Abstract
The human papillomavirus (HPV) infection may be associated with the development and progression of non-small cell lung cancer (NSCLC). However, the role of HPV-16 oncoproteins in the development and progression of NSCLC is not completely clear. Epithelial–mesenchymal transition (EMT), a crucial step for invasion and metastasis, plays a key role in the development and progression of NSCLC. Here we explored the effect of HPV-16 oncoproteins on EMT and the underlying mechanisms. NSCLC cell lines, A549 and NCI-H460, were transiently transfected with the EGFP-N1-HPV-16 E6 or E7 plasmid. Real-time PCR and Western blot analysis were performed to analyze the expression of EMT markers. A protein microarray was used to screen the involved signaling pathway. Our results showed that overexpression of HPV-16 E6 and E7 oncoproteins in NSCLC cells significantly promoted EMT-like morphologic changes, downregulated the mRNA and protein levels of EMT epithelial markers (E-cadherin and ZO-1), and upregulated the mRNA and protein levels of EMT mesenchymal markers (N-cadherin and vimentin) and transcription factors (ZEB-1 and Snail-1). Furthermore, the HPV-16 E6 oncoprotein promoted STAT3 activation. Moreover, WP1066, a specific signal transducer and activator of transcription 3 (STAT3) inhibitor, reversed the effect of HPV-16 E6 on the expression of ZO-1, vimentin, and ZEB-1 in transfected NSCLC cells. Taken together, our results suggest that overexpression of HPV-16 E6 and E7 oncoproteins enhances EMT, and the STAT3 signaling pathway may be involved in HPV-16 E6-induced EMT in NSCLC cells.
Key words: Human papillomavirus (HPV), Epithelial–mesenchymal transition (EMT), Signal transducer and activator of transcription 3 (STAT3), Non-small cell lung cancer (NSCLC)
INTRODUCTION
Non-small cell lung cancer (NSCLC) accounts for approximately 80% of lung cancer cases. It is well known that NSCLC is predominantly caused by cigarette smoking. However, global statistics in 2002 estimated that about 25% of patients with lung cancer all over the world were never-smokers1. Recently, more and more patients with NSCLC, especially women, have been found to be never-smokers2–5. Therefore, the role of nonsmoking factors in never-smokers with NSCLC should be investigated.
Human papillomavirus (HPV), a group of small nonenveloped DNA viruses, was first suggested to play a role in the development and progression of lung cancer in 19796. Recently, accumulating epidemiological evidence and meta-analyses have shown that the infection of high-risk HPV types, especially HPV types 16/18, may have an association with NSCLC7–11. The HPV 16/18 infection was found to increase the risk of squamous cell lung carcinoma7, and a higher frequency and increased significance of oncogenic HPV 16/18 were observed in never-smokers and women with lung cancer8–11. Specifically, HPV type 16 (HPV-16) was demonstrated to be the most frequent genotype of HPV in NSCLC12–14. Most recently, the HPV-16 infection was reported to act synergistically with environmental exposure to induce lung tumorigenesis in nonsmokers15. These reports indicated that the HPV-16 infection might contribute to NSCLC. However, different results have also been reported16,17, and the role of HPV-16 E6 and E7 oncoproteins in the development and progression of NSCLC is not completely clear. Therefore, more studies are essential to further explore the role of HPV-16 oncoproteins in the development and progression of NSCLC.
Invasion and metastasis play key roles in the development and progression of NSCLC. Epithelial–mesenchymal transition (EMT), a process by which epithelial cells lose the epithelial phenotype and gain the mesenchymal phenotype, is a crucial step for invasion and metastasis18. EMT is characterized by the downregulation of epithelial molecular markers such as E-cadherin and ZO-1, the upregulation of mesenchymal molecular proteins such as N-cadherin and vimentin, and the induction of EMT-related transcription factors such as ZEB-1 and Snail. Recently, Shen et al. found that EMT contributed to docetaxel resistance in human NSCLC19. Zhao et al. demonstrated that E-cadherin expression in pleural effusion cells was associated with EGFR mutation status and patient prognosis in lung adenocarcinoma patients in first-line chemotherapy20. Atmaca et al. reported that SNAI2/SLUG was prognostic of the outcome of NSCLC patients, and SNAI2/SLUG and estrogen receptor mRNA levels were inversely correlated21. These reports indicated that EMT played a crucial role in the development and progression of NSCLC. However, whether HPV-16 oncoproteins can promote the development and progression of NSCLC by interfering with EMT still remains unclear.
The signal transducers and activators of transcription (STATs) are a family of seven proteins with high homology including STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT622. STAT3, the most ubiquitous of the STATs, has been demonstrated to enhance tumorigenesis by mediating the expression of various target genes, including cell cycle regulators, antiapoptotic genes, and angiogenic factors22. Recently, STAT3 has been found to regulate the EMT program in various cancer cells23–25.
In this study, we analyzed the effect of HPV-16 oncoproteins on EMT in NSCLC cells and the role of STAT3 in that effect. To our knowledge, we found, for the first time, that HPV-16 E6 and E7 oncoproteins promoted EMT, and the STAT3 signaling pathway was involved in HPV-16 E6-induced EMT in both A549 and NCI-H460 NSCLC cells.
