Abstract
Emerging evidence suggests that the long noncoding RNA (lncRNA) plasmacytoma variant translocation 1 (PVT1) gene is involved in the pathogenesis of cervical cancer. However, the potential mechanism is rarely reported. Our study found that PVT1 was upregulated in cervical cancer tissue and cell lines. After transfecting PVT1 siRNA, the proliferation, migration, and invasion of cervical cancer cells were markedly decreased. miRNA expression profiles demonstrate that miR-424 was markedly downregulated in cervical cancer tissue. Bioinformatics analysis revealed that miR-424 was potentially targeted by PVT1, which was confirmed by dual-luciferase reporter assay. Pearson’s correlation analysis showed that PVT1 expression was negatively related to miR-424 expression in glioma cancer tissues. Finally, lowered expression of miR-424 could recover the tumor-suppressive effects of PVT1 knockdown in cervical cancer cell lines. Our results reveal a tumor-promoting role for PVT1, acting as a competing endogenous RNA (ceRNA) or a molecular sponge in negatively modulating miR-424, which might provide a novel therapeutic target for cervical cancer.
Key words: Plasmacytoma variant translocation 1 (PVT1), miR-424, Cervical cancer, Competing endogenous RNA (ceRNA)
INTRODUCTION
In developing countries, cervical cancer has the second highest morbidity and mortality rates of all gynecological malignancies. It is a leading cause of cancer-related deaths among women and seriously affects women’s health1. A series of research shows that the occurrence of cervical cancer is closely related to the persistent infection of high-risk human papilloma virus (HPV)2,3. Like in other tumors, the occurrence and development of cervical cancer involve a complex network system, including multiple gene expression regulation abnormalities, multiple signaling pathways, and abnormal activation or inhibition, which cause abnormal cell proliferation differentiation, precancerous lesions, tumor invasion, and metastasis of malignant change4,5.
With next-generation sequencing technology and the development of microarray technology based on the genome, more and more studies have focused on the function of long noncoding RNAs (lncRNAs) in cervical cancer6. lncRNAs are at least 200 bases in length and are involved in numerous physiological and pathological processes. Presently, only a small number of lncRNAs have been characterized in detail. Biological functions of lncRNAs include chromosome silencing, genomic imprinting, dosage compensation effect, protein activity regulation, and RNA alternative splicing7,8. Several types of lncRNAs have been tested to participate in the tumorigenesis of cervical cancer, including H19, TUG1, HOTAIR, and CCHE19. Plasmacytoma variant translocation 1 (PVT1) gene has been proven to play an oncogenic role in cervical cancer; however, the underlying molecular mechanisms remain unclear10,11.
In our previous research, we used a miRNA microarray to verify that miR-424 is downregulated in cervical cancer cell lines. Dysregulated expression of miR-424 has been identified in cervical cancers, and our study showed that miR-424 significantly increased radiation-induced DNA damage, cell apoptosis, and G2/M cell cycle arrest in radioresistant cervical cancer cells12. In addition, miR-424-5p may act as a novel antioncogene in cervical cancer by blocking cell growth through targeting the KDM5B–Notch pathway13. Generally, miRNAs could play a role in protein by combining with the 3′-untranslated region (3′-UTR) of target genes’ mRNA.
The effect and interaction of lncRNA PVT1 and miR-424 on tumor metastasis in cervical cancer are unknown. Here we investigate and describe the functional role of PVT1 and miR-424 and explore this novel therapeutic target for cervical cancer.
MATERIALS AND METHODS
Cell Lines and Tissues Samples
Human cervical cancer cell lines HeLa and SiHa, and one nonmalignant human cervical epithelial cell line (Ect1/E6E7) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS) at 37°C in an atmosphere of 95% air and 5% CO2. Cells were transfected with the indicated nucleotides or plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
This study was approved by the Research Ethics Review Committee of the Cangzhou Central Hospital. Patients were informed of the aims of this study, and informed consent was obtained from all patients. Human cervical cancer tissues and adjacent noncancerous tissues were surgically collected from patients. The tissue samples were immediately snap frozen in liquid nitrogen and stored at −80°C.
