Abstract
In this study, the putative genes involved in diterpenoid alkaloids biosynthesis in A. vilmorinianum roots were revealed by transcriptome sequencing. 59.39 GB of clean bases and 119,660 unigenes were assembled, of which 69,978 unigenes (58.48%) were annotated. We identified 27 classes of genes (139 candidate genes) involved in the synthesis of diterpenoid alkaloids, including the mevalonate (MVA) pathway, the methylerythritol 4-phosphate (MEP) pathway, the farnesyl diphosphate regulatory pathway, and the diterpenoid scaffold synthetic pathway. 12 CYP450 genes were identified. We found that hydroxymethylglutaryl-CoA reductase was the key enzyme in MVA metabolism, which was regulated by miR6300. Transcription factors, such as bHLH, AP2/EREBP, and MYB, used to synthesize the diterpenes were analyzed.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02646-6.
Keywords: Aconitum vilmorinianum, Transcriptome, Aconitine, Diterpene, Biosynthesis pathway, Traditional Chinese medicine
Introduction
The genus Aconitum (Ranunculaceae) is comprised of 350 species worldwide, with 211 species from China, and 76 species are used as aconitine medicinal plants in China (Li et al. 2012). Aconitine is a kind of terpenoid, which plays important roles in plant growth, development, and the synthesis of secondary metabolites. Many terpenoids with pharmacological activities are widely used for medical purposes (Li et al. 2013; Yang et al. 2013; Zhao et al. 2018a, b). Diterpenoid alkaloids are the main active components in Aconitum plants. The mevalerate (MVA) and methylerythritol 4-phosphate (MEP) pathways are upstream of diterpene alkaloid biosynthesis, and both form isopentenyl diphosphate (IPP), and then further synthesize terpenes. In the biosynthesis of diterpenoid alkaloids three IPP molecules are condensed into geranylgeranyl diphosphate (GGPP), to initiate biosynthesis of the diterpene alkaloid skeleton (Gao et al. 2009; Kai et al. 2010), and to form various diterpenoid compounds through CYP450-mediated oxidation and modification (Cherney and Baran 2011; Devkota and Sewald 2013) (Fig. S1).
The species Aconitum vilmorinianum is an important medicinal plant in the genus Aconitum and has been a focus because of its pharmacological action and trace toxicity. A. vilmorinianum is one of the main raw materials of Yunnan Baiyao, Bulleyaconitine A Tablets and other well-known Chinese medicines. Present research on A. vilmorinianum focuses on the chemical composition and pharmacological action. A. vilmorinianum contains more than 40 kinds of diterpenoid alkaloids, including yunaconitine, vilmorrianine A, and bulleyaconitine A, and other diterpenoid alkaloids, which have many medicinal functions including antirheumatic activity, improvement in human meridians, relief of swelling and pain, activation of the blood circulation, and inhibition of tumor growth (Yang et al. 1981; Li et al. 2016). The molecular mechanism of diterpenoid alkaloid synthesis in A. vilmorinianum and its regulation have not identified. Therefore, identity of the key genes in the A. vilmorinianum diterpenoid biosynthetic pathway remains an important question.
RNA-seq has been widely used in many kinds of medicinal plants, such as Glycyrrhizin biosynthesis, Lonicera japonica, Chamaemelum nobile, Dendrobium huoshanense, and Ocimum tenuiflorum. Mining of functional genes and construction of the gene regulatory networks of metabolic pathways play an important role in germplasm resource evaluations and molecular breeding, particularly in medicinal plants (Li et al. 2010; Yuan et al. 2012, 2018; Upadhyay et al. 2015; Liu et al. 2019). In this study, we used the BGISEQ-500 sequencing platform (Zhang et al. 2016; Huang et al. 2017) to identify the candidate genes involved in the biosynthesis of diterpene alkaloids. The CYP450 genes, transcription factors, and microRNAs regulating the biosynthesis of diterpene alkaloids were identified, through the transcriptomes of the initial, middle and final stages of root formation of Aconitum vilmorinianum. These results will contribute to the analysis of the molecular mechanism of alkaloid biosynthesis in A. vilmorinianum, lay a foundation for the study of functional genomics, metabonomics, and breeding of A. vilmorinianum, and provide a reference for the synthesis and metabolism of medicinal components of Aconitum.
Materials and methods
Material preparation and sample collection
The plants of Aconitum vilmorinianum from Malutang Town, Luquan County, Yunnan Province, China, was cultivated at the experimental base of the School of Life and Technology, Kunming University of Science and Technology (around N30.35°, E112.14°). The samples were collected during three periods of root formation (Fig. 1b). The initial stage of root formation was 6 months (R1). The middle stage of root formation was 8 months (R2), and the final stage of root formation was 10 months (R3). The samples from each period were set with three biological replicates (marked as R1-1, R1-2, R1-3; R2-1, R2-2, R2-3; R3-1, R3-2, R3-3). Then all samples were stored at − 80 °C for RNA extraction. The roots, stems, leaves, tubercles, flowers, capsules, and seed samples from the three stages were dried in an oven at 60 °C to constant weight, and all dried samples are used to determine the alkaloid content.
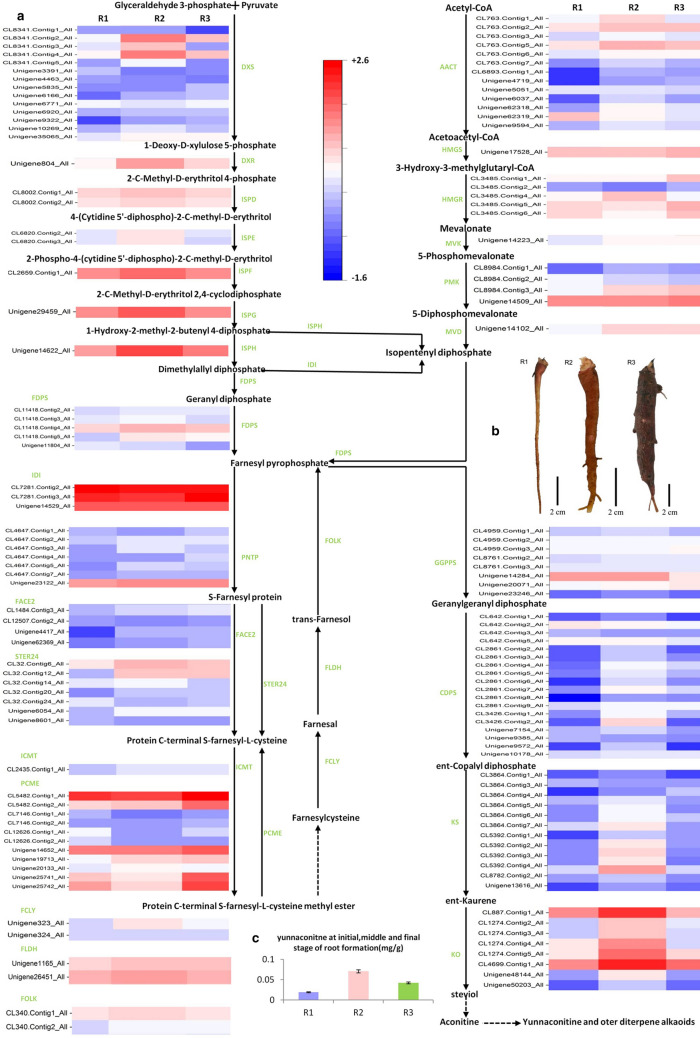
Fig. 1.
