Abstract
The long noncoding CPS1 intronic transcript 1 (lncRNA CPS1-IT1) is a recently identified tumor suppressor in the lncRNA family of proteins. Whether this lncRNA plays any functional role in solid tumors remains largely unknown. The present study aimed to investigate the role of lncRNA CPS1-IT1 in human lung cancer. Expression of lncRNA CPS1-IT1 was initially assessed in human lung cancer and in a series of lung cancer cell lines. The effects of CPS1-IT1 overexpression on cell proliferation, migration, and invasion were examined in lung cancer cell lines A549 and 95D. It was found that lncRNA CPS1-IT1 was significantly lower in cancerous tissues than in noncancerous tissues. lncRNA CPS1-IT1 was differentially expressed in lung cancer cell lines and expressed the least in two highly invasive cell lines, A549 and 95D. Overexpression of CPS1-IT1 slowed down cell proliferation by 35.7% in A549 cells and 30.8% in 95D cells on the fifth day. Cell migration was inhibited by 59% in A549 cells and 48% in 95D cells, and cell invasion was suppressed by 60% in both cell lines after overexpression of CPS1-IT1. While cell apoptosis was induced, CPS1-IT1 overexpression promoted the activities of caspase 3 and caspase 9 without affecting that of caspase 8. These observations were suggestive of the tumor-suppressive role of lncRNA CPS1-IT1 in lung cancer. Our data suggest that CPS1-IT1 may be used as a biomarker for early diagnosis and therapeutic targets against lncRNA and may be promising in the treatment of lung cancer.
Key words: Long noncoding RNA (lncRNA), CPS1-IT1, Lung cancer, Proliferation, Metastasis
INTRODUCTION
Lung cancer is the leading cause of cancer-related deaths among males in both developed and developing countries and has surpassed breast cancer as the leading cause of cancer deaths among females in more developed countries1. Patients suffering from this malignancy generally have a poor prognosis. The low 5-year survival rate may be attributed to the fact that affected patients have limited access to early detection and timely treatment2,3. Early detection and screening are therefore the best defense against lung cancer and are of vital importance in the development of therapeutic targets against this malignancy4.
Recent improvements in high-throughput screening of differentially expressed genes have led to the discovery of a novel category of RNAs that are referred to as noncoding RNAs5. These short or long noncoding RNAs (lncRNAs) come from <2% of the human genome with limited or no protein-coding ability because of a lack of open reading frames6–8. The number of lncRNAs is estimated to be 15,000, with most of them exhibiting a tissue-specific pattern. Unlike transcription factors, mounting evidence has indicated that lncRNAs regulate gene expression at different levels, including transcriptional processing, chromatin reprogramming, microRNA sponging, and other processes9. Increasing evidence has further shown that lncRNAs functionally serve as oncogenes or tumor suppressors and are critically involved in tumorigenesis, cell proliferation, migration, and apoptosis5,10. The lncRNA MEG3, for instance, is an identified tumor suppressor11 and has been associated with tumor grade in meningioma12, as well as regulated cell proliferation and apoptosis in non-small cell lung cancer13. In contrast, lncRNA MALAT1 serves as an oncoprotein that promotes tumor proliferation and metastasis in esophageal squamous cell carcinoma and glioma14,15. The identification of critical lncRNAs as oncoproteins or tumor suppressors has led to the discovery of biomarkers that are of value in the early diagnosis of cancer.
lncRNA CPS1 intronic transcript 1 (lncRNA CPS1-IT1) is a recently identified tumor suppressor in the lncRNA family of proteins. It was initially identified as a novel tumor suppressor and served as an independent predictor for overall survival and disease-free survival in hepatocellular carcinoma9. Overexpression of lncRNA CPS1-IT1 reduced tumor growth and metastasis by interacting with heat shock protein 90 (HSP90)9. However, whether this lncRNA plays any functional role in other solid tumors remains largely unknown. Of particular interest, the current study aimed to investigate the role of lncRNA CPS1-IT1 in human lung cancer. Expression of this lncRNA in clinical lung cancer tissues and in lung cancer cell lines was initially examined. The effects of CPS1-IT1 modulation on cell proliferation, metastasis, and apoptosis are fully discussed in this study.
MATERIALS AND METHODS
Human Samples
A total of 40 lung cancer patients underwent clinical surgeries in Tianjin Hospital (P.R. China) between 2013 and 2015. Before surgery, no chemotherapy or radiotherapy was performed on these patients. Once dissected, tumor tissues and adjacent noncancerous tissues were frozen in liquid nitrogen and subsequently used for real-time polymerase chain reaction (RT-PCR) analysis. All patients gave their full intention to participate in our study, and written consents were obtained. This study was approved by the ethics committee of the Tianjin Hospital.
