Abstract
Poly(3-hydroxybutyrate) (PHB) belongs to the family of polyhydroxyalkanoates, biopolymers used for agricultural, industrial, or even medical applications. However, scaling up the production is still an issue due to the myriad of parameters involved in the fermentation processes. The present work seeks, firstly, to scale up poly(3-hydroxybutyrate) (PHB) production by wild type C. necator ATCC 17697 from shaken flasks to a stirred-tank bioreactor with the optimized media and fructose as carbon source. The second purpose is to improve the production of PHB by applying both the batch and fed-batch fermentation strategies in comparison with previous works of wild type C. necator with fructose. Furthermore, thinking of biomedical applications, physicochemical, and cytotoxicity analyses of the produced biopolymer, are presented.
Fed-batch fermentation with an exponential feeding strategy enabled us to achieve the highest values of PHB concentration and productivity, 25.7 g/l and 0.43 g/(l h), respectively. The PHB productivity was 3.3 and 7.2 times higher than the one in batch strategy and shaken flask cultures, respectively. DSC, FTIR, 1H, and 13C NMR analysis led to determine that the biopolymer produced by C. necator ATCC 17697 has a molecular structure and characteristics in agreement with the commercial PHB. Additionally, the biopolymer does not induce cytotoxic effects on the NIH/3T3 cell culture.
Due to the improved fermentation strategies, PHB concentration resulted in 40 % higher of the already reported one for wild type C. necator using other fed-batch modes and fructose as a carbon source. Thus the produced PHB could be attractive for biomedical applications, which generate a rising interest in polyhydroxyalkanoates during recent years.
Keywords: Poly(3-hydroxybutyrate) (PHB), Cupriavidus necator, Fermentation strategies, Optimization, Polymer characterization, Cytotoxicity
Poly(3-hydroxybutyrate) (PHB); Cupriavidus necator; Fermentation strategies; Optimization; Polymer characterization; Cytotoxicity.
1. Introduction
Currently, polyhydroxyalkanoates (PHAs) are one of the most researched bioplastics because they are biodegradable, renewable, biocompatible and environmentally friendly [1]. They have similar physical characteristics (including molecular mass, brittleness, melting point, and glass transition temperature) to that of synthetic petrochemical polymers such as polypropylene [2]. These characteristics, together with the worldwide problem of the depletion of fossil fuels, place PHAs as potential substitutes for conventional plastics, the petroleum-based polymers, especially in short-lived industrial applications [3]. In addition, these polymers have a wide field of use in medicine due to their innocuousness [4]. Therefore, PHAs have immense industrial potential, as already shown by applications in tissue engineering, drug delivery, and packaging [5].
However, one drawback of PHAs is that the production cost is still not competitive with the conventional polymers. So, the target of the PHAs production focused on advanced medical applications and products for tissue engineering. In fact, applications such as polymer-based devices for controlled drug delivery and hormone release or 3D printed resorbable scaffolds for tissue regeneration [6] require biodegradable polymers, so that degradation products are not harmful to the body. Several PHAs fulfill these issues [7, 8] since their monomeric and oligomeric in vivo degradation products have no deleterious effect on living cells or tissues [9, 10].
Poly(3-hydroxybutyrate) (PHB) is the only homopolymer of this large family, produced on an industrial scale and also the most studied [11]. More than 300 different microorganisms including Eubacteria (Pseudomonas sp., Ralstonia sp., Bacillus sp., Vibrio sp., Azotobacter sp., Methylobacterium sp., Burkholderia sp.) and archaea (e.g. Haloarchaea), intracellularly produce PHA granules [12, 13, 14, 15, 16, 17, 18, 19], as storage of carbon and energy under limiting conditions of essential nutrients such as nitrogen or phosphate and in the presence of an excess of carbon source [13, 15, 20].
Cupriavidus necator (formerly known as Wautersia eutropha, Ralstonia eutropha, and Alcaligenes eutrophus) is the most promising producer of PHB due a remarkable capacity for accumulating PHB up to 90 % of the cellular dry weight using a wide range of substrates in a heterotrophic or autotrophic pathway [21, 22]. Several carbon sources have been tested as substrate for the PHB production by wild type C. necator: fructose, glucose, lactic acid, xylose, sucrose, molasses, sorbose, acetic acid, starch, sodium acetate, glycerol, lactose, propionic acid and different lignocellulosic biomass hydrolysates; fructose is the one that allows the highest PHB productivity [23, 24]. Recently the advantage of using fructose has been demonstrated in certain bacteria: Halomonas sp. and Bacillus sp, prefers fructose over other sources; in some cases increasing its size by using this carbon source [14, 25].
To enhance the fermentation process from Cupriavidus necator it is necessary to increase the cell concentration and the intracellular PHB accumulation. One way to improve the PHB production is by tuning the growing medium and the operating conditions [26, 27]. Since the specific growth rate for the PHB production could be inhibited by the substrate concentration, the fed-batch fermentation is an useful approach to improve the PHB productivity [28]. Different feeding sources with a limiting nutrient were used to improve the cell growth and the PHB-productivity. Furthermore, the aerobic dynamic feeding by pulses of the carbon source enables the increase of the PHB content [29, 30, 31]. A successful strategy is the three-stage fermentation: The first stage is a batch culture for adaptation of the bacteria, the second is a fed-batch culture where the total biomass further increases and the third stage under nitrogen limitation in the medium allows to increase the PHB accumulation [32]. Moreover, different feeding policies were used to improve PHB production in fed-batch cultivation [26, 33, 34].
Most researchers use the C. necator ATCC 17699 strain for the PHAs production using various carbon substrates and fermentation strategies [22, 32, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46], however C. necator ATCC 17697 have been studied only by one research group [23, 26, 47, 48, 49], so it is important to extend its research. Regarding this goal, Khanna and Srivastava have been studied the PHB production by C. necator using different fructose feeding strategies with limited nitrogen source [23, 26, 47, 48, 49, 50]. They produced PHB in a batch system using ATCC 17697 strain, where the dissolved oxygen concentration was maintained at 30 % saturation by manually adjusting the agitation speed and/or airflow rate [47, 50]. Another model based on fed-batch fermentation with nitrogen and fructose feeding by applying different constant rates, at different times, was also successfully performed [26, 48, 49]. Through these fed-batch strategies, the maximum concentration of PHB in the culture medium was 18.6 g/l. Nevertheless, in accordance to Blunt et al. [51] it is possible to further increase the PHB concentration by applying other fermentation strategies, particularly a two-step fermentation: first in batch mode to adapt the microorganism to the medium and then a fed-batch to maximize cell density at a high rate.
In our previous work, full factorial design and response surface analysis were carried out to optimize media parameters including the concentration of fructose, ammonium sulfate, potassium dihydrogen phosphate, microelement solution, and initial pH to increase PHB production by shaking cultures of wild type C. necator ATCC 17697 [52,53].
The present work seeks, firstly, to scale up the PHB production from shaken flasks to a stirred-tank bioreactor with the optimized media and fructose. Secondly, the aim is to improve the production of PHB both by batch and fed-batch fermentation strategies in comparison with previous works of wild type C. necator with fructose as carbon source. Furthermore, thinking of biomedical applications, physicochemical and cytotoxicity analyses of the produced biopolymer are presented.
