Abstract
Doxorubicin (DOX) is a commonly used antineoplastic agent for the treatment of various malignancies, and its use is associated with unpredictable cardiotoxicity. Susceptibility to DOX cardiotoxicity is largely patient dependent, suggesting genetic predisposition. We have previously found that individual sensitivity to DOX cardiotoxicity was associated with differential expression of genes implicated in inflammatory response and immune trafficking, which was consistent with the increasing number of reports highlighting the important role of human leukocyte antigen (HLA) complex polymorphism in hypersensitivity to drug toxicity. This pilot study aimed to investigate DNA from patients treated with DOX-based chemotherapy for breast cancer and to correlate the results with the risk for DOX-associated cardiotoxicity. We have identified 18 SNPs in nine genes in the HLA region (NFKBIL1, TNF-α, ATP6V1G2-DDX39B, MSH5, MICA, LTA, BAT1, and NOTCH4) and in the psoriasis susceptibility region of HLA-C as potential candidates for association with DOX cardiotoxicity. These results, albeit preliminary and involving a small number of patients, are consistent with reports showing the presence of susceptibility loci within the HLA gene region for several inflammatory and autoimmune diseases, and with our previous findings indicating that the increased sensitivity to DOX cardiotoxicity was associated with dysregulation of genes implicated both in inflammation and autoimmune disorders.
Key words: Doxorubicin (DOX), Cardiotoxicity, Genotyping, Breast cancer
INTRODUCTION
Doxorubicin (DOX) is a commonly used anthracycline anticancer agent for the treatment of various malignancies, and it may cause unpredictable cardiotoxicity1. The mechanism(s) of DOX cardiotoxicity is still uncertain; however, it is likely a multifactorial event involving diverse processes such as oxidative stress, inhibition of nucleic acid and protein synthesis, release of vasoactive amines, abnormalities in Ca2+ handling, activation of the ubiquitin–proteasome system, and impaired cardiac repair due to inhibition of bone marrow and cardiac progenitor cells2.
It has been well established that DOX cardiotoxicity is a cumulative dose-dependent process that begins with the first dose, suggesting that assessment of the cardiac function in patients prior to chemotherapy may be able to avoid permanent cardiac damage3. According to the American College of Cardiology guidelines, patients receiving chemotherapy are at an increased risk of developing cardiac dysfunction4. Evidence indicates that susceptibility to DOX cardiotoxicity is largely individual, with some patients developing cardiomyopathy at doses of 200–400 mg/m2( 3 ), and others tolerating much higher cumulative doses up to >800–1,000 mg/m2( 5 ), suggesting the presence of a genetic predisposition. Genetic variations in ABCB1, SLC22A16, and CBR1 genes were suggested to contribute to DOX adverse effects6. We have previously found that individual sensitivity to a low dose of DOX-based chemotherapy was associated with differential expression of genes implicated in inflammatory response and immune trafficking7, which was consistent with the increasing number of reports showing the important role of human leukocyte antigen (HLA) complex polymorphism in hypersensitivity to drug toxicity8.
This pilot study focused on obtaining further information about the genetic variations of cancer patients who develop cardiac abnormality after DOX-based chemotherapy and compare them with patients who maintain normal cardiac function.
MATERIALS AND METHODS
Ethical Statements
This study was carried out in accordance with the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the ethics committee of the University of Arkansas for Medical Sciences. All subjects signed an Institutional Review Board-approved informed consent where they were informed about the use of their blood samples and medical records for research purposes.
Patients
Thirty women were treated for breast cancer at the University of Arkansas for Medical Sciences between 2011 and 2015, and they were enrolled in an institutional review board-approved protocol and gave informed consent. The disease stage of all patients is presented in Table 1 under TNM stage. Sixteen patients were diagnosed with stage IIA stage disease, of which six patients had an abnormal decline of LVEF at completion of chemotherapy; eight patients had stage IIB breast cancer, of whom two developed abnormal LVEF decline; five patients had stage IIIA disease, of whom one developed abnormal LVEF decline; and one patient with abnormal LVEF had stage IIIC disease. All patients were treated with a combination of DOX (60 mg/m2) with cyclophosphamide (600 mg/m2) every 3 weeks for four cycles (cumulative dose of DOX 240 mg/m2 and cumulative dose of cyclophosphamide 2,400 mg/m2). Cardiac function of all subjects was assessed by multigated acquisition (MUGA) scan before the start of the DOX-containing chemotherapy and after the completion of DOX-based chemotherapy. A decline of LVEF by >10% or below 55% was considered abnormal.