Cell Culture and Transfection
Lung cancer cell lines A549, H1975, SPC-A-1, H125, and 95D were commercially purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in the recommended medium supplied with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher, Waltham, MA, USA) in a 5% CO2 atmosphere. Normal human lung cells (MRC-5) were purchased from the Cell Bank of the Chinese Academy of Sciences and used as a control. An expression plasmid for lncRNA CPS1-IT1 was constructed with pcDNA3.1 vector and transfected into A549 and D95 cells with Lipofectamine 2000 (Invitrogen, New Brunswick, NJ, USA) as per the manufacturer’s instructions.
Real-Time Polymerase Chain Reaction (RT-PCR)
Total RNAs from both clinical lung cancer tissues and cultured cells were extracted with TRIzol reagent (TaKaRa, Dalian, P.R. China) with a dose of 1 ml/well in six-well plates. RNAs were quantified with NanoDrop 2000 (Thermo Fisher Scientific, NJ, USA) by collecting the absorbance of 260 and 230 nm. Afterward, RNAs were reverse transcribed into cDNA with a TaKaRa kit. RT-PCR analysis was performed with the ABI 7900 (Applied Biosystems, Foster City, CA, USA). The primers were designed and synthesized by Sigma-Aldrich (St. Louis, MO, USA). GAPDH was included here as an internal control.
Colony Formation Assay
Both A549 and 95D cells in six-well plates were treated with control vector or CPS1-IT1 expression plasmid. Twenty-four hours after treatment, they were seeded into 12-well plates (100 cells/well) in triplicate. After incubation for 10 days, the colonies were fixed with pre-iced methanol and stained with crystal violet. Colonies containing more than 50 cells were considered as survivors and counted under a Nikon microscope (Minato, Tokyo, Japan) at a magnification of 200×. The following formula was used to calculate the rate of colony formation: colony formation rate = (number of colonies/number of seeded cells) × 100%.
Cell Viability Assay
A cell viability assay was performed with 3-(4,5-dimethyl thiazol-2yl)-2, 5-diphenyltetrazolium bromide (MTT). A total of 1 × 104 lung cancer cells were seeded in 96-well plates and transfected with CPS1-IT1 plasmid in triplicate. Cells were allowed to grow for an additional 48 h in a 37°C incubator, after which 10 μl of MTT (5 μg/ml) was mixed with the medium in each well and incubated for another 3 h at 37°C in the dark. Formazan crystal formed and was dissolved again in 100 μl of DMSO. The optical absorbance of each well was collected at 570 nM with a TECAN reader (Männedorf, Switzerland).
Transwell Assay
Cell migration and invasion were determined by Transwell assays. First, cells were seeded in six-well plates and transfected with CPS1-IT1 plasmid for 48 h. Second, the bottom chamber was filled with 600 μl of medium with 10% FBS. Third, cells were resuspended with the recommended medium without FBS, and 1 × 104 A549 or 95D cells were placed in the upper chamber (Corning, Corning, NY, USA) with a volume of 100 μl per well. After incubation for 12 h, cells were washed by PBS three times and fixed with methanol for 10 min, and then stained with crystal violet for 5 min. Cell migration was quantified by counting cells that migrated across the filter toward the lower surface of the chamber. Five random fields were calculated under a Nikon microscope (Minato). For invasion assays, the upper surface of the chamber was pretreated with Matrigel (BD Biosciences, San Jose, CA, USA) for 6 h in a 37°C incubator. All experiments were repeated at least three times in triplicate.
Wound-Healing Assay
Wound-healing assays were explored by creating identical wound areas for anchorage-dependent cells A549 with 10-μl sterile pipette tips. Cells were seeded into six-well plates and coincubated with plasmids in the presence or absence of CPS1-IT1 overexpression for 24 h. Afterward, cells were scraped in a cross in the center of each well, washed with PBS, and immediately replaced with fresh serum-free medium. After 24 h of growth, cells were observed and photographed under a Nikon microscope (Minato) at a magnification of 200× for each group.
Cell Apoptosis Assay
Cell apoptosis was examined with Hoechst 33258 (Beyotime, Nanjing, P.R. China) according to the manufacturer’s protocols. Both A549 and 95D cells were seeded into 12-well plates in triplicate and transfected with different plasmids for an additional 48 h. Afterward, Hoechst 33258 was mixed with the medium in each well and incubated for 15 min at 37°C. Fluorescence images were captured randomly with an inverted fluorescence microscope (Nikon; Minato). The percentages of apoptotic neurons were calculated with the following formula: apoptotic rate = (apoptotic cells/total cells) × 100%.