2. Materials and methods
2.1. Bacterial strain and media
Cupriavidus necator ATCC 17697 was obtained from American Type Culture Collection (Manassas, USA) and maintained on nutrient agar plates at 4 °C. Inoculum and bioreactor fermentations were performed in saline basal medium. It contained 20–40 g/l of fructose as the carbon source and 1.5–3.0 g/l of ammonium sulfate as the nitrogen source. These concentrations of carbon and nitrogen were changed depending on the fermentation strategy. The saline basal medium contained also potassium dihydrogen phosphate 4.35 g/l, sodium monobasic phosphate 4.35 g/l, magnesium sulfate heptahydrate 0.5 g/l and microelement solution 2 ml/l. The microelement solution contained FeSO4 2.0 g/l, MnCl2∙4 H2O 0.03 g/l, CaCl2∙2H2O 2.0 g/l, CuCl2∙2H2O 0.01 g/l, ZnSO4∙7 H2O 0.1 g/l, H3BO3 0.3 g/l, CoCl2∙6H2O 0.2 g/l, NiCl2∙6H2O 0.02 g/l and Na2MoO4∙2H2O 0.03 g/l in 0.1 N HCl solution [54]. The pH of the culture medium was adjusted to 7.0 ± 0.2 by the addition of 1 N HCl and 2 N NaOH.
2.2. PHB production by bioreactor fermentations
The production of PHB by C. necator ATCC 17697 was carried out by fermentation in a bioreactor using both batch and fed-batch strategies. Fermentations were performed in a 6 l stirred-tank bioreactor (BioFlo 110, New Brunswick Scientific; Edison, NJ), interfaced with Biocommand Bioprocessing software (New Brunswick Scientific) for parameter control and data acquisition. To obtain the bacterial inoculum, a volume of 200 ml of culture medium in a 1 l Erlenmeyer flask was inoculated with single colonies of C. necator ATCC 17697 and incubated at 30 ± 1 °C and 150 rpm for 24 h. This culture was employed to inoculate 2 l of culture medium contained in the bioreactor. The size of the inoculum (10%) is twice that used in the Erlenmeyer flask scale. The pH was measured in situ using a pH electrode (Mettler-Toledo GmbH, Germany). Dissolved oxygen concentration (dO2, % saturation) was measured with a polarographic probe (InPro6110/320, Mettler-Toledo GmbH) and modulated by controlling the agitation speed and adding filter-sterilized air (0.22 μm). Temperature was maintained at 30 ± 1 °C and foam formation was avoided by the addition of 0.3 % (v/v) antifoam 289 (Sigma-Aldrich; St. Louis, MO). Batch and fed-batch procedures are described below.
2.2.1. Batch strategy
At the beginning of the batch fermentation, the dO2 was set at 100 % saturation. When the dO2 reached to 20 %, the dO2 regulation was started using a cascade control strategy consisting of varying the agitation speed (800–1000 rpm) and air flow (1–2 l/min) to maintain dO2 saturation between 20 - 40 %.
The pH value was maintained at 7.0 ± 0.2 by the addition of 1 N HCl and 2 N NaOH. Cultures were withdrawn periodically during incubation for the quantification of fructose and ammonium concentration, cell biomass and PHB.
2.2.2. Fed-batch strategy
Two fed-batch fermentations were performed applying a three-stage procedure:
Stage 1: Batch culture: Cell growth phase.
Stage 2: Fed-batch culture with carbon and nitrogen supply: Cell growth and PHB accumulation phase.
Stage 3: Fed-batch culture without nitrogen supply: PHB accumulation phase.
In both fermentations, 1N HCl and 25 % (v/v) NH4OH were used to regulate the pH at 7.0 ± 0.2 from the beginning. The NH4OH solution also served as a source of nitrogen to avoid its limitation.
However, two fed-batch strategies with different modes of fructose feeding regulation were employed in Stage 2. In the first case (Fed-batch 1), fructose feeding was regulated by dO2 with a cut-off level of 70 % saturation. In the second case (Fed-batch 2), an exponential fructose feeding strategy was performed applying the exponential feeding equation (Eq. (1))
| (1) |
where F(t) is the substrate solution exponential flow rate (l/h); t denotes the time (h); μ0 is the specific growth rate (h−1); X0 and V0 are the biomass concentration (g/l) and the volume (l) at the beginning of the exponential feeding, respectively; YX/S is the biomass yield based on substrate consumption (gx/gs) and Sfeed is the substrate concentration in the feeding solution (gs/l) (600 g fructose/l). A series of batch experiments at the bioreactor level were performed with C. necator ATCC 17697 to determine YX/S and μ0, estimated as 0.44 g/g and 0.11 h−1 respectively [53].
The Stage 3, equal for both fermentations, consisted of a fed-batch culture without nitrogen supply. For this purpose, the fructose feeding (600 g/l) was controlled by dO2, with a cut-off level of 30–20 % saturation and the NH4OH solution was replaced by 2 N NaOH. Furthermore, in this stage, the agitation was reduced to 800-650 rpm and the aeration to 1 l/min, to improve the PHB production.
2.3. Analytical methods
Cell samples from bioreactor fermentations were harvested by centrifugation for 10 min at 3500 rpm (Presvac INS-DCA-300RTV), washed twice with distilled water and air-dried at 105 °C until constant weight (Numak DHG-9053A). Weights of the dried samples were considered as the dry cell weights and denoted as biomass (g/l). The recovered supernatant was used to determine the fructose and ammonium concentrations. Fructose concentration was determined by total organic carbon quantification through TOC-L Analyzer (Shimadzu) that operates in interface with TOC-Control L/V software.
The determination of ammonium concentration was performed according to the method of indophenol blue [55]. Quantitative estimation of PHB from biomass was carried out by the ultraviolet (UV) spectrophotometer method [56]. PHB was dissolved in 80 % (v/v) sulfuric acid solution and exposed at 100 °C for half an hour to be converted into crotonic acid. The sulfuric 80 % (v/v) acid solution without the polymer was used as blank. The absorbance of the solution was measured at 234 nm in a PerkinElmer Lambda 35 UV/Vis spectrometer. The commercially available Biocycle® 1000 - PHB polymer (PHB Industrial S/A, Brazil) was used as standard.
Biomass is composed of two components: i) the catalytically active component consists of proteins and nucleic acids (residual biomass), and ii) the inert component, the product PHB [47]. Henceforth, the residual biomass was defined as total biomass weight minus PHB weight.
2.4. Biopolymer extraction
Biomass from bioreactor fermentation was centrifuged for 30 min at 3500 rpm, supernatants removed and cell pellets freeze-dried and lyophilized at -83 °C and 3 × 10−3 mBar for 24 h (Labconco 7670030). PHB polymer was extracted from the lyophilized biomass with chloroform at 70 °C for 24 h using a Soxhlet extractor. The extract was then concentrated by rotary evaporation and the biopolymer was then precipitated in 10 volumes of ice-cold methanol at 4 °C. The precipitated PHB was filtered under vacuum using a ceramic filter and reprecipitated in chloroform and hexane to yield pure PHB as a white powder. Finally, PHB polymer was dried in an oven at 65 °C until reaching constant weight.
