Abstract
MicroRNA-200a (miR-200a) is frequently downregulated in most cancer types and plays an important role in carcinogenesis and cancer progression. In this study, we determined that miR-200a was downregulated in hepatocellular carcinoma (HCC) tissues and cell lines, consistent with the results of our previous study. Because a previous study suggested that downregulation of miR-200a is correlated with HCC metastasis, we aimed to elucidate the mechanism underlying the role of miR-200a in metastasis in HCC. Here we observed that overexpression of miR-200a resulted in suppression of HCC metastatic ability, including HCC cell migration, invasion, and metastasis, in vitro and in vivo. Furthermore, bioinformatics and luciferase reporter assays indicated that GAB1 is a direct target of miR-200a. Inhibition of GAB1 resulted in substantially decreased cell invasion and migration similar to that observed with overexpression of miR-200a in HCC cell lines, whereas restoration of GAB1 partially rescued the inhibitory effects of miR-200a. Taken together, these data provide novel information for comprehending the tumor-suppressive role of miR-200a in HCC pathogenesis through inhibition of GAB1 translation.
Key words: miR-200a, Hepatocellular carcinoma (HCC), Grb2-associated binding protein 1 (GAB1), Invasion and migration, Metastasis
INTRODUCTION
Hepatocellular carcinoma (HCC) is the sixth most prevalent and aggressive human malignancy and the third leading cause of cancer-related mortality worldwide1. The poor prognosis of HCC is attributed to tumor invasiveness, frequent intrahepatic spread, and extrahepatic metastasis at initial diagnosis2. Therefore, preventing the metastasis of cancer cells is a critical and effective therapeutic strategy for the successful management of HCC.
MicroRNAs (miRNAs) are a set of highly conserved noncoding small RNAs that regulate gene expression by binding to the 3′-untranslated region (3′-UTR) of target mRNAs3,4. Accumulating evidence suggests that miRNAs are involved in the development and progression of various human cancers3,5,6. Some miRNAs have been identified to function as tumor suppressors or oncogenes and participate in the regulation of various fundamental biological processes in HCC, including tumorigenesis, angiogenesis, proliferation, apoptosis, invasion, and metastasis7–13. Therefore, miRNAs may be potential therapeutic targets in HCC.
Emerging data have demonstrated that microRNA-200a (miR-200a) is frequently downregulated and may act as a tumor suppressor in many cancer types, including breast cancer, nasopharyngeal cancer, gastric cancer, colorectal cancer, and HCC14–18. Restoration of miR-200a expression has been proposed as a novel treatment strategy in HCC. We previously reported that miR-200a is downregulated in HCC tissues and cell lines and that reduced miR-200a expression is correlated with HCC metastasis and poor prognosis19. Experimental data suggest that miR-200a directly regulates several target mRNAs, including ZEB215, β-catenin20, CDK-621, HDAC418, IGF222, EPHA223, TGFB224, SIRT114, and Ap-2γ25. However, the potential mechanisms of miR-200a in affecting the malignant phenotype of HCC have not been completely elucidated. In the present study, we observed frequent downregulation of miR-200a in HCC tissues and cell lines, consistent with the results of our previous study19. In vitro and in vivo assays demonstrated that overexpression of miR-200a inhibits cell invasion and metastasis of HCC. Moreover, we determined that Grb2-associated binding protein 1 (GAB1), a novel tumor-suppressor gene, is a direct target of miR-200a. Silencing of GAB1 resulted in substantially decreased cell migration and invasion in HCC cell lines, whereas restoration of GAB1 reversed the inhibitory effects of miR-200a. Therefore, our results suggest a critical role for miR-200a in HCC pathogenesis and its possible application as a target for cancer therapy.
MATERIALS AND METHODS
Patients and Tissue Specimens
Forty-eight patients who were diagnosed with HCC at the Department of Hepatobiliary Surgery of Xijing Hospital of the Fourth Military Medical University between 2012 and 2015 were included in this study. The study protocol was approved by the Ethics Committee of Xijing Hospital. All participants provided informed written consent.
Cell Culture
The human HCC cell lines MHCC97-H, SMMC-7721, Huh7, and HepG2, the normal human hepatocyte cell line HL-7702, 293T cells (obtained from the Cell Bank of the Chinese Academy of Sciences) were cultivated in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin G, and 100 μg/ml streptomycin. In all experiments, cells were cultured at 37°C in a humidified 5% CO2/95% air atmosphere.