The diterpenoid alkaloid biosynthetic pathway and the differential expression of the unigenes involved in A. vilmorinianum. R1 is the early stage of root formation, R2 is the middle stage, and R3 is the final stage. a The expression levels of unigenes encoding enzymes from each step are shown. The columns are R1, R2, R3, corresponding to the early stage, middle stage and final stage of root formation, respectively, and the rows correspond to unigenes. Red and bule represent high and low expression levels, respectively. b The early stage (R1), middle stage (R2) and final stage (R3) of root formation. c Yunaconitine content of the early stage (R1), middle stage (R2) and final stage (R3) of root formation
Determination of alkaloids
Sample powder (0.20 g) was placed in a centrifuge tube with 1.5 ml of methanol and shaken. After pretreatment for 12 h and ultrasonic treatment for 30 min, the supernatant as the test solution was filtered through a microporous filter membrane (0.45 μm). The yunaconitine content during the initial, middle, and final stages of root formation of A. vilmorinianum was analyzed quantitatively by high-performance liquid chromatography (HPLC). The retention times of the standard markers under the specific column operating conditions were determined using Shimadzu LC-15C HPLC and Waters Xterra®RP18 (4.6 × 250 mm, 5 μm) columns (Xu et al. 2017). Acetonitrile (0.7%) and triethylamine aqueous solution (adjusted pH to 3 with phosphoric acid) were used as the mobile phase. Samples were eluted in a low pressure gradient at a flow rate of 1.0 ml/min under the detection wavelength for 260 nm and the sample volume for 10 μL. The yunaconitine content in roots, stems, leaves, tubercles flowers, capsules, and seeds was determined during three periods. Three replicates for each sample was used and the test was repeated three times. A standard curve was established using standards markers, and contents were quantified by comparing peak areas.
RNA extraction and transcriptome sequencing
Total RNA was extracted with a Trizol kit, and residual DNA was removed with RNase-free Dnase I. The quality of the extract was detected by an Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit; Agilent Technologies, Palo Alto, CA, USA), and the purity was detected by ultraviolet spectrophotometer, and determined by agarose gel electrophoresis. After the total RNA passed the quality inspection, magnetic beads with oligo DT were used to remove the mRNA with a Poly(A) tail, and RNA was segmented with the interrupt buffer. The random N6 primer was used for reverse transcription to synthesize the first cDNA chain. Then the buffer, dNTPs, RNase H, and DNA polymerase I were added to synthesize double-stranded cDNA. The end of the synthesized double-stranded DNA was flattened and phosphorylated to form a sticky end protruding from "A" and was then connected to a bubbly joint protruding from "T" at the end of 3′. The linked products were amplified by PCR with specific primers. The PCR products were thermally denatured into a single strand, and then the single-strand DNA library was obtained by cyclization of single-strand DNA with a bridge primer. The constructed library was sequenced with the BGISEQ-500 for mRNA and miRNA, and the size of the miRNA selected fragment was 18–30 nt.
Data assembly and functional annotation classification
Nine mRNA libraries were sent to BGI (Shenzhen, China) for sequencing to obtain the raw sequences (raw reads), to remove splices, unknown base N content > 5% and low-quality reads, to evaluate quality of the sequencing data, and to filter the sequencing data to obtain clean reads. Trinity was used for de novo assembly, and Tgicl was used for clustering and removing redundancy to obtain the unigenes. Blastx (http://blast.ncbi.nlm.nih.gov/Blast.cgi) or Diamond (https://github.com/bbuchfink/diamond) was used to annotate the unigenes with the NR, NT, COG, KEGG and SwissProt databases. The Blast2GO (https://www.blast2go.com) and NR annotation results were used for GO annotation. InterProScan 5 (https://code.google.com/p/interproscan/wiki/introduction) was used for InterProt annotation. Getorf (http://genome.csdb.cn/cgi-bin/boss/help/getorf) was used to detect the open reading frames (ORFs) of the unigenes, and then hmmsearch (http://hmmer.org) was used to compare the ORFs to the transcription factor protein domain (data from PlntFDB, plant transcription factor database). Unigenes were identified according to the characteristics of the transcription factor family described by plantFDB. MISA (http://pgrc.ipk-gatersleben.de/misa) was used to detect the simple sequence repeats (SSRs) of the unigene. Blast or Diamond was used to compare the unigenes to the plant resistance gene database (http://prgdb.crg.eu/) for annotation, and the possible resistance genes were obtained. We obtained the functional annotation information and classification information for all unigenes and sorted out all of the annotation information.
Differential gene expression analysis
Fragments per kilobase per million mapped fragments was calculated for each gene. RSEM (http://deweylab.biostat.wisc.edu/RSEM) (Li and Dewey 2011) was used to calculate the gene expression level of each sample. FDR (error detection rate) ≥ 0.001, absolute log2 ratio ≥ 1 as the threshold (fold change ≥ 4.00 and adjusted p value ≤ 0.001) were used to evaluate the differences in gene expression levels during the different periods. To reduce the false-positive error rate, three biological replicates were used during the initial, middle, and final stages of root formation.
microRNA sequencing and data processing
Nine samples were sent to BGI (Shenzhen, China) and sequenced using the BGISEQ-500. The length of the original RNA marker was 49 nt. By removing the low-quality reads, 5′ connector contaminated, over-inserted, Poly a containing, < 18 nt markers, and clean markers were obtained for the miRNA sample analysis. After removing rRNAs, scRNAs, snoRNAs, sNRNAs, and tRNAs, the remaining microRNAs was compared with the miRBase database to obtain mature microRNAs and new microRNAs. The predicted target gene was blasted with the NR, NT, GO, COG, KEGG, SwissProt, and InterProt databases to obtain the functional annotation information and classification information of the target genes.
