Abstract
NFBD1, a signal amplifier of the p53 pathway, is vital for protecting cells from p53-mediated apoptosis and the early phase of DNA damage response under normal culture conditions. Here we investigated its expression in patients with nasopharyngeal carcinoma (NPC), and we describe the biological functions of the NFBD1 gene. We found that NFBD1 mRNA and protein were more highly expressed in NPC tissues than in nontumorous tissues. To investigate the function of NFBD1, we created NFBD1-depleted NPC cell lines that exhibited decreased cellular proliferation and colony formation, an increase in their rate of apoptosis, and an enhanced sensitivity to chemotherapeutic agents compared with in vitro controls. However, N-acetyl cysteine (NAC) and downregulation of p53 expression could partially reverse the apoptosis caused by the loss of NFBD1. Further analysis showed that loss of NFBD1 resulted in increased production of intracellular reactive oxygen species (ROS) depending on p53, which subsequently triggered the mitochondrial apoptotic pathway. Using a xenograft model in nude mice, we showed that silencing NFBD1 also significantly inhibited tumor growth and led to apoptosis. Taken together, our data suggest that inhibition of NFBD1 in NPC could be therapeutically useful.
Key words: Nasopharyngeal carcinoma (NPC), NFBD1/MDC1, Apoptosis, Reactive oxygen species (ROS), Mitochondrial
INTRODUCTION
Nasopharyngeal carcinoma (NPC) is a nonlymphomatous carcinoma that occurs in the epithelial lining of the nasopharynx and is prevalent with increasing incidence in people of southern Chinese ancestry in southern China and Southeast Asia (1). Although radiotherapy is routinely used to treat patients with NPC, local recurrences and distant metastases often occur in 30%–40% of advanced stage NPC patients (2). Therefore, continued research on NPC is essential to expand our understanding of the disease mechanisms and to develop novel therapies.
Human nuclear factor with BRCT domain protein 1 (NFBD1), also known as mediator of DNA damage checkpoint protein (MDC1), regulates many aspects of the DNA damage response (DDR) pathway, including the intra-S phase checkpoint, G2/M checkpoint, spindle assembly checkpoint, and the formation of NBS/MRE/Rad50 (MRN complex), 53BP1, and BRCA1 foci (3–7). Previous studies have found that NFBD1−/− mice exhibit phenotypes such as retarded growth, male infertility, immune defects, chromosome instability, DNA repair defects, and radiation sensitivity (8). Also, our previous studies have found that silencing NFBD1 enhances the radiosensitivity and chemosensitivity of human nasopharyngeal cancer cells (9,10). However, the role of NFBD1 in the development of cancer is still relatively understudied. Recent studies have found that the expression of NFBD1 was aberrantly reduced in breast and lung cancers, but almost no loss of NFBD1 was found intesticular germ cell tumors and cervical cancer tissues (11,12). Thus, there are conflicting reports regarding the expression and role of NFBD1 in the different tumors.
Although much of what is known about NFBD1 was identified at sites of DNA damage, the cellular environment would also be expected to have an important supporting role in facilitating these responses and maintaining genomic stability. Numerous DNA breaks are generated by reactive oxygen species (ROS), which can also cause oxidative and mitochondrial stress (13–16). Cellular ROS homeostasis and ATP levels are required to support DNA repair and for the protection against the generation of double-strand breaks (DSBs); mitochondria are the primary mediators of the levels of both ROS and ATP (17–22). However, beyond a certain threshold, excess intracellular ROS can induce apoptosis in cancer cells (23,24). In addition, mitochondria act as central modulators of apoptotic cell death, which has an important role in the removal of cells with persistent or oncogenic DNA lesions (13,25). Thus, mitochondria would be expected to have a central role in the DDR. In this study, to further determine the role of the NFBD1 gene in NPC, we investigated the differential expression of NFBD1 in patients with NPC and detailed the biological functions of the NFBD1 gene.
MATERIALS AND METHODS
Clinical Specimens
A total of 66 NPC biopsy samples and 46 normal nasopharyngeal tissues used for quantitative real-time polymerase chain reaction (qRT-PCR) and Western blotting were obtained from The First Affiliated Hospital of Chongqing Medical University. These tissues were frozen in liquid nitrogen and stored during 2014. For immunohistochemical analysis, 120 paraffin-embedded NPC specimens and 20 normal nasopharyngeal tissues were collected in The First Affiliated Hospital of Chongqing Medical University from 2012 to 2014. All samples were pathologically confirmed by two pathologists. No patients received radiotherapy or chemotherapy before biopsy. Pathological stage was recorded according to the International Union Against Cancer (Union for International Cancer Control). An informed consent to participate in this study was obtained from each subject. All experiments were performed in accordance with the relevant guidelines and regulations of Chongqing Medical University, and this study was approved by the Ethical Committee of Chongqing Medical University.
Cell Culture and Reagents
Two human NPC cell lines, CNE1 and CNE2, were obtained from the Molecular Medicine and Cancer Research Center, Chongqing Medical University. All cells were grown in RMPI-1640 medium (HyClone, Logan City, UT, USA) with 10% fetal bovine serum (HyClone) at 37°C with 5% CO2. NAC was purchased from Beyotime (Beyotime Institute of Biotechnology, Nantong, P.R. China), 2′,7′-dichlorofluorescin diacetate (DCFH-DA), ADR, cisplatin, and 5-FU were purchased from Sigma-Aldrich (St. Louis, MO, USA). Lipofectamine RNAiMAX was purchased from Invitrogen (Carlsbad, CA, USA), and human p53 siRNA was purchased from GenePharma (Shanghai, P.R. China). The antibodies used in this study were anti-NFBD1 (Abcam, UK); anti-PARP, anti-cleaved caspase 3, anti-caspase 9, anti-Bcl-2, and anti-Bcl-xl (Cell Signaling Technology, Danvers, MA, USA); anti-cytochrome c, anti-SESN2, anti-GPX-1/2, and ALDH4A1 (Santa Cruz Biotechnology, USA); and anti-PUMA, anti-P53, anti-MDM2, anti-Bax, anti-SOD2, and anti-PA26 (Bioworld Technology, USA).
Immunohistochemistry
Immunohistochemistry (IHC) was performed to study alterations in the protein expression of human NPC tissues. In brief, paraffin-embedded specimens were cut into 5-μm sections and baked at 65°C for 30 min. The sections were deparaffinized with xylene and rehydrated. Sections were submerged into EDTA antigenic retrieval buffer and microwaved for antigen retrieval. The sections were treated with 3% hydrogen peroxide in methanol to quench the endogenous peroxidase activity, followed by incubation with 3% bovine serum albumin to block nonspecific binding. The sections were incubated with mouse monoclonal anti-NFBD1 (1:200) overnight at 4°C. Negative control staining for each tissue was performed in parallel in the absence of anti-NFBD1 primary antibody. After washing, the tissue sections were treated with a biotinylated anti-mouse secondary antibody (Cell Signaling Technology) followed by further incubation with a streptavidin–horseradish peroxidase complex (Cell Signaling Technology). The NFBD1 IHC scores ranged from 0 to 3+ as follows: 0 (no staining or <50% of tumor cells with any intensity); 1+ (≥50% of tumor cells with weak or higher staining intensity and <50% with moderate or higher intensity); 2+ (≥50% of tumor cells with moderate or higher staining intensity and <50% strong intensity); and 3+ (≥50% of tumor cells with strong staining intensity). Scores of 2+ and 3+ were defined as high NFBD1 expression, and scores of 1+ and 0 were defined as low NFBD1 expression. The IHC scores were independently evaluated by two pathologists who were blinded to the clinical and molecular characteristics of the patients.