Cell Transfection
All miRNA mimics and siRNAs were synthesized by GenePharma (Shanghai, P.R. China), including miR-424 mimic, miR-424 inhibitor, PVT1 siRNA, and the corresponding negative controls. The sequences of two siRNAs targeting PVT1 were listed as follows: si-PVT1-1: 5′-CACGCACAGACAUGAACAAUU-3′, si-PVT1-2: 5′-GGGUACUGGAAGUAGAAUAUU-3′. The sequence of the miR-424 inhibitor was listed as follows: 5′-AGCGGACGUUUGAGGGCCAGUU-3′. Oligonucleotide transfection was conducted using the Lipofectamine 2000 transfection reagent according to the manufacturer’s instructions.
Real-Time Quantitative Polymerase Chain Reaction (PCR)
RNA was extracted from samples and cell lines using TRIzol reagent (Invitrogen). Total RNA (2 μg) was extracted to synthesize first-strand cDNA reverse transcription using a reverse transcription reagent kit (Invitrogen) according to the manufacturer’s instructions. Real-time quantitative PCR (RT-qPCR) was performed using SYBR Green PCR Kit (TaKaRa, Dalian, P.R. China). All primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, P.R. China). Amplification conditions comprised an initial denaturation (95°C for 3 min), 30 cycles (denaturation at 95°C for 15 s, annealing at 60°C for 50 s, and elongation at 72°C for 55 s), and the final elongation (72°C for 3 min). β-Catenin acted as the internal standard used for all quantifications. The relative gene expression results were expressed as a fold change compared with the internal standard with the 2−ΔΔCT method.
Luciferase Assays
The sequences of PVT1 were predicted to contain target complementary-binding sequences with the 3′-UTR of miR-424. The sequences containing PVT1 3′-UTR, including wild-type (WT) or mutant (Mut) fragments, were amplified and cloned downstream of the pMIR-Report luciferase vector (Ambion, Cambridge, MA, USA) in order to construct the luciferase reporter vector. Cell lines (HeLa and SiHa) were transfected with luciferase reporter plasmids with Lipofectamine 2000 according to the manufacturer’s instructions. Forty-eight hours after transfection, luciferase activity was assessed with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Cell Proliferation and Viability Assay
Cells were transferred into 96-well plates (4,000 cells/well with the indicated nucleotides transfected). Forty-eight hours after transfection, cell viability was determined using a CCK-8 kit (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s protocol.
Matrigel Invasion Assay
Matrigel invasion assay was performed by BD Biocoat Matrigel Invasion Chamber (BD Biosciences, Bedford, MA, USA) with 24-well Transwell chambers and a polycarbonate membrane with 8-μm pore size according to the manufacturer’s protocol. Cells (1 × 105 cells/well) were seeded into the Matrigel-coated upper chamber, and serum-free medium (1% FBS) was added to the lower chamber. After incubation for 24 h, nonmigrating cells were removed and then fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 20 min. The stained cells were quantified by counting in five random areas under an inverted microscope.
Statistical Analysis
The results were expressed as mean ± standard deviation (SD) values. The Student’s t-test and one-way ANOVA were used to evaluate the difference. A value of p < 0.05 was considered to indicate a statistically significant result.
RESULTS
PVT1 Was Upregulated in Cervical Cancer Tissues and Cell Lines
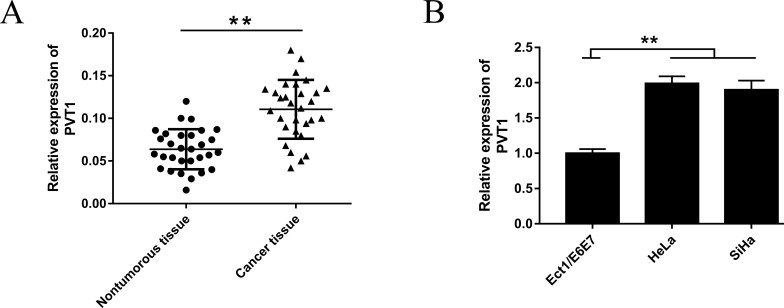
In the first step, we detected the PVT1 expression in cervical cancer tissues from patients and cell lines by real-time PCR. RT-qPCR analysis showed that PVT1 expression was distinctly increased in cervical cancer tissues compared to noncancerous normal tissues (Fig. 1A). Afterward, PVT1 expression in cell lines (HeLa and SiHa) demonstrated a similar overexpression (Fig. 1B). These results revealed that lncRNA PVT1 was significantly upregulated in cervical cancer, which suggests a potential oncogenicity.
