Abstract
Nasopharyngeal carcinoma (NPC) is a distinct head and neck cancer, which is occurring at a high frequency in Southern China. Emerging studies have shown that long noncoding RNAs (lncRNAs) play a critical role in carcinogenesis and progression. In this study, we established a comprehensive lncRNA profile in NPC and found that 35 lncRNAs were differentially expressed in NPC. We found that LINC0086 was decreased in NPC patient serum samples and tissues. The Kaplan–Meier survival curve showed that patients with high LINC0086 expression had a higher survival rate than those with low LINC0086 expression. LINC0086 expression was associated with NPC histological grade, lymph node metastasis, and clinical stage. Upregulation of LINC0086 inhibited cancer cell proliferation and promoted apoptosis. In addition, upregulation of LINC0086 dramatically decreased the expression of miR-214, an oncogene in several cancers, in C666-1 and HK-1 cells. An miR-214 binding site was found in the 3′-UTR of LINC0086. We also validated that both miR-214 and LINC0086 presented in the RISC complex, demonstrating that LINC0086 could decrease miR-214 expression by directly interacting with miR-214. Furthermore, the suppressive effects of LINC0086 on NPC cell growth were reversed by overexpression of miR-214 in vitro and in vivo. Thus, our study reports a novel mechanism underlying NPC carcinogenesis and provides a potential novel diagnosis and treatment biomarker for NPC.
Key words: Nasopharyngeal carcinoma (NPC), Long noncoding RNAs (lncRNAs), LINC0086, miR-214, Carcinogenesis
INTRODUCTION
Nasopharyngeal carcinoma (NPC) is a distinct head and neck cancer, which is occurring at a high frequency in Southern China1. It is estimated by the World Health Organization (WHO) that 40% of all NPC cases occurred in China2. Patients diagnosed with NPC at an early stage (stages I and II) had a satisfactory treatment result, which is significantly different from that of patients diagnosed at late stages (stages III and IV)3. NPC has a close association with the Epstein–Barr virus (EBV), a ubiquitous human herpes virus infecting about 95% of the world’s population and viral gene products that are expressed in all tumor cells4. However, the etiology of NPC is still unclear.
Emerging studies have shown that long noncoding RNAs (lncRNAs), a class of RNAs with 200-nucleotide, noncoding transcripts in length, play a critical role in carcinogenesis and progression5. lncRNAs function as an oncogene or tumor suppressor in various cancers6. For example, lncRNA Ewing sarcoma-associated transcript 1 (EWSAT1) promotes NPC cell growth in vitro through upregulating cyclin D1 partially via “sponging” micro-RNA (miR)-326/330-5p clusters7. lncRNA NEAT1 was significantly upregulated in NPC cell lines and tissues. Knockdown of NEAT1 could sensitize NPC cells to radiation by modulating the epithelial–mesenchymal transition (EMT) phenotype and by modulating the miR-204/ZEB1 axis in NPC8. EBV-miR-BART6-3p, an miRNA encoded by oncogenic EBV, inhibited EBV-associated cancer cell migration and invasion by targeting and downregulating a novel lncRNA LOC5531039. The function of many lncRNAs remains unknown.
In this study, we established a comprehensive lncRNA profile in NPC and also found that 35 lncRNAs were differentially expressed in NPC. We further investigated the role of LINC0086 in NPC. We found that LINC0086 was decreased in NPC patient serum samples and tissues. Upregulation of LINC0086 inhibited cancer cell proliferation and promoted apoptosis. We demonstrated that LINC0086 inhibited cell proliferation and promoted apoptosis by targeting and downregulating miR-214, an oncogene in several types of cancer. Thus, our study reports a novel mechanism underlying NPC carcinogenesis and provides a potential novel diagnosis and treatment biomarker for NPC.
MATERIALS AND METHODS
Serum and Tissue Samples
Twenty NPC and 20 healthy serum samples were used for microarray analysis and qPCR validation. Another cohort containing 112 NPC specimens with clinical TNM staging and survival information, and 56 adjacent normal nasopharyngeal epithelial tissue specimens were collected for in situ hybridization and for the associated analysis of LINC0086 expression with pathological and clinical data. This project was approved by the ethics committee of The Third Xiangya Hospital of Central South University.
miRNA Microarray Analysis
Total RNA was extracted using the Rneasy MiNi Kit (Qiagen, Valencia CA, USA) according to the manufacturer’s instructions. The RNA was then labeled by miRCURY LNA miRNA Power Labeling Kit (Exiqon Life Sciences, Vedbaek, Denmark) according to the manufacturer’s instructions. The test sample and the reference were labeled with Hy5 and Hy3, respectively, and cohybridized to the miRNA arrays. The hybridized arrays were scanned by illumina HiSeqTM2000/MiSeq (Novogene Corporation, Beijing, P.R. China). Expression profile clustering and visualization were performed with the Cluster and Treeview software (Ernest Orlando Lawrence Berkeley National Laboratory, Berkeley, CA, USA).