Lentivirus-Mediated shRNA Downregulation of Gene Expression
The shRNA oligonucleotide or a lentivirus-mediated shRNA (Genechem, Shanghai, P.R. China) construct was used to silence NFBD1. The sequence of shRNA oligonucleotide for the positive experiment group (NFBD1-shRNA) was 5′-GAGGCAGACUGUGGAUAAATT-3′; nonsilencing sequence 5′-TTCTCCGAACGTGTCACG T-3′ was used as a control, which was named negative control group (NC-shRNA).
NPC cells were transferred into six-well plates with a density of 2 × 104/well and divided into two groups: the NFBD1-shRNA group and the NC-shRNA group. The cells were cultured in 1 ml of medium (containing 4 μg/μl polybrene) and 2 μl of viral supernatants (the number of viruses is 2 × 105). Twenty-four hours later, the culture medium was replaced with normal medium. After 72 h, the cells were cultured in the medium with 1 μg/ml puromycin to select cells that have been transduced with the lentivirus containing NFBD1-shRNA or NC-shRNA. Seven days later, the live cells were collected and cultured for cell amplification. qRT-PCR, Western blotting, and immunofluorescence were used to detect the inhibition rate of lentivirus-mediated shRNA targeting NFBD1.
Real-Time Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions. Total RNA (500 ng) was reverse transcribed into cDNA using a One Step SYBR® PrimeScript™ RT-PCR Kit II (Takara Biotechnology, Dalian, P.R. China) following the supplied protocol. The qRT-PCR was performed using SYBR Premix Ex Taq (Takara Biotechnology) in a LightCycler 480 qRT-PCR system (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. The primers for NFBD1 were 5′-AGCAACCCCAGTTGTCATTC-3′ (forward) and 5′-TCAGATGTGCCAAAGTCAGC-3′ (reverse), and GAPDH was used as the internal standard in the qRT-PCR system. The primers for GAPDH were 5′-ACCTGACCTGCCGTCTAGAA-3′ (forward) and 5′-T CCACCACCCTGTTGCTGTA-3′ (reverse). The 2−ΔΔCt method was used to determine the relative quantification of the NFBD1 expression. Briefly, the threshold cycle (Ct) of fluorescence for each sample was determined. ΔCt indicates the difference in expression level with the Ct value between NFBD1 and GAPDH (ΔCt = CtNFBD1 − CtGAPDH), and ΔΔCt indicates the difference in the ΔCt value between tumor tissues and control tissues (ΔΔCt = ΔCttumor − ΔCtcontrol). The 2−ΔΔCt value (fold value) was also calculated.
Western Blotting
Total protein extracts from cells were prepared using RIPA buffer (Beyotime Institute of Biotechnology). Protein concentrations were determined using the BCA protein assay systems (Beyotime Institute of Biotechnology). Proteins were fractionated in SDS–polyacrylamide gels. Proteins were transferred to polyvinylidene fluoride (Millipore, Billerica, MA, USA), and Western blotting was performed using the appropriate antibody overnight at 4°C. This was followed by incubation with peroxidase-conjugated AffiniPure secondary antibodies for 1 h at 37°C. Antibody/antigen complexes were detected using ECL (Western Bright Sirius; Advansta, Inc., Menlo Park, CA, USA), and images were acquired using an enhanced chemifluorescence detection system (Amersham Biosciences) at room temperature.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde at room temperature for 30 min, permeablized with 0.5% Triton X-100 solution, washed three times in PBS for 5 min, blocked using 5% fetal bovine serum in PBS, and then primary antibodies were applied overnight at 4°C, and secondary antibodies (green; Invitrogen) were applied for 60 min at room temperature. Finally, the cells were washed three times in PBS, and the DNA was stained using DAPI (Sigma-Aldrich) at 50 ng/ml. The slides were observed under a Nikon Microphot-FX fluorescence microscope (Japan).
Cell Growth Assay
Cells were seeded in 96-well plates at 2 × 103 cells/well in 100 μl of medium. At 0 (after adherence), 24, 48, 72, and 96 h after seeding, cell proliferation was assessed with the use of a CCK-8 kit (Beyotime Institute of Biotechnology). Following the addition of 10 μl of CCK-8 to each well, the plates were incubated at 37°C for 2 h. The optical density (at 450 nm) in each well was measured by an enzyme-linked immunosorbent assay plate reader, and the viability rate was calculated.
Colony Formation Assay
Cells were seeded in six-well plates at 500 cells/well. Cells were left for 14 days at 37°C with 5% CO2 to allow colonies to form. Cells were fixed with 70% ethanol for 20 min and then stained with 0.5% crystal violet for 20 min. Colonies containing 50 or more cells were counted as survivors.
Flow Cytometric Analysis of Apoptosis
Cells were seeded in six-well plates at 1 × 105 cells/well. The cells were then incubated for 24 h and stained with FITC-conjugated annexin V and propidium iodide (PI) using the Annexin-V-FITC Apoptosis Detection Kit (Beyotime Institute of Biotechnology) and according to the manufacturer’s recommendation. Each sample was then subjected to analyses by flow cytometry (FCM) using a FACSVantage SE system (BD Biosciences, San Jose, CA, USA). The percentage of apoptotic cells was determined by FCM.
Hoechst 33342 Staining
Cells were seeded in 24-well plates at 2 × 104 cells/well, and 24 h after seeding, cells were fixed with 4% paraformaldehyde for 1 h at room temperature and then stained with Hoechst 33342 (Beyotime Institute of Biotechnology) in the dark for 30 min at 37°C. Images were acquired under a Nikon Microphot-FX fluorescence microscope. Normal cells were identified as having noncondensed chromatin throughout the nucleus, whereas apoptotic cells showed condensed chromatin that was contiguous with the nuclear membrane and/or fragmented nuclei.
Flow Cytometry and Immunofluorescence Analysis of Intracellular ROS
For the measurement of ROS production, cells were seeded in six-well plates at 1.0 × 105 cells/well and 24 h later treated with 10 μM of DCFH-DA at 37°C for 30 min followed by analysis by immunofluorescence (Nikon Microphot-FX fluorescence microscope) and FCM using a FACSVantage SE system (BD Biosciences) with excitation and emission wavelengths of 488 and 525 nm, respectively. Image-Pro Plus software (version 6.0; Media Cybernetics, Bethesda, MD, USA) was used for analysis of fluorescence intensity.
Mitochondrial Membrane Potential (MMP) Assay
The fluorescent, lipophilic, cationic probe JC-1 (Beyotime Institute of Biotechnology) was used to assess the MMP (Δψm) at 24 h after cells were seeded in six-well plates. Assays were carried out using the MMP assay kit with JC-1 according to the manufacturer’s instructions and then analyzed by FCM using a FACSVantage SE system (BD Biosciences).
Terminal Deoxynucleotidyl Transferase-Mediated Nick-End Labeling (TUNEL) Assays
Cells were seeded in 24-well plates at 2 × 104 cells/well. Twenty-four hours after seeding, the cells were fixed for 30 min with 4% paraformaldehyde at room temperature. TUNEL staining was carried out in accordance with the manufacturer’s protocol (In Situ Cell Death Detection Kit; Roche, Mannheim, Germany). Images were acquired under a Nikon Microphot-FX fluorescence microscope.
