Abstract
Hypopharyngeal cancer (HPC) frequently presents at an advanced stage and displays early submucosal spread, resulting in a poor prognosis. It is among the worst of all cancers in the head and neck subsites. Therefore, detection of HPC at an earlier stage would be beneficial to patients. In this study, we used differential in-gel electrophoresis (DIGE) and two-dimensional polyacrylamide gel electrophoresis (2-DE) proteomics analysis to identify the potential biomarkers for HPC. Among the differential proteins identified, calcium-binding protein S100A9 was overexpressed in HPC tissues compared with normal adjacent tissues, and S100A9 expression in metastatic tissues and advanced tumor tissues was higher than in nonmetastatic tissues and early tumor tissues. S100A9 expression was further confirmed in a large additional cohort. Our data showed that a higher S100A9 level was associated with a poor prognosis for HPC patients, and this may be an independent factor for predicting their prognosis. In addition, S100A9 protein expression was upregulated in human HPC cell lines compared with normal oral cavity epithelia. Knockdown of S100A9 induced significant inhibition of cell growth and their invasive ability. Mechanically, we found that downregulation of S100A9 significantly reduced the expression of NF-κB, phosphorylation of NF-κB and Bcl-2, as well as the expression of MMP7 and MMP2. Restoration of NF-κB expression sufficiently reversed the inhibitory effects on cell proliferation and invasion induced by S100A9 downregulation in vitro and in vivo. In conclusion, for the first time, we have identified S100A9 as an independent prognostic factor for HPC. Inhibiting S100A9 expression would be a potential novel diagnostic biomarker and therapeutic target for HPC treatment.
Key words: Hypopharyngeal cancer (HPC), Proteomics analysis, S100A9, Biomarker, NF-κB
INTRODUCTION
Hypopharyngeal cancer (HPC) mainly originates in the pyriform sinus, followed by sites not otherwise specified, and then the posterior hypopharyngeal wall1. These tumors frequently present at an advanced stage and display early submucosal spread, resulting in a poor prognosis and are among the worst in all head and neck subsites2. Surgery is difficult due to the multifocal nature of the disease and its early lymphatic spread. Although combining surgical therapy with chemoradiotherapy has improved the survival for patients with HPC over the past three decades, the mortality rate for patients with a late stage diagnosis of HPC is unsatisfactory3. Therefore, detection of HPC at an earlier stage would be beneficial to patients. Many molecules, such as p16, Bcl-23, microRNAs4, and cyclin D15, have been evaluated as candidate biomarkers for HPC, but none has been widely used in practice because each belongs to signaling pathways that have either multiple known or unknown proteins. Therefore, more effective biomarkers for the early diagnosis of HPC are necessary. In addition, a better understanding of the molecular mechanisms involved in HPC development, progression, and treatment response is also necessary.
To date, proteomics methods such as matrix-assisted laser desorption ionization-time of flight/time of flight mass spectrometry (MALDI-TOF/TOF MS), differential in-gel electrophoresis (DIGE), and two-dimensional polyacrylamide gel electrophoresis (2-DE) on cancer tissues have been used to search for biomarkers in numerous cancers, including HPC, followed by immortalized cell lines and malignantly transformed counterparts6–8. Tian et al. used 2D-DIGE to identify α-2-HS-glycoprotein as a plasma biomarker of hypopharyngeal squamous cell carcinoma9. Two-dimensional liquid chromatography–tandem mass spectrometry revealed that interactions among Sp1, c-Myc, and p53 may play a vital role in the carcinogenesis and metastasis of hypopharyngeal squamous cell carcinoma10.
In this study, we used 2D-DIGE proteomics analysis to identify potential biomarkers for HPC. Among the differential proteins identified, calcium-binding protein S100A9 was overexpressed in HPC tissues compared with normal adjacent tissues, and S100A9 expression in metastatic tissues and advanced tumor tissues was higher than in nonmetastatic tissues and early tumor tissues. We then validated the expression of S100A9 in additional cohorts. Furthermore, we investigated the role of S100A9 in HPC development and the underlying mechanisms in vitro and in vivo.