MATERIALS AND METHODS
Reagents
WP1066, a STAT3 inhibitor, was purchased from Merck China Ltd. (Shanghai, P.R. China). WP1066 was dissolved at a concentration of 50 mmol/L in 100% dimethyl sulfoxide (DMSO) as a stock solution and stored at −20°C. The final DMSO concentration did not exceed 0.1% throughout the study. Transfection reagent (Lipofectamine™ 2000) was obtained from Invitrogen Corporation (Carlsbad, CA, USA). TRIzol® reagent was purchased from Invitrogen. Reverse transcription and real-time PCR (SYBR Green) kits were purchased from Tiangen Biotech (Beijing, P.R. China). Lysis and blocking buffers were purchased from Beyotime Biotechnology Corporation (Shanghai, P.R. China). Rabbit anti-human E-cadherin, ZO-1, N-cadherin, vimentin, ZEB-1, and Snail-1 primary antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). Mouse anti-human β-actin antibody was purchased from Beyotime Biotechnology Corporation. Horseradish peroxidase (HRP)-conjugated secondary antibodies were from Cell Signaling Technology. The Human Phospho-Kinase Array Kit (No. ARY003) containing 46 kinase phosphorylation sites was obtained from RD Systems China Co. Ltd. (Shanghai, P.R. China).
Cell Lines and Cell Cultures
Human NSCLC cell lines, A549 and NCI-H460, were purchased from the American Type Culture Collection (Rockville, MD, USA) and the Chinese Academy of Sciences Cell Bank of Type Culture Collection (Shanghai, P.R. China), respectively. All NSCLC cells were grown in RPMI-1640 medium containing 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a humidified condition with 5% CO2.
Transient Transfection
EGFP plasmid vectors harboring HPV-16 E6, E7, or mutant cDNA were constructed by our lab. A549 and NCI-H460 cells at 70% to 80% confluence were transiently transfected for 4 h with pEGFP-N1-HPV-16 E6 or E7 plasmid using Lipofectamine™ 2000 transfection reagent. The cells transfected with empty vector or mutant plasmid were used as negative controls, and the cells exposed to Lipofectamine™ 2000 alone acted as mock transfection controls. Twenty-four hours after transfection, the transfected cells were harvested for further analysis. The green florescence signals in the transfected cells were observed under a fluorescence microscope, and the transfection efficiency was analyzed by flow cytometry (Epics-XL; Beckman Coulter, Brea, CA, USA). The expression of HPV-16 E6 and E7 oncoproteins in transfected cells was confirmed in a previous study26. E-cadherin, ZO-1, N-cadherin, vimentin, ZEB-1, and Snail-1 mRNA levels were analyzed by real-time PCR. The expression of E-cadherin, ZO-1, N-cadherin, vimentin, ZEB-1, and Snail-1 proteins in transfected cells was detected by Western blot analysis.
Real-Time PCR
Total RNA was extracted from transfected- and mock-transfected cells using the TRIzol® reagent. E-cadherin, ZO-1, N-cadherin, vimentin, ZEB-1, and Snail-1 mRNA relative levels were determined using a reverse transcription kit and real-time PCR (SYBR Green) kit according to the manufacturer’s instructions (Tiangen Biotech). The sequences of the primers were as follows: human E-cadherin, 5′-ttgctactggaacagggacac-3′ (forward) and 5′-cccgtgtgttagttctgctgt-3′ (reverse) (Genbank: NM_001317184.1); ZO-1, 5′-ggatgtttatcgtcgcattgta-3′ (forward) and 5′-aagagcccagttttccattgta-3′ (reverse) (Genbank: NM_001301025.1); N-cadherin, 5′-ttatccttgtgctgatgtttgtg-3′ (forward) and 5′-tcttcttctcctccaccttcttc-3′ (reverse) (Genbank: NM_001792.3); vimentin, 5′-tggcacgtcttgaccttgaa-3′ (forward) and 5′-ggtcatcgtgatgctgagaa-3′ (reverse) (Genbank: NM_003380.3); ZEB-1, 5′-tccccatcacctctaaacctt-3′ (forward) and 5′-ccctgttgctttggtagtgaa-3′ (reverse) (Genbank: NM_001174096.1); Snail-1, 5′-tccttcgtccttctcctctactt-3′ (forward) and 5′-tgttgcagtatttgcagttgaag-3′ (reverse) (Genbank: NM_005985.3); β-actin, 5′-tgacgtggacatccgcaaag-3′ (forward) and 5′-ctggaaggtggacagcgagg-3′ (reverse) (Genbank: NM_001101.3). All the primers were synthesized by Sangon Biotech (Shanghai, P.R. China). The reverse transcription conditions were as follows: 25°C for 10 min, 55°C for 30 min, and 85°C for 5 min. The real-time PCR conditions were as follows: 95°C for 5 min, followed by 40 cycles at 95°C for 10 s, 60°C for 20 s, and 72°C for 20 s. The size of the PCR product of E-cadherin, ZO-1, N-cadherin, vimentin, ZEB-1, Snail-1, and β-actin was 179, 158, 139, 176, 122, 155, and 186 bp, respectively. All relative mRNA levels were normalized to β-actin.
Western Blot Analysis
The method was described in our previous studies26–28. Briefly, transfected and mock-transfected cells were lysed with lysis buffer (Beyotime Biotechnology Corporation) and complete protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA). The proteins were run on a 10% SDS-PAGE gel and transferred to polyvinylidene difluoride membranes. After blocking with blocking buffer (Beyotime Biotechnology Corporation), the membranes were incubated with specific primary antibodies and HRP-conjugated secondary antibodies, respectively. Signals were visualized using enhanced chemiluminescence (ECL). The analysis of β-actin protein expression was used as a loading control.
Protein Microarray for Human Phosphor-Kinase
The levels of 46 kinase phosphorylation sites were analyzed using the Human Phospho-Kinase Array Kit (RD systems China Co. Ltd.) according to the manufacturer’s instructions. Briefly, two membranes spotting with capture and control antibodies were put into a well of the eight-well multidish and blocked with the blocking buffer on a rocking platform for 1 h. The total protein with reconstituted detection antibody cocktail was added into each well and incubated overnight at 4°C on a rocking platform. Afterward, the membranes were incubated with diluted streptavidin–HRP for 30 min at room temperature, followed by incubation with ECL. The signals were visualized after exposure to X-ray film.