Measurement of Cell Sensitivity to Chemotherapeutic Agents
Cells were seeded in 96-well plates at 5 × 103 cells/well, and 24 h after seeding the cells were treated with ADR, CDDP, or 5-FU. Forty-eight hours after treatment cell survival was assessed with a CCK-8 kit (Beyotime Institute of Biotechnology). The samples were measured in triplicate, and the percentages of cell survival were estimated by converting the absorbance [optical density (OD) values] of the CCK-8-stained cells to the percentage of the absorbance of untreated cells.
Tumor Xenografts
All animal husbandry and experiments were performed under a protocol approved by the Institutional Animal Care Committee at Chongqing Medical University. NC-shRNA and NFBD1-shRNA cells (CNE1: 6.0 × 106 cells, CNE2: 6.0 × 106) were resuspended in 0.1 ml of PBS. The cell suspension was subcutaneously injected into 4-week-old BALB/C nude mice (n = 5). Tumors were measured once a week with Vernier calipers, and tumor volumes were calculated as follows: volume = 1/2 × length × width2. Thirty-five days after injection, the mice were sacrificed, and tumor specimens were weighed and fixed. Histological examination was performed with hematoxylin and eosin staining. Apoptosis of the tumor tissues was examined with a TUNEL assay according to the manufacturer’s instructions. Brown staining was regarded as positive for apoptosis and blue was negative.
Statistical Analysis
Statistical comparison of mean values was performed using ANOVA, rank-sum tests (nonparametric statistics), or chi-square tests. Differences with a value of p < 0.05 were considered statistically significant.
RESULTS
Overexpression of NFBD1 in Tissues From NPC Patients
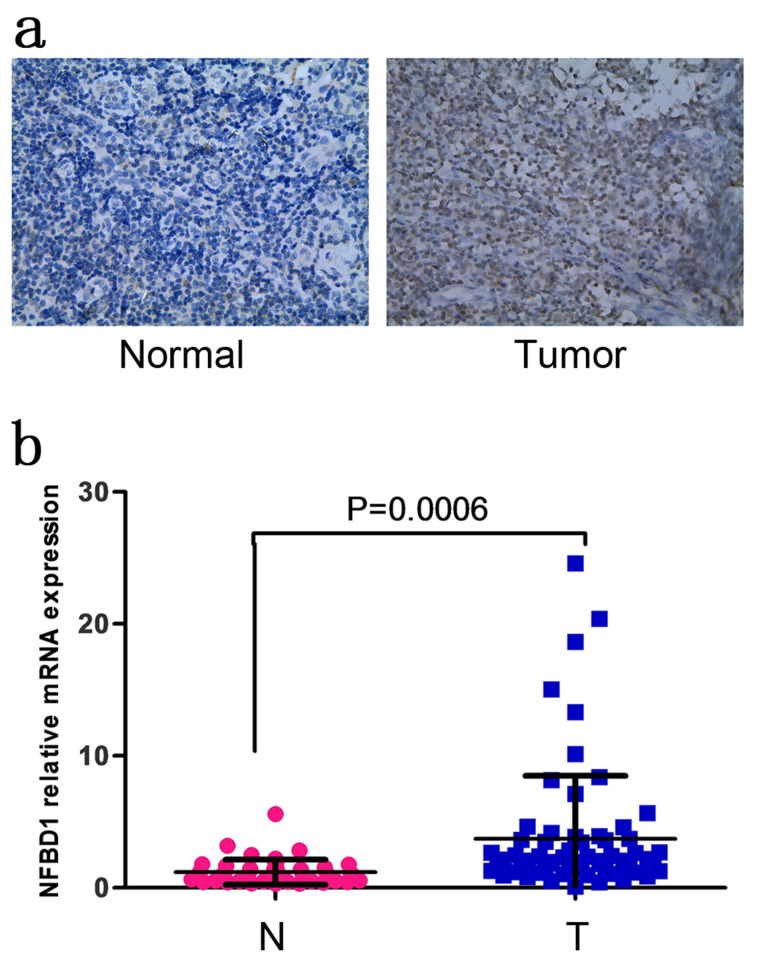
To reveal a potential role for NFBD1 in NPC, we performed immunohistochemical analysis of NFBD1 expression in patient-derived NPC biopsies. The NFBD1 protein was primarily detected in the nucleus of both normal nasopharyngeal epithelia and carcinoma cells (Fig. 1a). The positive rate of NFBD1 in tumor tissues is 71.7% (86/120) and that in normal nasopharyngeal tissues is 20% (4/20; χ2 = 16.523, p < 0.001) (Table 1). Furthermore, NFBD1 protein expression was not associated with clinical parameters of NPC patients, including gender, age, tumor type, lymph node metastasis, and TNM clinical stage (Table 2). Furthermore, we examined NFBD1 mRNA expression by qRT-PCR in 46 normal nasopharyngeal tissues and 66 NPC tissues, and high expression of NFBD1 mRNA level was found in NPC tissues (Fig. 1b).
Figure 1.
Human nuclear factor with BRCT domain protein 1 (NFBD1) expression in normal nasopharyngeal and NPC tissues. (a) Representative images of immunohistochemistry (IHC)-stained normal nasopharyngeal tissues (left) and NPC (right) showing levels of NFBD1 protein expression. (b) NFBD1 mRNA levels in normal nasopharyngeal tissues (N) and nasopharyngeal carcinoma (NPC) tissues (T). The y-axis represents the relative levels of NFBD1 mRNA expression.
Table 1.
The Expression of NFBD1 in Nasopharyngeal Carcinoma and Normal Nasopharyngeal Tissues
| Groups | NFBD1 High | NFBD1 Low | Chi-Square | p |
|---|---|---|---|---|
| Tumor | 86 (71.7%) | 34 (29.3%) | ||
| Control | 4 (20%) | 16 (80%) | 16.523 | 0.000 |
Table 2.
Relationship of NFBD1 Expression With Clinicopathological Characteristics in Nasopharyngeal Carcinoma
| Variable | NFBD1 Expression | Total | p | |
|---|---|---|---|---|
| Low | High | |||
| Age | ||||
| ≥45 | 20 | 50 | 70 | |
| <45 | 14 | 36 | 50 | 0.945 |
| Sex | ||||
| Male | 22 | 56 | 78 | |
| Female | 12 | 30 | 42 | 0.966 |
| Tumor type | ||||
| Undifferentiation | 16 | 36 | 52 | |
| Differentiation | 18 | 50 | 68 | 0.605 |
| Lymph node metastasis | ||||
| − | 11 | 30 | 41 | |
| + | 31 | 48 | 79 | 0.227 |
| TNM clinical stage | ||||
| I–II | 8 | 36 | 42 | |
| III–IV | 26 | 50 | 78 | 0.092 |
Silencing NFBD1 Inhibits NPC Cell Growth and Enhances Their Sensitivity to Chemotherapeutic Agents
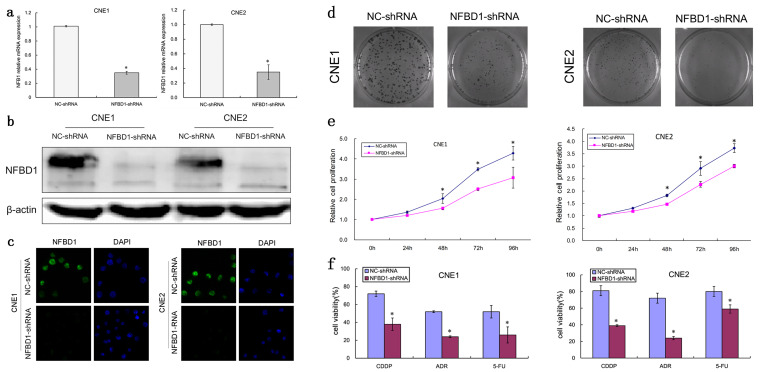
To investigate the biological function of NFBD1 in NPC, we established CNE1 and CNE2 cell lines with stable knockdown of NFBD1 and corresponding control cell lines (Fig. 2a–c). To determine a role for the NFBD1 gene in the regulation of NPC cell growth and proliferation, we performed cellular growth and colony formation assays. Both CNE1 and CNE2 cells with NFBD1 depletion showed decreased cellular proliferation and colony formation, compared with in vitro controls (Fig. 2d and e), indicating that the NFBD1 gene is involved in cellular growth. Most chemotherapy drugs target the DNA of tumor cells, and NFBD1 is an important player in the DDR pathway. Therefore, we next explored whether the depletion of NFBD1 affected the sensitivity of NPC cells to chemotherapeutic agents. As shown in Figure 2f, the knockdown of NFBD1 significantly enhanced the sensitivity of NPC cells to all tested chemotherapeutic agents (ADR, CDDP, and 5-FU).
