Abstract
The ATPase H+/K+ Transporting Beta Subunit (ATP4B) encodes the β subunit of the gastric H+, K+-ATPase, which controls gastric acid secretion and is therefore a target for acid reduction. Downregulation of ATP4B was recently observed in human gastric cancer (GC) without known mechanisms. In the present study, we demonstrated that ATP4B expression was decreased in human GC tissues and cell lines associated with DNA hypermethylation and histone hypoacetylation of histone H3 lysine 9 at its intragenic region close to the transcriptional start site. The expression of ATP4B was restored in GC cell lines by treatment with the DNA methyltransferase inhibitor, 5-aza-2′-deoxycytidine (5-AZA), or histone deacetylase inhibitor, trichostatin A (TSA), with further enhancement by combined treatment with both drugs. In contrast, 5-AZA had no effect on ATP4B expression in human hepatocellular carcinoma (HCC) and pancreatic cancer cell lines, in which ATP4B was silenced and accompanied by intragenic methylation. Chromatin immunoprecipitation (ChIP) showed that, in BGC823 GC cells, histone H3 lysine 9 acetylation (H3K9ac) was enhanced in the intragenic region of ATP4B upon TSA treatment, whereas 5-AZA showed a minimal effect. Additionally, ATP4B expression enhanced the inhibitory effects of chemotherapeutic mediation docetaxel on GC cell growth. Thus, as opposed to HCC and pancreatic cancer cells, the silencing of ATP4B in GC cells is attributable to the interplay between intragenic DNA methylation and histone acetylation of ATP4B, the restoration of which is associated with a favorable anticancer effect of docetaxel. These results have implications for targeting epigenetic alteration at the intragenic region of ATP4B in GC cells to benefit diagnosis and treatment of GC.
Key words: ATP4B, Gastric cancer (GC), Intragenic DNA methylation, Histone modification, Biomarker
INTRODUCTION
Gastric cancer (GC) is one of the leading malignancies and remains the second most lethal cancer worldwide, with increasing incidence in Asia1,2. It is highly desirable to develop specific molecular biomarkers for precise diagnosis and surveillance of GC3–9. Epigenetic regulation represented by cell type-specific DNA methylation and of histone modifications is essential for normal development and gene expression10–15. Epigenetic regulation is a progressive but reversible process, the alteration of which is involved in a variety of chronic inflammation-associated cancers, such as GC4,16–19. Silencing of the tumor suppressor gene in cancer cells caused by hypermethylation of promoter regions is a well-defined epigenetic hallmark and thus has significant implications for early detection, treatment, and prognostic assessment of GC4,17,18,20. Therefore, considerable effort has been expended to delineate the cell- and tissue-specific epigenetic alterations during tumorigenesis21–27.
The gastric H+, K+-ATPase (ATP4) is a dimeric heterodimer (〈αβ〉2), composed of two catalytic α-subunits and two regulatory β subunits, coded by the ATP4A and ATP4B genes, respectively28. This enzyme is expressed in parietal cells of the stomach as a gastric proton pump that exports H (+) in exchange for luminal K (+) and is also responsible for the last step in acid secretion. ATP4 hyperactivity may cause acid peptic disorders and gastric inflammation and thus is the major therapeutic target in the treatment of acid-related gastric diseases29.
In addition to high expression levels in the stomach, ATP4A is a widely expressed pattern in human tissues and is considered to play a “housekeeping” role, whereas ATP4B is expressed in a cell- and tissue-specific manner as a gene involved in gastric function30,31. Recent studies revealed the downregulation of ATP4B in GC5–7,32,33. We previously reported that ATP4B might act as a diagnostic biomarker because of abnormal expression changes in GC. ATP4B was also reported to be overexpressed in human hepatocellular carcinoma (HCC)34. However, the mechanism for the aberrant expression of ATP4B during gastric tumorigenesis remains elusive.
We therefore hypothesized that ATP4B is epigenetically silenced in human GC in a tissue-specific manner. In this study, we reported that epigenetic modifiers 5-aza-2′-deoxycytidine (5-AZA) and trichostatin A (TSA) restored ATP4B expression in GC cells but not in HCC and pancreatic cancer cells. Treatment of GC cells with docetaxel (DOC) showed an inhibitory effect on GC cell proliferation, which was synergistically enhanced when ATP4B was ectopically expressed in GC cells. Therefore, ATP4B is epigenetically silenced in GC, which may constitute a valuable target for diagnosis and chemotherapeutic intervention of GC.
MATERIALS AND METHODS
Cell Culture and Treatment
Human GC cell lines (BGC823, MGC803, SGC7901, NCI-N87, and AGS), immortalized human gastric mucosal epithelial cell line GES-1, human HCC cell lines (HepG2, M3, SMMC-7721, BEL-7402, and Huh7), immortalized human embryo liver cell line L02, and human pancreatic cancer cell lines (PANC 5.04, PANC 3.11, SW1990, MIAPACA-2, and PANC-1) were grown in 90% DMEM (Gibco, Life Technologies, Grand Island, NY, USA) with 10% fetal bovine serum (FBS; Gibco, Life Technologies). GC cells were split to low density (30% confluence) for overnight culture and then treated with 5 μM of DNA methyltransferase inhibitor, 5-AZA (Sigma-Aldrich, St. Louis, MO, USA) and/or 5 μM of histone deacetylase inhibitor, TSA (Sigma-Aldrich). Cells were exposed to 5-AZA for 96 h with the growth medium being changed every 24 h or to TSA for 24 h. For combined treatment, the cells were initially exposed to 5-AZA for 72 h and then TSA for 24 h. For cell viability assays, GC cells were exposed to 5-AZA for 3 days with the growth medium being changed every day and/or to TSA at day 1. GC cells were also treated with DOC (Taxotere, 20 mg; Aventis Pharma, Dagenham, UK) daily for 3 days at a concentration of 20 nM after transfection of ATP4B35.
DNA Construction and Transfection
The cDNA coding ATP4B was cloned into piRESneo vector (Clontech Laboratories, Inc., Mountain View, CA, USA) to construct recombinant plasmid piRESneo-ATP4B (ATP4B). In vitro transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instruction35,36. Transfection efficiency was evaluated by Western blotting of ectopic ATP4B expression.