Plasmids and Cell Transfection
Lentiviral plasmids encoding miR-200a (LV-miR-200a) and empty vector (LV-miR-NC) were designed and produced by Genechem (Shanghai, P.R. China). MHCC97-H and SMMC-7721 were plated in six-well plates and incubated overnight. The culture medium was replaced with transduction-enhancing solution containing 20 MOI lentivirus and 50 μg/ml polybrene. After 12 h, the medium was replaced with complete medium, and the cells were cultured for 72 h. The cells were transfected with miR-200a mimic, a mimic negative control, siRNA for GAB1 (si-GAB1), or siRNA-negative control (NC) (GenePharma, Shanghai, P.R. China) and pcDNA3.1-GAB1 plasmid lacking the 3′-UTR or pcDNA3.1-GAB1-NC (Genechem) using Lipofectamine 2000 (Invitrogen, USA) following the manufacturer’s instructions.
RNA Extraction and Real-Time PCR
Total RNA was isolated using RNAiso Plus reagent (TaKaRa, Dalian, P.R. China) according to the manufacturer’s instructions. miR-200a expression levels were quantified using a miScript PCR system (Qiagen, Hilden, Germany), a miScript II RT kit, miScript primer assays, and a miScript SYBR Green PCR kit. Small nuclear RNA U6 was employed for internal normalization.
For mRNA analysis, complementary DNA (cDNA) was generated with oligo-dT primers using the PrimeScript RT Reagent Kit (TaKaRa). The generated cDNA was amplified using SYBR Premix EX Taq II (TaKaRa) on a Bio-Rad IQTM5 detection system (Bio-Rad, Hercules, CA, USA). β-Actin was used as an internal control to normalize the mRNA expression levels of the target genes. All experiments were performed according to the manufacturer’s instructions. All reactions were assessed in triplicate, and data were analyzed with the comparative Ct (2−ΔΔCt) method.
Western Blot Analysis
The cells were washed three times with cold PBS and lysed in RIPA lysis buffer (Beyotime, Shanghai, P.R. China) containing fresh protease and phosphatase inhibitor cocktails (Sigma-Aldrich). The protein concentration was determined using the Bio-Rad assay system (Bio-Rad). The total protein extracts were separated by 10% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel electrophoresis, transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA), and sequentially incubated with primary antibodies against GAB1 (1:1,000; Cell Signaling Technology) and β-actin (1:2,000; Cell Signaling Technology) overnight at 4°C, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000; Abcam) at room temperature for 1 h. The blots were developed using enhanced chemiluminescence detection reagents (Pierce, Rockford, IL, USA) and scanned with a Molecular Imager System (Bio-Rad).
Migration and Invasion Assays
Cell migration ability was analyzed with non-Matrigel-coated Transwell cell culture chambers (8-μm pore size) (Millipore), and cell invasion ability was analyzed with Matrigel-coated Transwell cell culture chambers. A total of 3 × 104 cells were added to the top chamber on the noncoated or Matrigel-coated membrane (24-well insert) and allowed to migrate toward the serum-containing medium in the lower chamber. The cells were incubated for 24 h, then fixed with 4% formaldehyde for 10 min and stained with 0.5% crystal violet for 30 min. The invaded and migrated cells were counted at 200× magnification from five different fields of each sample.
In Vivo Pulmonary Metastasis Assays
All experimental procedures involving mice were performed in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by the Research Animal Care and Use Committee of the Fourth Military Medical University. For the in vivo pulmonary metastasis assay, 4- to 5-week-old female BALB/c nude mice (n = 8 mice per group) were injected through the lateral tail vein with MHCC97-H cells (2 × 106) transfected with miR-NC or miR-200a. After 5 weeks, the mice were sacrificed, and the lungs were collected and fixed in 10% formalin. After paraffin embedding, the lung samples were sectioned and stained with hematoxylin and eosin (H&E).
Luciferase Reporter Assay
The luciferase reporter assay was performed in 293T cells. The 293T cells were seeded into a 24-well plate and cotransfected with luciferase reporter constructs encoding the wild-type 3′-UTR region of GAB1 (GAB1-WT-3′-UTR) or a mutated GAB1 3′-UTR region (GAB1-MUT-3′-UTR) (RIBOBIO, Guangzhou, P.R. China) and miR-200a mimic or mi-NC using Lipofectamine 2000 (Invitrogen). After 48-h of incubation, the cells were washed with PBS and lysed with Passive Lysis Buffer (Promega). Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s protocol.
Statistical Analysis
Each experiment was repeated at least three times. All data are presented as the mean ± SD. Student’s t-test (two tailed) or the Student–Newman–Keuls test (SNK test, ANOVA) was employed to analyze the differences using SPSS 13.0 software (Chicago, IL, USA). A value of p < 0.05 was considered statistically significant.