Figure 2.
NFBD1 depletion suppresses the proliferation of NPC cells and enhances their sensitivity to chemotherapeutic agents. Lentiviruses expressing NFBD1 shRNA and control shRNA were transfected into CNE1 cells. Transfected cells that stably expressed NFBD1 shRNA and NC-shRNA were obtained under puromycin (1 μg/ml) selection. (a) The effects of the lentivirus on the expression of NFBD1 mRNA were determined by qRT-PCR. The relative expression level of NFBD1 mRNA was significantly downregulated in the NFBD1-shRNA group. *p < 0.01 compared with the NC-shRNA group. The effects of lentiviral infection on the expression of NFBD1 protein were determined by Western blotting (b) and immunofluorescence (c). (d) Colony formation assay for NPC cells. Cells were cultured for 14 days, and the surviving colonies were stained and counted. (e) Growth curves of CNE1 and CNE2 cells. A CCK-8 assay was performed, and absorbance values at 450 nm were recorded from 0 to 96 h after the cells were seeded. (f) The depletion of NFBD1 enhances the sensitivity of NPC cells to ADR (0.2 μg/ml), CDDP (0.2 μg/ml), and 5-FU (0.4 μg/ml). Cell viability was examined using a CCK-8 assay. *p < 0.05.
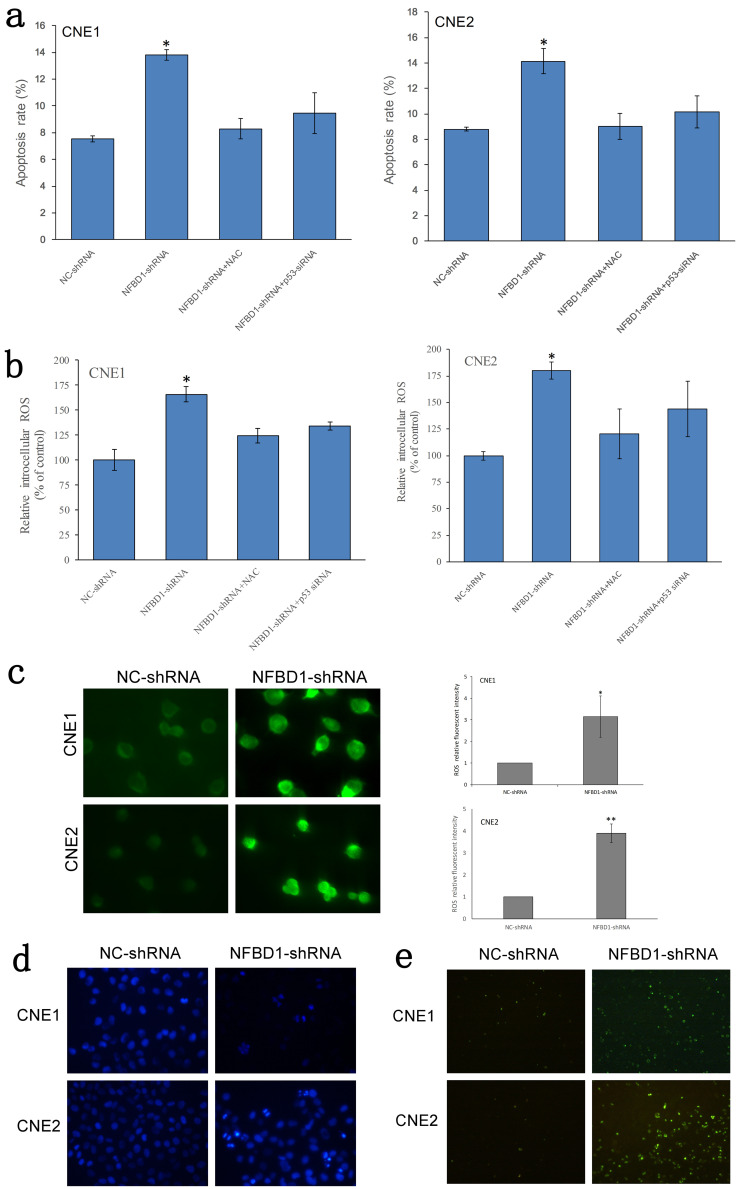
Silencing NFBD1 Induces the Apoptosis of NPC Cells in a p53-ROS-Dependent Manner
To determine whether the inhibition of NFBD1-deficient NPC cell growth was associated with apoptosis, we used FCM to detect the rate of apoptosis. The results showed that silencing NFBD1 increased the apoptotic rate in NPC cells; however, the apoptotic rate was significantly decreased in NFBD1-shRNA cells that were respectively pretreated with the ROS scavenger N-acetyl cysteine (NAC) and p53 siRNA (Fig. 3a). Next, we studied the changes in ROS levels in NFBD1-depleted cells. Cells were seeded overnight and then probed with DCFH-DA, a cell-permeable indicator that fluoresces when oxidized. The results showed that the inhibition of NFBD1 produced a marked increase in intracellular ROS (Fig. 3b and c). However, when NFBD1-shRNA cells were respectively treated with NAC and p53 siRNA, we observed a significant decrease in the levels of intracellular ROS (Fig. 3c). To further confirm that silencing NFBD1 induces apoptosis, we performed Hoechst 33342 and TUNEL staining. Consistent with our FCM results, the NFBD1-shRNA group demonstrated a greater number of fragmented nuclei and nuclear shrinkage (Hoechst 33342 staining) and more apoptotic bodies (TUNEL assay) compared with the NC-shRNA group (Fig. 3d and e). These results indicated that ROS might be the primary factor involved in apoptosis induced by NFBD1 depletion. It is widely accepted that the tumor suppressor p53 has been proposed as a critical regulator of intracellular ROS levels. Our study also showed that p53 is an important regulator of ROS levels (Fig. 3b). Therefore, we examined the link between p53 and NFBD1 by Western blotting, and we found that silencing NFBD1 has negative effects on the expression of p53, while the levels of MDM2 is decreased (Fig. 4a). We next determined if p53 expression affects the expression of antioxidants including ALDH4A1, SOD2, GPX-1/2, SESN2, and PA26. We observed an increase in these antioxidants by Western blotting (Fig. 4b). This observation suggests that silencing NFBD1 induces the apoptosis of NPC cells in a p53-ROS-dependent manner.