Patients and Specimens
Endoscopic biopsy specimens of primary GC were obtained from patients and stored at the Tissue Bank of Peking University Cancer Hospital according to the standard procedures of the ethics committee of Beijing Jiaotong University as previously described35,36.
RNA Isolation, Reverse Transcription (RT), and Polymerase Chain Reaction (PCR)
Cells were harvested for RNA isolation using TRIzol reagent (Invitrogen, Grand Island, NY, USA), and first-strand cDNA was synthesized with the SuperScript First-Strand Synthesis System (Invitrogen). PCR was performed using primers listed in Table 1. The mRNA expression levels of ATP4B and internal control glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were analyzed using 2× Taq PCR Mix (GenStar Biosolutions, Beijing, P.R. China) on Coyote Theater 4 × 4 PCR machine (Coyote, Beijing, P.R. China). Conventional PCR products were visualized on a 1.5% agarose gel. Real-time quantitative PCR (qPCR) was performed using 2× SYBR Green-based qPCR reagent on ABI 7500 qPCR machine (Applied Biosystems, Foster City, CA, USA). The relative expression level of each gene was normalized to the amount of the same cDNA using the 2−ΔΔCt method and was further compared to its own control.
Table 1.
Primer Sequences and PCR Conditions
| Accession No. | Target Gene (Size) | Primer ID | Sequence (5′ to 3′) | Oligo Use For |
|---|---|---|---|---|
| NM_000705 | ATP4B (183 bp) | F | AGGAGTTCCAGCGTTACTGC | RT-PCR |
| R | GGTCTTGGTAGTCCGGTGTG | |||
| NM_000704 | ATP4A (142 bp) | F | GCAATCGCTCTCATTGCTGTGGTT | RT-PCR |
| R | TCTGGAATTTGTCTCCATCGCGGA | |||
| NM_005953 | MT2A (318 bp) | F | AACCTGTCCCGACTCTAGC | RT-PCR |
| R | GGAATATAGCAAACGGTCACG | |||
| NM_002046 | GAPDH (472 bp) | F | ACCACAGTCCATGCCATCAC | RT-PCR |
| R | TCCACCACCCTGTTGCTGTA | |||
| NM_000705 | ATP4B (280 bp) | F | GGATCGTCAAGTTCCTCCCC | RT-qPCR |
| R | GGGTCGTGGGGATTGTTGAA | |||
| NM_002046 | GAPDH (189 bp) | F | GAGGCGGAGGAGAACAAACA | RT-qPCR |
| R | CCATGGAGAAGGCTGGG | |||
| NC_000013 REGION: complement (113648804..113658198) | ATP4B (70 bp) | MF | GAGTTTTAGCGTTATTGTTGGAATTCGGATAC | MSP |
| MR | GTACCCCACCGAAACAAAATACG | |||
| UF | GGAGTTTTAGTGTTATTGTTGGAATTTGGATATG | MSP | ||
| UR | CACACATACCCCACCAAAACAAAATACA | |||
| ATP4B (240bp) | F | GGAGGATTTTAGGTTAGGGA | BQ | |
| R | CACTATCAAAAACCCTCTACTC | |||
| ATP4B (125bp) | F | GCAACGGTGATTCCTTGGTC | ChIP-qPCR | |
| R | TTTACTCCCCACCTCC GCAT |
F, forward; R, reverse; M, methylated; U, unmethylated.
DNA Extraction, Bisulfite Modification, Methylation-Specific PCR (MSP-PCR), and Bisulfite Sequencing (BS)
Total DNA was extracted from cell lines and tissues using the genomic DNA rapid extraction kit (Aurora Biomed, Inc., Vancouver, BC, Canada). Bisulfite modification of DNA was performed using Zymo DNA Methylation Kit (Zymo Research, Irvine, CA, USA). MSP-PCR was performed using primer pairs that specifically amplify either methylated or unmethylated sequences of the ATP4B gene37 (Table 1). The in vitro methylation DNA (IVD) serving as the positive control was the A&D Human Methylated DNA Standard (A&D Technology, Beijing, P.R. China), and the negative control was the genomic DNA from normal human peripheral lymphocytes as previously described38. MSP products were analyzed using a 2% agarose gel electrophoresis. Bisulfite-treated DNA was amplified using BS primers as described in Table 1. PCR products were gel purified and cloned into pGEM-T vectors (Promega, Madison, WI, USA). Colonies were randomly selected for plasmid isolation using EasyPure Plasmid MiniPrep Kit (TransGen Biotech, Beijing, P.R. China) and subjected to sequencing with the SP6 reverse primer via automated sequencing (Beijing AUGCT Biotechnology Co. Ltd., Beijing, P.R. China) as previously described38.
Western Blotting
Total protein was prepared using RIPA lysis buffer (Beyotime Biotech, Jiangsu, P.R. China). Protein was measured using a BCA Protein Assay Kit (CWBIO, Beijing, P.R. China). Proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto PVDF membranes using a Bio-Rad Mini PROTEAN 3 system (Hercules, CA, USA). The membranes were blocked with PBS containing 5% milk and 0.1% Tween 20 at room temperature for 1 h. The membranes were then immunoblotted with the desired primary antibodies and incubated with the responding horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies. The primary antibodies were as follows: anti-ATP4B (mouse monoclonal; 1:1,000; ab2866), anti-H3K9ac (rabbit polyclonal; 1:1,000; ab4441), and H4K5ac (rabbit polyclonal; 1:1,000; ab114146) were purchased from Abcam (Cambridge, UK), and anti-HDAC1 (rabbit monoclonal; 1:800; AH379) and HDAC2 (rabbit monoclonal; 1:800; AH382) were purchased from Beyotime, Ltd. β-Actin (mouse monoclonal; 1:10,000; A4551) was from Sigma-Aldrich. Horseradish peroxidase-conjugated anti-mouse (1:2,500 dilution) or anti-rabbit (1:2,500 dilution) secondary antibodies were purchased from Bioworld Technology, Inc. (St. Louis, MO, USA). Immunoreactive bands were visualized using the Amersham ECL Western Blotting Detection Kit according to the manufacturer’s instructions. β-Actin served as a loading control.