2.5. Biopolymer characterization
2.5.1. Differential scanning calorimetry (DSC)
The thermal properties of the polymer were evaluated by the DSC method using a DSC-Q 2000 calorimeter (Thermo Analytics). The conditioned samples (10 mg) were sealed in aluminum pans, equilibrated at -90 °C, kept isothermally for 5 min, heated from -90 °C to 200 °C and kept at 200 °C for 5 min before cooling to -90 °C. The samples were maintained at -90 °C for 5 min and reheated to 200 °C. Both heating and cooling rates were 10 °C/min and this thermal analysis was performed under N2 flow at 80 ml/min. The first heating cycle was performed to erase the heat history of the polymer.
The degree of crystallinity of PHB (XC) was calculated by the following Eq. (2):
| (2) |
where ΔHm is the apparent melting enthalpy and ΔHom is the theoretical value for the thermodynamic melting enthalpy obtained from a 100 % crystalline polymer (146.6 J/g) [57].
2.5.2. Fourier transform infrared spectroscopy (FTIR)
FTIR analysis was performed in the Attenuated Total Reflection mode (ATR) by direct analysis of PHB powder on ZnSe crystal. Infrared spectra were obtained at 400-4000 cm−1 on an FT-IR Nicolet 8700 spectrophotometer with 32 scans, a resolution of 4 cm−1 and an interval of 2 cm−1. The infrared spectra were analyzed to identify side-chain and functional groups. The commercial PHB used as the standard was the same as in Analytical methods.
2.5.3. 1H and 13C Nuclear magnetic resonance spectroscopy (NMR)
The dried biopolymer was suspended in deuterated chloroform (15 mg/ml solvent) to determine the molecular structures of PHB. 1H and 13C NMR spectra were recorded using a Bruker BioSpin GmbH spectrometer at 500 MHz (1H) and 126 MHz (13C), respectively.
2.5.4. Indirect cytotoxicity testing
0.2 mg of control and test PHB samples were incubated in 1 ml of Dulbecco's Modified Eagle Medium (DMEM) medium for 24 h at 37 °C in a humidified atmosphere containing 5 % CO2, following ISO norms [58]. Latex rubber and polytetrafluoroethylene (PTFE) were used as the positive and negative controls, respectively. Aliquots of each preparation obtained after incubation were considered as controls pure extracts and test pure extracts. Besides, 1/16 dilutions were prepared from these pure extracts. Simultaneously, NIH/3T3 (ATCC® CRL-1658™) fibroblast cells were incubated at 37 °C in a 5% CO2 humidified incubator for 24 h in a 24-well plate (Corning Costar, MA) at a concentration of 1 × 105 cells per ml of DMEM. After 24 h of incubation, the NIH/3T3 culture medium was removed by a slightly vigorous inversion of the plates. Subsequently, each culture well-received 200 μl of the controls and test pure extracts and diluted extracts and was incubated for the next 24 h. The cytotoxicity of PHB was assessed qualitatively, for which cells were examined microscopically in a Nikon TE2000-U inverted microscope coupled to an ORCA-ER CCD camera (Hamamatsu). Changes in general morphology, vacuolization, detachment, and cell lysis were evaluated. All experiments were performed in triplicate.
3. Results and discussion
3.1. Development of the fermentation process to enhance PHB production
3.1.1. Enhanced batch production of PHB through the fermenter
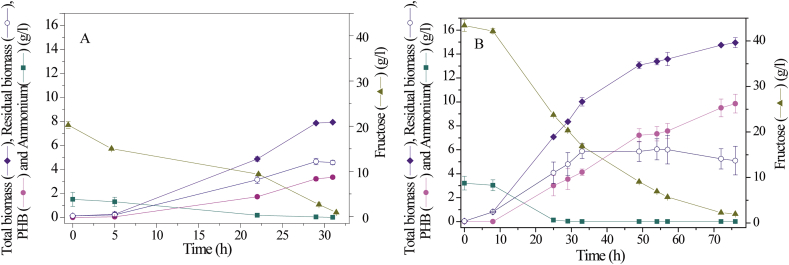
Two batch fermentations of C. necator ATCC 17697 were performed to determine the time evolution of total and residual biomass, fructose and ammonium consumption and PHB production. Figure 1A shows the parameters profile for a basal medium with 20 g/l fructose and 32 h incubation (Batch 1).
Figure 1.
Batch fermentation profiles of C. necator ATCC 17697 in culture medium with (A) 20 g/l fructose and 1.5 g/l ammonium sulfate for 32 h of incubation and (B) 40 g/l fructose and 3 g/l ammonium sulfate for 76 h of incubation.
PHB accumulation begins in the initial exponential phase and continues to increase until culture reaches the stationary phase. Fructose and ammonium consumptions correlate with bacterial growth and consequent polymer accumulation. Both cell growth and PHB accumulation are maximum at 31 h, 7.93 g/l, and 3.35 g/l, respectively. At this time, the PHB volumetric productivity (PPHB) is 0.11 g/(l h), the ammonium sulfate was already exhausted and fructose was consumed completely.
In this batch fermentation, the biomass concentration and polymer productivity were significantly higher, compared to the values obtained in our previous work [52] in shaken flask cultures during 72 h: 6.5 g/l and 0.06 g/(l h), respectively (Table 1). However, due to the depletion of the carbon source in the culture medium, the accumulation of PHB was stopped in the early stage of the stationary phase, without reaching higher values. The best medium chosen for shaken flask cultures is not always the best medium for bioreactor fermentations. This result has already been pointed out by Kennedy and Krouse [59].
Table 1.
Summary of PHB production and yield and its comparison with previous fermentation from fructose reported for C. necator ATCC 17697.
| Fermentation strategy | Biomass |
PHB |
PHB |
PPHB |
Reference |
|---|---|---|---|---|---|
| g/l | g/l | % | g/(l h) | ||
| Shaken flasks | 6.5 | 4.6 | 71 | 0.06 | [52] |
| Batch 1 | 7.9 | 3.4 | 42 | 0.11 | This work |
| Batch 2 | 14.4 | 9.9 | 69 | 0.13 | |
| Fed-batch 1 | 35.5 | 17.5 | 48 | 0.25 | |
| Fed-batch 2 | 50.8 | 25.7 | 51 | 0.43 | |
| Batch | 19.7–20.7 | 9.3–10.9 | 45–55 | 0.16–0.18 | [23, 47] |
| Fed-batch | 32–36 | 14–18.6 | 44–53 | 0.28–0.48 | [26, 48, 49] |
By this batch fermentation (Batch 1), higher biomass level was attained in less than half time compared to results obtained in our previous work using shaken flasks culture [52]; however, the PHB accumulation process stopped due to fructose depletion. Therefore, to optimize the production process in a bioreactor, a second batch fermentation (Batch 2) was carried out in a culture medium, doubling the concentrations of carbon and nitrogen sources (Figure 1B). In this fermentation, the exponential cell growth phase began after a lag phase of around 8 h. At 25 h all the ammonium sulfate in the culture medium was consumed, triggering PHB synthesis in the last phase of the fermentation process. The exponential growth finished after 33 h with a production of 10 g/l of biomass, where 4.13 g/l corresponds to the amount of accumulated polymer. Then, the residual biomass remained constant, while the total biomass increased exclusively by the intracellular accumulation of PHB. As a result, after 76 h of this batch fermentation 14.4 g/l of biomass and 9.90 g/l of PHB concentration were obtained, which represents a PPHB of 0.13 g/(l h) (Table 1). These results were consistent with those reported by Khanna and Srivastava [23], showing a slight improvement in PHB production.