CYP450 system development analysis
CYP450 family genes were detected according to the unigene annotation information, and the CYP450 family genes related to diterpene alkaloid synthesis were screened out. The homology of the genes with sequence length > 500 bps was blasted. The genes of other species with homology > 100 bps and CYP450 family involved in diterpene alkaloid synthesis were used for phylogenetic analysis. The amino acid sequences of the CYP450 family genes involved in diterpenoid synthesis were derived from their unigene sequences. A multisequence alignment was carried out using the ClustalW program together with the amino acid sequences of CYP450s related to diterpenoid biosynthesis in other known species, and a phylogenic tree was constructed by setting 1000 guiding values using Mega6 software (Tamura et al. 2013).
qRT-PCR analysis
Ten key genes encoding the MVA, MEP, FDP regulatory, diterpenoid skeleton synthetic pathways, CYP450 and Transcription factors were screened to verify gene expression. RNA of each sample was reverse transcribed into the first strand following the manufacturer’s instructions. Samples were run in triplicate and qRT-PCR analysis was performed using the PrimeScript RT kit. The PCR was carried out in 96-well plates under the following conditions: pre-denaturation at 95 °C for 5 min, 10 s at 95 °C, 20 s at 53 °C, 30 s at 72 °C, 15 s at 95 °C, and 40 cycle reactions using the CFX96TM Real-Time System (Bio-Rad Laboratories, Hercules, CA, USA). 18S RNA was used as the internal reference gene. Primers are listed (Table S1). The expression of the sample was calculated by the relative quantitative method (2−ΔΔCT), and the data were statistically analyzed (Manuka et al. 2013).
Results
Aconitine content
Yunaconitine is the main effective component of Aconitum vilmorinianum, which is generally collected after October. We performed HPLC of A. vilmorinianum root and found that yunaconitine content was the lowest (0.019%) during the initial stage (June) and highest (0.070%) during the middle stage (August) of root formation (Figs. S2, S3). Yunaconitine content in root decreased (0.022%) after the root completely formed (Fig. 1c). In addition, the content in flowers was highest during the entire three periods of root formation. Many alkaloids, including diterpenoids, accumulate in the reproductive organs. This includes yunaconitine which is enriched in the floral organs. Removing the inflorescences is an important cultivation for A. vilmorinianum, as it allows a large number of effective ingredients to accumulate in roots. Yunaconitine was also found in stems, leaves, tubercles, capsules, and seeds.
Sequencing analysis and de novo assembly
The sequencing results showed that 637.27 MB raw reads were obtained from nine libraries, with an average of 70.8 MB raw reads per library. After filtering, 593.76 MB clean reads and 59.39 GB clean bases were obtained, with an average of 6.6 GB clean bases per library. The sequences are uploaded to NCBI (Submission number: PRJNA667080). The Q20 and Q30 of the nine libraries were between > 97 and 90%. The proportion of clean reads ranged from 88.17 to 95.08%. The clean reads were used for subsequent analysis and research (Table S2). Using Trinity (Grabherr et al. 2011) to assemble the clean reads, we obtained 729,895 transcripts, with an average length of 705 nt and N50 length of 1163 nt (Table S3). We obtained unigenes using Tgicl (Pertea et al. 2003) to cluster and remove redundant transcripts. In total, we detected 119,660 unigenes with an average length of 1003 bases. The N50, N70, and N90 values were 1604, 1031, and 424 nt, respectively, and the GC ratio was between 41.15 and 42.38% (Table S4). We then blast the unigene sequences to the NR, NT, GO, COG, KEGG, SwissProt and InterProt databases. There are 69,978 unigenes with annotation information, accounting for 58.48% of total the unigenes, 63,070 unigenes (52.71%) in the NR database, 32,894 unigenes (52.71%) in the NT database, 37,836 unigenes (31.62%) in the GO database, 54,123 unigenes (45.23%) in the COG database, 48,383 unigenes (40.43%) in the KEGG database, 42,682 unigenes (35.67%) in the SwissProt database, and 55,535 unigenes (46.41%) in the InterProt database (Table S5).
NR annotation analysis
According to the similar sequence comparison of related species in the NR database, the similarity of unigenes to those from Macleaya cordata is the highest (17,999, 28.54%), followed by Nelumbo nucifera (10,467, 16.6%), Vitis vinifera (3426, 5.43%), Juglans regia (1,002, 1.59%), Hevea brasiliensis (707, 1.12%), and 11.84% for other species (Fig. S4a).
Functional classification of the unigenes
COG functional classification of unigene
The COG database classifies gene products using direct homology. We annotated 73,933 unigenes using the COG database, and 25 functional groups were annotated for classification and statistics (Fig. S4c). COG functional classification of the unigenes demonstrates the variety of gene activities in A. vilmorinianum roots. Among them, the most common is general function prediction (14,930, accounting for 20.2% of all unigenes). The second-most annotation is signal transmission mechanisms (8656, accounting for 11.7% of all unigenes). Post-translational modification, protein turnover, and chaperones are the third-most common (5817, 7.9% of all unigenes). In addition, there were 5231 unknown functions, accounting for 7.1% of all unigenes, and 4784 with transcription annotated functions, accounting for 6.5% of all unigenes. There were 2219 unigenes with a function of secondary metabolites biosynthesis, transport and catabolism, accounting for 3.0% of all unigenes. The smallest functional category was cell modifications with 115, accounting for only 0.16% of all unigenes.
Unigenes of GO classification
The GO database is an international standardized gene function database used to describe the biological characteristics of genes in different organisms. Blast2GO software (Conesa et al. 2005) was used to annotate all unigenes in A. vilmorinianum results in the NR database to the GO database. We annotated 170,089 unigenes using GO (Fig. S4b). The counts of the unigenes in three GO categories were biological processes (51,843 unigenes), cellular components (73,582 unigenes), and molecular functions (44,664 unigenes) with 57 sub categories. For biological processes, the top three categories were cell processes (14,806 unigenes), metabolic processes (13,995 unigenes), and biological regulation (4356 unigenes), according to the GO classification. Among the cell components unigenes, the top five categories were cell (14,191 unigenes), cell components (13,956 unigenes), cell membranes (12,198 unigenes), cell parts (11,230 unigenes), and organelles (9977 unigenes). In terms of molecular function, the top three categories were binding (19,628 unigenes), catalytic activity (19,351 unigenes), and carrier activity (2132 unigenes), while few unigenes were involved in protein tag and translation regulator activity. There were 13,995 unigenes related to the metabolism of diterpenoid alkaloids.
Unigene of KEGG classification
A total of 47,106 unigenes were annotated and divided into five major categories and 19 minor categories using KEGG classifications. The five categories include cellular processes (2121 unigenes), environmental information processing (3029 unigenes), genetic information processing (11,296 unigenes), metabolism (28,953 unigenes), and organismal systems (1707) (Fig. S4e). Among the 19 subgroups, 11,006 unigenes were included in global and overview maps, of these 4246 unigenes had translation annotations, 4137 had carbohydrate metabolism annotations, 3310 had folding, sorting and degradation annotationis, and 782 unigenes had membrane transport annotations. The unigenes related to diterpenoid alkaloids (1006 unigenes) were divided into terpenoids and polyketides. These annotations will be helpful in study the metabolism of diterpenoid alkaloids and to identify the genes involved in the metabolism of diterpenoid alkaloids. This information will be an important resource for molecular breeding of diterpenoid alkaloids in the future.