Figure 3.
The depletion of NFBD1 induces the reactive oxygen species (ROS)-dependent apoptosis of NPC cells. (a) Apoptotic cells were assessed by flow cytometry using annexin V and propidium iodide (PI) staining. (b) NPC cells were left untreated or treated with N-acetyl cysteine (NAC) (1.0 mM), and the level of ROS generation was assessed by flow cytometry. (c) ROS staining was performed using DCFH-DA. Left: Representative images of ROS staining. Right: ROS fluorescence intensity. (d) Cell morphology was observed under a fluorescence microscope by staining with Hoechst 33342. Fragmented nuclei and nuclear shrinkage were used as indicators of apoptotic cells (200×). (e) Apoptotic cells were evaluated by TUNEL assay (apoptotic bodies are shown in green) (100×). *p < 0.05, **p < 0.05.
Figure 4.
Silencing NFBD1 upregulates ROS generation through antioxidant effects depending on p53 levels. (a) Protein levels of p53 and MDM2 were examined by Western blotting analysis. (b) Expression of antioxidant-related proteins.
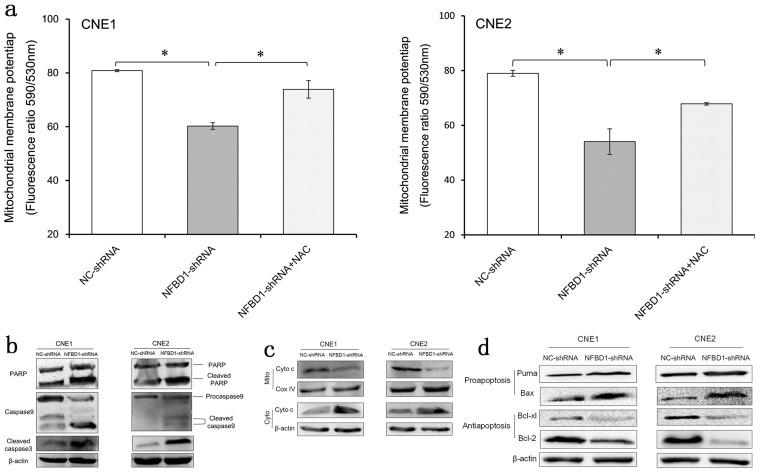
Silencing NFBD1 Activates the Mitochondrial Apoptosis Pathway
To determine whether the mitochondrial pathway is involved in apoptosis induced by NFBD1 depletion, we analyzed the role of the NFBD1 gene in mitochondria regulation. JC-1, which selectively enters the mitochondria, was used to assess the MMP; a reversible color change from red to green indicates a decrease in MMP as the release of JC-1 into depolarized mitochondria causes a reduction in red fluorescence intensity. The results showed a decrease in MMP in the NFBD1-shRNA group compared with the NC-shRNA group; however, pretreatment with NAC restored the MMP in the NFBD1-shRNA group (Fig. 5a). In addition, we also detected the expression of apoptotic factors and found that the cleavage of caspase 3, caspase 9, and PARP was significantly increased in the NFBD1-shRNA group compared with the NC-shRNA group (Fig. 5b). We further evaluated the expression of mitochondrial apoptosis-related proteins, and as shown in Figure 5c and d, NFBD1 depletion results in significant upregulation of several proapoptotic proteins including Bax and Puma, downregulation of antiapoptotic proteins including Bcl-2 and Bcl-xl, and release of cytochrome c into the cytosol. This observation suggests that the mitochondrial apoptotic pathway could mediate apoptosis resulting from the silencing of NFBD1.
Figure 5.
Depletion of NFBD1 activates the mitochondrial apoptosis pathway. (a) Measurement of mitochondrial membrane potential (MMP) using JC-1 by flow cytometry (FCM). (b) The expression of apoptotic factors, including the cleavage of caspase 3, caspase 9, and PARP. (c) Changes in mitochondrial and cytosolic cytochrome c levels. (d) Expression of apoptosis-related proteins.
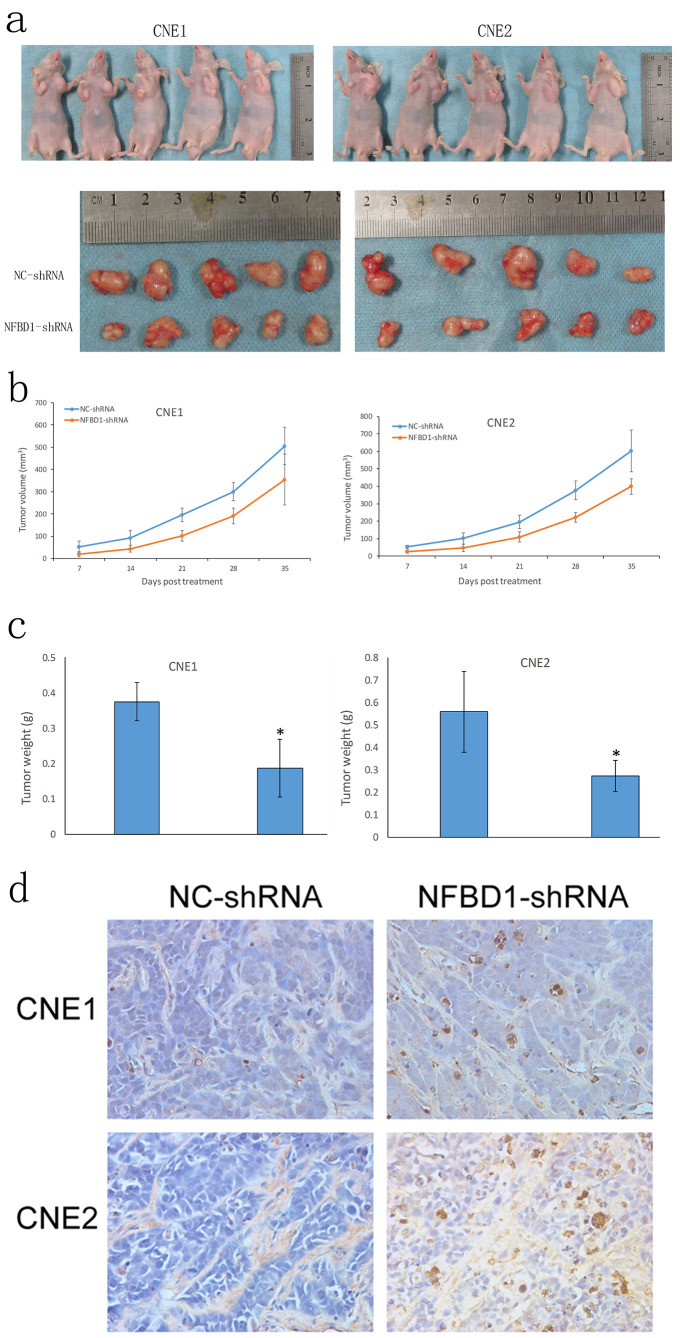
Silencing NFBD1 Inhibits the Tumorigenicity of NPC Cells and Induces Apoptosis In Vivo
To assess the effects of NFBD1 depletion on the tumorigenicity of NPC cells in vivo, we established a xenograft model in athymic nude mice. We noted a marked delay in the growth of tumors from NFBD1-shRNA cells compared with NC-shRNA cells, and the tumor weights from the NFBD1-depleted group decreased significantly compared with that from the controls (Fig. 6a–c). Tumor xenografts were sectioned, and apoptosis was assessed by TUNEL assay. Compared with the NC-shRNA group, tumors from the NFBD1-shRNA group demonstrated a greater number of brown apoptotic bodies (TUNEL+ cells) (Fig. 6d). Therefore, these results suggest that silencing NFBD1 results in the inhibition of tumor growth and induction of apoptosis in vivo.