Therefore, the second batch fermentation led to an increase of 10 % the PHB productivity and two and three times the biomass and PHB concentration, respectively, comparing to our results from shaken flask cultures. Since PHB productivity and concentration were not very high, the trial of fed-batch fermentation will be considered in the next section.
3.1.2. Improved fed-batch fermentation to enhance PHB production
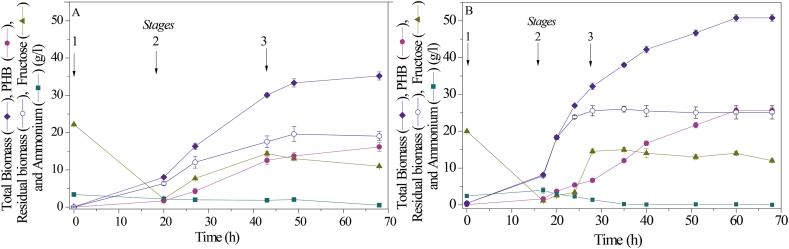
Two fed-batch fermentations of C. necator ATCC 17697 were performed: one by fructose feeding regulated with dO2 level (Figure 2A) and the other by exponential fructose feeding (Figure 2B).
Figure 2.
Fed-batch fermentation profiles of C. necator ATCC 17697 using fructose feeding regulated with dO2 level (A) and exponential fructose feeding (B). The arrows indicate the beginning of each stage.
The first fed-batch fermentation (Fed-batch 1) was developed using a three-stage production system, as shown in Figure 2A. The first stage was a batch culture for adaptation and biomass production. During 10 h of this stage corresponding to the lag phase of bacterial growth, the dO2 remained with a value higher than 90 %. Then, the dO2 began to decrease due to the exponential bacterial growth as well as the fructose concentration, which is completely depleted after 20 h. After this batch phase, total biomass and PHB concentrations were 8.0 and 1.6 g/l, respectively; this means that the carbon source was used mainly for biomass production.
The second stage was a fed-batch culture with fructose feeding regulated with dO2 level, in which the total biomass increased along with the PHB accumulation. The ammonium concentration, used to regulate pH and as a source of nitrogen remained relatively constant; thus, their supply and consumption rates were equal. Biomass and PHB concentrations increased to 30 g/l and 13.7 g/l, respectively, while the residual biomass reached a value of 16.3 g/l.
The third stage consisted of fed-batch culture with fructose feeding and without nitrogen supply, where only PHB production occurred. During the first 6 h, the ammonium concentration remained almost constant and then decreased from 1.4 g/l to 0.1 g/l; this condition of excess carbon is ideal to produce PHA [32, 47]. Throughout this stage, the residual biomass remained constant. At the end of this stage, 35.5 g/l of biomass and 17.5 g/l of PHB were achieved, with PHB productivity of 0.25 g/(l h) (Table 1). These results are slightly higher than those obtained by Khanna and Srivastava with the same strain but with a constant flow feeding strategy of 100 ml/h of 360 g/l fructose [26]. Therefore, using this fed-batch fermentation strategy it was possible to increase the PHB production in more than 3.5 and 1.5 fold, in comparison with shaken flask cultures and Batch 2 fermentation, respectively (Table 1).
The second fed-batch fermentation strategy (Fed-batch 2) was developed using exponential feeding to allow cells to grow at a constant specific growth rate to achieve high cell density in a short period [60, 61] and thus increase the amount of catalytic biomass.
After 17 h of the first batch stage, a biomass concentration of 8.17 g/l and PHB production of 0.30 g/l have been reached. In the second stage with exponential feeding from 17 h to 28 h, the bioreactor software determined the feeding flow rate according to Eq. (1), with V0 = 2.2 l and X0 = 7.5 g/l. Thus, the biomass raised to 32.2 g/l with a PHB content of 6.64 g/l (Figure 2B).
The main difference between the two fed-batch strategies in Stage 2 is the residual biomass concentration: 25.7 g/l with exponential fructose feeding, and only 16.4 g/l for fructose feeding with dO2 regulation. Also, due to the reduction of the second stage to less than half the time, the productivity of this phase highly improved. Therefore, the third stage of this fed-batch started earlier with more catalytic biomass for the biopolymer accumulation.
In the third stage —where only PHB accumulation takes place— the residual biomass remained constant and the total biomass reached its maximum value of 50.8 g/l with a PHB content of 25.7 g/l. The maximum volumetric productivity of this fed-batch process was 0.43 g/(l h) after 60 h of fermentation (Table 1).
By using the fed-batch fermentation with an exponential feeding strategy, we attained a PHB level (in g/l) 40% higher than that obtained by Khanna and Srivastava, who performed fed-batch fermentations using constant fructose feeding strategies [26, 48, 49].
Specific growth rates were computed by linear regression of natural log biomass concentration versus time; the slope of this line estimated the specific growth rates in the exponential phase (μ1) and the stationary phase (μ2) (Table 2).
Table 2.
Comparison of specific growth rates in the exponential phase (μ1) and the stationary phase (μ2) for the indicated fermentation strategies using C. necator ATCC 17697.
| Fermentation strategy | μ1 |
μ2 |
|---|---|---|
| h−1 | h−1 | |
| Shaken flasks | 0.115 | 0.006 |
| Batch 1 | 0.118 | 0.004 |
| Batch 2 | 0.114 | 0.011 |
| Fed-batch 1 | 0.101 | 0.013 |
| Fed-batch 2 | 0.119 | 0.011 |
Error ≤10 %
Cupriavidus necator is a model organism which has a strong ability to produce PHB in a non-growth-associated manner [62]. μ1 was determined in the growth-associated PHB production phase; its value is mainly due to the growth of biomass. μ2 was determined in non-growth-associated PHB production phase; this second rate is due exclusively to PHB production since the residual biomass remains constant. The occurrence of some growth-associated PHB production besides non growth-associated PHB production was demonstrated, although it is inhibited in the presence of nitrogen [63].
μ1 calculated for batch and fed-batch fermentations coincide with the value measured in shaken flask cultures, 0.115 h−1 (Table 2) and is similar to those obtained in fed-batch feeding of C. necator DSM 545 with glucose [64]. μ2 values calculated for shaken flasks were 0.006 h−1 which represents about 5 % of the maximum specific growth rate of this microorganism and could increase in large-scale fermentation where PHB production is higher. This feature, confirmed by comparing the μ2 values obtained at different fermentation scales, as shown in Table 1. Therefore, the residual biomass remains constant but the total biomass increased with the amount of accumulated biopolymer. In the case of batch 1 fermentation, the μ2 value was lower than that calculated in shaken flask cultures, since PHB accumulation stopped due to the depletion of the substrate in the culture medium.
3.2. Biopolymer characterization
Biopolymer samples produced by fermentation of C. necator ATCC 17697 were analyzed by differential scanning calorimetry, ATR-FTIR, 13C and 1H NMR spectroscopy, and indirect cytotoxicity testing.