Table 1.
Patient Demographics, Tumor Characteristics, Comorbid Conditions, and Concomitant Medications
| Characteristic | All Patients | Patients With Abnormal Decline of LVEF |
|---|---|---|
| Patients analyzed | 30 | 10 |
| Race | ||
| European American | 21 | 9 |
| African American | 9 | 1 |
| Age (years) | ||
| Median | 53.1 | 57.6 |
| Range | 35–76 | 47–66 |
| Tumor type: ductal | 30 | 10 |
| Tumor grade | ||
| I | 6 | 2 |
| II | 13 | 6 |
| III | 11 | 2 |
| TNM stage | ||
| IIA | 16 | 6 |
| IIB | 8 | 2 |
| IIIA | 5 | 1 |
| IIIC | 1 | 1 |
| Hormone receptor status | ||
| ER+, PR+/Her2-neu | 20 | 6 |
| ER−, PR− | 9 | |
| ER−/PR+ | 1 | 1 |
| HER2+ | 1 | 1 |
| Triple negative | 9 | 2 |
| Lymph node positive | 20 | 6 |
| Comorbidity and medications | ||
| Hypertension | 9 | 4 |
| Hydrochlorothiazide, 12.5 mg; metoprolol, 25 mg; candesartan, 16 mg; nebivolol, 10 mg; amlodipine, 10 mg; valsartan, 12.5 mg; lisinopril, 10 mg | ||
| Diabetes mellitus | 6 | 3 |
| Metformin, 1,000 mg; insulin glargine, 100 U/ml | ||
| Coronary artery disease | 1 | 1 |
| Atenolol, 25 mg | ||
| Autoimmune diseases | ||
| Asthma | 1 | |
| Albuterol; mometasone |
Methods and Data Analysis
DNA was extracted from peripheral blood using QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA). Samples were quality assessed and quantified by ultraviolet (UV) absorbance measured via the NanoDrop Technologies NanoDrop® ND-2000 Spectrophotometer (Wilmington, DE, USA) and software. DNA quality was evaluated by an allelic discrimination drug-metabolizing enzyme assay (Applied Biosystems Assay; Life Technologies, Carlsbad, CA, USA) with fluorescence detection on the 7900 Fast Real-Time PCR System (Applied Biosystems, Life Technologies) performed according to the manufacturer’s protocol. Genotyping was performed using the Illumina® HumanOmni5-4v1.1 BeadChip. Beadchips were imaged using the Illumina iScan System and analyzed with GenomeStudio V20111 software (Illumina), which has a built-in cnvPartition tool for copy number variants (CNVs). For whole-genome SNP genotyping analysis, individual samples with genotype call rates <93% and SNPs with call rates <95% were removed. The Cochran–Armitage test for trend was conducted with Purcell’s PLINK program (http://pngu.mgh.harvard.edu/purcell/plink). To map the SNPs-to-genes, containing genomic coordinates for all genes according to positions on the Genome Browser, hg18 (NCBI assembly GRCh36) were downloaded from the PLINK ftp server (http://pngu.mgh.harvard.edu/∼purcell/plink/res.shtml#hapmap) accessed in January 2015. SNPs were assigned to a gene if they located within its primary transcript (intragenic region) or 5 kilobases (kb) upstream or downstream of the gene start or end9. The small sample size precluded the ability to detect genome-wide associations; therefore, we used a candidate gene/SNP approach for analysis. We used a 5% level of significance criteria to identify the significant SNPs at first hand. Based on previous data showing significant alterations in the gene expression of genes associated with inflammation and immunity in patients with DOX-associated abnormal decline of LVEF, we manually selected SNPs (p < 0.05) that have been reported in autoimmune and inflammatory diseases.