Cell Lines and Cell Transfection
The two EBV+ NPC cell lines (C666-1 and HK-1) were obtained from the Cellbank of the Chinese Academy of Sciences. Cells were grown routinely in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA) and cultured in a 37°C humidified atmosphere with 5% CO2. Ectopic expression of miR-214 in cells was achieved by transfection with miR-214 mimic (GenePharma, Shanghai, P.R. China) using Lipofectamine 3000 (Invitrogen). Overexpression of LINC0086 was achieved by using lentivirus containing LINC0086-expressed plasmid (GeneCopoecia, Guangzhou, P.R. China). Cells were plated into 6-well clusters or 96-well plates and transfected for 24 or 48 h. Transfected cells were used in further assays.
In Situ Hybridization
The in situ hybridization (ISH) probe used for detecting LINC0086-labeled digoxin was designed and synthesized by Sangon Biotech Co., Ltd. (Shanghai, P.R. China). Slices were processed using the Enhanced Sensitive ISH Detection kit I (Cat. No. MK1030; Wuhan Boster Biological Technology, Ltd., Wuhan, P.R. China) according to the manufacturer’s protocol. The slides were visualized with 3,3′-diaminobenzidine (DAB; Fuzhou Maixin Biotech Co., Ltd., Fuzhou, P.R. China) for 5 min and counterstained with hematoxylin for 90 s. The slides were mounted and dried. Images of slides were captured with an Olympus BX51 microscope (magnification: 200× and 400×; Olympus Corporation, Tokyo, Japan).
RNA Extraction and SYBR Green Quantitative PCR Analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen). miR-214 expression in cells was detected using a Hairpin-it™ miRNAs qPCR kit (GenePharma) according to the manufacturer’s instructions. Expression of RNU6B was used as an endogenous control. The expression of LINC0086 was measured by SYBR green qPCR assay (Takara, Dalian, P.R. China) according to the manufacturer’s instructions. Expression of β-actin was used as an endogenous control. qPCR was performed at the following conditions: 95°C for 3 min, and 39 cycles of 95°C for 10 s and 60°C for 30 s. Data were processed using the 2−ΔΔCT method.
CCK-8 Cell Proliferation Assay
Cell proliferation rates were measured using a cell counting kit-8 (CCK-8; Beyotime, Hangzhou, P.R. China). Cells (0.5 × 104) were seeded into 96-well plates for 24 h, transfected with the indicated miRNA or lentivirus, and further incubated for 24, 48, and 72 h, respectively. CCK-8 reagents (10 μl) were added to each well 1 h before the endpoint of incubation. OD value at 570 nm in each well was determined with a microplate reader.
Flow Cytometric Analysis of Apoptosis With Annexin V/PI Double Staining
Annexin V apoptosis detection kit (Life Technologies, Grand Island, NY, USA) was used for analysis of apoptosis. After receiving the indicated treatment, C666-1 and HK-1 cells were trypsinized, collected, and resuspended. About 2 × 105 cells were harvested and washed twice with cold phosphate-buffered saline (PBS), and then resuspended in 500 μl of binding buffer. Annexin-V–FITC (10 μl) and 10 μl of propidium iodide were added to the solution and mixed well. After 15 min of incubation, the cells were analyzed using flow cytometric analysis (BD Biosciences, San Jose, CA, USA).
Luciferase Reporter Assay
The fragment of 3′-UTR of LINC0086 containing the putative BART-6 binding site was amplified by PCR. The PCR product was subcloned into a psiCHECK-2 vector (Promega, Madison, WI, USA) immediately downstream of the luciferase gene sequence. A psiCHECK-2 construct containing the 3′-UTR of LINC0086 with a mutant seed sequence of BART-6 was also synthesized by GenePharma. All constructs were verified by DNA sequencing. The cells were plated into 96-well clusters, and then cotransfected with 100 ng of constructs with or without the miR-BART-6 mimic. Forty-eight hours after transfection, luciferase activity was detected using a dual-luciferase reporter assay system (Promega) and normalized to Renilla activity.
RNA Immunoprecipitation (RIP)
RIP experiments were performed using the Magna RIP RNA-Binding Protein IP Kit (Millipore, Bedford, MA, USA) and the Ago2 antibody (Cell Signaling, Danvers, MA, USA) according to the manufacturer’s instructions. Finally, purified RNA in the precipitates was used to determine LINC0086 and miR-214 expression.
Tumor Xenograft in Nude Mice
Animal experiments were approved by the Ethical Committee for Animal Research of Central South University. All nude mice (4–5 weeks old, male, n = 5 per group) were purchased from The Central Animal Facility of Central South University. To assess tumor growth, 200 ml of C666-1 cells (1 × 106) was subcutaneously injected into the left side on the back of each mouse (five mice per group). Tumor size was measured regularly and calculated using the formula: 0.52 × L × W 2, where L and W are the long and short diameters of the tumor. The animals were euthanized on day 50 after injection, and the tumors were removed for immunohistochemical staining to evaluate the expression of Ki-67.