Caspase Activity Assay
The activity of caspase 3, caspase 8, and caspase 9 was determined by caspase activity kits (Beyotime), according to the manufacturer’s instructions. Briefly, both A549 and 95D cells were administered with CPS1-IT1-expressing plasmids for 24 h, and cell lysates were collected. Assays were performed in 96-well plates by incubating proteins of cell lysates and reaction buffers containing substrates for caspase 3, caspase 8, and caspase 9, respectively. After coincubation for 4 h, samples were measured with a TECAN reader at an absorbance of 405 nm. All experiments were repeated at least three times in triplicate.
Statistical Analysis
All data were presented as the mean ± standard deviation (SD). Each experiment was repeated at least three times in triplicate, except when stated otherwise. The Student’s t-test was included to compare the difference between groups. Any value of p < 0.05 was considered statistically significant.
RESULTS
The Transcript Levels of lncRNA CPS1-IT1 Were Decreased in Human Lung Cancers
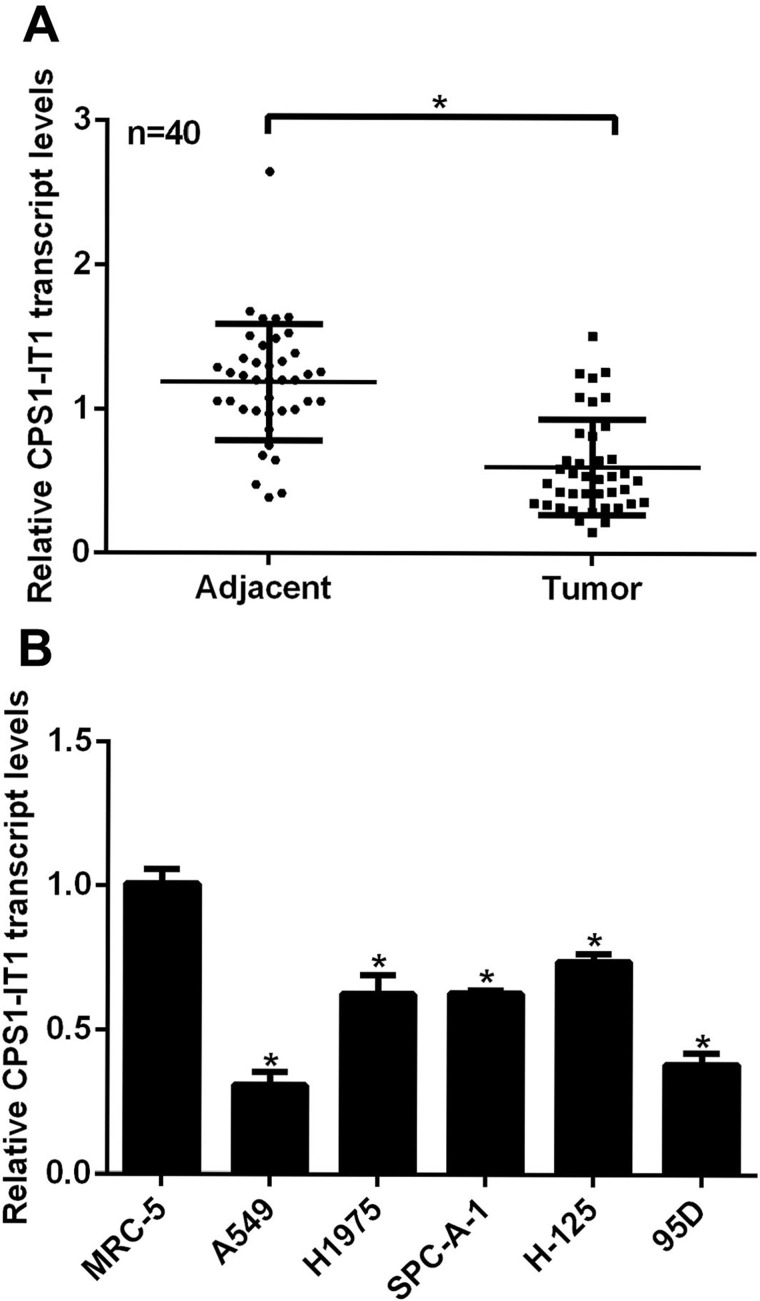
First, we examined the expression of lncRNA CPS1-IT1 in 40 clinical lung cancer patients with RT-PCR analysis. As shown in Figure 1A, the relative transcript levels of CPS1-IT1 in tumor tissues were notably decreased compared to their adjacent noncancerous counterparts. MRC-5 cells are derived from normal lungs and used here as a control. All five lung cancer cell lines showed significantly lower expressions of CPS1-IT1 when compared with MRC-5 cells, of which the relative transcript levels of CPS1-IT1 were the lowest in A549 and 95D cells (Fig. 1B); thus these two cell lines were selected for the subsequent overexpression assays. These data suggest that lncRNA CPS1-IT1 was downregulated in human lung cancers in vivo and in vitro.