Chromatin Immunoprecipitation-qPCR (ChIP-qPCR)
ChIP assay was performed by following the EpiTect ChIP One Day Kit protocol (QIAGEN, Valencia, CA, USA)35. BGC823 cells with different treatments were fixed with 1% formaldehyde. Chromatin was prepared by sonication of cell lysate and preclearing with protein A beads. Aliquots of precleared chromatin solution (named as “IP fractions”) were incubated with 2 μg of specific rabbit anti-H3K9ac or preimmune rabbit IgG on a rotation platform at 4°C overnight. IP fraction (1%) served as the “input control” for each ChIP assay. Protein A beads were added to precipitate the antibody-enriched protein–DNA complexes from the IP fractions. After washing, the immune complexes were subjected to reversal cross-link to release DNA fragments. Immunoprecipitated DNA fractions were purified using a QIAquick Purification Kit (QIAGEN) and analyzed by qPCR using 0.05% immunoprecipitated DNA as a template35.
Cell Viability
BGC823, MGC803, and SGC7901 cells were seeded in 96-well plates at a density of 2.5 × 103 cells/well in culture medium for 1 day and treated with 5-AZA, TSA, and DOC or in combination for 3 days as described above. Cell proliferation was measured every day after the indicated treatment using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay kit (Sigma-Aldrich). The absorbance at 570 nm was detected using a microplate reader (680 ELISA; Bio-Rad)35. All experiments were performed in triplicate.
Statistical Analysis
The data are expressed as means ± standard deviation (SD) of at least three independent experiments. The DNA methylation in human GC was analyzed by the Student’s t-test. Results were judged to be statistically significant at p < 0.05.
RESULTS
Decreased Expression of ATP4B in Human GC Cell Lines and Tissues
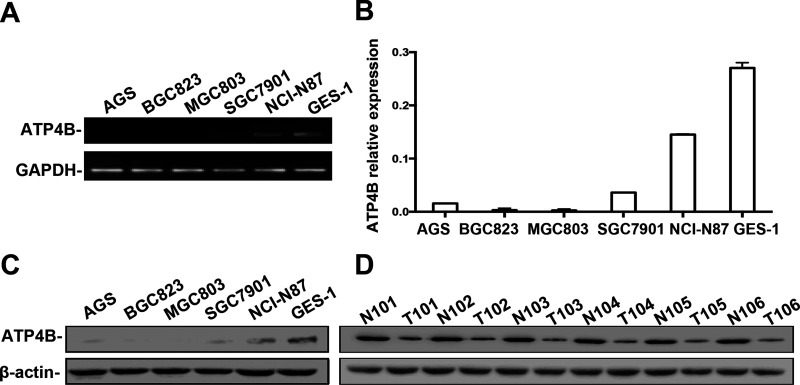
We first determined the expression of ATP4B in the human gastric cell lines BGC823, MGC803, AGS, SGC7901, and NCI-N87, as well as in the immortalized gastric cell line GES-1. Semiquantitative RT-PCR showed that ATP4B expression was absent in the BGC823, MGC803, and AGS cell lines, weakly expressed in the SGC7901 cell line, but highly expressed in the NCI-N87 cell line relative to GES-1 cells (Fig. 1A), which was confirmed by RT-qPCR (Fig. 1B). Western blotting showed that the expression of ATP4B was completely absent in BGC823 and MGC803 cells, but weakly detectable in AGS, SGC7901, and NCI-N87 cells (Fig. 1C). In specimens from GC patients, ATP4B was downregulated in tumor tissues compared with adjacent nontumorous tissues (Fig. 1D), confirming decreased expression of ATP4B in GC.
Figure 1.
ATP4B expression in GC cell lines and paired GC tumor and matched tumor-adjacent tissue samples. (A) Semiquantitative RT-PCR analysis of the mRNA level of ATP4B expression in human GC cell lines and an immortalized gastric cell line GES-1. (B) RT-qPCR was used to quantify the amount of ATP4B mRNA in the GC cell lines and GES-1 cells. Relative expression of ATP4B was related to GAPDH mRNA using the 2−ΔΔCt method and is presented as the mean ± standard deviation (SD). (C, D) Western blotting analysis of the protein level of ATP4B expression in GC cell lines and GES-1 cells (C) or the paired GC tumor and matched tumor-adjacent tissue samples (D).
Epigenetic Alterations in ATP4B in Human GC Cell Lines and Tissues
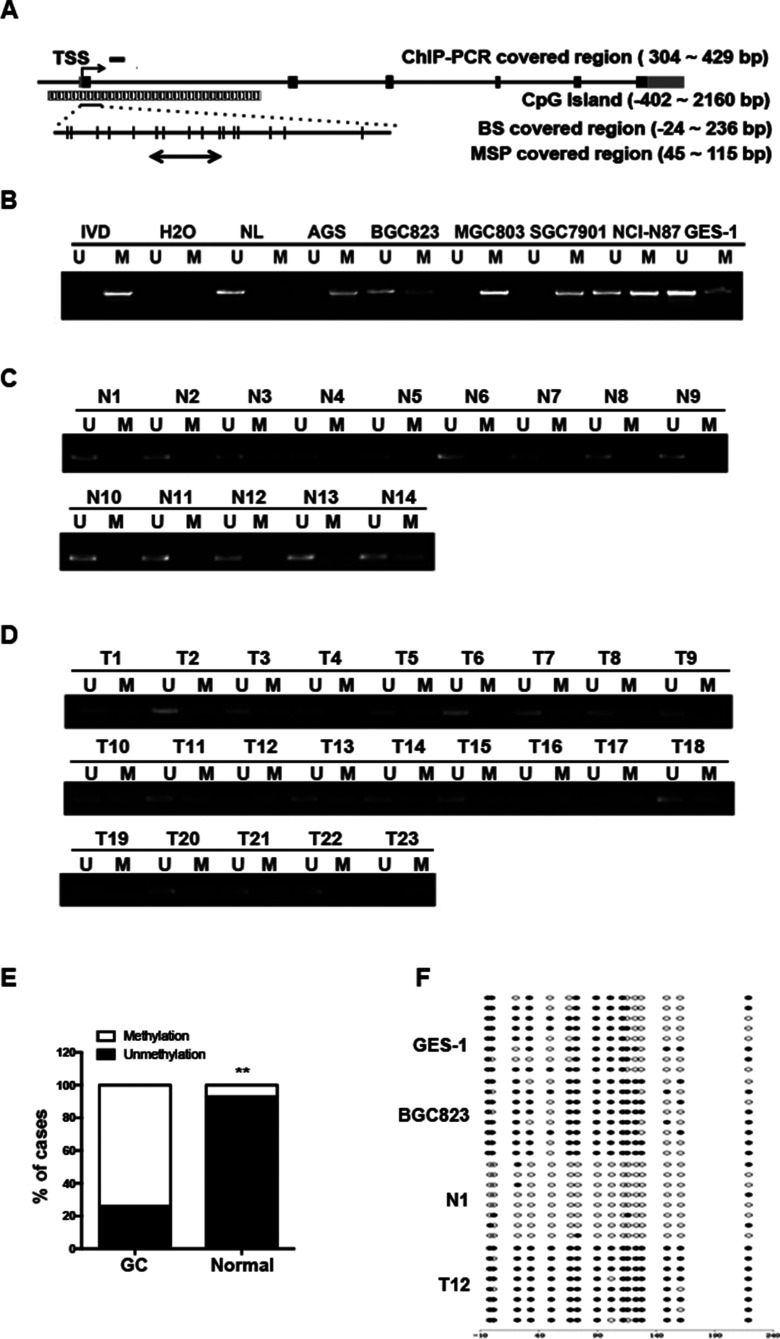