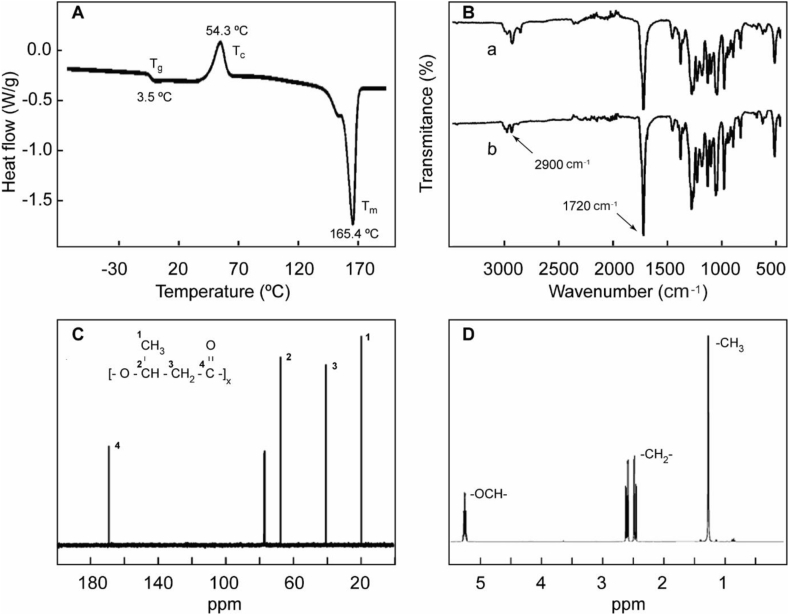
Figure 3 shows the polymer physicochemical characterization. The main properties of the produced PHB and their comparison with values from the literature are presented in Table 3.
Figure 3.
Polymer physicochemical characterization. DSC thermogram for PHB produced by C. necator ATCC 17697 (A). FTIR-ATR spectra of PHB produced by C. necator ATCC 17697 a) and commercially available b) (B). 13C NMR spectrum (C) and 1H NMR spectrum (D) of PHB produced by C. necator ATCC 17697.
Table 3.
NMR and DSC characterization of PHB produced by C. necator ATCC 17697 and comparison with PHB produced by other microorganisms.
| This work | Ref. [64] | Ref. [66] | Ref. [65] | Ref. [67] | ||||
|---|---|---|---|---|---|---|---|---|
| Bacterial strain | Cupriavidus necator | Cupriavidus necator | Cyanobacteria |
Bacillus cereus |
Cupriavidus necator | Bacillus megaterium | Standard PHB | |
| ATCC 17697 | DSM 545 | spp. | SPV | MTCC 8320 | MTCC 453 | |||
| Carbon source | fructose | glucose | glucose | glucose | fructose | fructose | ||
| Extraction method | solvent |
solvent |
solvent |
solvent |
solvent and ultrasonication |
solvent and ultrasonication |
||
| DSC analysis | ||||||||
| Tg (°C) | 3.5 | 6 | 6 | 2 | 6 | 6 | –8 | |
| Tm (°C) | 165.4 | 180 | 171 | 169.7 | 175 | 176 | 176 | |
| Tc (°C) | 54.3 | - | 78.8 | - | 84 | 104 | 90 | |
| Xc (%) | 56.0 |
64.6 |
56.8 |
57.7 |
44 |
23 |
- |
|
| NMR spectra (chemical shift in ppm) | ||||||||
| 13C NMR spectrum | -CH3 | 19.9 | - | - | 21.2 | 19.95 | 19.95 | 19.95 |
| -CH2- | 40.1 | - | - | 42.7 | 40.99 31.09 |
40.99 31.09 |
40.99 31.09 |
|
| -CH- | 67.7 | - | - | 68.5 | 67.81 | 67.80 | 67.81 | |
| -CO- | 169.3 | - | - | 169.7 | 169.32 | 169.32 | 169.32 | |
| 1H NMR spectrum | -CH- (m) | 5.26 | - | 5.22–5.28 | - | 5.26 | 5.26 | 5.26 |
| -CH2- (dq) | 2.45–2.63 | - | 2.43–2.64 | - | 2.17–2.60 | 2.17–2.62 | 2.17–2.60 | |
| -CH3 (d) | 1.27 | - | 1.27–1.29 | - | 1.28–1.60 | 1.26 | 1.28 | |
m: multiplet, dq:double quadruplet, d: doublet.
The thermal properties of PHB were determined using the DSC method (Figure 3A). The melting temperature (Tm), glass transition temperature (Tg) and crystallinity (XC) are key parameters to polymer processing and applications. Figure 3A shows the curve obtained from the second heating from which Tm was found at 165.4 °C and the crystallization temperature (Tc) at 54.3 °C. From these data, it was determined that the melting enthalpy and crystallinity were 81.3 J/g and 56 %, respectively. Tg was determined at 3.5 °C. Results obtained for the PHB produced by C. necator ATCC 17697 are similar to those reported in the literature for this biopolymer [64, 65, 66, 67]. ATR-FTIR spectra of the PHB samples from C. necator ATCC 17697 and the commercially available PHB are shown Figure 3B. The IR spectra revealed an intense band at 1720 cm−1 associated with the C=O bond stretching, which corresponds to the characteristic ester carbonyl group of polyhydroxyalkanoates [22, 68]. At 1181 cm−1 there is a band that is well known in the FTIR spectra of PHB due to the asymmetric stretching vibration of the C–O–C group. The C–H stretching from methyl and ethyl groups was assigned to the bands located in the spectral region around 2900 cm−1. The obtained ATR-FTIR spectrum of the polymer produced by fermentation was in agreement with the corresponding spectrum to the commercial PHB.
Figure 3C shows the 13C NMR spectrum of the polymer synthesized by C. necator ATCC 17697. Signals at chemical shifts (ppm): 169.3, 67.7, 40.1, 19.9 were assigned to –CO–, –CH–, –CH2- and –CH3, respectively. The signal at ca. 77 ppm is a triplet that corresponds to the solvent, CDCl3. 13C RMN analysis is in agreement with the expected values for the PHB chemical structure [65, 67]. Figure 3D shows the 1H-RMN of the polymer synthesized by C. necator ATCC 17697. The peaks observed correspond to those previously reported for the PHB structure [66, 67]. The doublet resonance signal at 1.27 ppm is attributed to the methyl –CH3 protons of the pendant chain in the PHB molecule. The doublet of quadruplet resonance signal at 2.45–2.63 ppm and the multiplet resonance signal at 5.26 ppm, corresponding to the methylene –CH2- and the methine –OCH– protons of the backbone chain, respectively.
The results obtained employing ATR-FTIR and NMR spectroscopy confirmed that the biopolymer produced by fermentation of C. necator ATCC17697 is the homopolyester PHB. The physico-chemical characterization is in the range expected for PHB obtained with the same extraction method but produced by different strains grown with different carbon sources (Table 3).
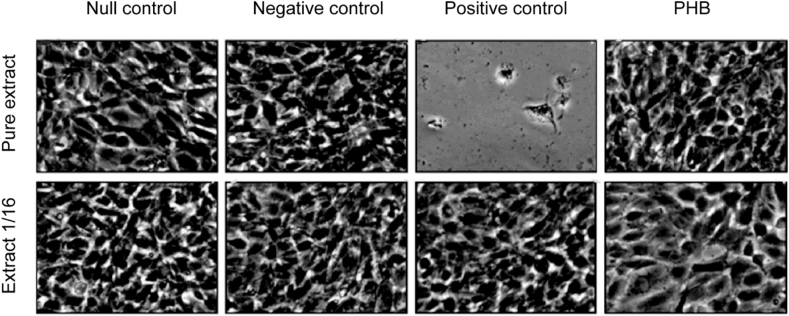
Indirect cytotoxicity assessment was carried out to establish the cytotoxic effect of PHB polymer produced by C. necator ATCC 17697. Microscopic photographs obtained from the NIH/3T3 fibroblast cells in the control and PHB extract assays are presented in Figure 4.