Statistical Analysis
All data in this study were expressed as mean ± SD for three independent experiments. One-way ANOVA and LSD methods were used as statistical analysis by SPSS 19.0 software. A value of p < 0.05 indicated the difference between two groups was statistically significant.
RESULTS
HPV-16 Oncoproteins Promoted EMT-Like Morphologic Changes in NSCLC Cells
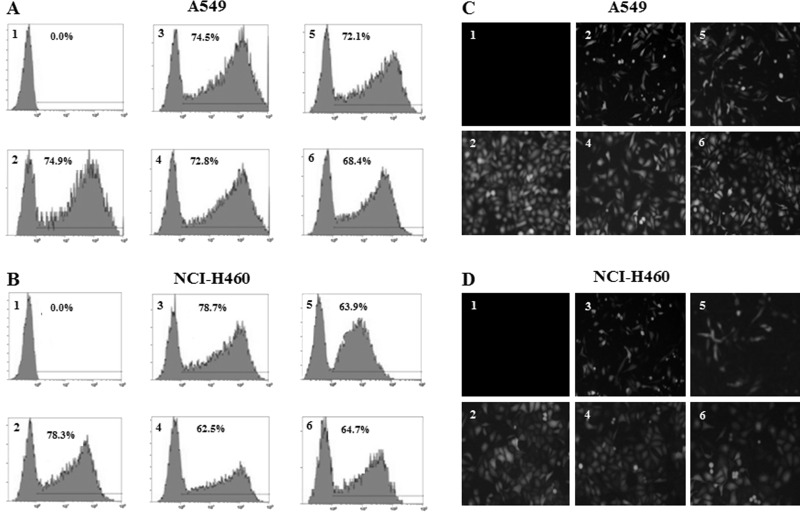
A549 and NCI-H460 NSCLC cells were transfected with EGFP plasmid vectors harboring HPV-16 E6, E7, or mutant cDNA. The transfection rates ranged from 62.5% to 78.7% (Fig. 1A and B). Moreover, 24 h after transfection, obvious EMT-like morphologic changes such as polarity were observed in HPV-16 E6- and E7-transfected NSCLC cells compared with empty vector or mutant controls (Fig. 1C and D).
Figure 1.
Analysis of transfection efficiency and morphologic changes. A549 and NCI-H460 cells were transiently transfected with EGFP plasmid vectors harboring HPV-16 E6, E7, or mutant cDNA. (A, B) The transfection efficiency was analyzed by flow cytometry (A: A549; B: NCI-H460). (C, D) The morphologic changes were observed under a fluorescence microscope (C: A549; D: NCI-H460). Original magnification: 200×. 1, Mock transfection control; 2, empty vector control; 3, HPV-16 E6; 4, HPV-16 E6 mutant (E6-mut) control; 5, HPV-16 E7; and 6, HPV-16 E7 mutant (E7-mut) control. The results are representative of three independent experiments.
HPV-16 Oncoproteins Regulated the mRNA Levels of EMT Markers in NSCLC Cells
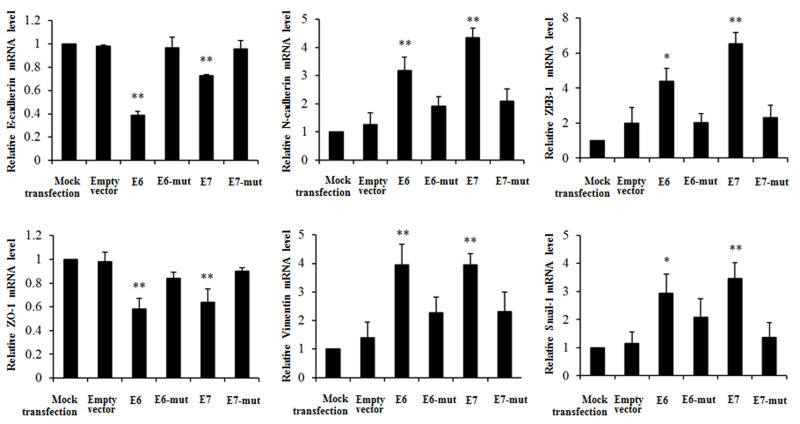
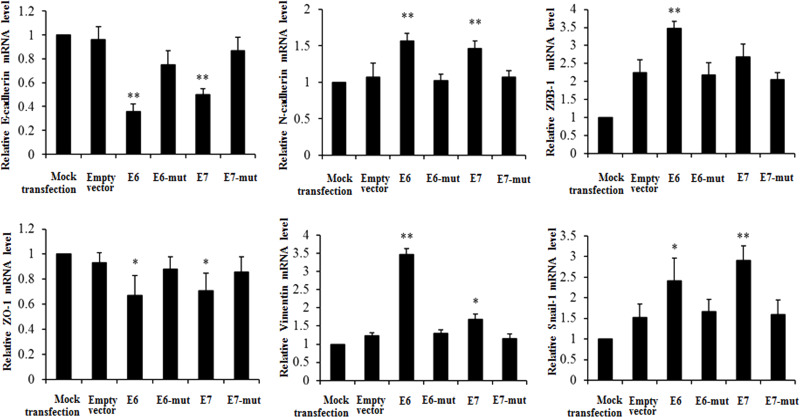
To study the effect of HPV-16 oncoproteins on EMT in NSCLC cells, A549 and NCI-H460 NSCLC cells were transfected with pEGFP-N1-HPV-16 E6 or E7 plasmid. Twenty-four hours after transfection, real-time PCR was performed to determine the mRNA levels of EMT epithelial markers (E-cadherin and ZO-1), mesenchymal markers (N-cadherin and vimentin), and transcription factors (ZEB-1 and Snail-1) in transfected NSCLC cells. We demonstrated that HPV-16 E6 and E7 oncoproteins significantly downregulated E-cadherin and ZO-1 mRNA levels and upregulated N-cadherin, vimentin, ZEB-1, and Snail-1 mRNA levels compared to the empty vector and mutant controls in A549 cells (Fig. 2). Similar results were found in NCI-H460 NSCLC cells (Fig. 3).