RESULTS
The Expression of miR-200a Is Downregulated in HCC Tissues and Cell Lines
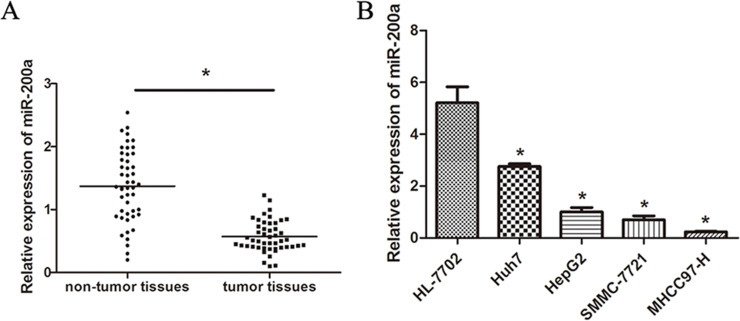
We previously demonstrated that miR-200a is greatly downregulated in HCC cell lines and tissues and is associated with HCC metastasis19. To investigate the expression of miR-200a in HCC tissues and cells, the expression of miR-200a in tumor specimens and four HCC cell lines (MHCC97-H, SMMC-7721, Huh7, and HepG2) was detected by real-time PCR. Our results demonstrated that the expression of miR-200a was significantly decreased in HCC tissues and cell lines (Fig. 1A and B). These data suggest that miR-200a may function as a tumor-suppressive gene.
Figure 1.
Expression of miR-200a in HCC tissues and cell lines. (A) Expression of miR-200a in HCC tissue and corresponding adjacent noncancerous tissues. (B) Relative expression of miR-200a in four HCC cell lines (Huh7, HepG2, SMMC-7721, and MHCC97-H) and a human normal liver cell line (HL-7702) as analyzed by RT-PCR. The data represent the mean ± SD. *p < 0.05.
miR-200a Inhibits HCC Cell Migration and Invasion In Vitro
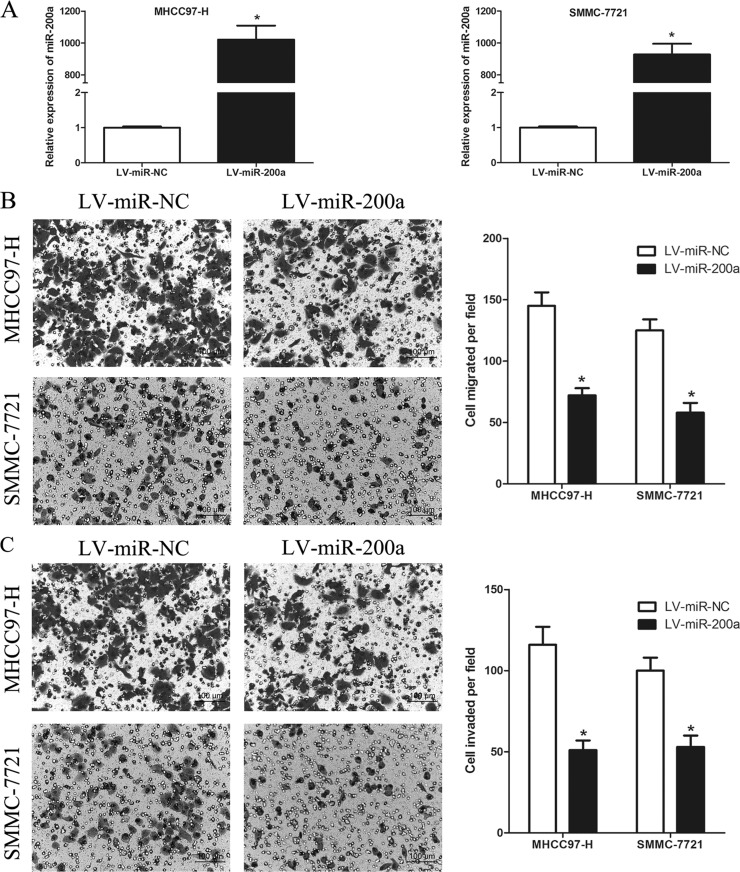
Because downregulation of miR-200a expression is correlated with HCC metastasis, we hypothesized that miR-200a might regulate this process. To test this hypothesis, we first transfected the lentiviral plasmid LV-miR-200a or a lentiviral control plasmid LV-miR-NC into the HCC cell lines MHCC97-H and SMMC-7721. As shown in Figure 2A, miR-200a was successfully overexpressed in the HCC cells transfected with LV-miR-200a. Then we investigated the role of miR-200a in HCC cell migration and invasion using in vitro Transwell assays. The Transwell assays revealed that overexpression of miR-200a dramatically suppressed cell migration and invasion in MHCC97-H and SMMC-7721 cells (p < 0.05) (Fig. 2B and C). These data imply that miR-200a can inhibit the metastatic ability of HCC cells by suppressing their migration and invasion.