Figure 4.
Indirect cytotoxicity test of PHB polymer synthesized by C. necator ATCC 17697. “Null control” (DMEM medium without polymer), “negative control” (PTFE), “positive control” (latex rubber) and “PHB” (PHB produced by C. necator ATCC 17697). Magnification 100x.
No cytotoxic effect of PHB polymer was observed on the NIH/3T3 fibroblast cells. There was neither alteration in cell morphology nor fibroblast monolayer detaching, both in PHB pure extract assay and diluted. The same results were observed in the negative and null control tests. Only in the positive control test, the fibroblast monolayer was not formed.
4. Conclusions
This work presents the PHB production by fermentation of wild type C. necator ATCC 17697 in a stirred-tank bioreactor. Different fermentation strategies were tested and the experimental results were compared to our previously achieved using shaken flask cultures. Variation in fermentation conditions has been explored to increase the parameters production of PHB. The sequential presentation allows assessing the effects of improvements in the batch and fed-batch fermentation strategies.
Firstly, it was necessary to increase the scale from 250 ml Erlenmeyer to a 5 l bioreactor with the culture medium previously optimized in shaken flasks. For that purpose, in batch mode fermentations several parameters were adjusted, such as aeration, agitation mode, and concentrations of carbon and nitrogen sources. In batch mode, a slight improvement on biomass, PHB productivity, and PHB concentration was achieved.
Secondly, fed-batch fermentation with an exponential feeding strategy enabled us to achieve the highest values of PHB concentration and productivity. The PHB productivity obtained by this fed-batch fermentation strategy was 3.3 and 7.2 fold higher than in batch strategy and shaken flask cultures and 40% higher than reported for wild type C. necator strains. Furthermore, the purified polymer was characterized by DSC, FTIR, H1 and C13 NMR techniques; the structure and characteristics of the biopolymer produced by C. necator ATCC 17697 correspond to the PHB. Finally, it was confirmed that the polymer does not induce cytotoxic effects on the NIH/3T3 cell culture which is one of the features to be fulfilled for biomedical application. Thus, the following steps of this work will intend to scale up the production of this biopolymer to produce filaments for 3D printing of resorbable scaffolds tailored to the patient.
Declarations
Author contribution statement
Daiana Nygaard: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Oxana Yashchuk: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Diego G. Noseda: Performed the experiments; Wrote the paper.
Beatriz Araoz: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Élida B. Hermida: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by CONICET (Argentine Council of Scientific and Technical Research) [PIP183].
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors wish to express their special thanks to Ph. D. Ana Gonzales Wusener and Mauro Fernandez for the indirect cytotoxicity test and DSC essay, respectively.
References
- 1.Sudesh K., Abe H., Doi Y. Synthesis, structure and properties of polyhydroxyalkonates: biological polyesters. Prog. Polym. Sci. 2000;25:1503–1555. [Google Scholar]
- 2.Taguchi S., Iwata T., Abe H., Doi Y., Aqida S.N. Poly(hydroxyalkanoate)s, Ref. Modul. Mater. Sci. Mater. Eng. 2016:1–28. [Google Scholar]
- 3.Jung H.R., Choi T.R., Han Y.H., Park Y.L., Park J.Y., Song H.S., Yang S.Y., Bhatia S.K., Gurav R., Park H.A., Namgung S., Choi K.Y., Yang Y.H. Production of blue-colored polyhydroxybutyrate (PHB) by one-pot production and coextraction of indigo and PHB from recombinant Escherichia coli. Dyes Pigments. 2020;173 [Google Scholar]
- 4.Lackner M. sixth ed. 2015. Bioplastics- Biobased Plastics as Renewable And/or Biodegradable Alternatives to Petroplastics. [Google Scholar]
- 5.Bhatia S.K., Yoon J.J., Kim H.J., Hong J.W., Gi Hong Y., Song H.S., Moon Y.M., Jeon J.M., Kim Y.G., Yang Y.H. Engineering of artificial microbial consortia of Ralstonia eutropha and Bacillus subtilis for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer production from sugarcane sugar without precursor feeding. Bioresour. Technol. 2018;257:92–101. doi: 10.1016/j.biortech.2018.02.056. [DOI] [PubMed] [Google Scholar]
- 6.Pereira T.F., Oliveira M.F., Maia I.A., Silva J.V.L., Costa M.F., Thiré R.M.S.M. 3D printing of poly(3-hydroxybutyrate) porous structures using selective laser sintering. Macromol. Symp. 2012;319:64–73. [Google Scholar]
- 7.Koller M. Biodegradable and biocompatible polyhydroxy-alkanoates (PHA): auspicious microbial macromolecules for pharmaceutical and therapeutic applications. Molecules. 2018;23:1–20. doi: 10.3390/molecules23020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luef K.P., Stelzer F., Wiesbrock F. Poly(hydroxyalkanoate)s in medical applications. Chem. Biochem. Eng. Q. 2015;29:287–297. doi: 10.15255/CABEQ.2014.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Q., Wang Y., Chen G.Q. Medical application of microbial biopolyesters polyhydroxyalkanoates, Artif. Cells, Blood Substitutes. Biotechnol. 2009;37:1–12. doi: 10.1080/10731190802664429. [DOI] [PubMed] [Google Scholar]
- 10.Singh A.K., Srivastava J.K., Chandel A.K., Sharma L. Biomedical applications of microbially engineered polyhydroxyalkanoates: an insight into recent advances, bottlenecks, and solutions. Appl. Microbiol. Biotechnol. 2019:1–26. doi: 10.1007/s00253-018-09604-y. [DOI] [PubMed] [Google Scholar]
- 11.Byrom D. Production of poly-3-hydroxybutyrate: poly-3-hydroxyvalerate copolymers. FEMS Microbiol. Rev. 1992;103:247–250. [Google Scholar]
- 12.Hong J.W., Song H.S., Moon Y.M., Hong Y.G., Bhatia S.K., Jung H.R., Choi T.R., yeon Yang S., Park H.Y., Choi Y.K., Yang Y.H. Polyhydroxybutyrate production in halophilic marine bacteria Vibrio proteolyticus isolated from the Korean peninsula. Bioproc. Biosyst. Eng. 2019;42:603–610. doi: 10.1007/s00449-018-02066-6. [DOI] [PubMed] [Google Scholar]
