e315–e329. doi: 10.1016/S1470-2045(17)30194-8. [DOI] [PubMed] [Google Scholar]
- 6.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM. The landscape of circular RNA in cancer. Cell. 2019;176:869–881. e813. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, Jiang B, Qin H, Guo X, Liu M, Li L, Guo H. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18:111. doi: 10.1186/s12943-019-1040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, Xie Z, Chen J, Chen J, Ni W, Ma Y, Huang K, Wang G, Wang J, Ma J, Shen S, Fan S. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18:73. doi: 10.1186/s12943-019-1007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, Xu Z, Zeng A, Zhang X, Zhang X, He Z, Li Q, Sun G, Wang S, Li Q, Wang L, Zhang L, Xu H, Xu Z. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18:71. doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Wang H, Chu J, Huang Q, Li G, Yan Y, Xu T, Chen J, Wang Y. Circular RNA hsa_circ_0008344 regulates glioblastoma cell proliferation, migration, invasion, and apoptosis. J Clin Lab Anal. 2018;32:e22454. doi: 10.1002/jcla.22454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Zhang D, Lv J, Wang S, Zhang Q. MiR-125a-5p suppresses bladder cancer progression through targeting FUT4. Biomed Pharmacother. 2018;108:1039–1047. doi: 10.1016/j.biopha.2018.09.100. [DOI] [PubMed] [Google Scholar]
- 17.Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, Gao J, Boothman DA. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr. 2014;24:15–28. doi: 10.1615/critreveukaryotgeneexpr.2013006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger W, Steiner E, Grusch M, Elbling L, Micksche M. Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell Mol Life Sci. 2009;66:43–61. doi: 10.1007/s00018-008-8364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hans MA, Muller M, Meyer-Ficca M, Burkle A, Kupper JH. Overexpression of dominant negative PARP interferes with tumor formation of HeLa cells in nude mice: evidence for increased tumor cell apoptosis in vivo. Oncogene. 1999;18:7010–7015. doi: 10.1038/sj.onc.1203178. [DOI] [PubMed] [Google Scholar]
- 21.Raval-Fernandes S, Kickhoefer VA, Kitchen C, Rome LH. Increased susceptibility of vault poly(ADP-ribose) polymerase-deficient mice to carcinogen-induced tumorigenesis. Cancer Res. 2005;65:8846–8852. doi: 10.1158/0008-5472.CAN-05-0770. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda Y, Kiyotani K, Yew PY, Kato T, Tamura K, Yap KL, Nielsen SM, Mester JL, Eng C, Nakamura Y, Grogan RH. Germline PARP4 mutations in patients with primary thyroid and breast cancers. Endocr Relat Cancer. 2016;23:171–179. doi: 10.1530/ERC-15-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szaflarski W, Sujka-Kordowska P, Pula B, Jaszczynska-Nowinka K, Andrzejewska M, Zawierucha P, Dziegiel P, Nowicki M, Ivanov P, Zabel M. Expression profiles of vault components MVP, TEP1 and vPARP and their correlation to other multidrug resistance proteins in ovarian cancer. Int J Oncol. 2013;43:513–520. doi: 10.3892/ijo.2013.1975. [DOI] [PubMed] [Google Scholar]
- 24.Jiang L, Huang Q, Zhang S, Zhang Q, Chang J, Qiu X, Wang E. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer. 2010;10:318. doi: 10.1186/1471-2407-10-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh TH, Hsu CY, Tsai CF, Long CY, Wu CH, Wu DC, Lee JN, Chang WC, Tsai EM. HDAC inhibitors target HDAC5, upregulate microRNA-125a-5p, and induce apoptosis in breast cancer cells. Mol Ther. 2015;23:656–666. doi: 10.1038/mt.2014.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishida N, Mimori K, Fabbri M, Yokobori T, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Mori M. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725–2733. doi: 10.1158/1078-0432.CCR-10-2132. [DOI] [PubMed] [Google Scholar]
- 27.Tang L, Zhou L, Wu S, Shi X, Jiang G, Niu S, Ding D. miR-125a-5p inhibits colorectal cancer cell epithelial-mesenchymal transition, invasion and migration by targeting TAZ. Onco Targets Ther. 2019;12:3481–3489. doi: 10.2147/OTT.S191247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao W, Chan JY, Wong TS. Curcumin exerts inhibitory effects on undifferentiated nasopharyngeal carcinoma by inhibiting the expression of miR-125a-5p. Clin Sci (Lond) 2014;127:571–579. doi: 10.1042/CS20140010. [DOI] [PubMed] [Google Scholar]