Figure 2.
miR-200a inhibits HCC cell migration and invasion in vitro. (A) Expression of miR-200a following transfection in MHCC97-H and SMMC-7721 cells was confirmed by RT-PCR. (B and C) Overexpression of miR-200a significantly inhibited cell migration and invasion in MHCC97-H and SMMC-7721 cells. Migrated and invaded cells were counted in five randomly selected areas under a 200× microscope field. *p < 0.05 compared to the control.
miR-200a Suppresses HCC Cells Metastasis In Vivo
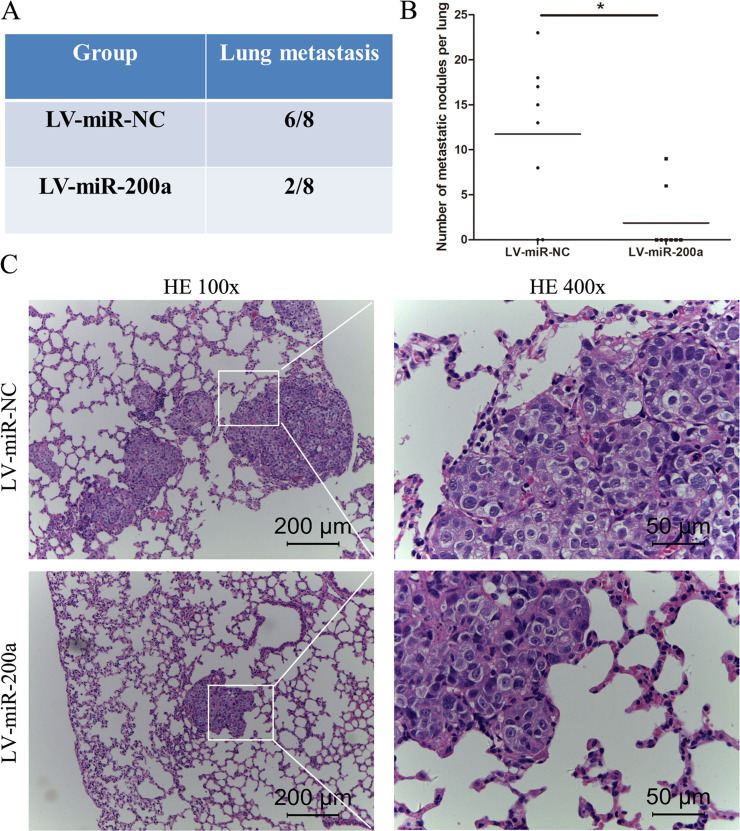
To further demonstrate the association between miR-200a and HCC cell metastasis, in vivo pulmonary metastasis assays of HCC cells were performed in nude mice. MHCC97-H cells transfected with LV-miR-200a or LV-miR-NC were transplanted into nude mice via lateral tail vein injection. After 5 weeks, the mice were sacrificed, and the lungs were dissected and fixed for H&E staining. As shown in Figure 3A and B, overexpression of miR-200a decreased the incidence of lung metastasis and the number of metastatic lung nodules. Representative H&E staining of metastatic lung nodules is shown in Figure 3C. These data indicate that miR-200a may act as a metastatic suppressor of HCC in vivo.
Figure 3.
miR-200a suppresses tumor metastasis in vivo. (A) Incidence of lung metastasis in mice injected with MHCC97-H cells transfected with miR-NC or miR-200a cells through the lateral tail vein. (B) The number of metastatic nodules on the surface of the lungs in mice from the different groups. (C) Representative H&E staining of lung metastatic nodules. *p < 0.05 compared to the control.