Figure 6.
Depletion of NFBD1 inhibits nasopharyngeal cancer tumor growth and induces apoptosis in vivo. NFBD1 shRNA and NC-shRNA cells were subcutaneously injected into two groups of athymic nude mice (n = 5 for each group). Tumor xenografts were measured once a week with Vernier calipers. (a) Images of tumor-bearing nude mice. (b) The growth curves of NPC cells in tumor xenografts plotted from tumor volumes recorded at different time points. Data are presented as mean ± SD from five mice. (c) Tumor weight was measured after the mice were killed. (d) Representative images of TUNEL staining from each group (400×). *p < 0.05.
DISCUSSION
NFBD1 plays an important role in DDR and is involved in a complex network of signaling pathways that regulate cell cycle checkpoints, DNA repair, and cell death (26–29). Previously, it was reported that NFBD1 is overexpressed in human cervical cancer and that the levels of NFBD1 might correlate with cancer grade (11). However, Bartkova et al. reported that NFBD1 expression was aberrantly reduced or lost in lung and breast cancers compared with normal tissues; in sharp contrast to both breast and lung tumors, no evidence for an aberrant lack of NFBD1 was found in any human testicular germ cell tumors (12). Additionally, in a cohort of early stage breast cancers, a decrease in the expression of NFBD1 was found to positively correlate with nodal failure; however, this did not correlate with ipsilateral breast relapse-free survival (IBRFS), distant metastasis-free survival (DMFS), or overall survival (OS) (30). Clearly, there have been conflicting reports about the role of NFBD1 in the malignancy of different tumors. To better understand the role of NFBD1 in NPC, we first determined the expression of NFBD1 in NPC and found that NFBD1 mRNA and protein levels were elevated in NPC tissues compared with normal nasopharyngeal tissues.
To further observe the detailed biological functions of the NFBD1 gene in NPC cells, we silenced the expression of NFBD1 using lentivirus-mediated shRNA. We found that silencing NFBD1 significantly inhibited the growth of NPC cell lines, reduced colony formation, and enhanced the apoptosis and sensitivity of NPC cells following exposure to chemotherapeutic agents. More importantly, the results of xenograft models in nude mice showed that silencing NFBD1 inhibited tumor growth and induced apoptosis in vivo. These findings suggest that NFBD1 might be a potential therapeutic target for improving the prognosis of patients with NPC.
NFBD1 is a signal amplifier of the p53 pathway and is vital for protecting cells from p53-mediated apoptosis under normal culture conditions and from the early phase of the DDR (31). The tumor suppressor p53 is a master regulator of the cellular response to stress, and it is activated in response to a wide range of cellular insults ranging from oncogene activation to DNA damage. These in turn drive diverse cellular outcomes such as cell cycle arrest and apoptosis (32–34). Previously, it was found that among the transcriptional targets of p53, there are several potential ROS-generating genes that presumably contribute to p53-mediated cell death (35–37). However, other p53-upregulated genes such as glutathione peroxidase (GPX-1) (38,39), Mn-superoxide dismutase (Mn-SOD) (39), and aldehyde dehydrogenase 4 (ALDH4) (40) would presumably act as antioxidants. In addition, the function of two p53-regulated sestrins (HI95 and PA26) is essential for the regeneration of overoxidized peroxiredoxins (41), which are enzymes involved in the decomposition of hydrogen peroxide (42). These findings suggest that p53 might play opposing roles in the regulation of ROS. We show here that silencing NFBD1 downregulates p53 expression. We also observed that downregulation of p53 increases the expression of its downstream antioxidant including ALDH4A1, SOD2, GPX-1/2, SESN2, and PA26. These results show that NFBD1 could negatively regulate intracellular ROS production via p53. Furthermore, ROS appear to be indispensable for the signal transduction pathways that regulate cell growth and the redox status. However, overproduction of ROS can damage lipids, proteins, and DNA (43–47). This study demonstrates that silencing NFBD1 leads to an increase in intracellular ROS production and a loss of MMP. An increase in ROS and a decrease in the MMP have been reported to be typical phenomena observed during mitochondria-dependent apoptosis (48–50). The decrease in MMP also induces apoptosis by causing the release of proapoptotic factors such as cytochrome c from the mitochondrial inner space to the cytosol (51). The release of cytochrome c from the mitochondria can activate caspase 9, which in turn activates executioner caspase 3 via cleavage induction (48). This hypothesis is strongly supported by our findings that the antioxidant NAC can effectively block the induction of apoptosis, the production of ROS, and the loss of MMP caused by the depletion of NFBD1.
Additionally, mitochondria have a central role in intrinsic apoptotic signaling by providing amplification factors such as cytochrome c, and this execution pathway is regulated by the Bcl-2 family (52). This is generally the function of a balance between proapoptotic members such as Bax and Puma and their antiapoptotic counterparts Bcl-2 and Bcl-xL. As a master regulator of the cellular response to stress, p53 controls a large transcriptional program that often activates genes with seemingly opposite functions such as proapoptotic genes, including Bax and Puma, and antiapoptotic genes, such as Bcl-2 and Bcl-xl. During apoptotic signaling, Bax translocates to the mitochondria and induces a loss of MMP, which facilitates the egress of proapoptotic factors and the assembly of the downstream apoptosome (53,54). By contrast, the overexpression of Bcl-2 prevents the oligomerization of Bax at the mitochondria and thereby inhibits cell death signaling (53). Interestingly, our results show that silencing NFBD1 triggers the loss of MMP in a ROS-dependent manner, whereas pretreatment with NAC or p53 siRNA prevented any significant effects of NFBD1 depletion on apoptotic activity. These data suggest that the silencing of NFBD1 could mediate its antitumor effects via the p53-ROS-mitochondria pathway.