Figure 1.
The transcript levels of lncRNA CPS1-IT1 were decreased in human lung cancers. (A) Tumor tissues and their adjacent noncancerous tissues from 40 lung cancer patients were subjected to RT-PCR analysis to explore the relative expression of lncRNA CPS1-IT1. *p < 0.05, Tumor versus Adjacent. (B) Normal lung cell MRC-5 and five lung cancer cell lines A549, H1975, SPC-A-1, H-125, and 95D were included for RT-PCR analysis, of which A549 and 95D cells showed the lowest CPS1-IT1 expression. *p < 0.05 versus MRC-5 cells.
Overexpression of CPS1-IT1 Inhibited Colony Formation and Cell Proliferation in Lung Cancer Cell Lines
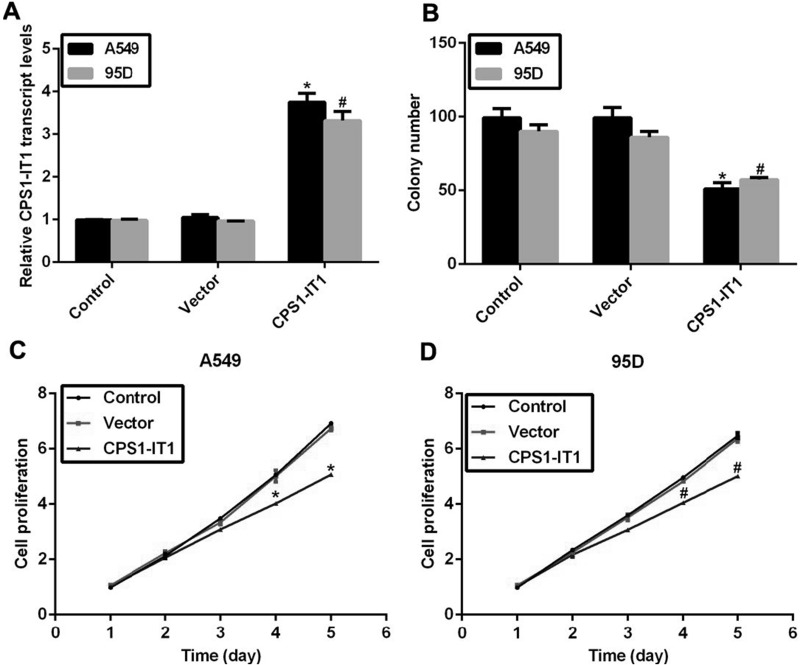
Next, we explored the detailed role of this downregulated lncRNA CPS1-IT1 in lung cancer. To this end, an expressing plasmid of CPS1-IT1 was constructed and transfected into A549 and 95D cells, after which the relative transcript levels of CPS1-IT1 were remarkably increased by approximately threefold compared to the control group in both cell lines (Fig. 2A), indicating the efficiency of the CPS1-IT1 expression plasmid. Afterward, a colony formation assay was performed in A549 and 95D cells. It was shown that the colony number in A549 cells was decreased from 100 to 50 upon CPS1-IT1 expression plasmid transfection, and the formed colonies were also decreased by approximately 40% in 95D cells (Fig. 2B). Cell viability assays were explored to assess the role of CPS1-IT1 in cell proliferation. No notable disparities were observed in the first 3 days for either cell line. However, the cell proliferative rate was suppressed by 21% in A549 cells and 18% in 95D cells on the fourth day, and the inhibitive effects of CPS1-IT1 were further increased on the fifth day in lung cancer cells (Fig. 2C and D). Our results showed that overexpression of lncRNA CPS1-IT1 in lung cancer cells inhibited colony formation and cell proliferation.
Figure 2.