GAB1 Is a Direct Downstream Target of miR-200a
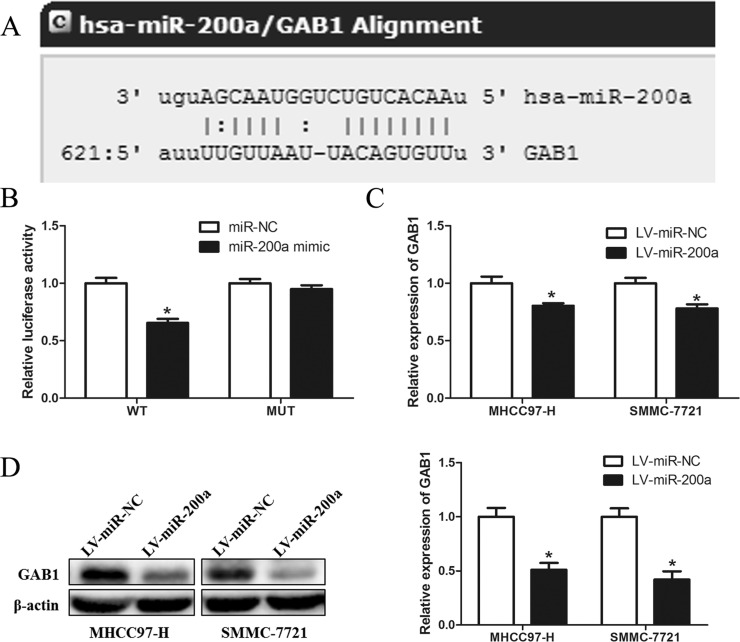
To further explore the mechanism of action of miR-200a in HCC metastasis suppression, we used bioinformatics tools to predict the putative target genes of miR-200a. Among the candidates, GAB1 was selected for further experimental validation. A predicted complementary sequence of miR-200a was identified in the 3′-UTR of GAB1 mRNA (Fig. 4A). As shown in Figure 4B, miR-200a significantly decreased the luciferase activity of GAB1 containing a wild-type 3′-UTR but did not decrease the activity of GAB1 with a mutant 3′-UTR in 293T cells. RT-PCR and Western blotting further demonstrated that overexpression of miR-200a markedly suppressed the mRNA and protein levels of GAB1 in MHCC97-H and SMMC-7721 cells (p < 0.05) (Fig. 4C and D). Taken together, these results strongly suggest that GAB1 is a target of miR-200a in HCC.
Figure 4.
GAB1 is a direct downstream target of miR-200a. (A) GAB1 was bioinformatically predicted as a target of miR-200a using online software (http://www.microrna.org/). (B) A miR-200a mimic or negative control and a luciferase vector encoding the wild-type or mutant GAB1 3′-UTR region were introduced into 239T cells, and the relative luciferase activity was measured. (C) Quantitation of GAB1 mRNA levels in MHCC97-H and SMMC-7721 cells by RT-PCR after transfection with LV-miR-NC or LV-miR-miR-200a. (D) Detection of GAB1 protein in MHCC97-H and SMMC-7721 cells by Western blot analysis after transfection with LV-miR-NC or LV-miR-200a. *p < 0.05 compared to the control.
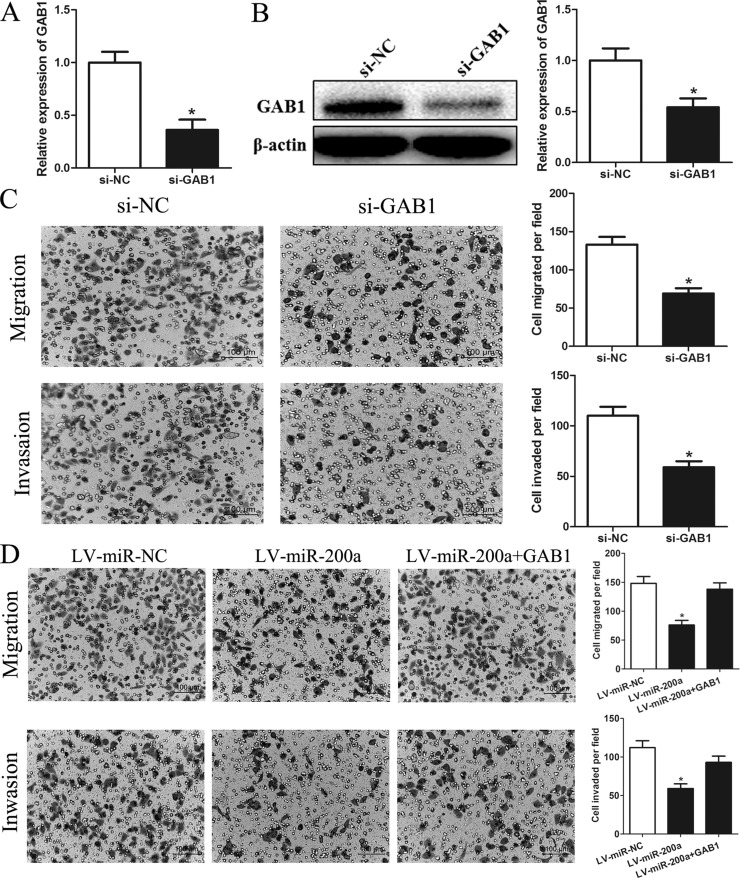
Alterations of GAB1 Levels Influence the Effects of miR-200a on HCC Cells
To determine if downregulation of GAB1 accounts for the inhibition of cell migration and invasion by miR-200a, we transfected SMMC-7721 cells with GAB1 siRNA to knock down endogenous GAB1 expression. As shown in Figure 5A and B, GAB1 was significantly downregulated in SMMC-7721 cells, and knockdown of GAB1 markedly inhibited cell migration and invasion (p < 0.05) (Fig. 5C). These results are similar to those induced by miR-200a. To further confirm that GAB1 is a functional target of miR-200a, we constructed a plasmid with GAB1 lacking its 3′-UTR and introduced the construct into miR-200a-overexpressing SMMC-7721 cells. The inhibitory effects of miR-200a on HCC cell migration and invasion were partially antagonized by restoring GAB1 expression (p < 0.05) (Fig. 5D). These data suggest that GAB1 is a downstream functional mediator of miR-200a.