- 13.Bhatia S.K., Gurav R., Choi T.R., Jung H.R., Yang S.Y., Song H.S., Jeon J.M., Kim J.S., Lee Y.K., Yang Y.H. Poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) production from engineered Ralstonia eutropha using synthetic and anaerobically digested food waste derived volatile fatty acids. Int. J. Biol. Macromol. 2019;133:1–10. doi: 10.1016/j.ijbiomac.2019.04.083. [DOI] [PubMed] [Google Scholar]
- 14.Yashchuk O., Hermida É.B. Influence of culture conditions on poly-3-hydroxybutyrate production by a newly isolated Bacillus cereus Y23, clean - soil. Air Water. 2020;48:1–7. [Google Scholar]
- 15.Koller Polyhydroxyalkanoate biosynthesis at the edge of water activitiy-Haloarchaea as biopolyester factories. Bioengineering. 2019 doi: 10.3390/bioengineering6020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chien C.C., Chen C.C., Choi M.H., Kung S.S., Wei Y.H. Production of poly-β-hydroxybutyrate (PHB) by Vibrio spp. isolated from marine environment. J. Biotechnol. 2007 doi: 10.1016/j.jbiotec.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Shrivastav A., Kim H.Y., Kim Y.R. Advances in the applications of polyhydroxyalkanoate nanoparticles for novel drug delivery system. BioMed Res. Int. 2013;2013 doi: 10.1155/2013/581684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zinn M., Bernard W., Thomas E. Occurrence, synthesis and medical application of bacterial polyhydroxyalkanoate. Adv. Drug Deliv. Rev. 2001;53:5–21. doi: 10.1016/s0169-409x(01)00218-6. [DOI] [PubMed] [Google Scholar]
- 19.Bonatto D., Matias F., Lisbôa M.P., Bogdawa H.M., Henriques J.A.P. Production of short side chain-poly[hydroxyalkanoate] by a newly isolated Ralstonia pickettii strain. World J. Microbiol. Biotechnol. 2004 [Google Scholar]
- 20.Anjum A., Zuber M., Zia K.M., Noreen A., Anjum M.N., Tabasum S. Microbial production of polyhydroxyalkanoates (PHAs) and its copolymers: a review of recent advancements. Int. J. Biol. Macromol. 2016;89:161–174. doi: 10.1016/j.ijbiomac.2016.04.069. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka K., Ishizaki A. Production of poly-D-3-hydroxybutyric acid from carbon dioxide by a two-stage culture method employing Alcaligenes eutrophus ATCC 17697. J. Ferment. Bioeng. 1994;77:425–427. [Google Scholar]
- 22.Aramvash A., Akbari Shahabi Z., Dashti Aghjeh S., Ghafari M.D. Statistical physical and nutrient optimization of bioplastic polyhydroxybutyrate production by Cupriavidus necator. Int. J. Environ. Sci. Technol. 2015;12:2307–2316. [Google Scholar]
- 23.Khanna S., Srivastava A.K. Statistical media optimization studies for growth and PHB production by Ralstonia eutropha. Process Biochem. 2005;40:2173–2182. [Google Scholar]
- 24.Bhatia S.K., Gurav R., Choi T.R., Jung H.R., Yang S.Y., Moon Y.M., Song H.S., Jeon J.M., Choi K.Y., Yang Y.H. Bioconversion of plant biomass hydrolysate into bioplastic (polyhydroxyalkanoates) using Ralstonia eutropha 5119. Bioresour. Technol. 2019;271:306–315. doi: 10.1016/j.biortech.2018.09.122. [DOI] [PubMed] [Google Scholar]
- 25.Park Y.L., Bhatia S.K., Gurav R., Choi T.R., Kim H.J., Song H.S., Park J.Y., Han Y.H., Lee S.M., Park S.L., Lee H.S., Kim Y.G., Yang Y.H. Fructose based hyper production of poly-3-hydroxybutyrate from Halomonas sp. YLGW01 and impact of carbon sources on bacteria morphologies. Int. J. Biol. Macromol. 2020;154:929–936. doi: 10.1016/j.ijbiomac.2020.03.129. [DOI] [PubMed] [Google Scholar]
- 26.Khanna S., Srivastava A.K. Productivity enhancement of poly-(β-hydroxybutyrate) by fed-batch cultivation of nutrients using variable (decreasing) nutrient rate by Wautersia eutropha. Chem. Eng. Commun. 2008;195:1424–1436. [Google Scholar]
- 27.Ertola R., Yantorno P., Mignone C. Microbiología industrial. Ser. Biol. 2006:1–103. [Google Scholar]
- 28.Grothe E., Chisti Y. Poly(β-hydroxybutyric acid) thermoplastic production by Alcaligenes latus: behavior of fed-batch cultures. Bioprocess Eng. 2000 [Google Scholar]
- 29.Patwardhan P.R., Srivastava A.K. Model-based fed-batch cultivation of R. eutropha for enhanced biopolymer production. Biochem. Eng. J. 2004 [Google Scholar]
- 30.Serafim L.S., Lemos P.C., Oliveira R., Reis M.A.M. Optimization of polyhydroxybutyrate production by mixed cultures submitted to aerobic dynamic feeding conditions. Biotechnol. Bioeng. 2004 doi: 10.1002/bit.20085. [DOI] [PubMed] [Google Scholar]
- 31.Wu W., Lai S.Y., Jang M.F., Chou Y.S. Optimal adaptive control schemes for PHB production in fed-batch fermentation of Ralstonia eutropha. J. Process Contr. 2013;23:1159–1168. [Google Scholar]
- 32.López-Cuellar M.R., Alba-Flores J., Rodríguez J.N.G., Pérez-Guevara F. Production of polyhydroxyalkanoates (PHAs) with canola oil as carbon source. Int. J. Biol. Macromol. 2011;48:74–80. doi: 10.1016/j.ijbiomac.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 33.López J.A., Bucalá V., Villar M.A. Application of dynamic optimization techniques for poly(b̃- hydroxybutyrate) production in a fed-batch bioreactor. Ind. Eng. Chem. Res. 2010 [Google Scholar]
- 34.Neddermeyer F., Rossner N., King R. Model-based control to maximise biomass and PHB in the autotrophic cultivation of Ralstonia eutropha. IFAC-Pap. Online. 2015;28:1100–1107. [Google Scholar]
- 35.Volova T.G., Kalacheva G.S., Steinbu A. Biosynthesis of multi-component polyhydroxyalkanoates by the bacterium Wautersia eutropha. Macromol. Symp. 2008;269:1–7. [Google Scholar]
- 36.Aramvash A., Hajizadeh-Turchi S., Moazzeni-zavareh F., Gholami-Banadkuki N., Malek-sabet N., Akbari-Shahabi Z. Effective enhancement of hydroxyvalerate content of PHBV in Cupriavidus necator and its characterization. Int. J. Biol. Macromol. 2016;87:397–404. doi: 10.1016/j.ijbiomac.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Jung Y., Lee Y. Utilization of oxidative pressure for enhanced production of poly-P-hydroxybutyrate and poly ( 3-hydroxybutyrate- 3-hydroxyvalerate) in Ralsto. Eutro. 2000;90:266–270. [PubMed] [Google Scholar]
- 38.Song J.Y., Kim B.S. Characteristics of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) production by Ralstonia eutropha NCIMB 11599 and ATCC 17699. Biotechnol. Bioproc. Eng. 2005;10:603–606. [Google Scholar]
- 39.Irorere V.U., Bagheriasl S., Blevins M., Kwiecień I., Stamboulis A., Radecka I. Electrospun fibres of polyhydroxybutyrate synthesized by ralstonia eutropha from different carbon sources. Int. J. Polym. Sci. 2014;2014 [Google Scholar]