Immunohistochemical Staining Assay
The expression of Ki-67 was evaluated using immunohistochemical staining. Briefly, the tissue sections cut at 4 μm were deparaffinized and hydrated and then were retrieved with citrate buffer at boiling water for 15 min. The sections were then incubated with primary antibodies (mouse monoclonal anti-Ki-67; 1:500; Sigma-Aldrich, St. Louis, MO, USA) overnight at 4°C. Afterward, the sections were incubated with secondary antibody for 60 min at 37°C. The signaling was visualized using substrate diaminobenzidine (DAB) and counterstained with hematoxylin.
Statistical Analysis
All data from three independent experiments were expressed as mean ± SD and processed using SPSS17.0 statistical software. The overall survival rate estimates over time were calculated using the Kaplan–Meier method with log-rank test. The clinical association between LINC0086 expression and clinicopathological variables in NPC patients was evaluated by the chi-square test. The difference among the groups was estimated by Student’s t-test or one-way ANOVA, depending on the conditions. A value of p < 0.05 was considered to be statistically significant.
RESULTS
LINC0086 Is Associated With Pathoclinical Features
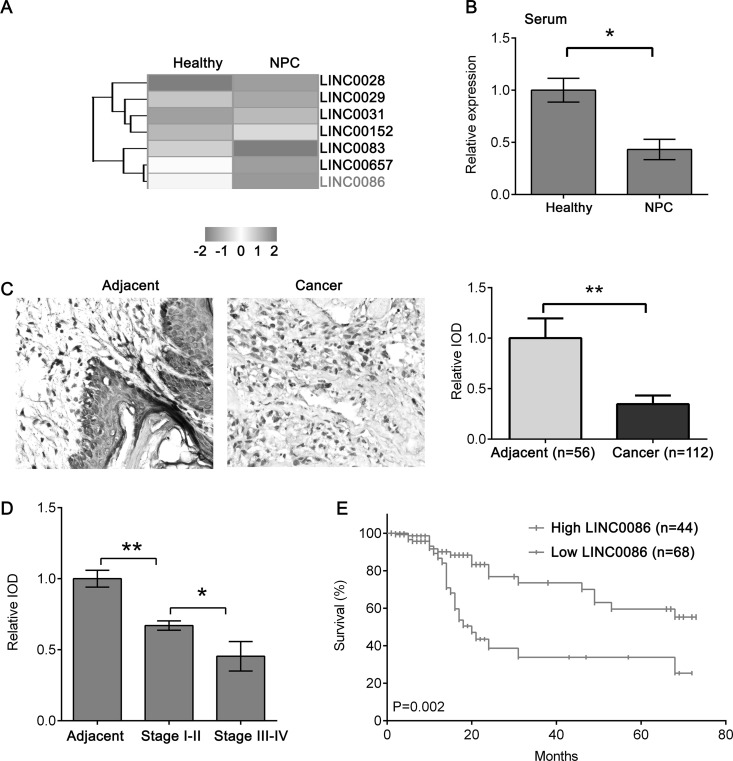
We performed miRNA expression profile microarray analysis of five NPC versus five healthy serum samples and identified 35 lncRNAs that were differentially expressed between NPC and healthy serum samples (Fig. 1A). Of those identified, 19 lncRNAs were upregulated and 16 lncRNAs were downregulated in NPC serum. Notably, LINC0086 was significantly downregulated in the top five, suggesting that LINC0086 may play a critical role in NPC. We also confirmed the LINC0086 expression in 20 NPC and 20 healthy serum samples (Fig. 1B). To investigate whether the LINC0086 expression is associated with pathoclinical features of NPC, we determined the expression of LINC0086 in another cohort of 112 NPC samples whose clinical stage and survival information using ISH were known. The expression of LINC0086 was dramatically decreased in NPC tissues compared with adjacent tissues (Fig. 1C) and reduced in stages III–IV compared with stages I–II (Fig. 1D). The Kaplan–Meier survival curve showed that patients with high LINC0086 expression had a higher survival rate than those with low LINC0086 expression (Fig. 1E). We also found that LINC0086 expression was associated with NPC histological grade (p = 0.012), lymph node metastasis (p = 0.007), and clinical stage (p = 0.006) (Table 1). Thus, the results suggest that LINC0086 might contribute to the development of NPC.
Figure 1.
Low LINC0086 is predictive of a worse clinical outcome. (A) Long noncoding RNA (lncRNA) expression profile microarray screening. Supervised hierarchical cluster analysis of 35 lncRNAs that were differentially expressed between five nasopharyngeal carcinoma (NPC) and five healthy serum samples (universal test p < 0.005, fold change: >1.5, false discovery rate: <0.05). (B) qPCR was performed to detect the expression of LINC0086 in 20 NPC and 20 healthy serum samples. (C) In situ hybridization (ISH) was performed to detect the expression of LINC0086 in NPC tissues and normal adjacent tissues. Right: Representative image of ISH staining of LINC0086. Magnification: 200×. Left: Quantification of ISH staining of LINC0086. IOD, integral optical density. (D) Quantification of ISH staining of LINC0086 in adjacent tissues, stages I–II, and stages III–IV. (E) Survival curve showed that patients with high LINC0086 had a better survival rate than those with low LINC0086 expression. *p < 0.05, **p < 0.01.