In summary, we report here that the NFBD1 protein is overexpressed in NPC tissues and that silencing NFBD1 can inhibit cell growth, induce apoptosis, increase the production of intracellular ROS, and enhance the sensitivity of NPC cells to chemotherapeutic agents. Furthermore, NFBD1 knockdown also inhibits the tumorigenicity of NPC cells in vivo. Mechanistically, silencing NFBD1 might trigger the p53-ROS-mitochondrial apoptotic pathway. These findings broaden our understanding of the role of NFBD1 with respect to the ROS network and highlight a functional interaction that could be exploited therapeutically.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81470676 and 81271061), Chongqing Postgraduate Research Innovation Project (CYB15097), and National Key Clinical Specialties Construction Program of China. Zhihai Wang conceived and designed the experiments, performed the experiments, analyzed the data, and wrote the manuscript. Quan Zeng, Kui Liao, and Xueliang Liu performed the experiments and analyzed the data. Zhili Qiu, Zhitao Gong, and Chuan Liu performed the experiments and assisted with writing the manuscript. Wenqi Zuo, Yi Qian, and Liang Jiang provided human NPC patient samples. Suling Hong and Youquan Bu analyzed the data. Guohua Hu conceived and designed the research, interpreted the results, wrote the manuscript, and oversaw the research project. All authors read and approved the final manuscript.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Wei W. I.; Sham J. S. Nasopharyngeal carcinoma. Lancet 365:2041–2054; 2005. [DOI] [PubMed] [Google Scholar]
- 2. Bensouda Y.; Kaikani W.; Ahbeddou N.; Rahhali R.; Jabri M.; Mrabti H.; Boussen H.; Errihani H. Treatment for metastatic nasopharyngeal carcinoma. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 128:79–85; 2010. [DOI] [PubMed] [Google Scholar]
- 3. Goldberg M.; Stucki M.; Falck J.; D’Amours D.; Rahman D.; Pappin D.; Bartek J.; Jackson S. P. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421:952–956; 2003. [DOI] [PubMed] [Google Scholar]
- 4. Stewart G. S.; Wang B.; Bignell C. R.; Taylor A. M. R.; Elledge S. J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961–966; 2003. [DOI] [PubMed] [Google Scholar]
- 5. Stiff T.; O’Driscoll M.; Rief N.; Iwabuchi K.; Lobrich M.; Jeggo P. A. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64:2390–2396; 2004. [DOI] [PubMed] [Google Scholar]
- 6. Eliezer Y.; Argaman L.; Kornowski M.; Roniger M.; Goldberg M. Interplay between the DNA damage proteins MDC1 and ATM in the regulation of the spindle assembly checkpoint. J. Biol. Chem. 289:8182–8193; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lou Z.; Minter-Dykhouse K.; Wu X.; Chen J. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421:957–961; 2003. [DOI] [PubMed] [Google Scholar]
- 8. Lou Z.; Minter-Dykhouse K.; Franco S.; Gostissa M.; Rivera M. A.; Celeste A.; Manis J. P.; van Deursen J.; Nussenzweig A.; Paull T. T. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21:187–200; 2006. [DOI] [PubMed] [Google Scholar]
- 9. Zeng Q.; Wang Z.; Liu C.; Gong Z.; Yang L.; Jiang L.; Ma Z.; Qian Y.; Yang Y.; Kang H.; Hong S.; Bu Y.; Hu G. Knockdown of NFBD1/MDC1 enhances chemosensitivity to cisplatin or 5-fluorouracil in nasopharyngeal carcinoma CNE1 cells. Mol. Cell. Biochem. 418:137–146; 2016. [DOI] [PubMed] [Google Scholar]
- 10. Wang Z.; Zeng Q.; Chen T.; Liao K.; Bu Y.; Hong S.; Hu G. Silencing NFBD1/MDC1 enhances the radiosensitivity of human nasopharyngeal cancer CNE1 cells and results in tumor growth inhibition. Cell Death Dis. 6:e1849; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yuan C.; Bu Y.; Wang C.; Yi F.; Yang Z.; Huang X.; Cheng L.; Liu G.; Wang Y.; Song F. NFBD1/MDC1 is a protein of oncogenic potential in human cervical cancer. Mol. Cell. Biochem. 359:333–346; 2012. [DOI] [PubMed] [Google Scholar]
- 12. Bartkova J.; Horejsi Z.; Sehested M.; Nesland J. M.; Rajpert-De Meyts E.; Skakkebaek N. E.; Stucki M.; Jackson S.; Lukas J.; Bartek J. DNA damage response mediators MDC1 and 53BP1: Constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene 26:7414–7422; 2007. [DOI] [PubMed] [Google Scholar]
- 13. Caputo F.; Vegliante R.; Ghibelli L. Redox modulation of the DNA damage response. Biochem. Pharmacol. 84:1292–1306; 2012. [DOI] [PubMed] [Google Scholar]
- 14. Hosoki A.; Yonekura S.; Zhao Q. L.; Wei Z. L.; Takasaki I.; Tabuchi Y.; Wang L. L.; Hasuike S.; Nomura T.; Tachibana A.; Hashiguchi K.; Yonei S.; Kondo T.; Zhang-Akiyama Q. M. Mitochondria-targeted superoxide dismutase (SOD2) regulates radiation resistance and radiation stress response in HeLa cells. J. Radiat. Res. 53:58–71; 2012. [DOI] [PubMed] [Google Scholar]
- 15. Ihrlund L. S.; Hernlund E.; Khan O.; Shoshan M. C. 3-Bromopyruvate as inhibitor of tumour cell energy metabolism and chemopotentiator of platinum drugs. Mol. Oncol. 2:94–101; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nair R. R.; Bagheri M.; Saini D. K. Temporally distinct roles of ATM and ROS in genotoxic-stress-dependent induction and maintenance of cellular senescence. J. Cell Sci. 128:342–353; 2015. [DOI] [PubMed] [Google Scholar]
- 17. Hekimi S. Enhanced immunity in slowly aging mutant mice with high mitochondrial oxidative stress. Oncoimmunology 2(4):e23793; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hopfner K. P.; Karcher A.; Shin D. S.; Craig L.; Arthur L. M.; Carney J. P.; Tainer J. A. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101:789–800; 2000. [DOI] [PubMed] [Google Scholar]
- 19. Sedelnikova O. A.; Redon C. E.; Dickey J. S.; Nakamura A. J.; Georgakilas A. G.; Bonner W. M. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat. Res. 704:152–159; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cadet J.; Ravanat J. L.; TavernaPorro M.; Menoni H.; Angelov D. Oxidatively generated complex DNA damage: Tandem and clustered lesions. Cancer Lett. 327:5–15; 2012. [DOI] [PubMed] [Google Scholar]
- 21. Kaneyuki Y.; Yoshino H.; Kashiwakura I. Involvement of intracellular reactive oxygen species and mitochondria in the radiosensitivity of human hematopoietic stem cells. J. Radiat. Res. 53:145–150; 2012. [DOI] [PubMed] [Google Scholar]
- 22. Chen J.; Liu B.; Yuan J.; Yang J.; Zhang J.; An Y.; Tie L.; Pan Y.; Li X. Atorvastatin reduces vascular endothelial growth factor (VEGF) expression in human non-small cell lung carcinomas (NSCLCs) via inhibition of reactive oxygen species (ROS) production. Mol. Oncol. 6:62–72; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu C.; Gong K.; Mao X.; Li W. Tetrandrine induces apoptosis by activating reactive oxygen species and repressing Akt activity in human hepatocellular carcinoma. Int. J. Cancer 129:1519–1531; 2011. [DOI] [PubMed] [Google Scholar]