For CNV analysis, the algorithm parameters in cnvPartition 2.4.4 were set at 25 minimum probes and 35 minimum confidence value threshold for CNV detection, and 250-kb minimal region size for LOH detection. Log R ratios (LRR) and B allele frequencies (BAF) were visualized for every CNV and CN-LOH call on each chromosome via Genome Studio’s chromosome browser. Each copy number event was assigned one of four possible calls based on the BAF, LRR, copy number values, and confidence scores: (1) LOH (BAF ≠ 0.5 and split into two components; LRR < 0; CNV = 1; confidence > 200), (2) homozygous deletion (BAF = SNPs scattered between 0 and 1; LRR < −1; CNV = 0; confidence > 200), (3) duplication (BAF split 0.65/0.35; LRR > 0; CNV > 2; confidence > 200), (4) CN-LOH (BAF split into two components, normally 0.95/0.5; LRR = 0; CNV = 2; confidence > 200; FISH negative). Long stretches of genomic DNA without CNAs have BAF = 0.5, LRR = 0, and CNV = 2. The mapping of chromosomal regions to genes was performed using the gene definitions and coordinates from UCSC Genome Build hg18 (the refFLAT file downloaded from UCSC database).
RESULTS AND DISCUSSION
We have analyzed and compared the genotype distribution of two distinct groups of breast cancer patients treated with DOX-based chemotherapy: 10 patients who developed abnormal LVEF decline at the completion of chemotherapy and 20 patients who did not. Patients’ demographics, tumor characteristics, comorbid conditions, and concomitant medications are presented in Table 1. All 30 patients were diagnosed with invasive ductal carcinoma (IDC). In this patient population, 9 patients were African American (AA) and 21 were European Americans (EA). One of the AA patients and nine of the EA patients presented with abnormal decline of LVEF at completion of chemotherapy. The patients with abnormal decline of LVEF were in the age range of 47–66 years, with mostly stage IIA disease (n = 6) and hormone receptor status mainly ER+/PR+ and Her2-neu+. Nine of the patients had hypertension; of these, three presented with abnormal LVEF decline, three of the patients with abnormal LVEF had diabetes (3:6), one had coronary artery disease (1:1), and one patient with normal LVEF had asthma. Table 2 presents the average changes of LVEF of the patients with abnormal decline of LVEF at completion of chemotherapy and patients who maintained normal LVEF. These results may suggest the possible involvement of hypertension and diabetes in the increased sensitivity to DOX cardiotoxicity, but the small number of patients does not allow definitive conclusions.
Table 2.
Cardiac Function of Women With Breast Cancer, Treated With DOX-Based Chemotherapy
| No. of Patients | Average LVEF (%) at Baseline (Mean ± SD) | Average LVEF (%) at Completion of Chemotherapy (Mean ± SD) | |
|---|---|---|---|
| Patients with abnormal LVEF (%) | 10 | 69.875 ± 5.436 | 57.875 ± 3.833 |
| 18–47 years | 2 | 68.5 ± 2.121 | 57.5 ± 0.707 |
| 48–55 years | 2 | 64.0 ± 0 | 54.0 ± 0 |
| >56 years | 4 | 73.5 ± 5.066 | 60.0 ± 4.242 |
| Patients with normal LVEF (%) | 20 | 61.583 ± 5.523 | 61.833 ± 6.919 |
| 18–47 years | 9 | 60.888 ± 6.050 | 62.222 ± 9.162 |
| 48–55 years | 9 | 60.888 ± 5.065 | 59.777 ± 5.118 |
| >56 years | 7 | 63.142 ± 5.080 | 57.714 ± 6.499 |
MUGA scan was performed before the start of DOX chemotherapy and at its completion. LVEF, left ventricle ejection fraction.