Table 1.
Clinical Association Between LINC0086 Expression and Clinicopathological Variables in Nasopharyngeal Carcinoma (NPC)
| Variable | LINC0086 Expression | Chi-Square Test p Value | |
|---|---|---|---|
| Low (n = 68) | High (n = 44) | ||
| Age | 0.841 | ||
| <60 | 25 | 15 | |
| ≥60 | 43 | 29 | |
| Gender | 0.847 | ||
| Male | 33 | 23 | |
| Female | 35 | 21 | |
| Histological grade | 0.012 | ||
| I | 28 | 29 | |
| II–III | 40 | 15 | |
| Lymph node metastasis | 0.007 | ||
| No | 25 | 28 | |
| Yes | 43 | 16 | |
| Clinical stage | 0.006 | ||
| I–II | 31 | 32 | |
| III–IV | 37 | 12 | |
Upregulation of LINC0086 Inhibits Cell Proliferation and Promotes Cell Apoptosis of NPC Cells
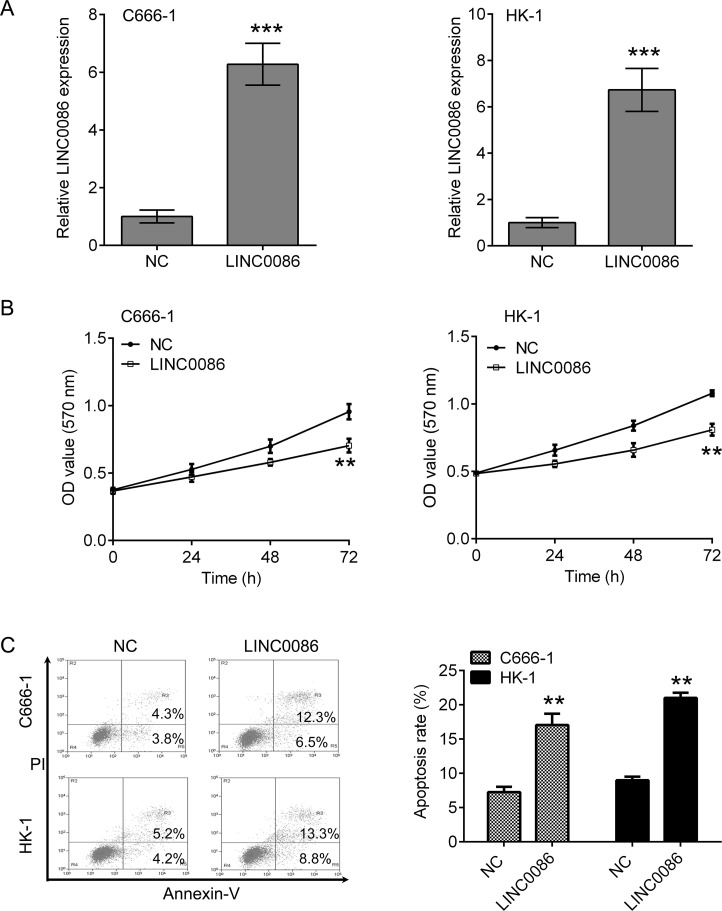
To investigate whether LINC0086 affected the growth of C666-1 and HK-1 cells, cells stably overexpressing LINC0086 were constructed (Fig. 2A). LINC0086 overexpression significantly impeded cell viability in C666-1 and HK-1 cells (Fig. 2B). A corresponding effect on apoptosis was also observed in a flow cytometry assay, which showed a significant induction of apoptosis in C666-1 and HK-1 cells compared with the respective NC group (Fig. 2C).
Figure 2.
Overexpression of LINC0086 inhibits cell proliferation and induces apoptosis in C666-1 and HK-1 cells. (A) qPCR was performed to detect the expression of LINC0086 in C666-1 and HK-1 cells after transfection with LINC0086 lentivirus. The cells treated with NC were used as control. (B) Cell counting kit-8 (CCK-8) was used for measuring cell proliferation. LINC0086 overexpression inhibited the proliferation of C666-1 and HK-1 cells. (C) Flow cytometry was used for measuring cell apoptosis. LINC0086 overexpression induced an increase in apoptosis of C666-1 and HK-1 cells. **p < 0.01, ***p < 0.001 versus NC group.