We next analyzed the methylation status of ATP4B in GC cell lines. The primers for MSP and BS were designed using the MethPrimer Program to cover the intragenic region close to the transcriptional start site (TSS) of the ATP4B (referred to as the intragenic region), where there is a CpG island (Fig. 2A). As shown in Figure 2B, the intragenic region of ATP4B was completely methylated in the AGS, MGC803, and SGC7901 cell lines and partially methylated in the BGC823 and NCI-N87 cell lines compared with GES-1 cells, which contained weak methylation in this region. BS39 showed a dense methylation pattern in BGC823 cells but partial methylation in GES-1 cells, confirming the MSP findings (Fig. 2F).
Figure 2.
The DNA methylation of ATP4B in human GC. (A) Schematic of ATP4B genomic structure indicating regions amplified in the methylation-specific PCR (MSP) and bisulfite sequencing (BS) as well as ChIP-PCR analysis. Each vertical bar represents one CpG site. The numbers indicate the position relative to the TSS. TSS, transcription start site; GC, gastric cancer; ChIP, chromatin immunoprecipitation. (B–D) MSP analysis of ATP4B methylation in GC cell lines and GES-1 cells (B), normal gastric tissue samples (n = 14) (C), and human primary GC (n = 23) (D). (E) Association of ATP4B methylation with GC. **p < 0.01, ATP4B methylation in GC versus ATP4B methylation in normal gastric tissue. (F) BS confirmation of ATP4B gene methylation. Representative results of BS in BGC823 cells, GES-1 cells, normal gastric tissue samples (N1), and human primary GC (T12). U, unmethylated; M, methylated; IVD, in vitro methylated DNA served as MSP-positive control; NL, normal blood lymphocyte DNA as negative control. Filled circles represent methylated CpG dinucleotides, and open circles indicate unmethylated sites.
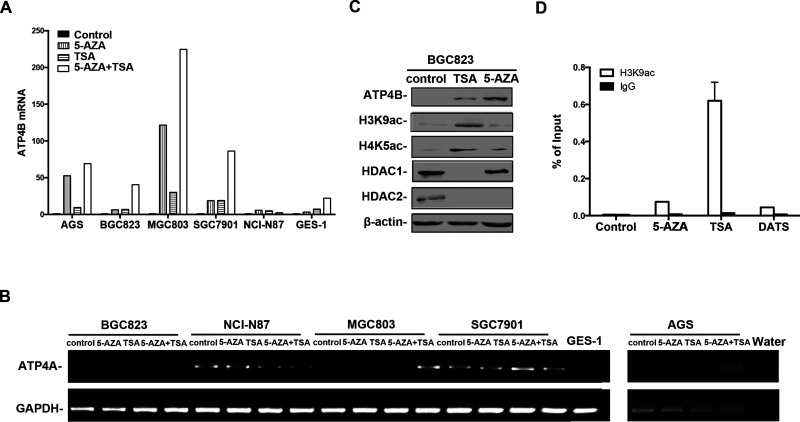
We further investigated histone acetylation status in BGC823 cells, in which ATP4B was silenced and accompanied by partial methylation in its intragenic region. Western blotting showed a relatively low level of global histone acetylation at H3 lysine 9 (H3K9ac) and H4 lysine 5 (H4K5ac) in BGC823 cells (Fig. 3C). ChIP-qPCR was further used to examine the specific association of histone acetylation with the ATP4B intragenic region (Fig. 2A). As shown in Figure 3D, there was no enrichment of H3K9ac in the intragenic region in BGC823 cells. Therefore, ATP4B was hypermethylated in the intragenic region accompanied by histone hypoacetylation.
Figure 3.
The expression of ATP4B and ATP4A and histone modifications in human GC cells upon epigenetic reagent treatment. (A) RT-qPCR analysis of the change of ATP4B expression following treatment with epigenetic reagents, 5-AZA and/or TSA (5-AZA: 5 μM, 96 h; TSA: 5 μM, 24 h). The relative expression of ATP4B mRNA in each cell line was normalized against GAPDH control using the 2−ΔΔCt and further compared to its untreated control. Results shown represent the mean ± SD. 5-AZA, 5-aza-2′-deoxycytidine; TSA, trichostatin A. (B) RT-PCR of ATP4A expression in human GC cell upon epigenetic reagent treatment. (C) Western blotting of BGC823 cells after 5-AZA or TSA treatment. (D) ChIP was performed on BGC823 cells treated with 5-AZA or TSA or DATS (40 μM, 12 h) using antibody against H3K9ac or control IgG. Precipitated ChIP DNA fractions were analyzed by qPCR for the enrichment of H3K9ac in the ATP4B intragenic region. Results are expressed as the percentage of input. DATS, diallyl trisulfide.