- 24. Zhu B.; Li X.; Zhang Y.; Ye C.; Wang Y.; Cai S.; Huang H.; Cai Y.; Yeh S.; Huang Z.; Chen R.; Tao Y.; Wen X. Cross-talk of alpha tocopherol-associated protein and JNK controls the oxidative stress-induced apoptosis in prostate cancer cells. Int. J. Cancer 132:2270–2282; 2013. [DOI] [PubMed] [Google Scholar]
- 25. Porcedda P.; Turinetto V.; Lantelme E.; Fontanella E.; Chrzanowska K.; Ragona R.; De Marchi M.; Delia D.; Giachino C. Impaired elimination of DNA double-strand break-containing lymphocytes in ataxia telangiectasia and Nijmegen breakage syndrome. DNA Repair (Amst.) 5:904–913; 2006. [DOI] [PubMed] [Google Scholar]
- 26. Nakanishi M.; Ozaki T.; Yamamoto H.; Hanamoto T.; Kikuchi H.; Furuya K.; Asaka M.; Delia D.; Nakagawara A. NFBD1/MDC1 associates with p53 and regulates its function at the crossroad between cell survival and death in response to DNA damage. J. Biol. Chem. 282:22993–23004; 2007. [DOI] [PubMed] [Google Scholar]
- 27. Bartek J.; Lukas J. DNA damage checkpoints: From initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19:238–245; 2007. [DOI] [PubMed] [Google Scholar]
- 28. Kastan M. B. Bartek J. Cell-cycle checkpoints and cancer. Nature 432:316–323; 2004. [DOI] [PubMed] [Google Scholar]
- 29. Burma S.; Chen B. P.; Murphy M.; Kurimasa A.; Chen D. J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462–42467; 2001. [DOI] [PubMed] [Google Scholar]
- 30. Patel A. N.; Goyal S.; Wu H.; Schiff D.; Moran M. S.; Haffty B. G. Mediator of DNA damage checkpoint protein 1 (MDC1) expression as a prognostic marker for nodal recurrence in early-stage breast cancer patients treated with breast-conserving surgery and radiation therapy. Breast Cancer Res. Treat. 126:601–607; 2011. [DOI] [PubMed] [Google Scholar]
- 31. Mitsuru N.; Toshinori O.; Hideki Y.; Takayuki H.; Hironobu K.; Kazushige F.; Masahiro A.; Domenico D.; Akira N. NFBD1/MDC1 associates with p53 and regulates its function at the crossroad between cell survival and death in response to DNA damage. J. Biol. Chem. 282:22993–23004; 2007. [DOI] [PubMed] [Google Scholar]
- 32. Vousden K. H.; Prives C. Blinded by the light: The growing complexity of p53. Cell 137:413–431; 2009. [DOI] [PubMed] [Google Scholar]
- 33. Bieging K. T., Mello S. S., and Attardi L. D. (2014) Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14:359–370; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sullivan K. D.; Gallant-Behm C. L.; Henry R. E.; Fraikin J. L.; Espinosa J. M. The p53 circuit board. Biochim. Biophys. Acta 1825:229–244; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Macip S.; Igarashi M.; Berggren P.; Yu J.; Lee S. W.; Aaronson S. A. Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol. Cell. Biol. 23:8576–8585; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sablina A. A.; Budanov A. V.; Ilyinskaya G. V.; Agapova L. S.; Kravchenko J. E.; Chumakov P. M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 11:1306–1313; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Polyak K.; Xia Y.; Zweier J. L.; Kinzler K. W.; Vogelstein B. A model for p53-induced apoptosis. Nature 389:300–305; 1997. [DOI] [PubMed] [Google Scholar]
- 38. Tan M.; Li S.; Swaroop M.; Guan K.; Oberley L. W.; Sun Y. Transcriptional activation of the human glutathione peroxidase promoter by p53. J. Biol. Chem. 274:12061–12066; 1999. [DOI] [PubMed] [Google Scholar]
- 39. Hussain S. P.; Amstad P.; He P.; Robles A.; Lupold S.; Kaneko I.; Ichimiya M.; Sengupta S.; Mechanic L.; Okamura S. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer Res. 64:2350–2356; 2004. [DOI] [PubMed] [Google Scholar]
- 40. Yoon K. A.; Nakamura Y.; Arakawa H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J. Hum. Gen. 49:134–140; 2004. [DOI] [PubMed] [Google Scholar]
- 41. Budanov A. V.; Sablina A. A.; Elena F.; Koonin E. V.; Chumakov P. M. Regeneration of peroxiredoxins by p53-regulated destrins, homologs of bacterial AhpD. Science 304:596–600; 2004. [DOI] [PubMed] [Google Scholar]
- 42. Jackson A. L.; Loeb L. A. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat. Res. 477:7–21; 2001. [DOI] [PubMed] [Google Scholar]
- 43. Gul N.; Grewal S.; Bogels M.; van der Bij G. J.; Koppes M. M.; Oosterling S. J.; Fluitsma D. M.; Hoeben K. A.; Beelen R. H.; van Egmond M. Macrophages mediate colon carcinoma cell adhesion in the rat liver after exposure to lipopolysaccharide. Oncoimmunology 1:1517–1526; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. An J.; Shi K.; Wei W.; Hua F.; Ci Y.; Jiang Q.; Li F.; Wu P.; Hui K.; Yang Y. The ROS/JNK/ATF2 pathway mediates selenite-induced leukemia NB4 cell cycle arrest and apoptosis in vitro and in vivo. Cell Death Dis. 4:e973; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Garrido-Urbani S.; Jaquet V.; Imhof B. A. [ROS and NADPH oxidase: Key regulators of tumor vascularisation]. Med. Sci. (Paris) 30:415–421; 2014. [DOI] [PubMed] [Google Scholar]
- 46. Soriani A.; Iannitto M. L.; Ricci B.; Fionda C.; Malgarini G.; Morrone S.; Peruzzi G.; Ricciardi M. R.; Petrucci M. T.; Cippitelli M. Reactive oxygen species- and DNA damage response-dependent NK cell activating ligand upregulation occurs at transcriptional levels and requires the transcriptional factor E2F1. J. Immunol. 193:950–960; 2014. [DOI] [PubMed] [Google Scholar]
- 47. Guo J.; Wu G.; Bao J.; Hao W.; Lu J.; Chen X. Cucurbitacin B induced ATM-mediated DNA damage causes G2/M cell cycle arrest in a ROS-dependent manner. PLoS One 9:e88140; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vaux D. L.; Korsmeyer S. J. Cell death in development. Cell 96:245–254; 1999. [DOI] [PubMed] [Google Scholar]
- 49. Park K.-R.; Nam D.; Yun H.-M.; Lee S.-G.; Jang H.-J.; Sethi G.; Cho S. K.; Ahn K. S. β-Caryophyllene oxide inhibits growth and induces apoptosis through the suppression of PI3K/AKT/mTOR/S6K1 pathways and ROS-mediated MAPKs activation. Cancer Lett. 312:178–188; 2011. [DOI] [PubMed] [Google Scholar]
- 50. Niu D.; Zhang J.; Ren Y.; Feng H.; Chen W. N. HBx genotype D represses GSTP1 expression and increases the oxidative level and apoptosis in HepG2 cells. Mol. Oncol. 3:67–76; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Loo G.; Saelens X.; Van Gurp M.; MacFarlane M.; Martin S.; Vandenabeele P. The role of mitochondrial factors in apoptosis: A Russian roulette with more than one bullet. Cell Death Diff. 9:1031–1042; 2002. [DOI] [PubMed] [Google Scholar]
- 52. Kluck R. M.; Bossy-Wetzel E.; Green D. R.; Newmeyer D. D. The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science 275:1132–1136; 1997. [DOI] [PubMed] [Google Scholar]
- 53. Ghibelli L.; Diederich M. Multistep and multitask Bax activation. Mitochondrion 10:604–613; 2010. [DOI] [PubMed] [Google Scholar]
- 54. Luo A.; Chen H.; Ding F.; Zhang Y.; Wang M.; Xiao Z.; Liu Z. Small proline-rich repeat protein 3 enhances the sensitivity of esophageal cancer cells in response to DNA damage-induced apoptosis. Mol. Oncol. 7:955–967; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]