Overexpression of CPS1-IT1 inhibited colony formation and cell proliferation in lung cancer cell lines. (A) An expressing plasmid for lncRNA CPS1-IT1 was constructed into pcDNA3.1 vector and transfected into A549 and 95D cells, after which RT-PCR analysis revealed that the relative transcript levels of CPS1-IT1 were upregulated in both cell lines. (B) Colony formation assays were performed in A549 and 95D cells upon CPS1-IT1 plasmid transfection. (C) A549 cells were transfected with or without CPS1-IT1-expressing plasmid and subjected to cell viability assays for a consecutive 5 days. (D) 95D cells were transfected with plasmids in the presence or absence of CPS1-IT1 overexpression and subjected to cell viability assays. Cell proliferative rate was detected each day for a consecutive 5 days. *p < 0.05 versus Control group in A549 cells. #p < 0.05 versus Control group in 95D cells.
Upregulated CPS1-IT1 Suppressed Cell Migration and Invasion in A549 and 95D Cells
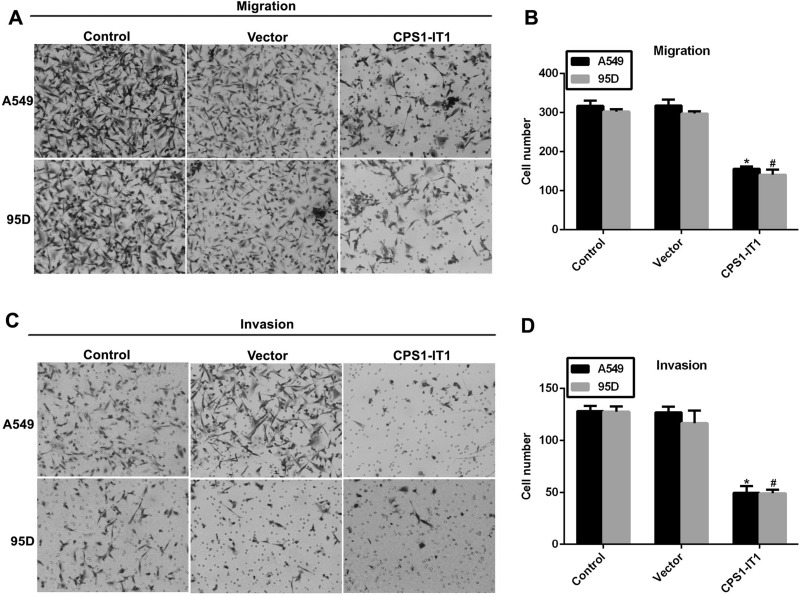
A549 and 95D cells showed the highest potential for migration and the lowest CPS1-IT1 expression among the five selected lung cancer cell lines; thus we assumed that downregulated CPS1-IT1 induced cell migration in these cells. Our results with the Transwell assays demonstrated this hypothesis (Fig. 3). The ability of cells to migrate was inhibited by approximately 50% when CPS1-IT1 was overexpressed in A549 and 95D cells (Fig. 3A and B). Similar observations were also made in cell-invasive assays. As shown in Figure 3C and D, about 150 A549 and 95D cells were counted on the lower surface of the membrane in the control group; however, only about 50 cells were observed to have invaded through the membrane upon CPS1-IT1 plasmid transfection in both cell lines.
Figure 3.
Overexpression of CPS1-IT1 inhibited cell migration and invasion in A549 and 95D cells. (A) Representative images of cell migration assays for A549 and 95D cells. Five random sites were photographed and quantified in each experimental group. (B) Quantification of cell migration assays revealed the inhibitive effects of CPS1-IT1 on cell migration in both cell lines. (C) Representative images of cell invasion assays for A549 and 95D cells. (D) Quantification of cell invasion assays revealed the inhibitive effects of CPS1-IT1 on cell invasion in both cell lines. *p < 0.05 versus Control group in A549 cells. #p < 0.05 versus Control group in 95D cells.