To ascertain the intragenic epigenetic alteration of ATP4B in GC, MSP was used to examine ATP4B methylation in 23 human GC tumor tissues and 14 normal gastric tissue samples. The intragenic region of ATP4B was moderately or heavily methylated in 73.91% (17 of 23) of GC tissues, but was unmethylated in 78.57% (11 of 14) of normal gastric tissues (p < 0.01) (Fig. 2C–E). Representative samples from tumor or normal tissues subjected to BS analysis showed heavily methylated or unmethylated status, respectively (Fig. 2F). These data support the epigenetic alterations in the intragenic region of ATP4B in GC.
Restoration of Silenced ATP4B in Human GC Cell Lines Through Epigenetic Regulations
To explore whether the decreased expression of ATP4B was attributable to epigenetic alteration, GC cells were treated with 5-AZA, or TSA, or in combination. As shown in Figure 3A, ATP4B expression was markedly increased in AGS, BGC823, MGC803, and SGC7901 cells, but less upregulated in NCI-N87 and GES-1 cells following 5-AZA or TSA treatment alone. Combined treatment with 5-AZA and TSA resulted in a synergistic effect on ATP4B expression in these cell lines, except NCI-N87. Similar results were observed in GC cells for the expression of ATP4A, which was previously identified to be epigenetically silenced in GC cells40 (Fig. 3B). Western blotting confirmed the upregulation of ATP4B in BGC823 cells treated with 5-AZA or TSA (Fig. 3C). Moreover, TSA treatment resulted in a marked upregulation of H3K9ac and H4K5ac, but a reduction in histone deacetylase 1 and 2 (HDAC1/2). 5-AZA treatment showed less effect on HDAC1 expression but was capable to mediate histone acetylation (Fig. 3C). Thus, epigenetic modifiers, 5-AZA and TSA, coordinately regulate the expression of ATP4B in GC cells.
The histone acetylation in the intragenic region was further analyzed in BGC823 cells, which showed that H3K9ac was significantly enriched upon TSA treatment. Additionally, treatment of BGC823 cells with 5-AZA caused a slight enrichment of H3K9ac in the intragenic region (Fig. 3D). These results suggest that histone acetylation provides the open chromatin status to be accessible to support active transcription of ATP4B in GC cells. Our results thus showed that ATP4B silencing in GC is attributable to DNA methylation and histone deacetylation in the intragenic region.
Specific Epigenetic Regulation of ATP4B in Human GC Cell Lines
To elucidate whether the epigenetic silencing of ATP4B gene in GC cells is cell and tissue type specific, we extended the study to other human cancer cell lines. RT-PCR showed that ATP4B was completely silenced in all human HCC (Fig. 4A) and pancreatic cancer cell lines (Fig. 4B). MSP analysis showed that the ATP4B gene was partially methylated in all HCC cell lines, including cancer cell lines, as well as an immortalized liver embryonic cell line L02 (Fig. 4C). The ATP4B gene was also completely or partially methylated in all pancreatic cancer cell lines (Fig. 4D). However, 5-AZA treatment failed to restore ATP4B expression in both HCC and pancreatic cancer cells (Fig. 4A and B). These data demonstrate that the silencing of ATP4B in HCC and pancreatic cancer cells is not due to epigenetic regulation, such as DNA methylation. These data indicate that epigenetic silencing of ATP4B in GC cells is cell/tissue type specific.
Figure 4.
The silencing of ATP4B in human hepatocellular carcinoma (HCC) and pancreatic cancer cell lines not due to epigenetic regulation. (A, B) Semiquantitative RT-PCR of the mRNA level of ATP4B expression in human HCC cell lines and immortalized liver embryonic cell line L02 (A) or in human pancreatic cancer cell lines (B). HCC, hepatocellular carcinoma. (C, D) MSP analysis of ATP4B methylation in HCC cell lines (C) or pancreatic cell lines (D). (E) RT-PCR of the expression of ATP4B and MT2A in human BGC823 cell line treated with DATS (40 μM, 12 h) as described. MT2A was used as a positive control for DATS treatment. MT2A, metallothionein 2A.
We recently demonstrated that a chemoprevention drug, diallyl trisulfide (DATS), acted as an HDAC inhibitor with the capability of inducing MT2A expression in BGC823 cells through histone acetylation35. We thus compared the effect of 5-AZA with DATS in BGC823 cells. As shown in Figure 4E, DATS treatment induced MT2A expression, but not ATP4B. In contrast, the expression of both MT2A and ATP4B was restored in BGC823 cells by 5-AZA or TSA35 (Fig. 3A). ChIP analysis revealed that DATS treatment was not able to enrich H3K9ac in the ATP4B intragenic region in GC cells (Fig. 3D), supporting the RT-PCR results (Fig. 4E). The results showed that the ATP4B gene is transcriptionally responsive to epigenetic drugs rather than the chemical reagent DATS.
Restoration of ATP4B Enhanced the Inhibitory Effect of Docetaxel on GC Cells
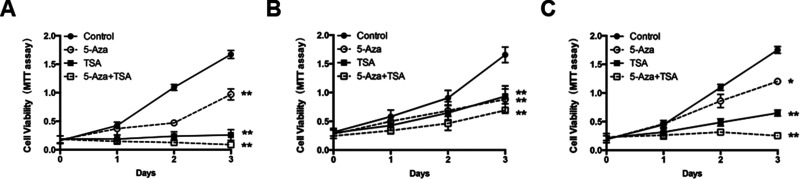
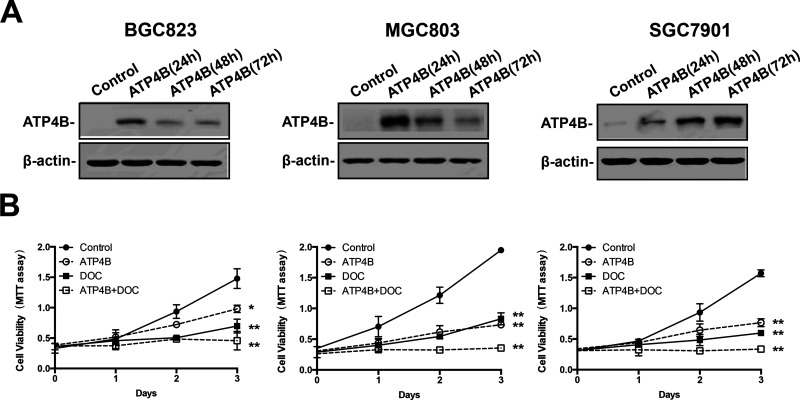
The specific epigenetic restoration of ATP4B by the epigenetic drugs in GC cells prompted us to explore the potential therapeutic relevance of ATP4B. Treatment of BGC823, MGC803, and SGC7901 cells with 5-AZA or TSA alone significantly reduced GC cell viability, and the combination treatment with two agents resulted in a synergistic inhibition of GC cell proliferation (Fig. 5A–C). To determine whether the antitumor activity of epigenetic drugs on GC cells is attributable to their capacity to restore ATP4B, a vector with ectopic expression of ATP4B was transfected into GC cells (Fig. 6A). MTT assays showed that restoration of ATP4B inhibited the growth of GC cells (Fig. 6B). We then used a chemotherapeutic drug, DOC, to test its anti-GC effects in conjunction with ATP4B restoration. MTT assays showed that the anti-GC effects of DOC were synergistically enhanced in GC cells transfected with ectopic ATP4B (Fig. 6B). These results indicate that ATP4B is a tumor suppressor that enhances the sensitivity of GC cells to the chemotherapeutic treatment by DOC.