MATERIALS AND METHODS
Ethics Approval
The present study was approved by the ethics committee of Xiangya Hospital of Central South University. All participants involved in this study provided written informed consent.
Study Population
A total of 189 cases of HPC tissue samples and 88 adjacent tissues were collected after surgery at the Department of Otorhinolaryngology Head and Neck Surgery. All tissues were formalin fixed, paraffin embedded, and stored at 4°C before usage. An additional five fresh HPC tissue samples and their matched adjacent tissues were immediately frozen at −150°C and used for proteomics analysis. The population of HPC patients included 103 males and 86 females ranging in age from 36 to 85 years. The clinical characteristics of these patients were collected from medical records. Before sample collection, none of the participants received any therapy.
Proteomics Analysis
Total proteins were extracted and purified from fresh tissues using a ReadyPrep™ Protein Extraction kit (GE Healthcare Bio-science, Fairfield, CT, USA) following the procedure recommended by the manufacturer. A 2-D Quant Kit (GE Healthcare Bio-science) was used to determine the concentrations of total proteins. Proteomics analysis was performed as previously described11. Protein samples were separated by 2-DE. The 20-cm IPG strips (pH 3–10) were loaded with samples and subjected to rehydration overnight. Samples containing 150 μg of protein for analytical gels and unlabeled samples containing 1 mg of protein were diluted to 450 μl with a rehydration solution and used for isoelectric focusing (IEF). After SDS-PAGE, each gel was stained with Coomassie brilliant blue dye and scanned with UMAXpowerlook 1120 (UMAX, Taipei, Taiwan). The DeCyder software version 6.5 (GE) was used to spot detect and determine quantity, intergel matching, and statistics. The statistical significance was assessed for each change in abundance using a one-way ANOVA. We selected protein spots for which the mean ratio was greater than 1.5-fold or less than −1.5-fold.
Immunohistochemical Staining Assay
The expression of S100A9, Ki-67, and MMP7 was evaluated using immunohistochemical staining. Briefly, the tissue sections cut at 4 μm were deparaffinized and hydrated, and then retrieved with citrate buffer in boiling water for 15 min. The sections were then incubated with primary antibodies (mouse monoclonal anti-S100A9, 1:200; mouse monoclonal anti-Ki-67, 1:500; rabbit polyclonal anti-MMP7, 1:500; Sigma-Aldrich, St. Louis, MO, USA) overnight at 4°C. The sections were then incubated with secondary antibody for 60 min at 37°C. Signaling was visualized using substrate diaminobenzidine (DAB) and counterstained with hematoxylin.
For evaluating the expression of S100A9 in tissues, the integral optical density (IOD) was obtained by ImageJ (National Institutes of Health, Bethesda, MD, USA). To compare the expression of S100A9 between adjacent and tumor tissues, the IOD of the S100A9 expression was normalized to the average score in normal tissues.
Cell Culture and Treatment
Normal epithelial cultures derived from oral cavity epithelia (HOK1) were obtained from ScienCell Research Laboratories (San Diego, CA, USA). Four human HPC cell lines, including Fadu, Tu212, Tu686, and Hep-2, were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were grown routinely in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and cultured in a 37°C humidified atmosphere of 5% CO2. Knockdown of S100A9 in Fadu cells was achieved by transfection with lentivirus containing S100A9 shRNA (shRNA-S100A9; GenePharma, Shanghai, P.R. China) using Lipofectamine 2000 (Invitrogen). Overexpression of NF-κB (p65) was achieved using a lentivirus containing NF-κB-expressed plasmid (GeneCopoeia, Guangzhou, P.R. China) using Lipofectamine 2000 (Invitrogen). Cells transfected with empty lentivirus were used as the control. Cells were plated into 6-well clusters or 96-well plates and transfected for 24 or 48 h. Transfected cells were used in further assays or protein extraction.
CCK-8 Cell Proliferation Assay
Cell proliferation rates were measured using a cell counting kit-8 (CCK-8; Beyotime, Hangzhou, P.R. China). Cells were seeded into each 96-well plate at a density of 0.5 × 104 for 24 h, transfected with the indicated plasmids, and further incubated for 24, 48, and 72 h, respectively. Ten microliters of the CCK-8 reagent was added to each well 1 h before the endpoint of incubation. OD 490 nm value in each well was determined by a microplate reader.