Notes from SwissProt and InterProt protein database
There are 42,682 unigenes (35.67%) and 55,535 unigenes (46.41%) annotated in the SwissProt and InterProt databases, respectively.
Notes from NR, COG, KEGG, SwissProt, and InterProt databases
The annotation results of the NR, COG, KEGG, SwissProt, and InterProt databases are displayed in a Venn diagram (Fig. S4d), and 32,222 unigenes were annotated in each database.
Transcription factor analysis and identification of the simple sequence repeat (SSR) markers
Transcription factors play an important role in the biosynthesis of secondary metabolites. We show that 2438 unigenes were predicted to be in 60 transcription factor families (Fig. S4f). Among them, the number of unigenes annotated as the MYB family was largest (299); 205 were from the MYB-related family, 153 from the bHLH family, 138 from the AP2/EREBP family, 135 from the C3H family, 113 from the NAC and mTERF family, and 106 from the C2H2 family. At least one unigene was annotated as the S1Fa-like and ULT family.
Among 119,660 unigenes in the transcriptome, 10,829 unigenes had SSRs, with a frequency of 9.49% (ratio of the number of unigenes containing a SSR to the total number of unigenes). The results show that 3098 unigenes are mono-nucleotides (24.67%), 2717 unigenes are di-nucleotides, accounting for 21.64%, and 5371 unigenes are tri-nucleotides, accounting for 42.77% (Fig. S4g). The quad-nucleotide and penta-nucleotide types are a very small proportion, accounting for 0.85 and 3.46%, respectively. There are 829 unigenes annotated as hexa-nucleotides, accounting for 6.60%. These results indicate that the main type of SSR in the root transcriptome of A. vilmorinianum is a tri-nucleotide, among which AAG/CTT is the most common (1848), accounting for 34.41% of the tri-nucleotides. These unique sequence markers provide valuable genetic resources for SSR mining and application.
Identification of microRNA and prediction of disease resistance genes
Using microRNA databases we discovered 54 mature microRNAs (Table S6), and 151 new microRNAs predictions from our data (Table S7). According to the KEGG functional analysis of the microRNA target genes, microRNAs regulating terpenoids and polyketides were found. The microRNAs involved in the regulation of polyketides are miR396a-5p, miR156k, and miR166d-5p-1, and the microRNA involved in the regulation of diterpene alkaloid synthesis is miR6300. The regulatory site is 3-hydroxy-3-methyl glutaryl-CoA reductase (HMGR), the key enzyme gene in the MVA metabolic pathway. Four unigenes in the resistance gene database (PRGDB) had annotation information (Table S8). These resistance genes’ information will provide an important reference for resistance breeding.
Unigene expression analysis
We analyzed the expression of the unigenes during the initial, middle, and final stages of root formation to analyze the specificity of unigene expression during the formation of roots of A. vilmorinianum. We found that a total of 55,271 unigenes are co-expressed among three samples from the initial stage of root formation, 66,965 unigenes are co-expressed among the three samples from the middle stage, and 56,769 unigenes are co-expressed among the three samples from the final stage (Fig. S5). According to the results of the gene expression level of each sample, we determined the differentially expressed genes (DEGs) during three stages of A. vilmorinianum root formation. The results show that 39,864 unigenes are upregulated and 11,228 unigenes were downregulated during the initial and middle stages, 26,789 unigenes are upregulated and 12,820 unigenes are downregulated during the initial and final stages, and 10,052 unigenes are upregulated and 22,950 unigenes are downregulated during the middle and final stages (Fig. S6).
Identification of candidate genes involved in diterpenoid alkaloid synthesis
According to the KEGG annotation results, the DEGs were classified into biological pathways. The results show that there are 24,165 DEGs during the initial and middle stages of root formation, among which 502 DEGs are involved in terpenoid and polyketide metabolism (Table S9), 162 DEGs were involved in terpenoid skeleton biosynthesis, and 87 DEGs are involved in diterpenoid synthesis (Table S10). Among the 17,656 DEGs detected during the initial and final stages of root formation, 423 are involved in terpenoid and polyketide metabolism (Table S11), 164 are involved in terpenoid skeleton synthesis, and 71 are involved in diterpenoid synthesis (Table S12). Among the 14,603 DEGs detected during the middle and final stages of root formation, 426 are involved in terpenoid and polyketide metabolism (Table S13), 127 are involved in terpenoid skeleton synthesis, and 71 are involved in diterpenoid synthesis (Table S14).
There are 27 enzyme genes (139 unigenes) regulated diterpenoid alkaloid synthesis (Fig. 1a). These candidate genes are involved in diterpenoid alkaloid synthesis of A. vilmorinianum (Table 1). There are six enzyme genes (25 unigenes) related to the MVA pathway, nine enzyme genes (31 unigenes) related to the MEP pathway, eight enzyme genes (36 unigenes) related to the FDP (farnesyl diphosphate) regulatory pathway, and four enzyme genes (48 unigenes) related to downstream of the diterpenoid skeleton synthetic pathway.
Table 1.