Figure 1.
Plasmacytoma variant translocation 1 (PVT1) was upregulated in cervical carcinoma tissues and cell lines. (A) Expression of PVT1 was significantly higher in cervical cancer tissues than in nontumorous tissues. (B) Expression of PVT1 was consistently upregulated in the cervical cell lines HeLa and SiHa. Data are presented as mean ± standard deviation (SD). **p < 0.01, calculated using Student’s t-test.
Knockdown of PVT1 Played a Growth-Inhibiting Role in Cervical Carcinoma Cells
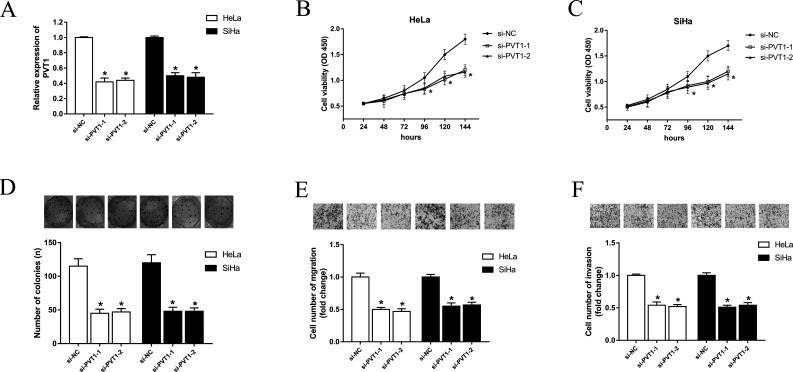
On the basis of the previous study, we found that lncRNA PVT1 was upregulated in cervical cancer tissues and cell lines. We then transfected PVT1 siRNA into cervical cancer cell lines (Fig. 2A) to examine the function of PVT1 in cervical cancer progression. Cell viability assay showed that PVT1 knockdown observably suppressed proliferation over time (Fig. 2B and C). The colony formation assay showed that the number of cell colonies was significantly decreased with siRNA-transfected PVT1 (Fig. 2D). PVT1 knockdown also markedly decreased migration and invasion abilities, as detected by the Transwell assay (Fig. 2E and F). The above results reveal that PVT1 knockdown inhibits proliferation, migration, and invasion in cervical cancer cells.
Figure 2.
Knockdown of PVT1 suppressed proliferation, migration, and invasion of cervical cancer cells. (A) Silencing of PVT1 with siRNAs in HeLa and SiHa cell lines. (B, C) Cell viability was determined at the indicated time point after seeding into 96-well plates using the CCK-8 assay. (D) Representative image of colony formation assay stained with 2% crystal violet. (E, F) Representative images of migration and invasion assays performed after transfection with siRNA, and randomly selected fields are shown. Data are presented as mean ± SD. *p < 0.05, calculated using Student’s t-test.
PVT1 Regulated miR-424 by Direct Targeting
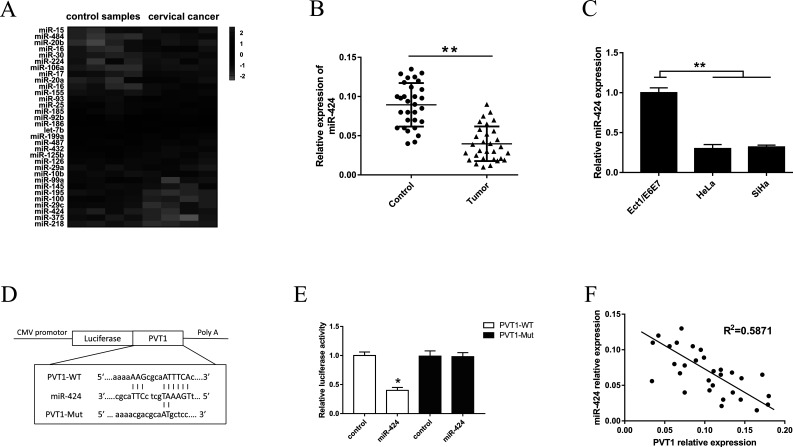
In our previous study, we screened the miRNA expression profiles in cervical cancer tissue compared with normal control tissue. Upregulated and downregulated expressions in miRNAs were demonstrated (Fig. 3A). Among the miRNAs with downregulated expression, we focused on the miR-424 that was observably downregulated in cervical cancer tissue and where the potential pathogenesis was unclear. miR-424 expression was consistently decreased in cancer tissue and cell lines (Fig. 3B and C). Putative complementary regions between miR-424 and PVT1 3′-UTR were demonstrated (Fig. 3D). Afterward, the alignment of complementary binding of miR-424 and PVT1 3′-UTR was verified by dual-luciferase reporter assay (Fig. 3E). Pearson’s correlation showed that PVT1 was negatively correlated to miR-424 expression in human cervical cancer tissues (Fig. 3F).