LINC0086 Interacts With miR-214
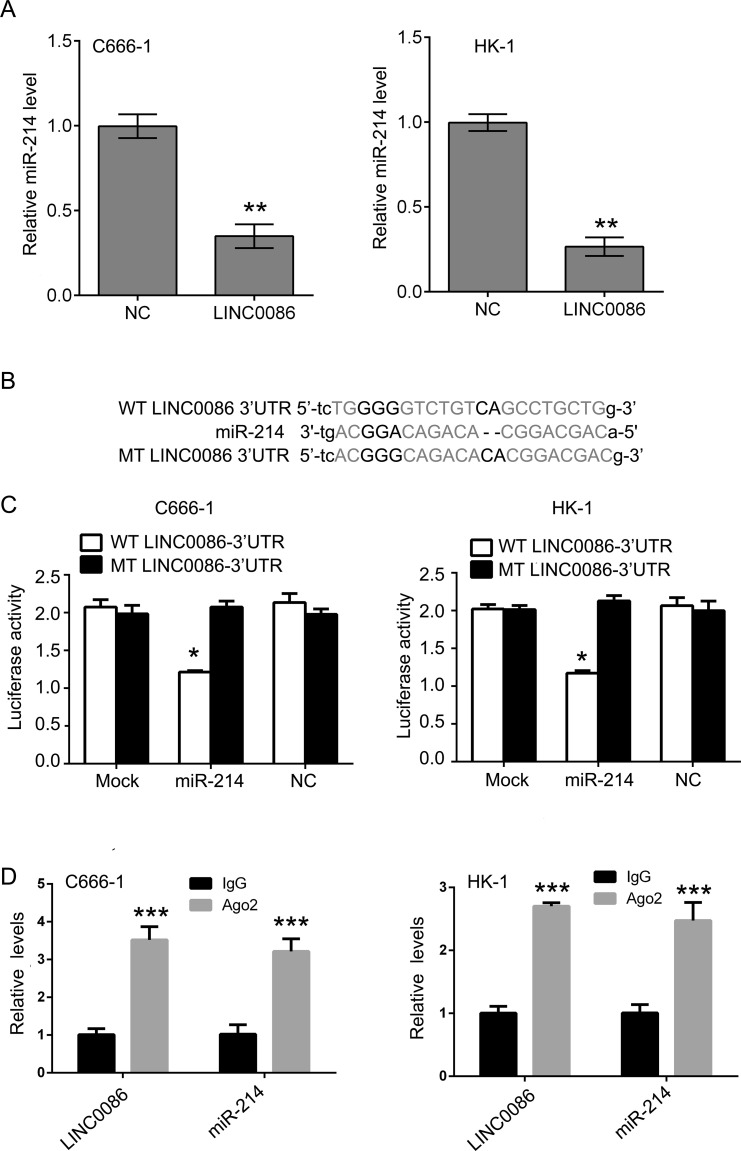
Bioinformatics analysis was used to search for the potential targeted miRNAs of LINC0086 (http://starbase.sysu.edu.cn/mirLncRNA.php)10. An miR-214 binding site was found in the LINC0086 transcript, and LINC0086 was a predicted gene of miR-214. Upregulation of LINC0086 dramatically decreased the expression of miR-214 in C666-1 and HK-1 cells (Fig. 3A). To investigate whether the predicted binding site of miR-214 to the 3′-UTR of LINC0086 was responsible for this regulation, we cloned the 3′-UTR of LINC0086 downstream to a luciferase reporter gene (WT-LINC0086 3′-UTR). Its mutant version (MT-LINC0086 3′-UTR), by binding site mutagenesis, was also constructed (Fig. 3B). The luciferase activity of cells cotransfected with miR-214 mimics and WT-LINC0086 was significantly reduced compared to the scramble control cells. Moreover, miR-214-mediated repression of luciferase activity was abolished by the mutant putative binding site in C666-1 and HK-1 cells (Fig. 3C). The RNA-induced silencing complex (RISC) is a major mechanism of miRNAs to silence target genes, and Ago2 protein is a key constituent of the RISC complex. To validate the presence of both miR-214 and LINC0086 in the RISC complex, an RIP experiment was conducted using Ago2 antibody to confirm that both miR-214 and LINC0086 were found in the Ago2 pellet (Fig. 3D).
Figure 3.
LINC0086 interacts with miR-214 and regulates its expression. (A) qPCR was performed to detect the expression of miR-214 in C666-1 and HK-1 cells after LINC0086 transfection. (B) The wild type (WT) and mutant type (MT) of LINC0086 3′-UTR, and the binding sites were shown. (C) The relative luciferase activities were inhibited in the C666-1 and HK-1 cells cotransfected with wild-type LINC0086 3′-UTR and miR-214, and not with the mutant type. Firefly luciferase activity was normalized to Renilla luciferase. (D) Association of LINC0086 and miR-214 with Ago2 in C666-1 and HK-1 cells. Cellular lysates from C666-1 and HK-1 cells were used for RNA immunoprecipitation (RIP) with antibody against Ago2. LINC0086 and miR-214 expression levels were detected using qRT-PCR. *p < 0.05, **p < 0.01, ***p < 0.001 versus NC group.