The unigenes involved in terpenoid backbone and diterpenoid biosynthesis
| Pathways | Abbreviation of gene | Name of the gene | Enzyme no. | Number of genes |
|---|---|---|---|---|
| MVA | AACT | Acetyl-CoA C-acetyltransferase | K00626 | 14 |
| HMGS | Hydroxymethylglutaryl-CoA synthase | K01641 | 1 | |
| HMGR | Hydroxymethylglutaryl-CoA reductase (NADPH) | K00021 | 5 | |
| MVK | Mevalonate kinase | K00869 | 1 | |
| PMK | Phosphomevalonate kinase | K00938 | 3 | |
| MVD | Diphosphomevalonate decarboxylase | K01597 | 1 | |
| MEP | DXS | 1-Deoxy-D-xylulose-5-phosphate synthase | K01662 | 14 |
| DXR | 1-Deoxy-D-xylulose-5-phosphate reductoisomerase | K00099 | 1 | |
| ISPD | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | k00991 | 2 | |
| ISPE | 4-Diphosphocytidyl-2-C-methyl-D-erythritol kinase | K00919 | 2 | |
| ISPF | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | K01770 | 1 | |
| ISPG | (E)-4-Hydroxy-3-methylbut-2-enyl-diphosphate synthase | K03526 | 1 | |
| ISPH | 4-Hydroxy-3-methylbut-2-en-1-yl diphosphate reductase | K03527 | 1 | |
| IDI | Isopentenyl-diphosphate delta-isomerase | K01823 | 3 | |
| FDPS | Farnesyl diphosphate synthase | K00787 | 5 | |
| FDP regulatory pathway | FNTP | Protein farnesyl transferase subunit beta | K05954 | 7 |
| FACE2 | Prenyl protein peptidase | K08658 | 4 | |
| STE24 | STE24 endopeptidase | K06013 | 7 | |
| ICMT | Protein-S-isoprenylcysteine O-methyltransferase | K00587 | 1 | |
| PCME | Prenylcysteine alpha-carboxyl methylesterase | K15889 | 11 | |
| FCLY | Prenylcysteine oxidase/farnesylcysteine lyase | K05906 | 2 | |
| FLDH | NAD + -dependent farnesol dehydrogenase | K15891 | 2 | |
| FOLK | Farnesol kinase | K15892 | 2 | |
| GGPPS | Geranylgeranyl diphosphate synthase, type II | K13789 | 8 | |
| Diterpenoid skeleton synthetic pathway | CDPS | ent-Copalyl diphosphate synthase | K04120 | 18 |
| KS | ent-Kaurene synthase | K04121 | 14 | |
| KO | ent-Kaurene oxidase | K04122 | 8 | |
| Total | 139 |
Phylogenetic analysis of the CYP450s family
After sequencing the transcriptome, we found 504 genes that are annotated as the CYP450s family, including CYP4, CYP19, CYP26, CYP71A1, CYP71A3, CYP71A4, CYP71A26, CYP71D10, CYP82C4, CYP81B25, CYP81E8, CYP85A1, CYP710A1, CYP90B1, CYP86B1, CYP97B1, and other families. Among the CYP450s genes, 146 CYP450s genes are related to terpenoid metabolism, 13 of which have annotation information, indicating that they are related to the ent-kaurenoic acid oxidase synthesized by diterpenoids (Table S15). We analyzed the phylogeny of these 13 genes together with 42 known CYP450s involved in diterpenoid synthesis in different plants (Fig. S7). We identified unigene19493-all, unigene19495-all, CL9995-contig1-all, unigene19497-all, unigene19496-all, unigene19498-all, unigene3912-all, unigene5986-all, unigene8612-all, and CL12054-contig3-All belongs to the CYP88A1 family, unigene17135-all belongs to the CYP97C family, and unigene 28628-all belongs to the CYP76 family.
Validation of key genes involved in A. vilmorinianum alkaloid synthesis
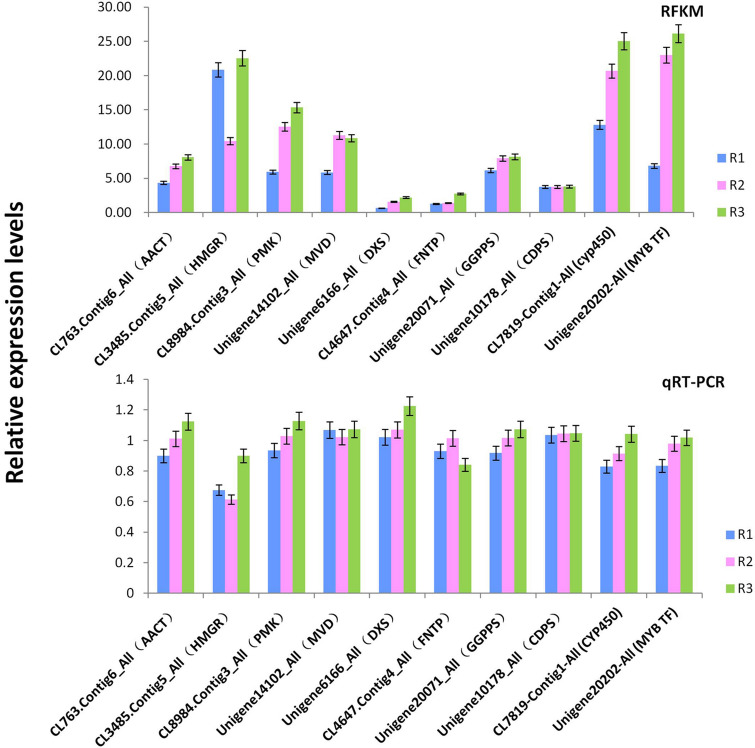
Using qRT-PCR we confirmed the expression patterns of ten key genes encoding genes in the MVA metabolic pathway, the MEP metabolic pathway, the FDP regulatory pathway, the diterpenoid skeleton synthetic pathway, CYP450 and important transcription factors (Fig. 2). The ten genes are CL763-Contig6-All (acetyl-CoA C-acetyl transferase, AACT), CL3485-Contig5-All (hydroxymethylglutaryl-CoA reductase, HMGR), CL8984-Contig3-All (phosphomevalonate kinase, PMK), Unigene14102-All (diphosphomevalonate decarboxylase, MVD), Unigene6166-All (1-deoxy-D-xylulose-5-phosphate synthase, DXS), CL4647-Contig4-All (protein farnesyltransferase subunit beta, FNTP), Unigene20071-All (geranylgeranyl diphosphate synthase, GGPPS), Unigene10178-All (ent-copalyl diphosphate synthase, CDPS), CL7819-Contig1-All (CYP450) and Unigene20202-All (MYB Transcription factors). We found that the changes in gene expression during the initial, middle, and final stages of root formation were consistent with the transcriptome expression trend, with the exception of Unigene14102-All (diphosphomevalonate decarboxylase, MVD) and CL4647-Contig4-All (protein farnesyltransferase subunit beta, FNTP).
Fig. 2.
Relative expression levels and FPKM of transcriptome data of key genes involved in A. vilmorinianum alkaloid synthesis. R1 is the early stage of root formation, R2 is the middle stage of root formation, and R3 is the final stage of root formation
Discussion
Many functional genes related to the biosynthesis of plant secondary metabolites have been predicted and screened by transcriptome sequencing (Hemmerlin et al. 2003; Laule et al. 2003; Dudareva et al. 2005). In this paper, the biosynthesis and metabolism pathway of the main active components diterpenoid alkaloids in A. vilmorinianum and its related key candidate genes are obtained by transcriptome sequencing (Fig. 1a). We obtained 59.39 gb of clean bases totally and 119,660 unigenes were assembled, in which 69,978 unigenes are annotated. There are 139 candidate genes involved in the synthesis of diterpenoid alkaloids, including hydroxymethylglutaryl-CoA reductase (HMGR) which is the key enzyme in MVA metabolism, and regulated by miR6300. Additionally, we identified 13 CYP450 genes and transcription factors, used to synthesize the diterpenes, such as bHLH, AP2/EREBP, and MYB. These results provide a scientific reference for studying the diterpenoid metabolic pathway of Aconitum and other plants.
The diterpenoid aconitine and its derivatives are the main active components in A. vilmorinianum, including diterpenoid alkaloids, such as yunaconitine, vilmorrianine A, vilmorrianine B, vilmorrianine C, vilmorrianine D, and bulleyaconitine A (Yang et al. 1981; Li et al. 2016). The complete diterpenoid alkaloid biosynthetic pathway of A. vilmorinianum has not been studied. According to the KEGG differential gene expression analysis, we found 27 enzyme genes that are involved in diterpenoid biosynthesis (Fig. 1, Table 1). There are six enzyme genes in the MVA pathway, nine in the MEP pathway, eight in the FDP regulatory pathway, and four downstream of the diterpenoid skeleton synthetic pathway.