Cell Invasion Assay
The invasive and migratory potential of cells was evaluated using Transwell inserts with 8-μm pores (Corning, Corning NY, USA). For the invasion assay, 24 h after transfection, 3.0 × 105 cells in serum-free medium were added to each upper insert precoated with Matrigel matrix (BD Biosciences, Franklin Lakes, NJ, USA). Five hundred microliters of 10% FBS medium was added to the matched lower chamber. After 48 h of incubation, cells that did not invade were removed from the upper surface of the Transwell membrane with a cotton swab, and cells on the lower membrane surface were fixed in methanol, stained with 0.1% crystal violet, and photographed. The staining was then dissolved by 5% acetic acid, and the OD 490 nm value in each well was determined by a microplate reader.
Western Blot Analysis
Cultured or transfected cells were lysed in RIPA buffer with 1% PMSF. Western blotting was performed on 10% SDS-PAGE using Mini-PROTEAN® Tetra Cell Systems (Bio-Rad, Hercules, CA, USA). Proteins were transferred onto polyvinylidine difluoride (PVDF) membranes (Immobilon; Millipore, Boston, MA, USA). Membranes were incubated with S100A9 rabbit monoclonal antibody (Cell Signaling, Danvers, MA, USA), NF-κB (p65) mouse monoclonal antibody (Cell Signaling), phospho-NF-κB (p65) (Ser536) rabbit monoclonal antibody (Cell Signaling), Bcl-2 rabbit monoclonal antibody (Cell Signaling), MMP7 rabbit monoclonal antibody (Cell Signaling), MMP2 rabbit monoclonal antibody (Cell Signaling) at 1:1,000 dilution, or GAPDH-specific antibody (Sigma-Aldrich) at 1:5,000 dilution at 4°C overnight. Signals were visualized using ECL Substrates (Millipore).
Tumor Xenograft in Nude Mice
Animal experiments were approved by the Ethical Committee for Animal Research of Central South University (Protocol No. 2014-030). All nude mice (4–5 weeks old, male) were purchased from the Central Animal Facility of Central South University. To assess tumor growth, 200 ml of Fadu cells (1 × 106) was subcutaneously injected into the left side of the back of each mouse (five mice per group). The tumor sizes were measured regularly and calculated using the formula: 0.52 × L × W 2, where L and W are the long and short diameters of the tumor, respectively. The animals were euthanized on day 50 after injection, and the tumors were removed for immunohistochemical staining to evaluate the expression of Ki-67 and MMP7.
Statistical Analysis
Data were expressed as means ± standard deviation (SD). SPSS 16.0 software (SPSS Inc., Chicago, IL, USA) was used to perform the statistical analysis. Student’s t-test was used to analyze the differential expression of S100A9 between HPC patients and adjacent controls. Chi-square test was used to analyze the association between the level of S100A9 and clinicopathological parameters of HPC. Kaplan–Meier analysis with the log-rank test was used to examine the association between the serum level of S100A9 and the overall survival (OS). Cox proportional hazard regression model was used to estimate the independent predictors for the prognosis of HPC patients. Values of p < 0.05 were considered statistically significant.