Figure 3.
Validation of PVT1 as a direct target of miR-424. (A) miRNA expression profiles detected with miRNA microarray in cervical cancer tissue and normal control tissue. (B) miR-424 relative expression was assessed by real-time quantitative PCR (RT-qPCR) in cervical cancer tissues from 30 patients (Tumor) compared to normal tissues (Control). (C) Similarly, the relative expression of miR-424 was assessed by RT-qPCR in cervical cancer cell lines (HeLa and SiHa). (D) A putative complementary sequence alignment between miR-424 and PVT1 3′-untranslated region (3′-UTR). (E) The wild-type (WT)/mutant (Mut)-PVT1 vectors and miR-424 NC/mimics were cotransfected into HeLa cells. Dual-luciferase reporter assay showed the probability of binding of miR-424 with the 3′-UTR of PVT1 in HeLa cell lines. (F) Pearson’s correlation showed the correlations between PVT1 and miR-424 expression in cervical cancer tissues (R 2 = 0.5871). All data are expressed as mean ± SD. *p < 0.05, **p < 0.01.
In conclusion, the above results revealed that miR-424 was downregulated in cervical cancer tissues compared to PVT1.
Effect of PVT1 and miR-424 on Cervical Cancer Cell Lines
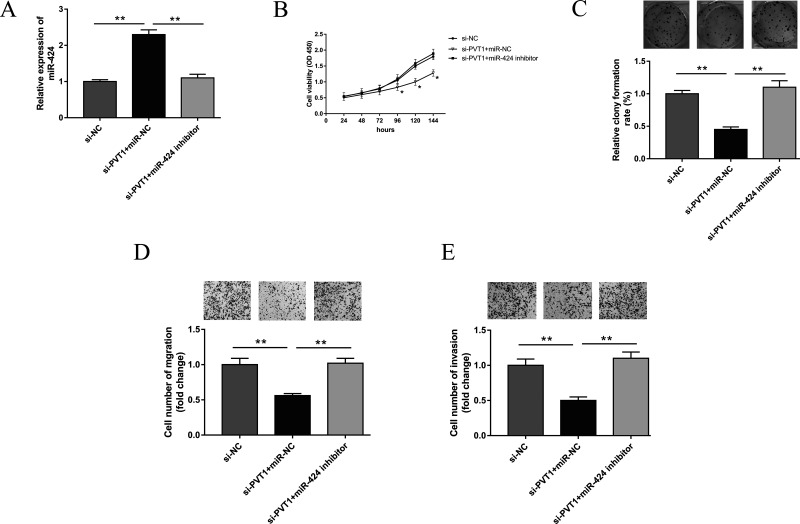
A previous study has shown that PVT1 was upregulated in cervical cancer tissues and cell lines. Nevertheless, miR-424 was distinctly downregulated. In addition, knockdown of PVT1 played a growth-inhibiting role in cervical carcinoma cells. Subsequently, we continued to verify the tumorigenetic effect of PVT1 and miR-424 on cervical cancer cell lines. Anti-miR-424 (miR-424 inhibitors), anti-miR-NC, and si-PVT1 were cotransfected. The cells cotransfected with anti-miR-424 could rescue the inhibiting effect of si-PVT1 on proliferation, colony formation, migration, and invasion (Fig. 4). The above results indicate that lowered expression of miR-424 could recover the tumor-suppressive effects of PVT1 knockdown in cervical cancer cell lines.
Figure 4.
PVT1 knockdown and miR-424 inhibitor mediated cervical cancer progression in HeLa cell. (A) Expression of miR-424 in HeLa cells respectively cotransfected with miR-424 inhibitor and si-PVT1. (B) Cell viability of HeLa cells detected by CCK-8 assay. (C) Cell colony formation. (D, E) Transwell assays were performed to evaluate potential migration and invasion. All data are expressed as mean ± SD. **p < 0.01.