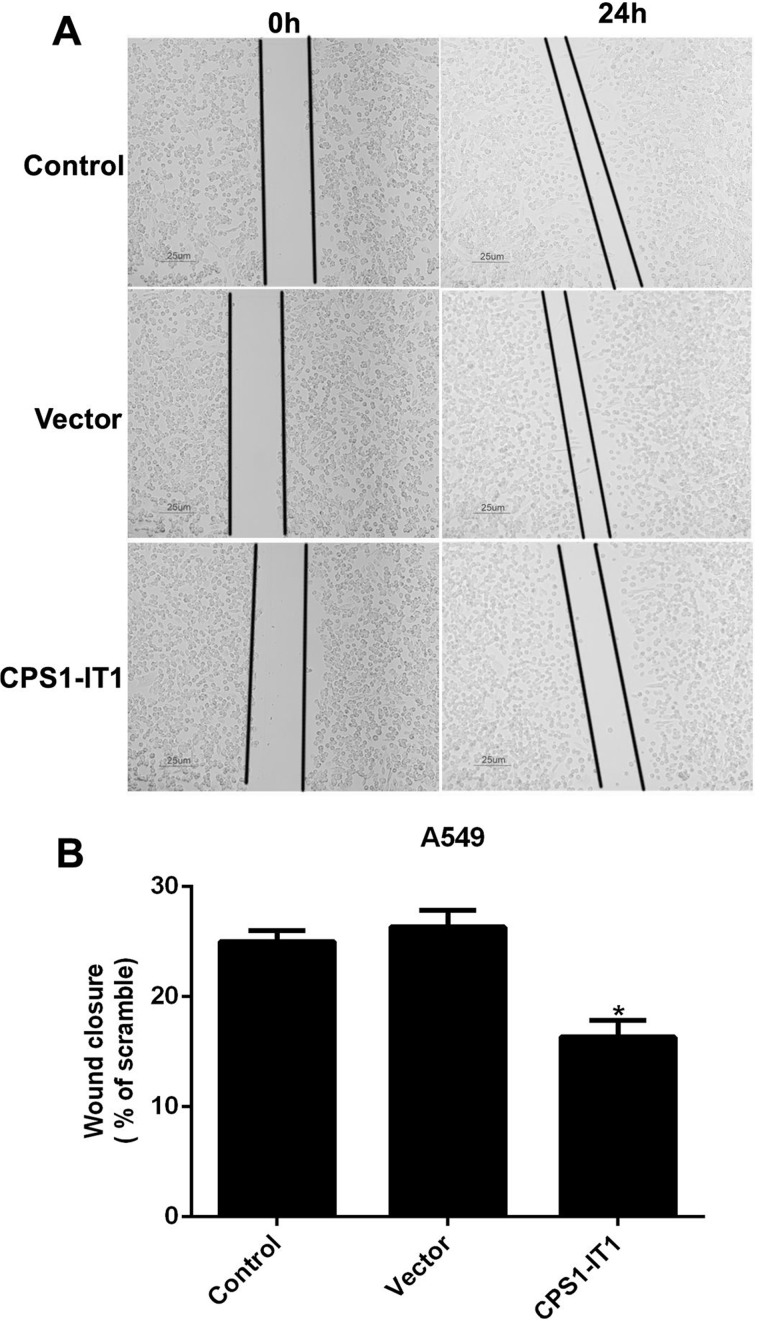
Afterward, a wound-healing assay was also explored in A549 cells to further assess the effects of CPS1-IT1 on cell metastasis. A549 cells were transfected with CPS1-IT1-expressing plasmid and allowed to grow for an additional 24 h. As shown in Figure 4, the wound closure ability was remarkably decreased in A549 cells treated with CPS1-IT1 when compared with the control counterpart. All of these results showed that overexpression of CPS1-IT1 in lung cancer cells significantly suppressed cell metastasis in vitro.
Figure 4.
Upregulated CPS1-IT1 in A549 cells retarded wound closure abilities in wound-healing assays. (A) Representative images of wound-healing assays for A549 cells. Five random sites were photographed and quantified in each experimental group. (B) Quantification of wound-healing assays showed the inhibitive role of CPS1-IT1 in cell migration. *p < 0.05 versus Control cells.