A total of 1,859 autosomal SNPs passed quality thresholds (p < 0.05) for association analysis. Potential candidate genes in chromosome 6p32 and 6p33 were identified (Table 3). These included 15 SNPs in nine genes in the HLA region (NFKBIL1, TNF-α, ATP6V1G2-DDX39B, MSH5, MICA, LTA, BAT1, NOTCH4), and 3 SNPs in the psoriasis susceptibility region of HLA-C (rs9264942, rs2523619, and rs10484554) (Table 1). Six of the SNPs in the BAT1-NFKBIL1-LTA region (rs2071591, rs3093949, rs2071592, rs2239527, rs909253, and rs1041981) have been reported in association with various inflammatory and autoimmune disorders including rheumatoid arthritis (RA)10, myocardial infarction11, and Grave’s disease12. TNF-α rs1800629 polymorphism was reported to be an associated risk for coronary heart disease and myocardial infarction13 and Crohn’s disease14. C6orf10 rs2050190 was associated with coronary artery disease15, MSH5 rs3131379 and rs3131378 in systemic lupus erythematosus (SLE)16, and MICA rs2523451 was reported as a marker for RA17. NOTCH4 rs3134931 was identified in coronary artery diseases18. HLA-C rs9264942, rs2523619, and rs10484554 were associated with psoriasis and psoriatic arthritis19, and rs9264942 was reported in patients with inflammatory bowel disease20. The telomeric class III region of HLA bordering the class I region is particularly gene dense, containing at least 10 genes in addition to TNF within an 82-kb interval: BAT1, ATP6V1G2, NFkBIL1, LTA, TNF, LTB, LST1, NCR3, AIF-1, BAT3, and BAT221. The function of most of these molecules remains poorly characterized, although evidence suggests their role in immune and inflammatory responses does exist for several22. NF-kB, a transcription factor that modulates the transcription of a variety of genes, including cytokines and growth factors, adhesion molecules, immune receptors, and acute phase proteins, is considered an important factor in inflammation and has been implicated in RA, atherosclerosis, asthma, multiple sclerosis (MS), inflammatory bowel disease, and ulcerative colitis23.
Table 3.
SNPs in chr6p Associated With Abnormal Decline of LVEF in Patients Treated With DOX-Based Chemotherapy
| SNP | Gene Symbol | Base Position | Minor Allele | Major Allele | Min | Maj | All | Abnormal LVEF | Normal LVEF | OR | p Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs9264942 | HLA-C | 31274380 | G | A | 0.6 | 0.15 | 4/10/16 | 4/10/16 | 0/6/14 | 8.61 | 0.01 |
| rs2523619 | HLA-C | 31318144 | G | A | 0.55 | 0.15 | 4/9/17 | 4/9/17 | 0/6/14 | 6.56 | 0.01 |
| rs10484554 | HLA-C | 31274555 | A | G | 0.35 | 0.07 | 2/6/22 | 2/6/22 | 0/3/17 | 5.41 | 0.04 |
| rs2071591 | NFKBIL1 | 31515799 | A | G | 0.6 | 0.25 | 5/12/13 | 5/12/13 | 1/8/11 | 6.83 | 0.02 |
| rs3093949 | NFKBIL1 | 31525184 | A | G | 0.6 | 0.22 | 5/11/14 | 5/11/14 | 1/7/12 | 8.87 | 0.01 |
| rs2071592 | NFKBIL1 | 31515340 | A | T | 0.6 | 0.22 | 5/11/14 | 5/11/14 | 1/7/12 | 7.99 | 0.01 |
| rs2071594 | ATP6V1G | 31512720 | C | G | 0.6 | 0.25 | 5/12/13 | 5/12/13 | 1/8/11 | 6.83 | 0.02 |
| rs3130059 | ATP6V1G | 31509284 | G | C | 0.6 | 0.25 | 5/12/13 | 5/12/13 | 1/8/11 | 6.83 | 0.02 |
| rs11796 | ATP6V1G | 31501212 | T | A | 0.6 | 0.27 | 6/11/13 | 6/11/13 | 2/7/11 | 4.12 | 0.03 |
| rs2050190 | C6orf10 | 32339076 | G | A | 0.6 | 0.2 | 6/8/16 | 6/8/16 | 1/6/13 | 3.89 | 0.02 |
| rs1800629 | TNF-α | 31543031 | A | G | 0.35 | 0.07 | 2/6/22 | 2/6/22 | 0/3/17 | 5.67 | 0.03 |
| rs3131379 | MSH5 | 31721033 | A | G | 0.2 | 0.05 | 0/6/24 | 0/6/24 | 0/2/18 | 11.58 | 0.04 |
| rs3131378 | MSH5 | 31725285 | G | A | 0.2 | 0.05 | 0/6/24 | 0/6/24 | 0/2/18 | 11.58 | 0.04 |
| rs2523451 | MICA | 31369151 | A | G | 0.5 | 0.22 | 3/13/14 | 3/13/14 | 0/9/11 | 4.50 | 0.04 |
| rs909253 | LTA | 31540313 | G | A | 0.6 | 0.25 | 5/12/13 | 5/12/13 | 1/8/11 | 6.83 | 0.02 |
| rs1041981 | LTA | 31540784 | A | C | 0.6 | 0.25 | 5/12/13 | 5/12/13 | 1/8/11 | 6.83 | 0.02 |
| rs2239527 | BAT1 | 31509779 | G | C | 0.5 | 0.25 | 4/12/14 | 4/12/14 | 1/8/11 | 4.13 | 0.05 |
| rs3134931 | NOTCH4 | 32190620 | G | A | 0.2 | 0.45 | 3/16/11 | 3/16/11 | 3/12/5 | 0.22 | 0.05 |
Min, minor allele frequency; Maj, major allele frequency; All, distribution of genotypes among all subjects treated with Dox-based chemotherapy (genotypes are ordered as mm/Mm/MM, where “m” and “M” are the minor and major alleles, respectively); abnormal LVEF, distribution of genotypes among subjects who developed abnormal LVEF decline; normal LVEF, distribution of genotypes among subjects who maintained normal LVEF; OR, estimated odds ratio for development of DOX cardiotoxicity (for minor allele; major allele is a reference).