DISCUSSION
Emerging studies have revealed that lncRNAs are important regulators in cell biology14, and increasing evidence has revealed the internal relation of aberrant expression and tumorigenesis15,16. lncRNA PVT1, downstream of MYC, has been indicated to frequent coamplification with MYC in a series of tumors17. It is illuminated that PVT1 is overexpressed in multiple types of tumors and plays a tumor-promoting role.
In our study, we detected the expression of PVT1 in cervical cancer tissues and cell lines using real-time PCR. PVT1 is significantly upregulated in tumor tissue and cell lines, which is quite fitting with the existing literature (Fig. 1). We then assessed the regulation of PVT1 on the tumorigenesis of cervical cancer. Results revealed that PVT1 knockdown markedly decreased the proliferation, migration, and invasion abilities of cervical cancer cells (Fig. 2). Research has indicated that non-small cell lung cancer patients with a higher expression of PVT1 showed poorer overall survival than those with lower PVT1 expression18. Subsequent studies demonstrated that PVT1 is increased in asthmatic patients and is associated with the aberrant phenotype19. Therefore, PVT1 expression could be identified as an independent prognostic marker of overall survival in a multivariate analysis.
Our previous study screened the miRNA expression profiles in cervical cancer tissue compared with normal control tissue. miR-424 is observably downregulated in cervical cancer tissue and cell lines (Fig. 3). Xu et al. reported that miR-424 might be a candidate for prognostic predictor and plays a crucial role of tumor suppression in the progression of cervical cancer via upregulating the expression of Chk1 and p-Chk120. Zhou et al. reported that miR-424-5p blocks cell growth via inhibiting the KDM5B–Notch pathway, which could be a novel anti-oncogene in cervical cancer13. Bioinformatics analysis predicts the potential binding domains within miR-424 and PVT1 3′-UTR, which was verified by dual-luciferase reporter assay. It could be described that miR-424 inhibits PVT1 expression and reduces 3′-UTR luciferase activity; thus, PVT1 is identified as a direct target of miR-424. In addition, Pearson’s correlation showed that PVT1 was negatively correlated to miR-424 expression in human cervical cancer tissues. Because the dual-luciferase reporter assay showed the opposite expression of PVT1 and miR-424, we assume that an antagonistic relationship exists within PVT1 and miR-424.
To investigate the interaction of PVT1 and miR-424 on tumor-suppressive effects, oligonucleotide interference sequences were cotransfected into cervical cancer lines. The cells cotransfected with anti-miR-424 could rescue the inhibiting effect of si-PVT1 on proliferation, colony formation, migration, and invasion (Fig. 4). In gastric cancer, PVT1 could directly interact with miR-186, and this interaction leads to the suppression of downstream HIF-1α expression21. Zhang et al. reported that PVT1 promotes cervical cancer cell proliferation, cell cycle progression, and migration through silencing miR-200b11. Our study first illustrates the interactive relationship between PVT1 and miR-424 in the progression of cervical cancer tumorigenesis. The results manifest an obvious intrinsic regulation pathway, which provides remarkable therapeutic significance.
In conclusion, lncRNA PVT1 and miR-424 play a key role in cervical cancer progression. Our study reveals that lncRNA PVT1 acts as a sponge or ceRNA for miR-424, and lncRNA PVT1 could be an important molecular marker and a significant therapeutic target for cervical cancer.