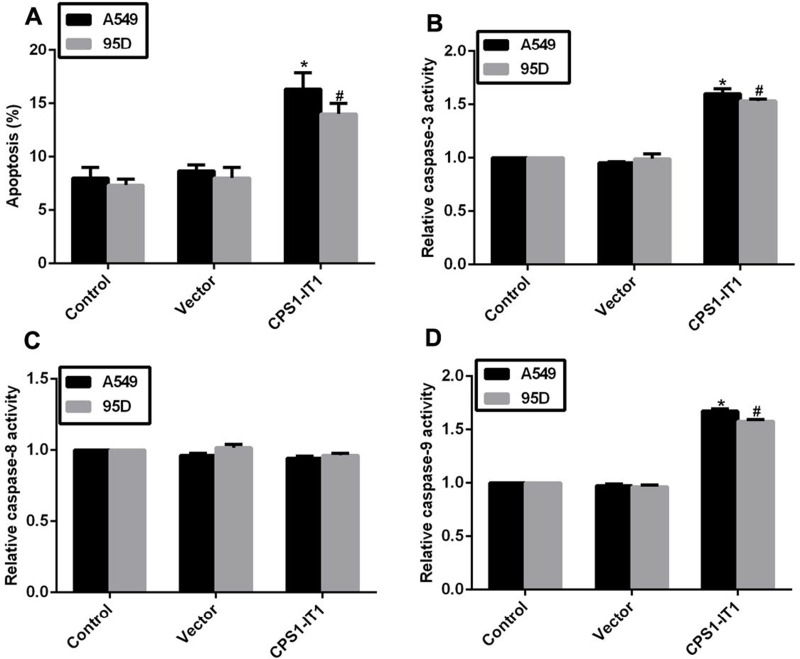
Overexpression of CPS1-IT1 Promoted Cell Apoptosis and Changed the Activities of Related Caspases
Cell proliferation and cell metastasis are two main manifestations of human cancer, while the apoptotic rate of cells varies in tumorigenesis. Next, we explored the potential role of CPS1-IT1 in cell apoptosis in lung cancer cells. As shown in Figure 5A, the apoptotic rates were increased by 7% in A549 cells and 6% in 95D cells upon treatment with CPS1-IT1 plasmid. Afterward, the relative activities of caspase 3, caspase 8, and caspase 9 were examined with a specific caspase activity kit. When cells were transfected with CPS1-IT1 plasmids, the relative activities of caspase 3 and caspase 9 were prominently increased by approximately 50% in both cell lines, while that of caspase 8 remained unchanged (Fig. 5B–D). These data revealed that the auxo action of lncRNA CPS1-IT1 on cell apoptosis is associated with the intrinsic apoptosis signaling pathway.
Figure 5.
Overexpression of CPS1-IT1 promoted cell apoptosis and the activities of related caspase. (A) Cell apoptotic rate was examined in A549 and 95D cells when cells were transfected with CPS1-IT1 plasmids. (B) The relative activity of caspase 3 was explored in A549 and 95D cells upon CPS1-IT1 transfection. (C) Overexpression of CPS1-IT1 in A549 and 95D cells caused no notable change in the relative activity of caspase 8. (D) The relative activity of caspase 9 was upregulated in A549 and 95D cells upon CPS1-IT1 transfection. *p < 0.05 versus Control group in A549 cells. #p < 0.05 versus Control group in 95D cells.
DISCUSSION
Lung cancer remains a great health threat for patients worldwide. Although the 5-year survival rate varies across countries, it is consistently low due to late-stage diagnosis and the paucity of timely intervention2,3,16. Therefore, it is imperative to develop strategies that could serve as sensitive and specific biomarkers for early detection of this malignancy.
The emerging discovery of lncRNAs has broadened our knowledge of human tumorigenesis. Evidence to date links lncRNA dysregulation to diverse human diseases, including cancer17,18. However, the role of lncRNA in human lung cancer remains to be elucidated. In the present study, we investigated the role of lncRNA CPS1-IT1 in lung cancer and found that expression of CPS1-IT1 was significantly lower in cancerous tissues relative to the adjacent noncancerous tissues. This finding was consistent with a previous pioneer study that identified CPS1-IT1 as a novel tumor suppressor that was lowly expressed in hepatocellular carcinoma9. Further in vitro functional assays demonstrated that CPS1-IT1 significantly inhibited the capacity for cell proliferation, migration, and invasion in lung cancer A549 and 95D cells. Moreover, cell apoptosis was induced by lncRNA CPS1-IT1 overexpression. CPS1-IT1 overexpression interestingly promoted the activities of caspase 3 and caspase 9 without affecting that of caspase 8. Caspase 8 is a known initiator of apoptosis involved in the extrinsic apoptosis pathway, whereas caspase 9 is an initiator in the intrinsic pathway19,20. Caspase 3 cleaves the inhibitor of caspase-activated DNase (ICAD) and is the most significant executioner caspase19. Hence, it could be concluded that lncRNA CPS1-IT1 induced cell apoptosis mainly through an intrinsic pathway. Since the induction of apoptotic activity is a good basis for anticancer proliferation21, the apoptosis promotion effect materially supports the antiproliferation effect of CPS1-IT1. All these data might suggest that lncRNA CPS1-IT1 is a tumor suppressor in lung cancer and serves as a useful biomarker for early diagnosis of lung cancer.