The MVA pathway is an indispensable metabolic pathway in the cytoplasm and main products are terpenes, including sterol, coenzyme Q, carotene, astaxanthin, and other synthetic precursors(Roberts 2007; Lange and Ahkami 2013; Pal et al. 2015). The expression of six related genes in the MVA pathway is lower during the initial stage of root formation, higher during the middle stage, and highest during the final stage in A. vilmorinianum. HMGR, is the rate-limiting enzyme gene in the MVA pathway, is an important regulatory site in the terpene metabolic pathway (Kai et al. 2012; Liao et al. 2016). HMGR expression is high during the initial stage of root formation, lowest during the middle stage, and highest during the final stage. Diphosphomevalonate decarboxylase (MVD) expression is lowest during the initial stage of root formation and highest during the middle and final stages. We found that HMGR and MVD genes are highly expressed in roots of Aconitum heterophyllum and are key genes in alkaloid biosynthesis (Rai et al. 2017); HMGR genes are highly expressed in roots of A. carmichaelii (Pal et al. 2015). The expression of HMGR has an obvious effect on products downstream in the MVA pathway, and is also regulated by miR6300 (Cuperus et al. 2011; Ng et al. 2011; Kai et al. 2012). The functional expression of HMGR and miR6300 is one of the key points in future studies of the regulation of diterpenoid biosynthesis in A. vilmorinianum.
In the MEP pathway, the expression of nine related genes is lowest during the initial stage of root formation, highest during the middle stage, and lower during the final stage. The changes in expression of these genes are consistent with the change in yunaconitine content. 1-Deoxy-d-xylulose-5-phosphate synthase (DXS) is the first rate-limiting enzyme in the MEP pathway (Enfissi et al. 2005; Morris et al. 2006; Rai et al. 2017), which catalyzes the production of 1-deoxy-d-xylulose-5-phosphate (DXP) from pyruvate and glyceraldehyde-3-phosphate. 1-Deoxy-d-xylulose-5-phosphate reductoisomerase (DXR) is the second rate-limiting enzyme gene in the MEP pathway (Botella-Pavia et al. 2004; Carretero-Paulet et al. 2006). DXP plays a key role in this pathway under the action of DXR, reducing DXP to 2-C-methyl-D-phosphate-erythritol (MEP). DXR expression is lowest during the initial stage of root formation, highest during the middle stage, and lower during the final stage. The change of gene expression is consistent with that of yunaconitine content. In the MEP pathway, the gene expression of DXS and DXR, as rate-limiting enzyme genes, is consistent with the change in yunaconitine content. These genes will be key in future studies of diterpenoid alkaloid synthesis of A. vilmorinianum.
The functional characteristics of the CYP450s family have generated great interest in the scientific community. There are numerous studies of the CYP450s involved in terpene synthesis (Banerjee and Hamberger 2017; Liao et al. 2017). 124 CYP450s are found in A. carmichaelii, 21 of which belong to the CYP450s family, and are related to the synthesis of diterpenoid alkaloids (Rai et al. 2017). 326 potential CYP450s have been identified in Panax ginseng, including the CYP716A47 family related to ginsenoside biosynthesis (Li et al. 2013). In Taxus, Salvia miltiorrhiza, Artemisia annua, and other species, CYP450s family genes are involved in the biosynthesis of the diterpenoid compounds paclitaxel, tanshinone, and artemisinin. The functions of these CPY450s have been characterized (Jennewein et al. 2001; Yang et al. 2013; Liu et al. 2018). The terpenoid synthetase gene and CYP450s are key to diterpenoid biosynthesis in Aconitum vilmorinianum. CYP450s play a decisive role in diterpenoid diversity (Pateraki et al. 2015), but little is known about the CYP450s involved in the synthesis of diterpenoid alkaloids. We identified 13 genes involved in the synthesis of diterpenoid alkaloids. Nine genes, such as unigene19493-all, unigene19495-all, and CL9995-contig1-all, have high homology with CYP88A1 from Tripterygium wilfordii, Salvia miltiorrhiza, and Papaver somniferum and other plants, and are related to ent-kaurenoic acid oxidase during diterpenoid biosynthesis. Unigene-17135 has high homology with CYP97C of Lycium ruthenicum, Medicago truncate, and other plants, and its function is related to oxygenases and hydroxylases. Unigene-28628-all has a high homology with CYP76 in Radish sativus, Lactuca sativa, and Vitis vinifera, and is related to monooxygenases. These CYP450 genes, particularly those related to the CYP88A1 family, may be of great significance for further understanding the target diterpenoid biosynthetic pathway.
Conclusion
In this study, transcriptome and metabolism analyses were performed to identify the genes and transcription factors related to diterpenoid alkaloid synthesis (Fig. 3). 119,660 unigenes in total were identified, in which 69,978 unigenes were annotated. We identified 27 enzyme genes (139 candidate genes) in the diterpenoid biosynthetic pathway and involved in its biosynthesis. The transcription factors, microRNA, and CYP450 genes were also identified. These results lay a foundation for the functional identification of the candidate genes for diterpenoid aconitine biosynthesis in the future and provide a scientific reference for the molecular mechanisms and metabolic pathways of diterpenoid synthesis in Aconitum and other plants.
Fig. 3.
The hypothetical model of diterpenoid synthesis regulatory network associated with A. vilmorinianum. In cytoplasm, acetyl CoA forms IPP through MVA pathway. In MVA pathway, HMGS gene is regulated by mir6300. In plastids, glycolide 3-phosphate and pyruvic acid form IPP through MEP pathway. IPP molecules are condensed into farnesyl pyrophosphate, and farnesyl pyrophosphate is regulated by FDP regulatory pathway to complete terpene skeleton synthesis. GGPPS enters the diterpene metabolic pathway through the interaction of CDPS, KS and KO genes, and forms aconitine and its derivatives through the interaction of transcription factors and CYP450. Finally, aconitine and other diterpenoid alkaloids were synthesized
Supplementary Information
Below is the link to the electronic supplementary material.