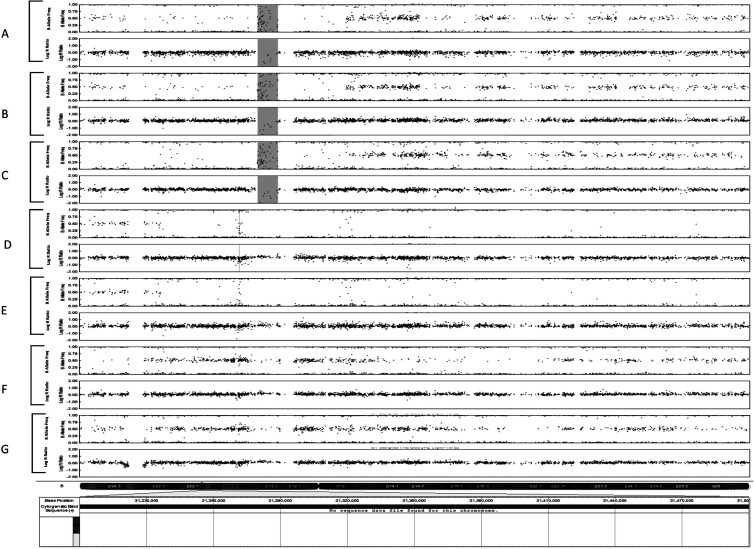
The CNV analysis identified a total of 311 CNVs among the patients in both examined groups, with abnormal and normal LVEF. Most of the CNVs were loss of heterozygosity (LOH) and duplications, without correlation with the heart function. Three of the patients with abnormal decline of LVEF showed LOH of 22 kb in the region of the HLA-DRB5 gene (chr6p.32), and three other patients with abnormal decline of LVEF showed homozygous deletion of 9 kb in chr6p.32 in the area of HLA-B (Fig. 1).
Figure 1.
Homozygous deletion in chr6p.32 in the area of HLA-B.
In conclusion, our data, although preliminary and involving a small number of cancer patients, are consistent with reports that have identified the presence of susceptibility loci within the HLA gene region for several inflammatory and autoimmune diseases, including RA, SLE, MS, and Grave’s diseases24. Autoimmune features and rheumatic manifestations have been reported in cancer patients after chemotherapy25, as well as the association between chemotherapy and development of rheumatism, and SLE has been reported in breast cancer26, although the mechanism of this syndrome was not established. In addition, evidence indicates that systemic inflammatory diseases are associated with increased coronary artery disease morbidity and mortality27. Moreover, our data are in line with our previous findings showing that the increased susceptibility to DOX cardiotoxicity was associated with dysregulation of genes implicated both in inflammation and immunity5. To confirm our finding, a targeted SNPs study is warranted.
ACKNOWLEDGMENTS
This research was supported by grants to V.K.T. from the Arkansas Breast Cancer Research Program and the University of Arkansas for Medical Sciences (1UL1RR029884) and National Institutes of Health/National Institute on Aging (NIH/NIA) Claude Pepper Center (P30AG028718).