- 29.Leotta M, Biamonte L, Raimondi L, Ronchetti D, Di Martino MT, Botta C, Leone E, Pitari MR, Neri A, Giordano A, Tagliaferri P, Tassone P, Amodio N. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J Cell Physiol. 2014;229:2106–2116. doi: 10.1002/jcp.24669. [DOI] [PubMed] [Google Scholar]
- 30.Fu Y, Cao F. MicroRNA-125a-5p regulates cancer cell proliferation and migration through NAIF1 in prostate carcinoma. Onco Targets Ther. 2015;8:3827–3835. doi: 10.2147/OTT.S92314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akcakaya P, Caramuta S, Ahlen J, Ghaderi M, Berglund E, Ostman A, Branstrom R, Larsson C, Lui WO. microRNA expression signatures of gastrointestinal stromal tumours: associations with imatinib resistance and patient outcome. Br J Cancer. 2014;111:2091–2102. doi: 10.1038/bjc.2014.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan J, Xiao G, Peng G, Liu D, Wang Z, Liao Y, Liu Q, Wu M, Yuan X. MiRNA-125a-5p inhibits glioblastoma cell proliferation and promotes cell differentiation by targeting TAZ. Biochem Biophys Res Commun. 2015;457:171–176. doi: 10.1016/j.bbrc.2014.12.078. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, Xiang D, Jia H, He X, Sheng J, Long Y, Zhu S, Wang K, Liu Q. The lncRNA BCYRN1 functions as an oncogene in human glioma by downregulating miR-125a-5p in vitro. Cancer Manag Res. 2020;12:1151–1161. doi: 10.2147/CMAR.S227327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cortez MA, Nicoloso MS, Shimizu M, Rossi S, Gopisetty G, Molina JR, Carlotti C Jr, Tirapelli D, Neder L, Brassesco MS, Scrideli CA, Tone LG, Georgescu MM, Zhang W, Puduvalli V, Calin GA. miR-29b and miR-125a regulate podoplanin and suppress invasion in glioblastoma. Genes Chromosomes Cancer. 2010;49:981–990. doi: 10.1002/gcc.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng L, Luo S, Jin C, Ma H, Zhou H, Jia L. FUT family mediates the multidrug resistance of human hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell Death Dis. 2013;4:e923. doi: 10.1038/cddis.2013.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, Liu S, Yan Q. Role of fucosyltransferase IV in epithelial-mesenchymal transition in breast cancer cells. Cell Death Dis. 2013;4:e735. doi: 10.1038/cddis.2013.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu B, Ma H, Liu Q, Xiao Y, Pan S, Zhou H, Jia L. MiR-29b/Sp1/FUT4 axis modulates the malignancy of leukemia stem cells by regulating fucosylation via Wnt/beta-catenin pathway in acute myeloid leukemia. J Exp Clin Cancer Res. 2019;38:200. doi: 10.1186/s13046-019-1179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang L, Gao C, Li Y, Sun M, Xu J, Li H, Jia L, Zhao Y. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis. 2017;8:e2968. doi: 10.1038/cddis.2017.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu K, Tang X, Tang G, Yao S, Yao B, Wang H, Nie D, Liang X, Tang C, He S. 18F-FP-PEG2-beta-Glu-RGD2: a symmetric integrin alphavbeta3-targeting radiotracer for tumor PET imaging. PLoS One. 2015;10:e0138675. doi: 10.1371/journal.pone.0138675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu P, Xu L, Su H. HULC functions as an oncogene in ovarian carcinoma cells by negatively modulating miR-125a-3p. J Physiol Biochem. 2019;75:163–171. doi: 10.1007/s13105-019-00669-5. [DOI] [PubMed] [Google Scholar]
- 41.Kim KJ, Lee KH, Kim HS, Moon KS, Jung TY, Jung S, Lee MC. The presence of stem cell marker-expressing cells is not prognostically significant in glioblastomas. Neuropathology. 2011;31:494–502. doi: 10.1111/j.1440-1789.2010.01194.x. [DOI] [PubMed] [Google Scholar]
- 42.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Read TA, Fogarty MP, Markant SL, McLendon RE, Wei Z, Ellison DW, Febbo PG, Wechsler-Reya RJ. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell. 2009;15:135–147. doi: 10.1016/j.ccr.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.