Figure 5.
The effects of epigenetic drugs on GC cell proliferation. MTT assays monitoring the cell variability of GC cells treated with 5 μM 5-AZA daily for 3 days, or with 5 μM TSA for 1 day, or with 5-AZA daily for 3 days and TSA at day 1. (A) BGC 823. (B) MGC803. (C) SGC7901. 5-AZA, 5-aza-2′-deoxycytidine; TSA, trichostatin A. Results shown represent the mean ± SD (n = 3). *p < 0.05, **p < 0.005, significant differences from the control.
Figure 6.
The antiproliferative effects of docetaxel on GC cells. (A) Ectopic ATP4B expression vector (ATP4B) was constructed and transfected into BGC823, MGC803, and SGC7901 cells for transient expression. The expression of ATP4B was examined daily by Western blotting. (B) MTT assays were performed to monitor the variability of GC cells treated with docetaxel daily for 3 days at a concentration of 20 nM. ATP4B, ectopic ATP4B expression vector; DOC, docetaxel. Data are presented as the mean ± SD (n = 3). *p < 0.05, **p < 0.005, significant differences from the control.
DISCUSSION
Human ATP4B is mainly expressed in the parietal cells of the stomach and represents a gastric function gene, which was downregulated in human GC tissues5–7,32,33, and associated with poor survival of patients with gastric noncardia adenocarcinoma cancer7. In agreement with this, we showed that the expression of ATP4B is significantly decreased in human GC tumor tissues and cell lines. We further found that ATP4B was silenced in HCC and pancreatic cancer cell lines, consistent with recent studies41. However, in HCC, overexpression of ATP4B was reported as more frequent in HCC tissues at the stages of severe fibrosis and cirrhosis than in non-HCC tissues34.
Cell and tissue type-specific gene expression is regulated epigenetically in the patterns of genomic DNA methylation and histone modifications11,12,23,24. The disruption is associated with cancer development, as shown by prevalent tissue-specific CpG island methylation in gene bodies, regulated by intragenic DNA methylation24. Cell type-specific methylation is most frequently present in intragenic CpG islands. Intragenic DNA methylation surrounding TSS is thought to be tightly associated with transcriptional silencing regulated during cell differentiation and cancer in a cell and tissue type-specific manner15,21,22. In support of this, we provided evidence that intragenic epigenetic alteration in ATP4B is critical for its silencing in GC tissues and cell lines as shown by the significant restoration of ATP4B expression in GC cells by 5-AZA or TSA, alone or in combination. Moreover, we confirmed that the suppressive effect of TSA on HDAC activity resulted in hyperacetylation in the ATP4B intragenic region to provide accessible chromatin in response to 5-AZA treatment. In contrast, in both HCC and pancreatic cancer cells, although ATP4B is also silenced and accompanied by DNA methylation, its transcription may be regulated through nonepigenetic mechanisms, since 5-AZA failed to alter ATP4B expression. A recent study showed that both ATP4B and ATP4A were repressed in gastric tumor tissues and associated with intragenic DNA methylation. However, the data shown are technically questionable since a pair of the same primers of ATP4A was used in RT-PCR for both ATP4A and ATP4B40.
Garlic-derived component DATS was capable of inducing transcriptional expression of another tumor suppressor gene MT2A in GC through an epigenetic mechanism35. MT2A belongs to the metallothionein family, functioning as a heavy metal-binding protein for metal storage and transport42. The expression of MT2A is thought to be through a tissue-specific manner, and thus is considered to be a parallel control of ATP4B in GC upon drug treatment35. Interestingly, we found that ATP4B in GC cells appears to respond more favorably to epigenetic drugs rather than DATS since DATS failed to alter ATP4B expression at the transcription level, although DATS acting as an HDAC inhibitor induced MT2A expression in GC35. These data suggest that ATP4B in GC may be regulated by the interplay between intragenic DNA methylation and histone modifications.
ATP4B is a critical component of gastric H+, K+-ATPase and is essential for homeostasis of gastric tissue28. ATP4B was reported in a mouse model to be required for normal development and function of mouse parietal cells43. Gastric H+, K+-ATPase is the major target for treatment of peptic and duodenal ulcers and gastroesophageal reflux disease by acid-suppressive drugs such as proton pump inhibitors (PPIs), as well as treatment of parietal cell autoantibodies associated with autoimmune gastritis44. However, recently, the increased risk of GC associated with acid-suppressive drugs has been reported45–50. A meta-analysis of 11 studies by pooling 94,558 participants (5,980 GC patients and 88,578 controls) showed that acid-suppressive drug use was associated with an increased risk of GC46. Nevertheless, there has been no study so far to show whether functional disruption of ATP4B is associated with the development of GC. Therefore, it is important to clarify whether the incidence of GC is increased due to acid suppression using PPIs to block the gastric H+, K+-ATPase.