RESULTS
Expression of S100A9 Is Higher in HPC Tissues Than in the Adjacent Controls
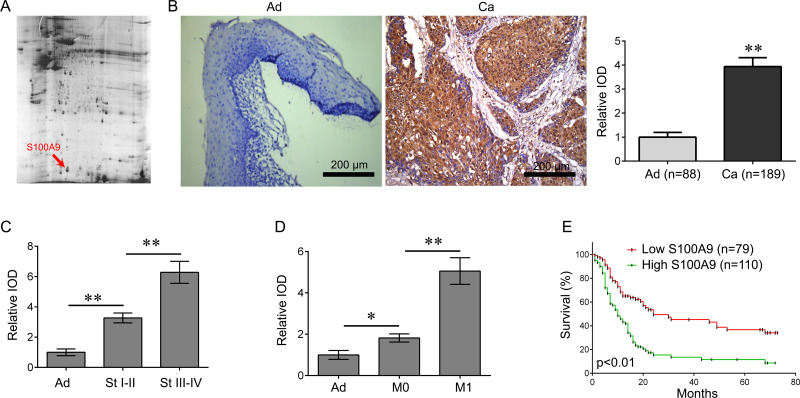
The proteins from five matched human HPC samples and their adjacent tissues were run in duplicate using DIGE together with an internal pool sample on each gel. Using DeCyder software version 6.5 (GE Healthcare Bio-science), positional deviation of the protein spot was 1.67 ± 0.24 mm for IEF and 1.25 ± 0.13 mm for 2-DE. Fully automated spot detection and quantification were further carried out using the Decyder software (GE; >1.5-fold) (Fig. 1A). Among the differential proteins identified, S100A9 was overexpressed in cancer tissues compared with normal adjacent tissues, and S100A9 expression in metastatic tissues and advanced tumor tissues was higher than in nonmetastatic tissues and early tumor tissues, respectively (Table 1). Furthermore, immunohistochemical staining analysis was performed to identify the expression of S100A9 in the 189 cases of HPC and adjacent tissues in 88 cases. Our data showed that the S100A9 levels were significantly higher in HPC tissues than in adjacent tissues. (p < 0.01) (Fig. 1B). In addition, the S100A9 levels increased with clinical stages (Fig. 1C), and its expression in metastatic tissues was higher than in nonmetastatic tissues (Fig. 1D).
Figure 1.
S100A9 is increased in human hypopharyngeal cancer (HPC) tissues. (A) Three differential protein spots from two-dimensional polyacrylamide gel electrophoresis (2-DE) gels were identified as S100A9 by HPC tissues. (B) S100A9 was further identified as positively expressed by immunohistochemical (IHC) staining analysis, and its expression was significantly higher in human HPC tissues than in adjacent tissues (**p < 0.01). Scale bar: 200 μm. (C) The expression of S100A9 in different clinical stages (**p < 0.01). (D) The expression of S100A9 in metastatic and nonmetastatic tissues (*p < 0.05, **p < 0.01). (E) The relationship between S100A9 expression and overall survival rate of HPC patients. Patients with a high S100A9 expression have a shorter survival time than those with a low S100A9 expression (p < 0.01).
Table 1.
Identification of the Differentially Expressed Protein in Laryngeal Cancer
| Gene Name | Protein | Fold Changes | ||
|---|---|---|---|---|
| Cancer versus Adjacent | Nonmetastasis versus Metastasis | Early Tumor versus Advanced Tumor | ||
| FGB | Fibrinogen fragment D | 3 | 2 | 1.5 |
| PKM | Pyruvate kinase isozymes M1/M2 | 2.3 | 4 | 1.9 |
| PSMB5 | Proteasome subunit α type 5 | 1.8 | 2.2 | 1.1 |
| FABP5 | Fatty acid-binding protein 5 | 1.9 | 3.2 | 2.1 |
| S100A9 | Protein S100-A9 | 4.5 | 6.8 | 5.1 |
| ANXA2 | Annexin A2 isoform 2 | 3.1 | 3.4 | 2.5 |
| LGALS7 | Chain A, galectin-7 | 2.5 | 2.8 | 2.4 |
| VIM | Vimentin | 1.9 | 1.7 | 2.3 |
| KRT1 | Keratin, type II cytoskeletal 1 | −1.5 | −1.7 | −2.2 |
| HSPB1 | Heat shock protein 27 | −4.1 | −2.1 | −1.7 |
| GLRX3 | Thioredoxin-like protein 2 | −2.3 | −3.1 | −3.2 |
| PRDX4 | Peroxiredoxin 4 | −2.6 | −1.5 | −3.2 |
Expression of S100A9 Is Significantly Associated With Tumor Size, Clinical Stage, and Lymph Node Metastasis as Well as Poor 5-Year Survival Rates in HPC Patients
We further studied the association between S100A9 levels and clinical characteristics in HPC. All HPC patients were divided into two groups relating to the mean of IOD of adjacent tissues: high S100A9 level group (n = 110) and low S100A9 level group (n = 79). S100A9 levels were not correlated with age (p = 0.657), gender (p = 0.632), or histological grade (p = 0.360) (Table 2). However, levels were significantly associated with tumor size (p = 0.001), lymph node metastasis (p = 0.002), distant metastasis (p = 0.002), and clinical stage (p = 0.038). Thus, our data suggest that S100A9 levels may be used as a prognostic biomarker for evaluating the malignant progression of HPC.