REFERENCES
- 1. Gianni L, Herman EH, Lipshultz SE, Minotti G, Sarvazyan N, Sawyer DB. Anthracycline cardiotoxicity: From bench to bedside. J Clin Oncol. 2008;26:3777–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi Y, Moon M, Dawood S, McManus B, Liu PP. Mechanisms and management of doxorubicin cardiotoxicity. Herz 2011;36:296–305. [DOI] [PubMed] [Google Scholar]
- 3. Floyd JD, Nguyen DT, Lobins RL, Bashir Q, Doll DC, Perry MC. Cardiotoxicity of cancer therapy. J Clin Oncol. 2005;23:7685–96. [DOI] [PubMed] [Google Scholar]
- 4. Bonow RO, Bennett S, Casey DE Jr, Ganiats TG, Hlatky MA, Konstam MA, Lambrew CT, Normand SL, Piña IL, Radford MJ, Smith AL, Stevenson LW, Bonow RO, Bennett SJ, Burke G, Eagle KA, Krumholz HM, Lambrew CT, Linderbaum J, Masoudi FA, Normand SL, Ritchie JL, Rumsfeld JS, Spertus JA; American College of Cardiology; American Heart Association Task Force on Performance Measures (Writing Committee to Develop Heart Failure Clinical Performance Measures); Heart Failure Society of America. ACC/AHA Clinical Performance Measures for Adults with Chronic Heart Failure: A report of the American College of Cardiology/American Heart Association Task Force on Performance Measures (Writing Committee to Develop Heart Failure Clinical Performance Measures): Endorsed by the Heart Failure Society of America. Circulation 2005;112:1853–87. [DOI] [PubMed] [Google Scholar]
- 5. VonHoff DD, Layard M, Basa P, Davis HL Jr, Von Hoff AL, Rozencweig M, Muggia FM. Risk factors for doxorubicin-induced congestive heart failure. Ann Intern Med. 1979;91:710–7. [DOI] [PubMed] [Google Scholar]
- 6. Bray J, Sludden J, Griffin MJ, Cole M, Verrill M, Jamieson D, Boddy AV. Influence of pharmacogenetics on response and toxicity in breast cancer patients treated with doxorubicin and cyclophosphamide. Br J Cancer 2010;102:1003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Todorova VK, Makhoul I, Siegel ER, Wei J, Stone A, Carter W, Beggs ML, Owen A, Klimberg VS. Biomarkers for presymptomatic doxorubicin-induced cardiotoxicity in breast cancer patients. PLoS One 2016;11:e0160224. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8. Alfirevic A, Pirmohamed M. Drug induced hypersensitivity and the HLA complex. Pharmaceuticals (Basel) 2011;4:69–74. [Google Scholar]
- 9. Petersen A, Alvarez C, DeClaire S, Tintle NL. Assessing methods for assigning SNPs to genes in gene-based tests of association using common variants. PLoS One 2013;8:e62161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okamoto K, Makino S, Yoshikawa Y, Takaki A, Nagatsuka Y, Takaki A, Nagatsuka Y, Ota M, Tamiya G, Kimura A, Bahram S, Inoko H. Identification of IkBL as the second major histocompatibility complex-linked susceptibility locus for rheumatoid arthritis. Am J Hum Genet. 2003;72:303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koch W, Hoppmann P, Michou E, Jung V, Pfeufer A, Mueller JC, Gieger C, Wichmann HE, Meitinger T, Schömig A, Kastrati A. Association of variants in the BAT1-NFKBIL1-LTA genomic region with protection against myocardial infarction in Europeans. Hum Mol Genet. 2007;16:1821–7. [DOI] [PubMed] [Google Scholar]
- 12. Kurylowicz A, Miśkiewicz P, Bar-Andziak E, Nauman J, Bednarczuk T. Association of polymorphism in genes encoding κB inhibitors (IκB) with susceptibility to and phenotype of Graves’ disease: A case-control study. Thyroid Res. 2009;2:10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chu H, Yang J, Mi S, Bhuyan SS, Li J, Zhong L, Liu S, Tao Z, Li J, Chen H. Tumor necrosis factor-alpha G-308 A polymorphism and risk of coronary heart disease and myocardial infarction: A case-control study and meta-analysis. J Cardiovasc Dis Res. 2012;3:84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mao YQ, Dong SQ, Gao M. Association between TNF-α rs1799724 and rs1800629 polymorphisms and the risk of Crohn’s disease. Gen Mol Res. 2015;14:15811–8. [DOI] [PubMed] [Google Scholar]