Figure 5.
Alterations of GAB1 levels influence the effects of miR-200a on HCC cells. (A) Quantitation of GAB1 mRNA levels in SMMC-7721 cells by RT-PCR after transfection with siRNA targeting GAB1 (si-GAB1) or a negative control siRNA (si-NC). (B) Detection of GAB1 protein in SMMC-7721 cells by Western blot analysis after transfection with GAB1 siRNAs or si-NC. (C) GAB1 knockdown inhibited SMMC-7721 cell migration and invasion. (D) GAB1 reintroduction into SMMC-7721 cells partially rescued the miR-200a-mediated inhibition of cell migration and invasion. Migrated and invaded cells were counted in five randomly selected areas under a 200× microscope field. *p < 0.05 compared to the control.
DISCUSSION
miR-200a suppresses different biological processes such as proliferation, invasion, metastasis, and tumorigenesis in HCC18,21,26,27; however, the molecular mechanisms by which miR-200a inhibits HCC remain unclear. Therefore, in this study, we aimed to elucidate the biological functions and mechanism of miR-200a in HCC. Our results demonstrated that miR-200a is frequently downregulated in HCC tissues and cell lines, consistent with the results of our previous study. Furthermore, we elucidated the role of miR-200a deregulation in tumor invasion and metastasis using in vitro and in vivo assays. We observed that overexpression of miR-200a suppressed cell migration and invasion in vitro and restrained tumor metastasis in vivo. These results suggest that miR-200a is a novel tumor suppressor that plays an important role in the regulation of invasion and metastasis in HCC.
We also explored the exact molecular mechanism of miR-200a in suppressing invasion and metastasis in HCC. Luciferase reporter assays, RT-PCR, and Western blotting demonstrated that GAB1 is a direct target of miR-200a. More importantly, the effects of miR-200a modulation on cell migration and invasion were accompanied by alteration of GAB1 levels and activities. Overexpression of GAB1 abolished the effects induced by miR-200a. Thus, we confirmed that miR-200a plays a critical role in the inhibition of invasion and metastasis in HCC, partially by downregulating the expression of GAB1.
GAB1, which belongs to the Grb2-associated binder (Gab) family, has been reported to be involved in tumor occurrence, invasion, and metastasis, particularly in gastrointestinal tumors28–31. GAB1 is most commonly and widely distributed in mammals and plays a crucial role in transmitting key signals that control cell growth, differentiation, and function from multiple receptors28,32,33. Zhang et al.34 demonstrated that GAB1 protein expression is significantly associated with aggressive tumor progression and poor prognosis in patients with HCC. Felic et al.35 and Oka et al.36 observed that Gab1 upregulates proto-oncogene expression by amplifying signaling pathways related to tumor biological behaviors. Downregulation of GAB1 inhibits cell proliferation, invasion, and migration in intrahepatic and hilar cholangiocarcinoma and colorectal cancer30,31,37. Interestingly, we also observed that knockdown of GAB1 using siRNA can inhibit cell invasion and migration in HCC. Recent studies have reported that GAB1 and FOXP1 expression are modulated by miR-150, resulting in proficient B-cell receptor signaling in chronic lymphocytic leukemia38, and are also regulated by microRNA-409-3p, resulting in the suppression of colorectal cancer invasion and metastasis in colorectal cancer37. Our study identified GAB1 as the target of miR-200a in HCC and revealed an important role for GAB1 dysregulation in HCC progression. Thus, the posttranscriptional regulation of GAB1 expression by miRNA might be a novel mechanism by which GAB1 regulates cancer progression.
In summary, our results demonstrated that miR-200a is downregulated in HCC cell lines. Overexpression of miR-200a inhibits cell invasion and migration in vitro and metastasis of HCC in vivo partly by targeting GAB1. This study might provide new insight into the molecular mechanisms of HCC progression and metastasis and suggests that the miR-200a/GAB1 axis is a possible therapeutic strategy for the treatment of HCC.