Biosynthetic pathway of diterpenoid compounds (TIF 9291 KB)
High performance liquid chromatography (TIF 943 KB)
Overview of unigenes annotation (TIF 14937 KB)
Venn diagram analysis of nine sample (TIF 530 KB)
Phylogenetic analysis of annotated as CYP450s genes (TIF 13848 KB)
Primer sequences for real time qPCR (DOCX 15 KB)
Quality of sample of clean reads (DOCX 14 KB)
Quality metrics of sample squencing (DOCX 15 KB)
Overview of assembly of sample (DOCX 14 KB)
Unigenes functional annotation with 7 functional database (DOCX 12 KB)
Result of plant disease resistance unigene detection (DOCX 13 KB)
Number of differential genes annotated by KEGG pathway( R1-vs-R2) (DOCX 14 KB)
Function enrichment results of DEG by KEGG pathway(R1-VS-R2) (DOCX 14 KB)
Number of differential genes annotated by KEGG pathway( R1-vs-R3) (DOCX 14 KB)
Function enrichment results of DEG by KEGG pathway(R1-VS-R3) (DOCX 14 KB)
Number of differential genes annotated by KEGG pathway( R2-vs-R3) (DOCX 14 KB)
Function enrichment results of DEG by KEGG pathway(R2-VS-R3) (DOCX 14 KB)
The P450 unigene of annotation to ent-kaurenoic acid oxidase (DOCX 13 KB)
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (No. 31560351; No. 31760349).
Author contributions
All authors read and approved the final manuscript. Y-GL conceived and designed the experiments, performed the experiments, wrote the paper, read and approved the final manuscript. F-JM contributed reagents, materials, analysis tools, read and approved the final manuscript. K-ZL conceived and designed the experiment, read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Yi-Guo Li, Email: liyiguo73@126.com.
Feng-Juan Mou, Email: moufengjuan@126.com.
Kun-Zhi Li, Email: likzkm@163.com.
References
- Banerjee AB, Hamberger B. P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem Rev. 2017;17(1):81–111. doi: 10.1007/s11101-017-9530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella-Pavía P, Besumbes Ó, Phillips MA, Carretero-Paulet L, Boronat A, Rodríguez-Concepción M. Regulation of carotenoid biosynthesis in plants: evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J. 2004;40(2):188–199. doi: 10.1111/j.1365-313x.2004.02198.x. [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Cairó A, Botella-Pavía P, Besumbes O, Campos N, Boronat A, Rodríguez-Concepción M. Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol Biol. 2006;62(4–5):683–695. doi: 10.1007/s11103-006-9051-9. [DOI] [PubMed] [Google Scholar]
- Cherney EC, Baran PS. Terpenoid-alkaloids: Their biosynthetic twist of fate and total synthesis. Isr J Chem. 2011;51:391–405. doi: 10.1002/ijch.201100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of miRNA genes. Plant Cell. 2011;23(2):431–442. doi: 10.1105/tpc.110.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota KP, Sewald N. Terpenoid alkaloids derived by amination reaction. In: Ramawat KG, Mérillon JM, editors. Natural products: phytochemistry, botany and metabolism of alkaloids, phenolics and terpenes. Berlin/Heidelberg: Springer; 2013. pp. 923–951. [Google Scholar]
- Dudareva N, Andersson S, Orlova I, Gatto N, Reichelt M, Rhodes D, Boland W, Gershenzon J. The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. PNAS. 2005;102(3):933–938. doi: 10.1073/pnas.0407360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enfissi EM, Fraser PD, Lois LM, Boronat A, Schuch W, Bramley PM. Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotech J. 2005;3(1):17–27. doi: 10.1111/j.1467-7652.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Gao W, Hillwig ML, Huang LQ, Cui GH, Wang JQ, Bin Y, Peters RJ. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett. 2009;11:5170–5173. doi: 10.1021/ol902051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng QD, Chen ZH, Mauceli E, Hacohen N, Gnirke A, Rhind N, Palma FD, Birren BW, Nusbaum C, Kerstin LT, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerlin A, Hoeffler JF, Meyer O, Tritsch D, Kagan IA, Grosdemange-Billiard C, Rohmer M, Bach TJ. Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J Biol Chem. 2003;278(29):26666–26676. doi: 10.1074/jbc.m302526200. [DOI] [PubMed] [Google Scholar]
- Huang J, Liang XM, Xuan YK, Geng CY, Li YX, Lu HR, Qu SF, Mei XL, Chen HB, Yu T, Sun N, Rao JH, Wang JH, Zhang WW, Chen Y, Liao S, Jiang H, Liu X, Yang ZP, Mu F, Gao SX. A reference human genome dataset of the BGISEQ-500 sequencer. GigaScience. 2017;6(5):1–9. doi: 10.1093/gigascience/gix024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein S, Rithner CD, Williams RM, Croteau RB. Taxol biosynthesis: taxane 13-hydroxylase is a cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA. 2001;98(24):13595–13600. doi: 10.1073/pnas.251539398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai GY, Liao P, Zhang T, Zhou W, Wang J, Xu H, Yuanyun Liu YY, Zhang L. Characterization, expression profiling, and functional identification of a gene encoding geranylgeranyl diphosphate synthase from Salvia miltiorrhiza. Biotech Bioproc Eng. 2010;15:236–245. doi: 10.1007/s12257-009-0123-y. [DOI] [Google Scholar]
- Kai GY, Li SS, Wang W, Lu Y, Wang J, Liao P, Cui LJ. Molecular cloning and expression analysis of a gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Camptotheca acuminata. Rus J Plant Physiol. 2012;60(1):131–138. doi: 10.1134/s102144371206009x. [DOI] [Google Scholar]
- Lange BM, Ahkami A. Metabolic engineering of plant monoterpenes, sesquiterpenes and diterpenes-current status and future opportunities. Plant Biotech J. 2013;11(2):169–196. doi: 10.1111/pbi.12022. [DOI] [PubMed] [Google Scholar]
- Laule O, Furholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange BM. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci. 2003;100(11):6866–6871. doi: 10.1073/pnas.1031755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Luo HM, Sun C, Song JY, Sun YZ, Wu Q, Wang N, Yao H, Steinmetz A, Chen SL. EST analysis reveals putative genes involved in glycyrrhizin biosynthesis. BMC Genomics. 2010;11:268. doi: 10.1186/1471-2164-11-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Zhu YJ, Guo X, Sun C, Luo HM, Song JY, Li Y, Wang LZ, Qian J, Shilin Chen SL. Transcriptome analysis reveals ginsenosides biosynthetic genes, microRNAs and simple sequence repeats in Panax ginseng C. A. Meyer. BMC Genomics. 2013;14:245. doi: 10.1186/1471-2164-14-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Guo LN, Zheng J, Ma SC. Reaserch progress of medicinal genus Aconitum. Chin J Pharm Anal. 2016;36(7):1129–1149. [Google Scholar]