The antitumor effects of epigenetic drugs have been observed in cultured and primary cancer cells by altering epigenetic pathways and affecting sensitivity of cancer cells to chemotherapeutic drugs51. We showed that the inhibitory effects of 5-AZA and TSA on GC cells might be attributable to their capacity to induce ATP4B expression, since restoration of ATP4B by ectopic expression displayed a similarly tumor-suppressive role in GC cells. Importantly, ectopic expression of ATP4B enhanced the inhibitory effect of DOC on GC cell growth. Docetaxel is one of the widely used chemotherapeutic medications approved by the FDA for the treatment of many cancers, including GC52. Our results therefore suggest that restoration of ATP4B may enhance the sensitivity of GC cells to chemotherapeutic treatments, which provides new insights into gastric carcinogenesis and a biomarker for diagnosis, as well as a target for treatment of GC.
In conclusion, we reported in this study that ATP4B is epigenetically silenced in GC, attributable to the intragenic DNA methylation and histone deacetylation in a tissue-specific manner. Our findings of epigenetic alteration of ATP4B in the intragenic region may therefore represent a promising target for future efforts to develop diagnostic biomarkers as well as chemotherapeutic intervention for GC.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research funding of Beijing Jiaotong University (S12RC00030) and the Open Project funded by Key Laboratory of Carcinogenesis and Translational Research Ministry of Education (2014 Open Project-8). S.L., J.H., and J.M.W. were also funded in part by federal funds from the National Cancer Institute, National Institutes of Health (NCI, NIH), under Contract No. HHSN261200800001E and were supported in part by the Intramural Research Program of the NCI, NIH.
REFERENCES
- 1. Bray F, Jemal A, Grey N, Ferlay J, Forman D. Global cancer transitions according to the Human Development Index (2008–2030): A population-based study. Lancet Oncol. 2012;13(8):790–801. [DOI] [PubMed] [Google Scholar]
- 2. Mahipal A, Choi M, Kim R. Second-line treatment of advanced gastric cancer: Where do we stand? J Natl Compr Canc Netw. 2015;13(10):1281–91; quiz 1292. [DOI] [PubMed] [Google Scholar]
- 3. Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10(11):643–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lim B, Kim JH, Kim M, Kim SY. Genomic and epigenomic heterogeneity in molecular subtypes of gastric cancer. World J Gastroenterol. 2016;22(3):1190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hasegawa S, Furukawa Y, Li M, Satoh S, Kato T, Watanabe T, Katagiri T, Tsunoda T, Yamaoka Y, Nakamura Y. Genome-wide analysis of gene expression in intestinal-type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res. 2002;62(23):7012–7. [PubMed] [Google Scholar]
- 6. Kim B, Bang S, Lee S, Kim S, Jung Y, Lee C, Choi K, Lee SG, Lee K, Lee Y, Kim SS, Yeom YI, Kim YS, Yoo HS, Song K, Lee I. Expression profiling and subtype-specific expression of stomach cancer. Cancer Res. 2003;63(23):8248–55. [PubMed] [Google Scholar]
- 7. Wang G, Hu N, Yang HH, Wang L, Su H, Wang C, Clifford R, Dawsey EM, Li JM, Ding T, Han XY, Giffen C, Goldstein AM, Taylor PR, Lee MP. Comparison of global gene expression of gastric cardia and noncardia cancers from a high-risk population in China. PLoS One 2013;8(5):e63826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513(7517):202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lordick F, Janjigian YY. Clinical impact of tumour biology in the management of gastroesophageal cancer. Nat Rev Clin Oncol. 2016;13(6):348–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659(1–2):40–8. [DOI] [PubMed] [Google Scholar]
- 11. Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, Bird A. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21(7):1074–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Novak P, Stampfer MR, Munoz-Rodriguez JL, Garbe JC, Ehrich M, Futscher BW, Jensen TJ. Cell-type specific DNA methylation patterns define human breast cellular identity. PLoS One 2012;7(12):e52299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bloushtain-Qimron N, Yao J, Snyder EL, Shipitsin M, Campbell LL, Mani SA, Hu M, Chen H, Ustyansky V, Antosiewicz JE, Argani P, Halushka MK, Thomson JA, Pharoah P, Porgador A, Sukumar S, Parsons R, Richardson AL, Stampfer MR, Gelman RS, Nikolskaya T, Nikolsky Y, Polyak K. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci USA 2008;105(37):14076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31(2):175–9. [DOI] [PubMed] [Google Scholar]
- 15. Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11(11):1068–75. [DOI] [PubMed] [Google Scholar]
- 16. Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17(3):330–9. [DOI] [PubMed] [Google Scholar]
- 17. Schneider BG, Peek RM Jr. Gastric cancer prevention by demethylation. Cancer Prev Res. (Phila) 2013;6(4):253–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oh JH, Jung SH, Hong SJ, Rhyu MG. DNA methylation as surrogate marker for gastric cancer. J Cancer Prev. 2015;20(3):172–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, Tamura G, Saito D, Sugimura T, Ichinose M, Ushijima T. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12(3 Pt 1):989–95. [DOI] [PubMed] [Google Scholar]
- 20. Baylin SB, Jones PA. A decade of exploring the cancer epigenome—Biological and translational implications. Nat Rev Cancer 2011;11(10):726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kulis M, Queiros AC, Beekman R, Martin-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta 2013;1829(11):1161–74. [DOI] [PubMed] [Google Scholar]