ACKNOWLEDGMENT
This study was supported by the National Natural Science Foundation of China (Grant No. 81172061).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–55. [DOI] [PubMed] [Google Scholar]
- 2. Huo TI, Lin HC, Huang YH, Wu JC, Chiang JH, Lee PC, Lee SD. The model for end-stage liver disease-based Japan Integrated Scoring system may have a better predictive ability for patients with hepatocellular carcinoma undergoing locoregional therapy. Cancer 2006;107:141–48. [DOI] [PubMed] [Google Scholar]
- 3. Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–79. [DOI] [PubMed] [Google Scholar]
- 4. Iorio MV, Croce CM. MicroRNA involvement in human cancer. Carcinogenesis 2012;33:1126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiemer EA. The role of microRNAs in cancer: No small matter. Eur J Cancer 2007;43:1529–44. [DOI] [PubMed] [Google Scholar]
- 6. Mizuguchi Y, Takizawa T, Yoshida H, Uchida E. Dysregulated microRNAs in progression of hepatocellular carcinoma: A systematic review. Hepatol Res. 2016;46(5):391–406. [DOI] [PubMed] [Google Scholar]
- 7. Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 2012;55:1852–62. [DOI] [PubMed] [Google Scholar]
- 8. Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. MiR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA 2010;107:264–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang X, Zhang XF, Lu X, Jia HL, Liang L, Dong QZ, Ye QH, Qin LX. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology 2014;59:1874–85. [DOI] [PubMed] [Google Scholar]
- 10. Zhang Z, Zhang Y, Sun XX, Ma X, Chen ZN. MicroRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol Cancer 2015;14:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang L, Wong CM, Ying Q, Fan DN, Huang S, Ding J, Yao J, Yan M, Li J, Yao M, Ng IO, He X. MicroRNA-125b suppressed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology 2010;52:1731–40. [DOI] [PubMed] [Google Scholar]
- 12. Fornari F, Milazzo M, Chieco P, Negrini M, Marasco E, Capranico G, Mantovani V, Marinello J, Sabbioni S, Callegari E, Cescon M, Ravaioli M, Croce CM, Bolondi L, Gramantieri L. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21 PTEN, AKT3 and TIMP2. J Pathol. 2012;227:275–85. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Takahashi S, Tasaka A, Yoshima T, Ochi H, Chayama K. Involvement of microRNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:565–75. [DOI] [PubMed] [Google Scholar]
- 14. Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. MiR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem. 2011;286:25992–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xia H, Ng SS, Jiang S, Cheung WK, Sze J, Bian XW, Kung HF, Lin MC. MiR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth migration and invasion. Biochem Biophys Res Commun. 2010;391:535–41. [DOI] [PubMed] [Google Scholar]
- 16. Shinozaki A, Sakatani T, Ushiku T, Hino R, Isogai M, Ishikawa S, Uozaki H, Takada K, Fukayama M. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res. 2010;70:4719–27. [DOI] [PubMed] [Google Scholar]
- 17. Pichler M, Ress AL, Winter E, Stiegelbauer V, Karbiener M, Schwarzenbacher D, Scheideler M, Ivan C, Jahn SW, Kiesslich T, Gerger A, Bauernhofer T, Calin GA, Hoefler G. MiR-200a regulates epithelial to mesenchymal transition-related gene expression and determines prognosis in colorectal cancer patients. Br J Cancer 2014;110:1614–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuan JH, Yang F, Chen BF, Lu Z, Huo XS, Zhou WP, Wang F, Sun SH. The histone deacetylase 4/SP1/microRNA-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology 2011;54:2025–35. [DOI] [PubMed] [Google Scholar]
- 19. Yang X, Wang J, Qu S, Zhang H, Ruan B, Gao Y, Ma B, Wang X, Wu N, Li X, Dou K, Li H. MicroRNA-200a suppresses metastatic potential of side population cells in human hepatocellular carcinoma by decreasing ZEB2. Oncotarget 2015;6:7918–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su J, Zhang A, Shi Z, Ma F, Pu P, Wang T, Zhang J, Kang C, Zhang Q. MicroRNA-200a suppresses the Wnt/beta-catenin signaling pathway by interacting with beta-catenin. Int J Oncol. 2012;40:1162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiao F, Zhang W, Zhou L, Xie H, Xing C, Ding S, Chen K, Zheng S. MicroRNA-200a is an independent prognostic factor of hepatocellular carcinoma and induces cell cycle arrest by targeting CDK6. Oncol Rep. 2013;30:2203–10. [DOI] [PubMed] [Google Scholar]