The molecular mechanisms underlying lncRNA functions are diverse, including (i) as a decoy to locate transcription factors; (ii) as a regulatory signal for transcription; (iii) as a scaffold to bridge different proteins; (iv) as a “sponge” to sequester microRNAs; (v) as a guide for proteins to reach their targets; and (vi) as a modified protein that can allosterically alter the functions of other proteins9,22. The mechanism of how lncRNA CPS1-IT1 suppresses lung cancer proliferation and metastasis needs to be elucidated in the future. However, several clues are valuable. In human hepatocellular carcinoma, CPS1-IT1 is found to interact with HSP90, which is a required chaperone for the activation and stabilization of numerous proteins involved in essential cellular processes such as signal transduction pathways23. Interaction of CPS1-IT1 with HSP90 inhibited its binding affinity to HIF-1α, thereby leading to decreased expression of mesenchymal proteins9. In addition, the tumor suppressor lncRNA MEG3 inhibits the activity of DNA methyltransferase and reduces the transcription of the Meg3 gene24. Since CPS1-IT1 also works as a tumor suppressor, as evidenced in hepatocellular carcinoma9 and in our present study, it could be hypothesized that CPS1-IT1 might play a critical role in silencing target genes via DNA methylation modifications. More exploration would be needed as to the detailed mechanisms of lncRNA CPS1-IT1 in lung cancer.
In all, we identified a long noncoding RNA, CPS1-IT1, as a tumor suppressor in lung cancer. To the best of our knowledge, this is the first study to address the function of CPS1-IT1 in human lung cancer. Our results demonstrate that CPS1-IT1 reduces the capacity for cell proliferation, migration, and invasion in lung cancer. Our study provides novel insights into the role of lncRNAs in the development of human lung cancer and suggests that CPS1-IT1 may serve as a biomarker for the early diagnosis of lung cancer. Therapeutic molecules targeting lncRNA CPS1-IT1 may be promising for the treatment of lung cancer.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. [DOI] [PubMed] [Google Scholar]
- 3. van Rens MT, Zanen P, Brutel de La Riviere A, Elbers HR, van Swieten HA, van Den Bosch JM. Survival in synchronous vs. single lung cancer: Upstaging better reflects prognosis. Chest 2000;118:952–8. [DOI] [PubMed] [Google Scholar]
- 4. Peng G, Tisch U, Adams O, Hakim M, Shehada N, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A, Haick H. Diagnosing lung cancer in exhaled breath using gold nanoparticles. Nat Nanotechnol. 2009;4:669–73. [DOI] [PubMed] [Google Scholar]
- 5. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014;157:77–94. [DOI] [PubMed] [Google Scholar]
- 6. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 2011;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–23. [DOI] [PubMed] [Google Scholar]
- 8. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. [DOI] [PubMed] [Google Scholar]
- 9. Wang TH, Yu CC, Lin YS, Chen TC, Yeh CT, Liang KH, Shieh TM, Chen CY, Hsueh C. Long noncoding RNA CPS1-IT1 suppresses the metastasis of hepatocellular carcinoma by regulating HIF-1alpha activity and inhibiting epithelial-mesenchymal transition. Oncotarget 2016;7(28):43588–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang TH, Lin YS, Chen Y, Yeh CT, Huang YL, Hsieh TH, Shieh TM, Hsueh C, Chen TC. Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transition. Oncotarget 2015;6:23342–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88:5119–26. [DOI] [PubMed] [Google Scholar]
- 12. Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, Cheunsuchon P, Louis DN, Klibanski A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70:2350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 2013;13:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, Wu JW. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015;36:3355–9. [DOI] [PubMed] [Google Scholar]
- 16. Marcus MW, Raji OY, Field JK. Lung cancer screening: Identifying the high risk cohort. J Thorac Dis. 2015;7(Suppl 2):S156–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget 2014;5:8027–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: A new player in cancer. J Hematol Oncol. 2013;6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene 2008;27:6194–206. [DOI] [PubMed] [Google Scholar]
- 20. Spencer SL, Sorger PK. Measuring and modeling apoptosis in single cells. Cells 2011;144:926–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luscan A, Shackleford G, Masliah-Planchon J, Laurendeau I, Ortonne N, Varin J, Lallemand F, Leroy K, Dumaine V, Hivelin M, Borderie D, De Raedt T, Valeyrie-Allanore L, Larousserie F, Terris B, Lantieri L, Vidaud M, Vidaud D, Wolkenstein P, Parfait B, Bièche I, Massaad C, Pasmant E. The activation of the WNT signaling pathway is a Hallmark in neurofibromatosis type 1 tumorigenesis. Clin Cancer Res. 2014;20:358–71. [DOI] [PubMed] [Google Scholar]
- 22. Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta 2012;1823:624–35. [DOI] [PubMed] [Google Scholar]
- 24. Zhi H, Ning S, Li X, Li Y, Wu W, Li X. A novel reannotation strategy for dissecting DNA methylation patterns of human long intergenic non-coding RNAs in cancers. Nucleic Acids Res. 2014;42:8258–70.1. [DOI] [PMC free article] [PubMed] [Google Scholar]