ACKNOWLEDGMENTS
This work was supported by the Department of Gynecology and Obstetrics, Cangzhou Central Hospital. The authors want to thank Hebei Medical University for its technical help and laboratory equipment support.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2. Grat K, Grat M, Wronka KM, Pietrzak B, Suchonska B, Walter de Walthoffen S, Mlynarczyk G, Krawczyk M, Wielgos M. Cervical human papilloma virus infection in the early postoperative period after liver transplantation: Prevalence, risk factors, and concordance with anal infections. Clin Transplant. 2017;31(3). [DOI] [PubMed] [Google Scholar]
- 3. Teymouri M, Pirro M, Johnston TP, Sahebkar A. Curcumin as a multifaceted compound against human papilloma virus infection and cervical cancers: A review of chemistry, cellular, molecular, and preclinical features. Biofactors 2017;43(3):331–46. [DOI] [PubMed] [Google Scholar]
- 4. Peralta-Zaragoza O, Deas J, Meneses-Acosta A, De la OGF, Fernandez-Tilapa G, Gomez-Ceron C, Benitez-Boijseauneau O, Burguete-Garcia A, Torres-Poveda K, Bermudez-Morales VH, Madrid-Marina V, Rodríguez-Dorantes M, Hidalgo-Miranda A, Pérez-Plasencia C. Relevance of miR-21 in regulation of tumor suppressor gene PTEN in human cervical cancer cells. BMC Cancer 2016;16:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao X, Li X, Ren Q, Tian J, Chen J. Calycosin induces apoptosis in colorectal cancer cells, through modulating the ERbeta/MiR-95 and IGF-1R, PI3K/Akt signaling pathways. Gene 2016;591(1):123–8. [DOI] [PubMed] [Google Scholar]
- 6. Kobayashi R, Miyagawa R, Yamashita H, Morikawa T, Okuma K, Fukayama M, Ohtomo K, Nakagawa K. Increased expression of long non-coding RNA XIST predicts favorable prognosis of cervical squamous cell carcinoma subsequent to definitive chemoradiation therapy. Oncol Lett. 2016;12(5):3066–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015;521(7551):232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Richard JL, Ogawa Y. Understanding the complex circuitry of lncRNAs at the X-inactivation Center and its implications in disease conditions. Curr Top Microbiol Immunol. 2016;394:1–27. [DOI] [PubMed] [Google Scholar]
- 9. Xia S, Wang C, Ni X, Ni Z, Dong Y, Zhan W. NONHSAT076754 aids ultrasonography in predicting lymph node metastasis and promotes migration and invasion of papillary thyroid cancer cells. Oncotarget 2017;8(2):2293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 contributes to the cervical cancer phenotype and associates with poor patient prognosis. PLoS One 2016;11(5):e0156274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang S, Zhang G, Liu J. Long noncoding RNA PVT1 promotes cervical cancer progression through epigenetically silencing miR-200b. APMIS 2016;124(8):649–58. [DOI] [PubMed] [Google Scholar]
- 12. Wang X, Li Q, Jin H, Zou H, Xia W, Dai N, Dai XY, Wang D, Xu CX, Qing Y. miR–424 acts as a tumor radiosensitizer by targeting aprataxin in cervical cancer. Oncotarget 2016;7(47):77508–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou Y, An Q, Guo RX, Qiao YH, Li LX, Zhang XY, Zhao XL. MiR424-5p functions as an anti-oncogene in cervical cancer cell growth by targeting KDM5B via the Notch signaling pathway. Life Sci. 2017;171:9–15. [DOI] [PubMed] [Google Scholar]
- 14. Degirmenci U, Lei S. Role of lncRNAs in cellular aging. Front Endocrinol. (Lausanne) 2016;7:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He R, Hu Z, Wang Q, Luo W, Li J, Duan L, Zhu YS, Luo DX. The role of long non-coding RNAs in nasopharyngeal carcinoma: A systemic review. Oncotarget 2017;8(9):16075–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pan Y, Li C, Chen J, Zhang K, Chu X, Wang R, Chen L. The emerging roles of long noncoding RNA ROR (lincRNA-ROR) and its possible mechanisms in human cancers. Cell Physiol Biochem. 2016;40(1–2):219–29. [DOI] [PubMed] [Google Scholar]
- 17. Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 2016;7(8):8601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7(10):6929–35. [PMC free article] [PubMed] [Google Scholar]
- 19. Austin PJ, Tsitsiou E, Boardman C, Jones SW, Lindsay MA, Adcock IM, Chung KF, Perry MM. Transcriptional profiling identifies the long noncoding RNA plasmacytoma variant translocation (PVT1) as a novel regulator of the asthmatic phenotype in human airway smooth muscle. J Allergy Clin Immunol. 2017;139(3):780–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, Xie X, Zhou C, Lu W. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene 2013;32(8):976–87. [DOI] [PubMed] [Google Scholar]
- 21. Huang T, Liu HW, Chen JQ, Wang SH, Hao LQ, Liu M, Wang B. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomed Pharmacother. 2017;88:302–8. [DOI] [PubMed] [Google Scholar]