Table 2.
Clinical Association Between S100A9 Expression and Clinicopathological Variables in Laryngeal Cancer Patients
| Variable | S100A9 Expression | Chi-Square Test p Value | |
|---|---|---|---|
| Low (n = 79) | High (n = 110) | ||
| Ag | 0.657 | ||
| <60 | 35 | 45 | |
| ≥60 | 44 | 65 | |
| Gender | 0.632 | ||
| Male | 53 | 78 | |
| Female | 26 | 32 | |
| Tumor size | 0.001 | ||
| <5cm | 58 | 55 | |
| ≥5cm | 21 | 55 | |
| Histological grade | 0.360 | ||
| I | 47 | 73 | |
| II–III | 32 | 37 | |
| Lymph node metastasis | 0.002 | ||
| No | 53 | 48 | |
| Yes | 26 | 62 | |
| Distant metastasis | 0.002 | ||
| No | 56 | 52 | |
| Yes | 23 | 58 | |
| Clinical stage | 0.038 | ||
| I–II | 43 | 42 | |
| III–IV | 36 | 68 | |
We then investigated the association between S100A9 levels and the survival rate in HPC patients using the Kaplan–Meier method. We found that HPC patients having higher S100A9 levels showed a shorter 5-year OS (p < 0.01) (Fig. 1E) when compared to those with lower S100A9 levels. Therefore, a higher S100A9 level is associated with a poor prognosis for HPC patients.
In addition, we investigated the factors that could predict the prognosis of HPC patients using univariate and multivariate analyses. Univariate analysis data indicated that the S100A9 level (p = 0.021), as well as the tumor size (p = 0.025), lymph node metastasis (p = 0.015), distant metastasis (p = 0.008), and clinical stage (p = 0.023), was significantly associated with survival (Table 3). Moreover, the S100A9 level (p = 0.011), tumor size (p = 0.029), lymph node metastasis (p = 0.017), distant metastasis (p = 0.007), and clinical stage (p = 0.023) were found to be independent factors for predicting the prognosis of HPC patients (Table 4).
Table 3.
Univariate Analysis of Prognostic Factors of Laryngeal Cancer
| Variable | Hazard Ratio | p Value |
|---|---|---|
| Age (≥60/<60) | 1.591 | 0.571 |
| Gender (male/female) | 1.325 | 0.653 |
| Tumor size (≥5 cm/<5 cm) | 2.543 | 0.025 |
| Histological grade (II–III/I) | 1.862 | 0.055 |
| Lymph node metastasis (yes/no) | 3.356 | 0.015 |
| Distant metastasis (yes/no) | 4.131 | 0.008 |
| Clinical stage (III–IV/I–II) | 2.821 | 0.023 |
| S100A9 expression (high/low) | 2.653 | 0.021 |
Table 4.