- 22. Saha S, Choudhury J, Ain R. MicroRNA-141-3p and miR-200a-3p regulate insulin-like growth factor 2 during mouse placental development. Mol Cell Endocrinol. 2015;414:186–93. [DOI] [PubMed] [Google Scholar]
- 23. Tsouko E, Wang J, Frigo DE, Aydogdu E, Williams C. MiR-200a inhibits migration of triple-negative breast cancer cells through direct repression of the EPHA2 oncogene. Carcinogenesis 2015;36:1051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu R, Ji Z, Li X, Qin J, Cui G, Chen J, Zhai Q, Zhao C, Zhang W, Yu Z. Tumor suppressive microRNA-200a inhibits renal cell carcinoma development by directly targeting TGFB2. Tumour Biol. 2015;36:6691–700. [DOI] [PubMed] [Google Scholar]
- 25. Gao SL, Wang LZ, Liu HY, Liu DL, Xie LM, Zhang ZW. MiR-200a inhibits tumor proliferation by targeting AP-2gamma in neuroblastoma cells. Asian Pac J Cancer Prev. 2014;15:4671–6. [DOI] [PubMed] [Google Scholar]
- 26. Hung CS, Liu HH, Liu JJ, Yeh CT, Chang TC, Wu CH, Ho YS, Wei PL, Chang YJ. MicroRNA-200a and-200b mediated hepatocellular carcinoma cell migration through the epithelial to mesenchymal transition markers. Ann Surg Oncol. 2013;20(Suppl 3):S360–8. [DOI] [PubMed] [Google Scholar]
- 27. Feng J, Wang J, Chen M, Chen G, Wu Z, Ying L, Zhuo Q, Zhang J, Wang W. MiR-200a suppresses cell growth and migration by targeting MACC1 and predicts prognosis in hepatocellular carcinoma. Oncol Rep. 2015;33:713–20. [DOI] [PubMed] [Google Scholar]
- 28. Yamasaki S, Nishida K, Yoshida Y, Itoh M, Hibi M, Hirano T. Gab1 is required for EGF receptor signaling and the transformation by activated ErbB2. Oncogene 2003;22:1546–56. [DOI] [PubMed] [Google Scholar]
- 29. Seiden-Long I, Navab R, Shih W, Li M, Chow J, Zhu CQ, Radulovich N, Saucier C, Tsao MS. Gab1 but not Grb2 mediates tumor progression in Met overexpressing colorectal cancer cells. Carcinogenesis 2008;29:647–55. [DOI] [PubMed] [Google Scholar]
- 30. Sang H, Li T, Li H, Liu J. Down-regulation of Gab1 inhibits cell proliferation and migration in hilar cholangiocarcinoma. PLoS One 2013;8:e81347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sang H, Li T, Li H, Liu J. Gab1 regulates proliferation and migration through the PI3K/Akt signaling pathway in intrahepatic cholangiocarcinoma. Tumour Biol. 2015;36:8367–77. [DOI] [PubMed] [Google Scholar]
- 32. Gu H, Neel BG. The “Gab” in signal transduction. Trends Cell Biol. 2003;13:122–30. [DOI] [PubMed] [Google Scholar]
- 33. Nishida K, Hirano T. The role of Gab family scaffolding adapter proteins in the signal transduction of cytokine and growth factor receptors. Cancer Sci. 2003;94:1029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y, Li Z, Yang M, Wang D, Yu L, Guo C, Guo X, Lin N. Identification of GRB2 and GAB1 coexpression as an unfavorable prognostic factor for hepatocellular carcinoma by a combination of expression profile and network analysis. PLoS One 2013;8:e85170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Felici A, Giubellino A, Bottaro DP. Gab1 mediates hepatocyte growth factor-stimulated mitogenicity and morphogenesis in multipotent myeloid cells. J Cell Biochem. 2010;111:310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oka M, Kikkawa U, Nishigori C. Protein kinase C-betaII represses hepatocyte growth factor-induced invasion by preventing the association of adapter protein Gab1 and phosphatidylinositol 3-kinase in melanoma cells. J Invest Dermatol. 2008;128:188–95. [DOI] [PubMed] [Google Scholar]
- 37. Bai R, Weng C, Dong H, Li S, Chen G, Xu Z. MicroRNA-409-3p suppresses colorectal cancer invasion and metastasis partly by targeting GAB1 expression. Int J Cancer 2015;137:2310–22. [DOI] [PubMed] [Google Scholar]
- 38. Mraz M, Chen L, Rassenti LZ, Ghia EM, Li H, Jepsen K, Smith EN, Messer K, Frazer KA, Kipps TJ. MiR-150 influences B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1. Blood 2014;124:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]