Figure 2.
Effects of HPV-16 oncoproteins on the mRNA levels of EMT markers in A549 cells. Real-time PCR was performed to analyze the mRNA levels of E-cadherin, ZO-1, N-cadherin, vimentin, ZEB-1, and Snail-1 in transfected A549 cells. All data were expressed as mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, compared with empty vector control or mutant controls (E6-mut and E7-mut).
Figure 3.
Effects of HPV-16 oncoproteins on the mRNA levels of EMT markers in NCI-H460 cells. Real-time PCR was performed to analyze the mRNA levels of E-cadherin, ZO-1, N-cadherin, vimentin, ZEB-1, and Snail-1 in transfected NCI-H460 cells. All data were expressed as mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, compared with empty vector control or mutant controls (E6-mut and E7-mut).
HPV-16 Oncoproteins Regulated the Protein Levels of EMT Markers in NSCLC Cells
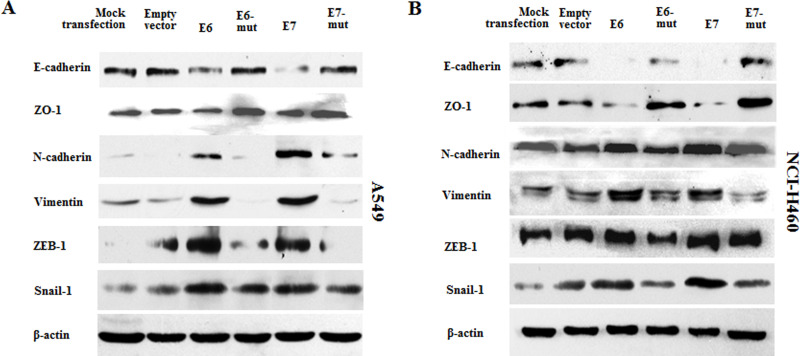
To further observe the effect of HPV-16 oncoproteins on the translational levels of EMT markers in NSCLC cells, Western blot analysis was performed to analyze the expression of E-cadherin, ZO-1, N-cadherin, vimentin, ZEB-1, and Snail-1 proteins in transfected NSCLC cells. Our results showed that the protein expression of E-cadherin and ZO-1 was decreased, while the protein expression of N-cadherin, vimentin, ZEB-1, and Snail-1 was increased by overexpression of HPV-16 E6 and E7 oncoproteins in A549 cells (Fig. 4A). Similar results were found in NCI-H460 NSCLC cells (Fig. 4B).
Figure 4.
Effects of HPV-16 oncoproteins on the protein expression of EMT markers in NSCLC cells. Western blot analysis was performed to analyze the expression of E-cadherin, ZO-1, N-cadherin, vimentin, ZEB-1, and Snail-1 proteins in transfected A549 (A) and NCI-H460 (B) cells. The results are representative of three independent experiments.
STAT3 Signaling Pathway Was Involved in HPV-16 E6-Induced EMT
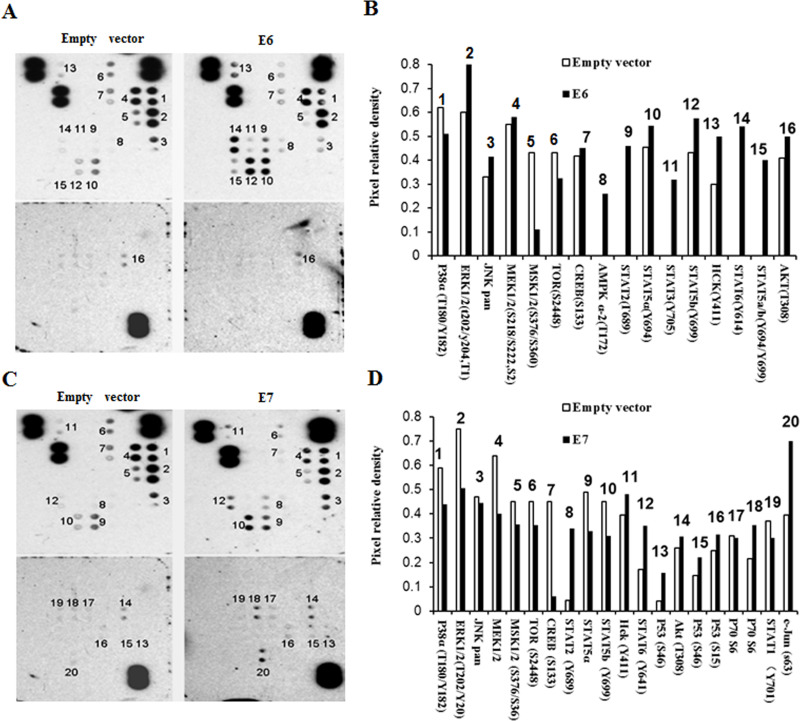
Multiple signaling pathways have been found to mediate EMT. To investigate signaling pathways that can regulate HPV-16 oncoprotein-induced EMT, we analyzed the phosphorylation levels of 46 proteins using protein microarray. Our results showed that the HPV-16 E6 oncoprotein interfered with the phosphorylation levels of 16 proteins, especially the HPV-16 E6 oncoprotein upregulated the phosphorylation levels of STAT3, STAT5a, STAT5b, and STAT6 (Fig. 5A and B). The HPV-16 E7 oncoprotein regulated the phosphorylation levels of 20 proteins. The HPV-16 E7 oncoprotein especially upregulated the phosphorylation levels of STAT1, STAT2, STAT5a, STAT5b, STAT6, and c-Jun (Fig. 5C and D). These results indicated that HPV-16 oncoproteins activated STAT signaling.