- 15. Tang W, Schwienbacher C, Lopez LM, Ben-Shlomo Y, Oudot-Mellakh T, Johnson AD, Samani NJ, Basu S, Gögele M, Davies G, Lowe GD, Tregouet DA, Tan A, Pankow JS, Tenesa A, Levy D, Volpato CB, Rumley A, Gow AJ, Minelli C, Yarnell JW, Porteous DJ, Starr JM, Gallacher J, Boerwinkle E, Visscher PM, Pramstaller PP, Cushman M, Emilsson V, Plump AS, Matijevic N, Morange PE, Deary IJ, Hicks AA, Folsom AR. Genetic associations for activated partial thromboplastin time and prothrombin time, their gene expression profiles, and risk of coronary artery disease. Am J Hum Genet. 2012;91:152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN)1, Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Snir O, Gomez-Cabrero D, Montes A, Perez-Pampin E, Gómez-Reino JJ, Seddighzadeh M, Klich KU, Israelsson L, Ding B, Catrina AI, Holmdahl R, Alfredsson L, Klareskog L, Tegnér J, Gonzalez A, Malmström V, Padyukov L. Non-HLA genes PTPN22, CDK6 and PADI4 are associated with specific autoantibodies in HLA-defined subgroups of rheumatoid arthritis. Arthritis Res Ther. 2014;16:414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reiner AP, Hartiala J, Zeller T, Bis JC, Dupuis J, Fornage M, Baumert J, Kleber ME, Baldus S, Bielinski SJ, Fontes JD, Illig T, Keating BJ, Lange LA, Ojeda F, Müller-Nurasyid M, Munzel TF, Psaty BM, Rice K, Rotter JI, Schnabel RB, Tang WH, Thorand B, Erdmann J, CARDIoGRAM Consortium, Jacobs DR Jr, Wilson JG, Koenig W, Tracy RP, Blankenberg S, März W, Gross MD, Benjamin EJ, Hazen SL, Allayee H. Genome-wide and gene-centric analyses of circulating myeloperoxidase levels in the charge and care consortia. Hum Mol Genet. 2013;22:3381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Y, Helms C, Liao W, Zaba LC, Duan S, Gardner J, Wise C, Miner A, Malloy MJ, Pullinger CR, Kane JP, Saccone S, Worthington J, Bruce I, Kwok PY, Menter A, Krueger J, Barton A, Saccone NL, Bowcock AM. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4:e1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D’Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H; International IBD Genetics Consortium (IIBDGC), Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neville MJ, Campbell RD. A new member of the Ig superfamily and a V-ATPase G subunit are among the predicted products of novel genes close to the TNF locus in the human MHC. J Immunol. 199;162:4745–9. [PubMed] [Google Scholar]
- 22. Greetham D, Ellis CD, Mewar D, Fearon U, Ultaigh SN, Veale DJ, Guesdon F, Wilson AG. Functional characterization of NF-kappaB inhibitor-like protein 1 (NFkappaBIL1), a candidate susceptibility gene for rheumatoid arthritis. Hum Mol Genet. 2007;16:3027–31. [DOI] [PubMed] [Google Scholar]
- 23. Tak PP, Firestein GS. NF-κB: A key role in inflammatory diseases. J Clin Invest. 2001;107:7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fernando MMA, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, Vyse TJ, Rioux JD. Defining the role of the MHC in autoimmunity: A review and pooled analysis. PLoS Genet. 2008;4:e1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abu-Shakra M, Buskila D, Ehrenfeld M, Conrad K, Shoenfeld Y. Cancer and autoimmunity: Autoimmune and rheumatic features in patients with malignancies. Ann Rheum Dis. 2001;60:433–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Michl I, Zielinski CC. More postchemotherapy rheumatism. J Clin Oncol. 1993;11:2051. [DOI] [PubMed] [Google Scholar]
- 27. Kriszbacher I, Koppan M, Bodis J. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;429:353–7. [PubMed] [Google Scholar]