- 22. Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One 2011;6(1):e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiench M, John S, Baek S, Johnson TA, Sung MH, Escobar T, Simmons CA, Pearce KH, Biddie SC, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, Hager GL. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30(15):3028–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010;466(7303):253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jjingo D, Conley AB, Yi SV, Lunyak VV, Jordan IK. On the presence and role of human gene-body DNA methylation. Oncotarget 2012;3(4):462–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Avraham A, Cho SS, Uhlmann R, Polak ML, Sandbank J, Karni T, Pappo I, Halperin R, Vaknin Z, Sella A, Sukumar S, Evron E. Tissue specific DNA methylation in normal human breast epithelium and in breast cancer. PLoS One 2014;9(3):e91805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y, Jadhav RR, Liu J, Wilson D, Chen Y, Thompson IM, Troyer DA, Hernandez J, Shi H, Leach RJ, Huang TH, Jin VX. Roles of distal and genic methylation in the development of prostate tumorigenesis revealed by genome-wide DNA methylation analysis. Sci Rep. 2016;6:22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gumz ML, Lynch IJ, Greenlee MM, Cain BD, Wingo CS. The renal H+-K+-ATPases: Physiology, regulation, and structure. Am J Physiol Renal Physiol. 2010;298(1):F12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sachs G, Shin JM, Vagin O, Lambrecht N, Yakubov I, Munson K. The gastric H,K ATPase as a drug target: Past, present, and future. J Clin Gastroenterol. 2007;41(Suppl 2):S226–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Herrmann M, Selige J, Raffael S, Sachs G, Brambilla A, Klein T. Systematic expression profiling of the gastric H+/K+ ATPase in human tissue. Scand J Gastroenterol. 2007;42(11):1275–88. [DOI] [PubMed] [Google Scholar]
- 31. Ordonez GR, Hillier LW, Warren WC, Grutzner F, Lopez-Otin C, Puente XS. Loss of genes implicated in gastric function during platypus evolution. Genome Biol. 2008;9(5):R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yan Z, Luke BT, Tsang SX, Xing R, Pan Y, Liu Y, Wang J, Geng T, Li J, Lu Y. Identification of gene signatures used to recognize biological characteristics of gastric cancer upon gene expression data. Biomark Insights 2014;9:67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rajkumar T, Vijayalakshmi N, Gopal G, Sabitha K, Shirley S, Raja UM, Ramakrishnan SA. Identification and validation of genes involved in gastric tumorigenesis. Cancer Cell Int. 2010;10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shao RX, Hoshida Y, Otsuka M, Kato N, Tateishi R, Teratani T, Shiina S, Taniguchi H, Moriyama M, Kawabe T, Omata M. Hepatic gene expression profiles associated with fibrosis progression and hepatocarcinogenesis in hepatitis C patients. World J Gastroenterol. 2005;11(13):1995–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pan Y, Lin S, Xing R, Zhu M, Lin B, Cui J, Li W, Gao J, Shen L, Zhao Y, Guo M, Wang JM, Huang J, Lu Y. Epigenetic upregulation of metallothionein 2A by diallyl trisulfide enhances chemosensitivity of human gastric cancer cells to docetaxel through attenuating NF-kappaB activation. Antioxid Redox Signal. 2016;24(15):839–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pan Y, Huang J, Xing R, Yin X, Cui J, Li W, Yu J, Lu Y. Metallothionein 2A inhibits NF-kappaB pathway activation and predicts clinical outcome segregated with TNM stage in gastric cancer patients following radical resection. J Transl Med. 2013;11:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002;18(11):1427–31. [DOI] [PubMed] [Google Scholar]
- 38. Jia Y, Yang Y, Liu S, Herman JG, Lu F, Guo M. SOX17 antagonizes WNT/beta-catenin signaling pathway in hepatocellular carcinoma. Epigenetics 2010;5(8):743–9. [DOI] [PubMed] [Google Scholar]
- 39. Grabsch HI, Tan P. Gastric cancer pathology and underlying molecular mechanisms. Dig Surg. 2013;30(2):150–8. [DOI] [PubMed] [Google Scholar]
- 40. Raja UM, Gopal G, Rajkumar T. Intragenic DNA methylation concomitant with repression of ATP4B and ATP4A gene expression in gastric cancer is a potential serum biomarker. Asian Pac J Cancer Prev. 2012;13(11):5563–8. [DOI] [PubMed] [Google Scholar]
- 41. Kong SC, Giannuzzo A, Novak I, Pedersen SF. Acid-base transport in pancreatic cancer: Molecular mechanisms and clinical potential. Biochem Cell Biol. 2014;92(6):449–59. [DOI] [PubMed] [Google Scholar]
- 42. Babula P, Masarik M, Adam V, Eckschlager T, Stiborova M, Trnkova L, Skutkova H, Provaznik I, Hubalek J, Kizek R. Mammalian metallothioneins: Properties and functions. Metallomics 2012;4(8):739–50. [DOI] [PubMed] [Google Scholar]
- 43. Scarff KL, Judd LM, Toh BH, Gleeson PA, Van Driel IR. Gastric H(+),K(+)-adenosine triphosphatase beta subunit is required for normal function, development, and membrane structure of mouse parietal cells. Gastroenterology 1999;117(3):605–18. [DOI] [PubMed] [Google Scholar]
- 44. Toh BH, Sentry JW, Alderuccio F. The causative H+/K+ ATPase antigen in the pathogenesis of autoimmune gastritis. Immunol Today 2000;21(7):348–54. [DOI] [PubMed] [Google Scholar]
- 45. Garcia Rodriguez LA, Lagergren J, Lindblad M. Gastric acid suppression and risk of oesophageal and gastric adenocarcinoma: A nested case control study in the UK. Gut 2006;55(11):1538–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ahn JS, Eom CS, Jeon CY, Park SM. Acid suppressive drugs and gastric cancer: A meta-analysis of observational studies. World J Gastroenterol. 2013;19(16):2560–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Waldum HL, Hauso O, Fossmark R. Letter: Proton pump inhibitors, hypergastrinaemia and the risk of gastric neoplasia. Aliment Pharmacol Ther. 2015;42(3):389. [DOI] [PubMed] [Google Scholar]
- 48. Waldum HL, Qvigstad G. Proton pump inhibitors and gastric neoplasia. Gut 2007;56(7):1019–20; author reply 1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kuipers EJ. Proton pump inhibitors and gastric neoplasia. Gut 2006;55(9):1217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Waldum HL, Brenna E, Sandvik AK. Long-term safety of proton pump inhibitors: Risks of gastric neoplasia and infections. Expert Opin Drug Saf. 2002;1(1):29–38. [DOI] [PubMed] [Google Scholar]
- 51. Azad N, Zahnow CA, Rudin CM, Baylin SB. The future of epigenetic therapy in solid tumours—Lessons from the past. Nat Rev Clin Oncol. 2013;10(5):256–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kang BW, Kwon OK, Chung HY, Yu W, Kim JG. Taxanes in the treatment of advanced gastric cancer. Molecules 2016;21(5). [DOI] [PMC free article] [PubMed] [Google Scholar]