Figure 5.
Effects of HPV-16 oncoproteins on the phosphorylation levels of 46 proteins. Protein microarray was performed to analyze the effects of HPV-16 oncoproteins on the phosphorylation levels of 46 proteins. (A, C) Results of protein microarray (A: E6; C: E7) and (B, D) density results (B: E6; D: E7).
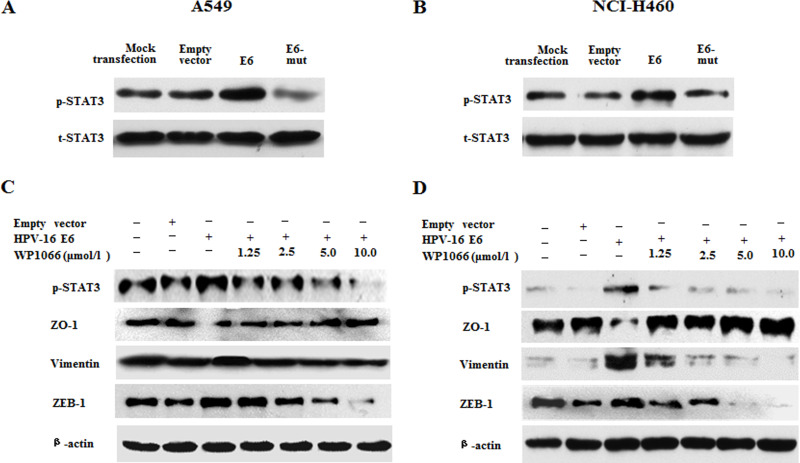
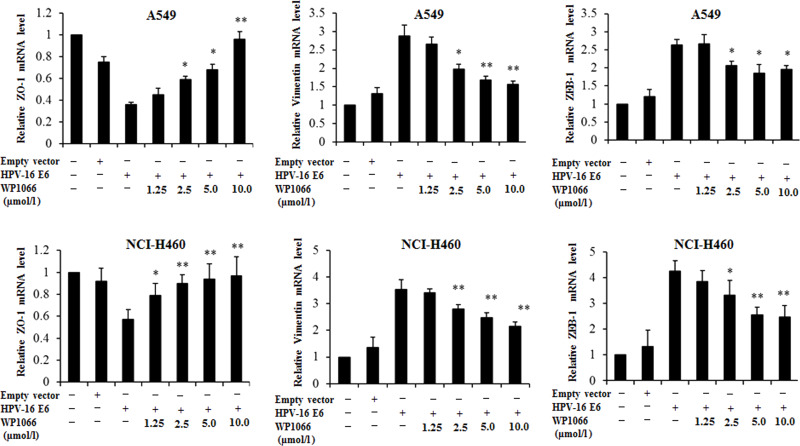
Previous studies have demonstrated that STAT3 can mediate EMT23–25, so we further analyzed the role of STAT3 signaling in HPV-16 E6 oncoprotein-induced EMT in NSCLC cells. The results from the Western blot analysis further confirmed that HPV-16 E6 upregulated the phosphorylated STAT3 (p-STAT3) protein level in both A549 (Fig. 6A) and NCI-H460 cells (Fig. 6B), indicating that HPV-16 E6 promoted the activation of STAT3 in two types of NSCLC cells. Next, NSCLC cells were pretreated with different concentrations of WP1066, a specific STAT3 inhibitor, followed by transfection with the pEGFP-N1-HPV-16 E6 plasmid. Our results showed that p-STAT3 protein expression was inhibited by WP1066. As expected, the inhibition of p-STAT3 expression reversed the effect of HPV-16 E6 on ZO-1, vimentin, and ZEB-1 protein expression in A549 (Fig. 6C) and NCI-H460 (Fig. 6D) cells. Moreover, the inhibition of p-STAT3 expression dramatically abrogated the effect of HPV-16 E6 on ZO-1, vimentin, and ZEB-1 mRNA expression in two types of NSCLC cells (Fig. 7). Taken together, our results suggest that STAT3 signaling is involved in HPV-16 E6 oncoprotein-induced EMT in NSCLC.
Figure 6.
WP1066 reversed the effect of HPV-16 E6 on the expression of ZO-1, vimentin, and ZEB-1 proteins in NSCLC cells. (A, B) Western blot analysis was performed to analyze the expression of total-STAT 3 (t-STAT) and phosphorylated-STAT3 (p-STAT3) in transfected A549 (A) and NCI-H460 (B) cells. (C, D) Transfected A549 (C) and NCI-H460 (D) cells were pretreated with different concentrations of WP1066 for 24 h, followed by Western blot analysis for the expression of STAT3, ZO-1, vimentin, and ZEB-1 proteins.
Figure 7.
WP1066 reversed the effect of HPV-16 E6 on ZO-1, vimentin, and ZEB-1 mRNA expression in NSCLC cells. Transfected A549 and NCI-H460 cells were pretreated with different concentrations of WP1066 for 24 h, followed by real-time PCR analysis for STAT3, ZO-1, vimentin, and ZEB-1 mRNA levels. All data were expressed as mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, compared with HPV-16 E6-transfected cells.