Upregulation of miR-214 Reversed LINC0086-Mediated Inhibitory Effects In Vitro and In Vivo
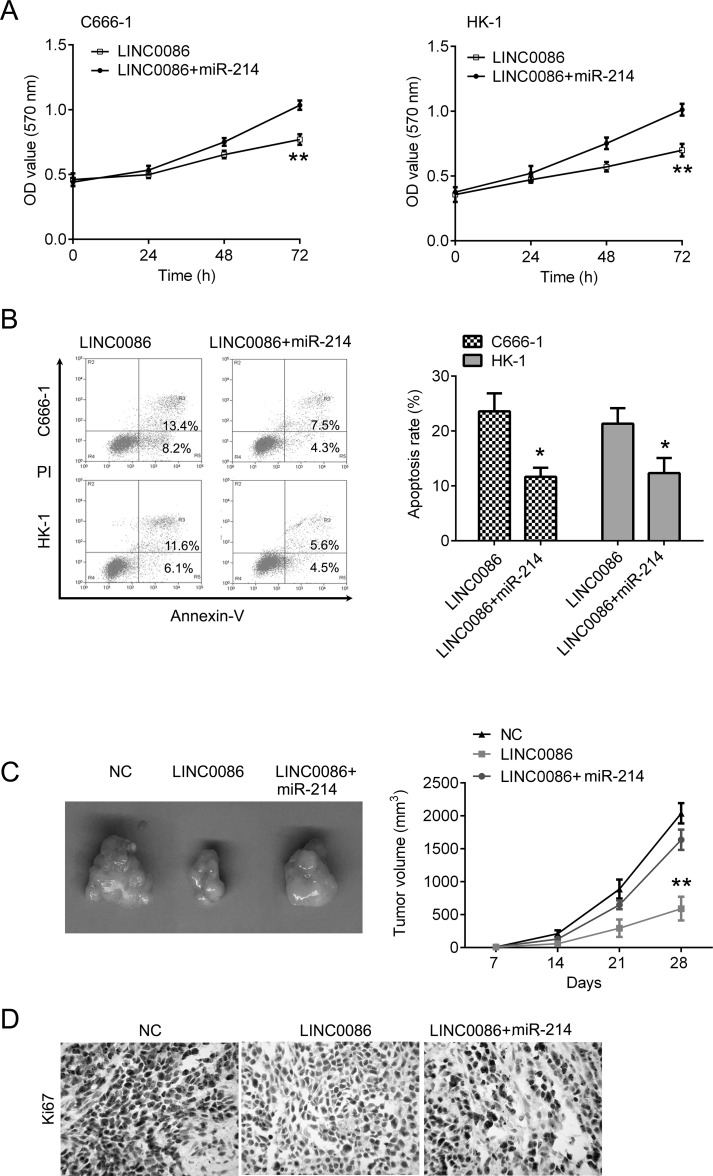
Because of the regulatory effect between LINC0086 and miR-214, we further tested the biological function between LINC0086 and miR-214. C666-1 and HK-1 cells were transfected with LINC0086 alone or together with the miR-214 mimic. We found that miR-214 upregulation largely attenuated LINC0086-induced cell growth inhibition and apoptosis in C666-1 and HK-1 cells (Fig. 4A and B). Moreover, we confirmed these results in vivo. Compared with the NC control group, overexpression of LINC0086 significantly inhibited tumor growth. However, these inhibitory effects were reversed by miR-214 upregulation (Fig. 4C). Additionally, the IHC staining of Ki-67 in tumor sections showed that overexpression of LINC0086 significantly inhibited Ki-67 expression, which was reversed by miR-214 upregulation (Fig. 4D). These results indicate that LINC0086 regulates the cell proliferation and apoptosis of NPC cells via targeting miR-214.
Figure 4.
Upregulation of miR-214 reversed LINC0086-mediated inhibitory effects in vitro and in vivo. (A) CCK-8 was used for measuring cell proliferation. miR-214 overexpression reversed LINC0086-mediated inhibition of cell proliferation in C666-1 and HK-1 cells. (B) Flow cytometry was used for measuring cell apoptosis. miR-214 overexpression reversed LINC0086-mediated promotion of cell apoptosis in C666-1 and HK-1 cells. (C) The C666-1 cells transfected with LINC0086 or together with miR-214 were injected into nude mice. The cells transfected with empty plasmid were used as negative control. Right: Tumors obtained from mice on day 28 after injection. Left: Tumor volumes were calculated every week. (D) Representative image of IHC staining of Ki-67 in xenografted tumor section. Magnification: 200×. *p < 0.05, **p < 0.01.
DISCUSSION
In this study, we first identified LINC0086 as a novel tumor suppressor in NPC. LINC0086 is markedly downregulated in NPC patient serum samples and tissues, and the expression of LINC0086 was associated with tumor histological grade, lymph node metastasis, and clinical stage. Patients with low LINC0086 expression had a lower survival rate than those with high LINC0086 expression. Overexpression of LINC0086 expression exhibited an inhibitory effect on cell proliferation both in NPC cells and the xenograft model. These results illustrate that LINC0086 acts as a tumor-suppressive lncRNA in NPC development.