- 40.Kunioka M., Nakamura Y., Doi Y. New bacterial copolyesters produced in Alcaligenes eutrophus from organic acids. Polym. Commun. Guildf. 1988 [Google Scholar]
- 41.Yang Y.H., Brigham C.J., Budde C.F., Boccazzi P., Willis L.B., Hassan M.A., Yusof Z.A.M., Rha C., Sinskey A.J. Optimization of growth media components for polyhydroxyalkanoate (PHA) production from organic acids by Ralstonia eutropha. Appl. Microbiol. Biotechnol. 2010;87:2037–2045. doi: 10.1007/s00253-010-2699-8. [DOI] [PubMed] [Google Scholar]
- 42.Chuah J.A., Yamada M., Taguchi S., Sudesh K., Doi Y., Numata K. Biosynthesis and characterization of polyhydroxyalkanoate containing 5-hydroxyvalerate units: effects of 5HV units on biodegradability, cytotoxicity, mechanical and thermal properties. Polym. Degrad. Stabil. 2013;98:331–338. [Google Scholar]
- 43.Franz A., Song H.S., Ramkrishna D., Kienle A. Experimental and theoretical analysis of poly(β-hydroxybutyrate) formation and consumption in Ralstonia eutropha. Biochem. Eng. J. 2011 [Google Scholar]
- 44.Wang J., Yue Z.B., Sheng G.P., Yu H.Q. Kinetic analysis on the production of polyhydroxyalkanoates from volatile fatty acids by Cupriavidus necator with a consideration of substrate inhibition, cell growth, maintenance, and product formation. Biochem. Eng. J. 2010;49:422–428. [Google Scholar]
- 45.Yan Q., Du G., Chen J. Biosynthesis of polyhydroxyalkanoates (PHAs) with continuous feeding of mixed organic acids as carbon sources by Ralstonia eutropha. Process Biochem. 2003;39:387–391. [Google Scholar]
- 46.Azhar A., Abdelhady H.M., Hafez A.M.A., Khodair T.A. Batch production of polyhydroxybutyrate (PHB) by ralstonia eutropha and Alcaligenes latus using bioreactor different culture. Strategies. 2009;5:556–564. [Google Scholar]
- 47.Khanna S., Srivastava A.K. A simple structured mathematical model for biopolymer (PHB) production. Biotechnol. Prog. 2005;21:830–838. doi: 10.1021/bp0495769. [DOI] [PubMed] [Google Scholar]
- 48.Khanna S., Srivastava A.K. Computer simulated fed-batch cultivation for over production of PHB: a comparison of simultaneous and alternate feeding of carbon and nitrogen. Biochem. Eng. J. 2006;27:197–203. [Google Scholar]
- 49.Khanna S., Srivastava A.K. Optimization of nutrient feed concentration and addition time for production of poly(β-hydroxybutyrate) Enzym. Microb. Technol. 2006;39:1145–1151. [Google Scholar]
- 50.Khanna S., Srivastava A.K. Recent advances in microbial polyhydroxyalkanoates. Process Biochem. 2005;40:607–619. [Google Scholar]
- 51.Blunt W., Levin D.B., Cicek N. Bioreactor operating strategies for improved polyhydroxyalkanoate (PHA) productivity. Polymers. 2018;10 doi: 10.3390/polym10111197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nygaard D., Yashchuk O., Hermida É.B. Evaluation of culture medium on poly(3-hydroxybutyrate) production by Cupriavidus necator ATCC 17697: application of the response surface methodology. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nygaard D. International Market Book Service Ltd.; 2020. Producción escalable del biopolímero polihidroxibutirato a partir de Cupriavidus necator ATCC 17697. [Google Scholar]
- 54.Barbosa M., Espinosa Hernández A., Malagón Romero D., Moreno Sarmiento N. Producción de poli-β-hidroxibutirato (PHB) por Ralstonia eutropha ATCC 17697. Univ. Sci. Rev. La Fac. Ciencias. Pontif. Univ. Javeriana. 2005;10:45–54. [Google Scholar]
- 55.Grasshoff K., Kremling K., Ehrhardt M. third ed. 2007. Methods of Seawater Analysis. [Google Scholar]
- 56.Repaske R., Repaske A.C. Quantitative requirements for exponential growth of Alcaligenes eutrophus. Appl. Environ. Microbiol. 1976;32:585–591. doi: 10.1128/aem.32.4.585-591.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lüpke T., Radusch H.J., Metzner K. Solid-state processing of PHB-powders. Macromol. Symp. 1998;127:227–240. [Google Scholar]
- 58.ISO 10993-5, Biological Evaluation of Medical Devices – Part 5 - Tests for Cytotoxicity: in Vitro Methods. 2009. [Google Scholar]
- 59.Kennedy M., Krouse D. Strategies for improving fermentation medium performance: a review. J. Ind. Microbiol. Biotechnol. 1999;23:456–475. [Google Scholar]
- 60.Ienczak J.L., Quines L.K., de Melo A.A., Brandellero M., Mendes C.R., Schmidell W., Aragão G.M.F. High cell density strategy for poly(3-hydroxybutyrate) production by Cupriavidus necator. Braz. J. Chem. Eng. 2011;28:585–596. [Google Scholar]
- 61.Lee J., Lee S.Y., Park S., Middelberg A.P.J. Control of fed-batch fermentations. Biotechnol. Adv. 1999;17:29–48. doi: 10.1016/s0734-9750(98)00015-9. [DOI] [PubMed] [Google Scholar]
- 62.Doran P.M. second ed. 2012. Bioprocess Engineering Principles. [Google Scholar]
- 63.Mozumder M.S.I., Goormachtigh L., Garcia-Gonzalez L., De Wever H., Volcke E.I.P. Modeling pure culture heterotrophic production of polyhydroxybutyrate (PHB) Bioresour. Technol. 2014;155:272–280. doi: 10.1016/j.biortech.2013.12.103. [DOI] [PubMed] [Google Scholar]
- 64.Koller M., Bona R., Hermann C., Horvat P., Martinz J., Neto J., Pereira L., Varila P., Braunegg G. Biotechnological production of poly(3-hydroxybutyrate) with Wautersia eutropha by application of green grass juice and silage juice as additional complex substrates. Biocatal. Biotransform. 2005;23:329–337. [Google Scholar]
- 65.Valappil S.P., Misra S.K., Boccaccini A.R., Keshavarz T., Bucke C., Roy I. Large-scale production and efficient recovery of PHB with desirable material properties, from the newly characterised Bacillus cereus SPV. J. Biotechnol. 2007;132:251–258. doi: 10.1016/j.jbiotec.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 66.Ansari S., Fatma T. Cyanobacterial polyhydroxybutyrate (PHB): screening, optimization and characterization. PloS One. 2016;11:1–20. doi: 10.1371/journal.pone.0158168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pradhan S., Dikshit P.K., Moholkar V.S. Production, ultrasonic extraction, and characterization of poly (3-hydroxybutyrate) (PHB) using Bacillus megaterium and Cupriavidus necator. Polym. Adv. Technol. 2018 [Google Scholar]
- 68.Bayarı S., Severcan F. FTIR study of biodegradable biopolymers: P(3HB), P(3HB-co-4HB) and P(3HB-co-3HV) J. Mol. Struct. 2005;747:529–534. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.