DISCUSSION
EMT is accompanied by degradation of the extracellular matrix and gain of mesenchymal cytoskeletal proteins18. HPV-16 E6 and E7 oncoproteins were found to induce EMT-like processes in the epithelial MDCK cell line via induction of the EMT transcription factors including Slug, Twist, ZEB-1, and ZEB-2, especially ZEBs29. HPV-16 E5 expression was also reported to cause switching from fibroblast growth factor receptor (FGFR) 2b to FGFR2c and EMT in cell models of transfected human keratinocytes as well as in cervical epithelial cells30. E5 and E6/E7 of high-risk HPVs were reported to cooperate in enhancing cancer progression through initiation of EMT31. Most recently, HPV-16 E6/E7 was found to promote cell migration and invasion in cervical cancer via regulating cadherin switch in vitro and in vivo32. However, HPV+ vulvar cancers did not exhibit EMT-like events and had a better prognosis33. These reports indicate that the relationship between HPV and EMT is not completely clear. Moreover, the effect of HPV-16 oncoproteins on EMT in NSCLC has not been reported.
Our previous studies demonstrated that overexpression of HPV-16 E6 and E7 oncoproteins promoted angiogenesis in A549 and NCI-H460 NSCLC cells via enhancing the expression of hypoxia-inducible factor-1α (HIF-1α) and vascular endothelial growth factor (VEGF)26, and PI3K/Akt and c-Jun signaling pathways were involved in HPV-16 oncoprotein-induced HIF-1α, VEGF, and IL-8 expression and in vitro angiogenesis27. Accumulating evidence has demonstrated that HIF-1α, VEGF, and IL-8, the important angiogenic factors, can promote EMT34–37. HIF-1α was found to promote EMT and metastasis through the direct regulation of ZEB-1 in colorectal cancer34 and mediate hypoxia-induced EMT in peritoneal mesothelial cells35. HIF-1α and HIF-2α were reported to enhance the migratory and neoplastic capacities of hepatocellular carcinoma cells by promoting EMT36. Additionally, increased expression of VEGF was demonstrated to lead to EMT in prostate intraepithelial neoplasia-like cells through an autocrine loop37. IL-8 was also found to inhibit E-cadherin expression in nasopharyngeal carcinoma cells by enhancing E-cadherin promoter DNA methylation38. Moreover, overexpression of HPV-16 E6 and E7 oncoproteins was demonstrated to mediate HIF-1α upregulation of GLUT1 expression in lung cancer cells39 and enhance the abilities of migration and invasion by promoting the expression of MMP-2 and MMP-9 in NSCLC cells40,41. A growing body of evidence has demonstrated that EMT plays a crucial role in invasion and metastasis18,42. Therefore, in this study, we further investigated the effect of HPV-16 oncoproteins on EMT in NSCLC cells. Interestingly, we found that overexpression of HPV-16 E6 and E7 oncoproteins in A549 and NCI-H460 NSCLC cells significantly promoted EMT-like morphologic changes, inhibited the expression of EMT epithelial markers (E-cadherin and ZO-1), and enhanced the expression of EMT mesenchymal markers (N-cadherin and vimentin) and transcription factors (ZEB-1 and Snail-1) at both transcriptional and translational levels, indicating that HPV-16 oncoproteins can promote EMT in NSCLC cells. EMT and angiogenesis play a key role in the development and progression. Therefore, taken together, our previous26,27 and present studies suggest that HPV-16 oncoproteins can promote NSCLC development and progression by enhancing angiogenesis and EMT.
STAT3, a key transcriptional factor, is involved in a wide variety of essential cellular functions related to proliferation, survival, and angiogenesis43. Recently, STAT3 has been demonstrated to mediate EMT23–25. STAT3 was found to play an important role in Notch1-induced EMT in breast cancer cells23 and cooperate with Twist to mediate EMT in human hepatocellular carcinoma cells24. Recently, it was reported that NANOG mediated EMT and drug resistance through the activation of the STAT3 pathway in epithelial ovarian cancer25. In the present study, we found that HPV-16 E6 enhanced the activation of STAT3 in NSCLC cells, and WP1066, a specific STAT3 inhibitor, significantly reversed the effects of HPV-16 E6 on the expression of EMT markers including ZO-1, vimentin, and ZEB-1 at both mRNA and protein levels in NSCLC cells. Taken together, our results indicate that STAT3 is involved in HPV-16 E6-induced EMT in NSCLC cells, suggesting that STAT3 may play a role in HPV-16 E6-mediated development and progression of NSCLC.
To summarize, in this study we first demonstrated that overexpression of HPV-16 E6 and E7 oncoproteins enhanced EMT in NSCLC cells, contributing to the development and progression of NSCLC. Furthermore, the STAT3 signaling pathway is involved in HPV-16 E6-induced EMT in NSCLC cells.
ACKNOWLEDGMENTS
This work was supported by the grants from the National Natural Science Foundation of China (81372511; to X. Tang), the Guangdong Natural Science Foundation (S2012010008232; to X. Tang), and the Science and Technology of Guangdong Province (2013B031100002; to X. Tang). Zhanjiang Municipal Governmental Specific Financial Fund Allocated for Competitive Scientific and Technological Projects (2012C0303-56; to X. Tang).