Increasing evidence has demonstrated a significant role for lncRNAs in tumorigenesis. lncRNAs may serve as a diagnostic marker, tumor suppressor, or oncogene, depending on cancer type. Microarray analysis for the expression profile of lncRNAs in high and low metastatic NPC cell lines showed that 167 lncRNAs were differentially expressed. The analysis also identified that lncRNA ENST00000470135 was the most upregulated lncRNA in high metastatic cell lines and was significantly higher in NPC cell lines and tissues with lymph node metastasis. Furthermore, knocking down ENST00000470135 suppressed the migration, invasion, and proliferation of NPC cells in vitro11. In addition, by comparison between radioresistant CNE-2-Rs and parental CNE-2 cells with next-generation deep sequencing, Li et al. identified a total of 781 known lncRNAs and 2,054 novel lncRNAs. Among them, 7 of 10 known lncRNAs and 3 of 10 novel lncRNAs were demonstrated to have significant differential expression trends that were the same as those predicted by deep sequencing, including lncRNA n373932 and SLITRK5, n409627 and PRSS12, and n386034 and RIMKLB12. Ectopic expression of lncRNA MEG3 in NPC cell lines resulted in repression of in vitro anchorage-independent growth, and in vivo tumorigenicity may be via p53 signaling cascade13. Zou et al. reported that lncRNA ANRIL serves as an independent predictor of overall survival and is generally upregulated in NPC, which was highly expressed in advanced-stage cancer. Knockdown of lncRNA ANRIL significantly repressed NPC cell proliferation and transformation14. Gong et al. found that long noncoding RNA LOC401317 was directly regulated by p53, and that ectopic expression of LOC401317 inhibited NPC cell proliferation in vitro and in vivo by inducing cell cycle arrest and apoptosis15. Another lncRNA LINC00312 was significantly downregulated in NPC tissues compared with noncancerous nasopharyngeal epithelium tissues, which could distinguish noncancerous patients from NPC patients and serve as independent contributors to nasopharyngeal carcinogenesis. Low expression of LINC00312 was an independent risk factor for overall survival16. These findings suggest that lncRNAs regulate wide aspects of biofunction in NPC development.
Recent evidence indicates that lncRNAs can interact with miRNAs, and these interactions play a crucial role in cancer cell proliferation and apoptosis through regulating critical events17. lncRNA X inactivate-specific transcript (XIST), an oncogenic gene, was upregulated in NPC tissues, and higher expression of XIST contributed to a markedly poorer survival time. XIST overexpression enhanced cell growth in NPC by upregulating the expression of the miR-34a-5p-targeted gene E2F3 through acting as a competitive sponge of miR-34a-5p18. Mechanically, we demonstrated that LINC0086 could directly interact with miR-214 and decrease its expression. The suppressive effect of LINC0086 on NPC cell growth was reversed by overexpression of miR-214. miR-214 is a double-faced gene that functions as a tumor suppressor or oncogene in different cancers. miR-214 functions as a tumor suppressor in breast cancer and colon cancer19,20. However, several studies showed an oncogenic role for miR-214 by targeting PTEN in gastric cancer21, by targeting β-catenin in esophageal cancer22, and by promoting EMT and metastasis in lung adenocarcinoma by targeting the suppressor-of-fused protein23. In addition, it was found that miR-214 could interact with lncRNA DANCR, which was overexpressed in stem-like hepatocellular carcinoma (HCC) cells and served as a prognostic biomarker for HCC patients24. Our findings in this study indicate that miR-214 functions as an oncogene in NPC because it can reverse the inhibitory role of LINC0086 on NPC cell growth.