- Liao P, Hemmerlin A, Bach TJ, Chye ML. The potential of the mevalonate pathway for enhanced isoprenoid production. Biotech Adv. 2016;34(5):697–713. doi: 10.1016/j.biotechadv.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Liao WF, Zhao SY, Zhang M, Dong K, Chen Y, Fu CH, Yu LJ. Transcriptome assembly and systematic identification of novel cytochrome P450s in Taxus chinensis. Front Plant Sci. 2017;8:1468. doi: 10.3389/fpls.2017.01468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MM, Zhu JH, Wu SB, Wang CK, Guo XY, Wu JW, Zhou MQ. De novo assembly and analysis of the Artemisia argyi transcriptome and identification of genes involved in terpenoid biosynthesis. Sci Rep. 2018;8(1):5824. doi: 10.1038/s41598-018-24201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XM, Wang XH, Chen ZX, Ye JB, Liao YL, Zhang WW, Chang J. De novo assembly and comparative transcriptome analysis: novel insights into terpenoid biosynthesis in Chamaemelum nobile L. Plant Cell Rep. 2019;38(1):101–116. doi: 10.1007/s00299-018-2352-z. [DOI] [PubMed] [Google Scholar]
- Morris W, Ducreux L, Hedden P, Millam S, Taylor M. Overexpression of a bacterial 1-deoxy-D-xylulose 5-phosphate synthase gene in potato tubers perturbs the isoprenoid metabolic network: Implications for the control of the tuber life cycle. J Exp Bot. 2006;57(12):3007–3018. doi: 10.1093/jxb/erl061. [DOI] [PubMed] [Google Scholar]
- Ng DW-K, Zhang CQ, Miller M, Palmer G, Whiteley M, Tholl D, Chen ZJ. Cis- andtrans-regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell. 2011;23(5):1729–1740. doi: 10.1105/tpc.111.083915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal T, Malhotra N, Chanumolu SK, Chauhan RS. Next-generation sequencing (NGS) transcriptomes reveal association of multiple genes and pathways contributing to secondary metabolites accumulation in tuberous roots of Aconitum heterophyllum Wall. Planta. 2015;242(1):239–258. doi: 10.1007/s00425-015-2304-6. [DOI] [PubMed] [Google Scholar]
- Pateraki I, Heskes A, Hamberger B. Cytochromes P450 for terpene functionalisation and metabolic engineering. Adv Biochem Eng Biotechnol. 2015;148:107–139. doi: 10.1007/10-2014-301. [DOI] [PubMed] [Google Scholar]
- Pertea G, Huang XQ, Liang F, Antonescu V, Sultana R, Karamycheva S, Karamycheva S, Lee YD, White J, Cheung F, Parvizi B, Tsai J, Quackenbush J. TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19(5):651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- Rai M, Rai A, Kawano N, Yoshimatsu K, Takahashi H, Suzuki H, Kawahara N, Saito K, Yamazaki M. De novo RNA sequencing and expression analysis of Aconitum carmichaelii to analyze key genes involved in the biosynthesis of diterpene alkaloids. Molecules. 2017;22(12):2155. doi: 10.3390/molecules22122155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SC. Production and engineering of terpenoids in plant cell culture. Nature Chem Biol. 2007;3(7):387–395. doi: 10.1038/nchembio.2007.8. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay AK, Chacko AR, Gandhimathi A, Ghosh P, Harini K, Joseph AP, Sowdhamini R. Genome sequencing of herb Tulsi (Ocimum tenuiflorum) unravels key genes behind its strong medicinal properties. BMC Plant Biol. 2015;15:212. doi: 10.1186/s12870-015-0562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Qin B, Cao HY, Zhang XN, Liu H, You Y. Simultaneous determination of yunaconitine and bulleyaconitine A in Aconitum vilmorinianum by HPLC. J Yunnan Trad Chin Med. 2017;38(9):70–72. [Google Scholar]
- Yang CR, Hao XJ, Wang DZ, Zhou J. The alkaloids of Aconitum vilmorrianum Kom. I: the structures of valmorrianine A and C. Acta Chim Sinica. 1981;39(2):147–152. [Google Scholar]
- Yang L, Ding GH, Lin HY, Cheng HN, Kong Y, Wei YK, Fang X, Liu RY, Wang LG, Chen XY, Yang CQ. Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS ONE. 2013;8(11):e80464. doi: 10.1371/journal.pone.0080464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Song LP, Li MH, Liu GM, Chu YN, Ma LY, Zhou YY, Wang X, Gao W, Qin SS, Yu J, Wang XM, Luqi Huang LQ. Genetic variation and metabolic pathway intricacy govern the active compound content and quality of the Chinese medicinal plant Lonicera japonica Thunb. BMC Genomics. 2012;13:195. doi: 10.1186/1471-2164-13-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Yu M, Jia Z, Song X, LiangY ZJ. Analysis of Dendrobium huoshanense transcriptome unveils putative genes associated with active ingredients synthesis. BMC Genomics. 2018;19:978. doi: 10.1186/s12864-018-5305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhang W, En R, Li WZ, Segraves KA, Yang XK, Xue HJ. Comparative transcriptome analysis of chemosensory genes in two sister leaf beetles provides insights into chemosensory speciation. Insect Biochem Mol Biol. 2016;79:108–118. doi: 10.1016/j.ibmb.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Zhao D, Shen Y, Shi YN, Shi XQ, Qiao Q, Zi SH, Zhao EQ, Yu DQ, Kennelly EJ. Probing the transcriptome of Aconitum carmichaelii reveals the candidate genes associated with the biosynthesis of the toxic aconitine-type C19-diterpenoid alkaloids. Phytochemistry. 2018;152:113–124. doi: 10.1016/j.phytochem.2018.04.022. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Li R, Zhang Y, Huang KJ, Wang WG, Li J. Transcriptome analysis reveals in vitro-cultured regeneration bulbs as a promising source for targeted Fritillaria cirrhosa steroidal alkaloid biosynthesis. 3 Biotech. 2018;8:191. doi: 10.1007/s13205-018-1218-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biosynthetic pathway of diterpenoid compounds (TIF 9291 KB)
High performance liquid chromatography (TIF 943 KB)
Overview of unigenes annotation (TIF 14937 KB)
Venn diagram analysis of nine sample (TIF 530 KB)
Phylogenetic analysis of annotated as CYP450s genes (TIF 13848 KB)
Primer sequences for real time qPCR (DOCX 15 KB)
Quality of sample of clean reads (DOCX 14 KB)
Quality metrics of sample squencing (DOCX 15 KB)
Overview of assembly of sample (DOCX 14 KB)
Unigenes functional annotation with 7 functional database (DOCX 12 KB)
Result of plant disease resistance unigene detection (DOCX 13 KB)
Number of differential genes annotated by KEGG pathway( R1-vs-R2) (DOCX 14 KB)
Function enrichment results of DEG by KEGG pathway(R1-VS-R2) (DOCX 14 KB)
Number of differential genes annotated by KEGG pathway( R1-vs-R3) (DOCX 14 KB)
Function enrichment results of DEG by KEGG pathway(R1-VS-R3) (DOCX 14 KB)
Number of differential genes annotated by KEGG pathway( R2-vs-R3) (DOCX 14 KB)
Function enrichment results of DEG by KEGG pathway(R2-VS-R3) (DOCX 14 KB)
The P450 unigene of annotation to ent-kaurenoic acid oxidase (DOCX 13 KB)