REFERENCES
- 1. Parkin DM, Bray F, Ferley J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- 2. Huang Y, Wang R, Pan Y, Zhang Y, Li H, Cheng C, Zheng D, Zheng S, Li Y, Shen X, Hu H, Cai D, Wang S, Zhang Y, Xiang J, Sun Y, Zhang J, Chen H. Clinical and genetic features of lung squamous cell cancer in never-smokers. Oncotarget 2016;7:35979–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clément-Duchêne C, Stock S, Xu X, Chang ET, Gomez SL, West DW, Wakelee HA, Gould MK. Survival among never-smokers with lung cancer in the Cancer Care Outcomes Research and Surveillance Study. Ann Am Thorac Soc. 2016;13:58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pirie K, Peto R, Green J, Reeves GK, Beral V. Lung cancer in never-smokers. Int J Cancer 2016;139:347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bae JM. Modifiable risk factors of lung cancer in “never-smoker” women. Epidemiol Health 2015;37:e2015047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Syrjanen KJ. Condylomatous changes in neoplastic bronchial epithelium. Report of a case. Respiration 1979;38:299–304. [DOI] [PubMed] [Google Scholar]
- 7. Zhai K, Ding J, Shi HZ. HPV and lung cancer risk: A meta-analysis. J Clin Virol. 2015;63C:84–90. [DOI] [PubMed] [Google Scholar]
- 8. Zhang EY, Tang XD. Human papillomavirus type 16/18 oncoproteins: Potential therapeutic targets in non-smoking associated lung cancer. Asian Pac J Cancer Prev. 2012;13:5363–9. [DOI] [PubMed] [Google Scholar]
- 9. Bae JM, Kim EH. Human papillomavirus infection and risk of lung cancer in never-smokers and women: An ‘adaptive’ meta-analysis. Epidemiol Health 2015;37:e2015052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin FC, Huang JY, Tsai SC, Nfor ON, Chou MC, Wu MF, Lee CT, Jan CF, Liaw YP. The association between human papillomavirus infection and female lung cancer: A population-based cohort study. Medicine (Baltimore) 2016;95:e3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Freitas AC, Gurgel AP, de Lima EG, de França São Marcos B, do Amaral CM. Human papillomavirus and lung carcinogenesis: An overview. J Cancer Res Clin Oncol. 2016;142:2415–27. [DOI] [PubMed] [Google Scholar]
- 12. Ciotti M, Giuliani L, Ambrogi V, Ronci C, Benedetto A, Mineo TC, Syrjänen K, Favalli C. Detection and expression of human papillomavirus oncogenes in non-small cell lung cancer. Oncol Rep. 2006;16:183–9. [PubMed] [Google Scholar]
- 13. Baba M, Castillo A, Koriyama C, Yanagi M, Matsumoto H, Natsugoe S, Shuyama KY, Khan N, Higashi M, Itoh T, Eizuru Y, Aikou T, Akiba S. Human papillomavirus is frequently detected in gefitinib-responsive lung adenocarcinomas. Oncol Rep. 2010;23:1085–92. [DOI] [PubMed] [Google Scholar]
- 14. Sarchianaki E, Derdas SP, Ntaoukakis M, Vakonaki E, Lagoudaki ED, Lasithiotaki I, Sarchianaki A, Koutsopoulos A, Symvoulakis EK, Spandidos DA, Antoniou KM, Sourvinos G. Detection and genotype analysis of human papillomavirus in non-small cell lung cancer patients. Tumour Biol. 2014;35:3203–9. [DOI] [PubMed] [Google Scholar]
- 15. Cheng YW, Lin FC, Chen CY, Hsu NY. Environmental exposure and HPV infection may act synergistically to induce lung tumorigenesis in nonsmokers. Oncotarget 2016;7:19850–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang SY, Keeney M, Law M, Donovan J, Aubry MC, Garcia J. Detection of human papillomavirus in non-small cell carcinoma of the lung. Hum Pathol. 2015;46:1592–7. [DOI] [PubMed] [Google Scholar]
- 17. Isa SI, Kurahara Y, Yamamoto S, Tamiya A, Omachi N, Asami K, Okishio K, Utsumi T, Ito N, Yoon HE, Matsumura A, Atagi S, Kawaguchi T. Molecular analysis of human papillomavirus in never-smokers with non-small cell lung cancer. Oncol Lett. 2015;9:927–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871–90. [DOI] [PubMed] [Google Scholar]
- 19. Shen W, Pang H, Liu J, Zhou J, Zhang F, Liu L, Ma N, Zhang N, Zhang H, Liu L. Epithelial-mesenchymal transition contributes to docetaxel resistance in human non-small cell lung cancer. Oncol Res. 2014;22:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao C, Li X, Su C, Li J, Cheng N, Ren S, Chen X, Zhou C. High expression of E-cadherin in pleural effusion cells predicts better prognosis in lung adenocarcinoma patients. Int J Clin Exp Pathol. 2015;8:3104–9. [PMC free article] [PubMed] [Google Scholar]
- 21. Atmaca A, Wirtz RW, Werner D, Steinmetz K, Claas S, Brueckl WM, Jäger E, Al-Batran SE. SNAI2/SLUG and estrogen receptor mRNA expression are inversely correlated and prognostic of patient outcome in metastatic non-small cell lung cancer. BMC Cancer 2015;15:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wake MS, Watson CJ. STAT3 the oncogene—Still eluding therapy? FEBS J. 2015;282:2600–11. [DOI] [PubMed] [Google Scholar]
- 23. Zhang X, Zhao X, Shao S, Zuo X, Ning Q, Luo M, Gu S, Zhao X. Notch1 induces epithelial-mesenchymal transition and the cancer stem cell phenotype in breast cancer cells and STAT3 plays a key role. Int J Oncol. 2015;46:1141–8. [DOI] [PubMed] [Google Scholar]