In conclusion, for the first time we demonstrated the tumor-suppressive role of LINC0086 in NPC. Low expression of LINC0086 is associated with a poor prognosis of NPC patients. Upregulation of LINC0086 significantly inhibited NPC cancer cell growth and caused apoptosis in vitro and in vivo by targeting miR-214. Our study facilitates the understanding of LINC0086 function in the tumorigenesis of NPC and provides a novel diagnostic marker and therapeutic target for NPC treatment.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Bruce JP, Yip K, Bratman SV, Ito E, Liu FF. Nasopharyngeal cancer: Molecular landscape. J Clin Oncol. 2015;33:3346–55. [DOI] [PubMed] [Google Scholar]
- 2. Wang Q, Fan H, Liu Y, Yin Z, Cai H, Liu J, Wang Z, Shao M, Sun X, Diao J, Liu Y, Tong L, Fan Q. Curcumin enhances the radiosensitivity in nasopharyngeal carcinoma cells involving the reversal of differentially expressed long non-coding RNAs. Int J Oncol. 2014;44:858–64. [DOI] [PubMed] [Google Scholar]
- 3. Fu WM, Lu YF, Hu BG, Liang WC, Zhu X, Yang HD, Li G, Zhang JF. Long noncoding RNA hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget 2016;7:4712–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lo AK, Dawson CW, Jin DY, Lo KW. The pathological roles of BART miRNAs in nasopharyngeal carcinoma. J Pathol. 2012;227:392–403. [DOI] [PubMed] [Google Scholar]
- 5. Yang QQ, Deng YF. Genome-wide analysis of long non-coding RNA in primary nasopharyngeal carcinoma by microarray. Histopathology 2015;66:1022–30. [DOI] [PubMed] [Google Scholar]
- 6. Gao W, Chan JY, Wong TS. Differential expression of long noncoding RNA in primary and recurrent nasopharyngeal carcinoma. Biomed Res Int. 2014;2014:404567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Song P, Yin SC. Long non-coding RNA EWSAT1 promotes human nasopharyngeal carcinoma cell growth in vitro by targeting miR-326/-330-5p. Aging (Albany NY) 2016;8:2948–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu Y, Li T, Wei G, Liu L, Chen Q, Xu L, Zhang K, Zeng D, Liao R. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016;37:11733–41. [DOI] [PubMed] [Google Scholar]
- 9. He B, Li W, Wu Y, Wei F, Gong Z, Bo H, Wang Y, Li X, Xiang B, Guo C, Liao Q, Chen P, Zu X, Zhou M, Ma J, Li X, Li Y, Li G, Xiong W, Zeng Z. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wen X, Tang X, Li Y, Ren X, He Q, Yang X, Zhang J, Wang Y, Ma J, Liu N. Microarray expression profiling of long non-coding RNAs involved in nasopharyngeal carcinoma metastasis. Int J Mol Sci. 2016;17:1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li G, Liu Y, Liu C, Su Z, Ren S, Wang Y, Deng T, Huang D, Tian Y, Qiu Y. Genome-wide analyses of long noncoding RNA expression profiles correlated with radioresistance in nasopharyngeal carcinoma via next-generation deep sequencing. BMC Cancer 2016;16:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chak WP, Lung RW, Tong JH, Chan SY, Lun SW, Tsao SW, Lo KW, To KF. Downregulation of long non-coding RNA MEG3 in nasopharyngeal carcinoma. Mol Carcinog. 2017;56:1041–54. [DOI] [PubMed] [Google Scholar]
- 14. Zou ZW, Ma C, Medoro L, Chen L, Wang B, Gupta R, Liu T, Yang XZ, Chen TT, Wang RZ, Zhang WJ, Li PD. LncRNA ANRIL is up-regulated in nasopharyngeal carcinoma and promotes the cancer progression via increasing proliferation, reprogramming cell glucose metabolism and inducing side-population stem-like cancer cells. Oncotarget 2016;7:61741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gong Z, Zhang S, Zeng Z, Wu H, Yang Q, Xiong F, Shi L, Yang J, Zhang W, Zhou Y, Zeng Y, Li X, Xiang B, Peng S, Zhou M, Li X, Tan M, Li Y, Xiong W, Li G. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS One 2014;9:e110674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang W, Huang C, Gong Z, Zhao Y, Tang K, Li X, Fan S, Shi L, Li X, Zhang P, Zhou Y, Huang D, Liang F, Zhang X, Wu M, Cao L, Wang J, Li Y, Xiong W, Zeng Z, Li G. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44:545–54. [DOI] [PubMed] [Google Scholar]
- 17. Su Y, Wu H, Pavlosky A, Zou LL, Deng X, Zhang ZX, Jevnikar AM. Regulatory non-coding RNA: New instruments in the orchestration of cell death. Cell Death Dis. 2016;7:e2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene 2016;592:8–14. [DOI] [PubMed] [Google Scholar]
- 19. Liu B, Tian Y, Li F, Zhao Z, Jiang X, Zhai C, Han X, Zhang L. Tumor-suppressing roles of miR-214 and miR-218 in breast cancer. Oncol Rep. 2016;35:3178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Long LM, He BF, Huang GQ, Guo YH, Liu YS, Huo JR. microRNA-214 functions as a tumor suppressor in human colon cancer via the suppression of ADP-ribosylation factor-like protein 2. Oncol Lett. 2015;9:645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xin R, Bai F, Feng Y, Jiu M, Liu X, Bai F, Nie Y, Fan D. MicroRNA-214 promotes peritoneal metastasis through regulating PTEN negatively in gastric cancer. Clin Res Hepatol Gastroenterol. 2016;40:748–54. [DOI] [PubMed] [Google Scholar]
- 22. Xu Y, Lu S. Regulation of beta-catenin-mediated esophageal cancer growth and invasion by miR-214. Am J Transl Res. 2015;7:2316–25. [PMC free article] [PubMed] [Google Scholar]
- 23. Long H, Wang Z, Chen J, Xiang T, Li Q, Diao X, Zhu B. microRNA-214 promotes epithelial-mesenchymal transition and metastasis in lung adenocarcinoma by targeting the suppressor-of-fused protein (Sufu). Oncotarget 2015;6:38705–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, Fan J, Liu L, Sun SH, Zhou WP. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology 2016;63:499–511. [DOI] [PubMed] [Google Scholar]