- 24. Zhang C, Guo F, Xu G, Ma J, Shao F. STAT3 cooperates with Twist to mediate epithelial-mesenchymal transition in human hepatocellular carcinoma cells. Oncol Rep. 2015;33:1872–82. [DOI] [PubMed] [Google Scholar]
- 25. Liu S, Sun J, Cai B, Xi X, Yang L, Zhang Z, Feng Y, Sun Y. NANOG regulates epithelial-mesenchymal transition and chemoresistance through activation of the STAT3 pathway in epithelial ovarian cancer. Tumour Biol. 2016;37:9671–80. [DOI] [PubMed] [Google Scholar]
- 26. Li G, He L, Zhang E, Shi J, Zhang Q, Le AD, Zhou K, Tang X. Overexpression of human papillomavirus (HPV) type 16 oncoproteins promotes angiogenesis via enhancing HIF-1α and VEGF expression in non-small cell lung cancer cells. Cancer Lett. 2011;311:160–70. [DOI] [PubMed] [Google Scholar]
- 27. Zhang E, Feng X, Liu F, Zhang P, Liang J, Tang X. Roles of PI3K/Akt and c-Jun signaling pathways in human papillomavirus type 16 oncoprotein-induced HIF-1α, VEGF, and IL-8 expression and in vitro angiogenesis in non-small cell lung cancer cells. PLoS One 2014;9:e103440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi J, Liu F, Zhang W, Liu X, Lin B, Tang X. Epigallocatechin-3-gallate inhibits nicotine-induced migration and invasion by the suppression of angiogenesis and epithelial–mesenchymal transition in non-small cell lung cancer cells. Oncol Rep. 2015;33:2972–80. [DOI] [PubMed] [Google Scholar]
- 29. Jung YS, Kato I, Kim HR. A novel function of HPV16-E6/E7 in epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2013;435:339–44. [DOI] [PubMed] [Google Scholar]
- 30. Ranieri D, Belleudi F, Magenta A, Torrisi MR. HPV16 E5 expression induces switching from FGFR2b to FGFR2c and epithelial-mesenchymal transition. Int J Cancer. 2015;137:61–72. [DOI] [PubMed] [Google Scholar]
- 31. Al Moustafa AE. E5 and E6/E7 of high-risk HPVs cooperate to enhance cancer progression through EMT initiation. Cell Adh Migr. 2015;9:392–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu D, Zhou J, Wang F, Shi H, Li Y, Li B. HPV-16 E6/E7 promotes cell migration and invasion in cervical cancer via regulating cadherin switch in vitro and in vivo. Arch Gynecol Obstet. 2015;292:1345–54. [DOI] [PubMed] [Google Scholar]
- 33. Rodrigues IS, Lavorato-Rocha AM, de M Maia B, Stiepcich MM, de Carvalho FM, Baiocchi G, Soares FA, Rocha RM. Epithelial-mesenchymal transition-like events in vulvar cancer and its relation with HPV. Br J Cancer 2013;109:184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang W, Shi X, Peng Y, Wu M, Zhang P, Xie R, Wu Y, Yan Q, Liu S, Wang J. HIF-1α promotes epithelial-mesenchymal transition and metastasis through direct regulation of ZEB1 in colorectal cancer. PLoS One 2015;10:e0129603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morishita Y, Ookawara S, Hirahara I, Muto S, Nagata D. HIF-1α mediates hypoxia-induced epithelial-mesenchymal transition in peritoneal mesothelial cells. Ren Fail. 2016;38:282–9. [DOI] [PubMed] [Google Scholar]
- 36. Liu Y, Liu Y, Yan X, Xu Y, Luo F, Ye J, Yan H, Yang X, Huang X, Zhang J, Ji G. HIFs enhance the migratory and neoplastic capacities of hepatocellular carcinoma cells by promoting EMT. Tumour Biol. 2014;35:8103–14. [DOI] [PubMed] [Google Scholar]
- 37. Gonzalez-Moreno O, Lecanda J, Green JE, Segura V, Catena R, Serrano D, Calvo A. VEGF elicits epithelial-mesenchymal transition (EMT) in prostate intraepithelial neoplasia (PIN)-like cells via an autocrine loop. Exp Cell Res. 2010;316:554–67. [DOI] [PubMed] [Google Scholar]
- 38. Zhang RL, Peng LX, Yang JP, Zheng LS, Xie P, Wang MY, Huang BJ, Zhao HR, Bao YX, Qian CN. IL-8 suppresses E-cadherin expression in nasopharyngeal carcinoma cells by enhancing E-cadherin promoter DNA methylation. Int J Oncol. 2016;48:207–14. [DOI] [PubMed] [Google Scholar]
- 39. Fan R, Hou WJ, Zhao YJ, Liu SL, Qiu XS, Wang EH, Wu GP. Overexpression of HPV16 E6/E7 mediated HIF-1α upregulation of GLUT1 expression in lung cancer cells. Tumour Biol. 2016;37:4655–63. [DOI] [PubMed] [Google Scholar]
- 40. Shiau MY, Fan LC, Yang SC, Tsao CH, Lee H, Cheng YW, Lai LC, Chang YH. Human papillomavirus upregulates MMP-2 and MMP-9 expression and activity by inducing interleukin-8 in lung adenocarcinomas. PLoS One 2013;8:e54423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu Q, Hu Q, Liu F, Feng Y, Tang X. Effects of HPV-16 oncoproteins on migration and invasion in A549 lung cancer cells and their mechanisms. Mod Oncol. 2015;23:2553–7. [in Chinese] [Google Scholar]
- 42. Smith BN, Bhowmick NA. Role of EMT in metastasis and therapy resistance. J Clin Med. 2016;5:pii:E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yuan J, Zhang F, Niu R. Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci Rep. 2015;5:17663. [DOI] [PMC free article] [PubMed] [Google Scholar]