Multivariate Analysis of Independent Prognostic Factors of Laryngeal Cancer
| Variable | Hazard Ratio | p Value |
|---|---|---|
| Tumor size | 2.153 | 0.029 |
| Lymph node metastasis | 2.618 | 0.017 |
| Distant metastasis | 3.616 | 0.007 |
| Clinical stage | 2.418 | 0.023 |
| S100A9 expression | 3.343 | 0.011 |
Knockdown of S100A9 Inhibits HPC Cell Proliferation and Invasion
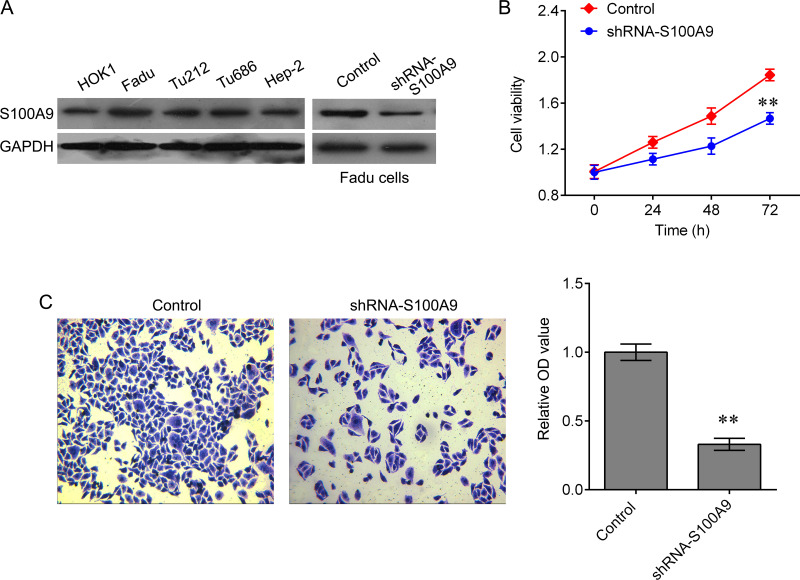
To further investigate the biological role of S100A9 in HPC progression, we detected the expression of S100A9 in one normal epithelial culture derived from oral cavity epithelia (HOK1) and four human HPC cell lines, including Fadu, Tu212, Tu686, and Hep-2. A total of three cell lines showed a notable upregulation of S100A9 protein expression, particularly in Fadu cells, in comparison with the control HOK1 cell (Fig. 2A). The Fadu cells transfected with lentivirus containing S100A9 shRNA or NF-κB-overexpressed plasmid were used for further analysis (Fig. 2A). Results showed that the knockdown of S100A9 induced significant inhibition of cell growth (Fig. 2B) and invasion ability (Fig. 2C).
Figure 2.
Knockdown of S100A9 represses Fadu cell growth and invasion. (A) Western blot detected the expression of S100A9 in HPC cell lines, including Fadu, Tu212, Tu686, and Hep-2, and the HOK1. (B) Cell counting kit-8 (CCK-8) was used to measure cell proliferation after shRNA-S100A9 treatment (**p < 0.01 vs. negative control). (C) Transwell assay was used to measure invasion ability after shRNA-S100A9 treatment. Data are expressed as mean ± standard deviation (SD) (**p < 0.01 vs. negative control).
Restoration of NF-κB Abolishes S100A9 Knockdown-Mediated Inhibition of Tumor Growth In Vitro and In Vivo
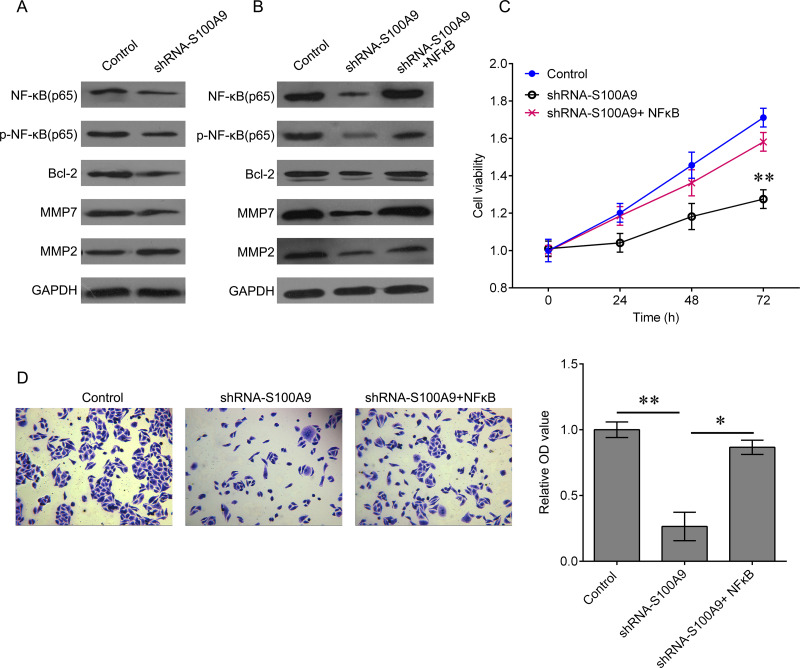
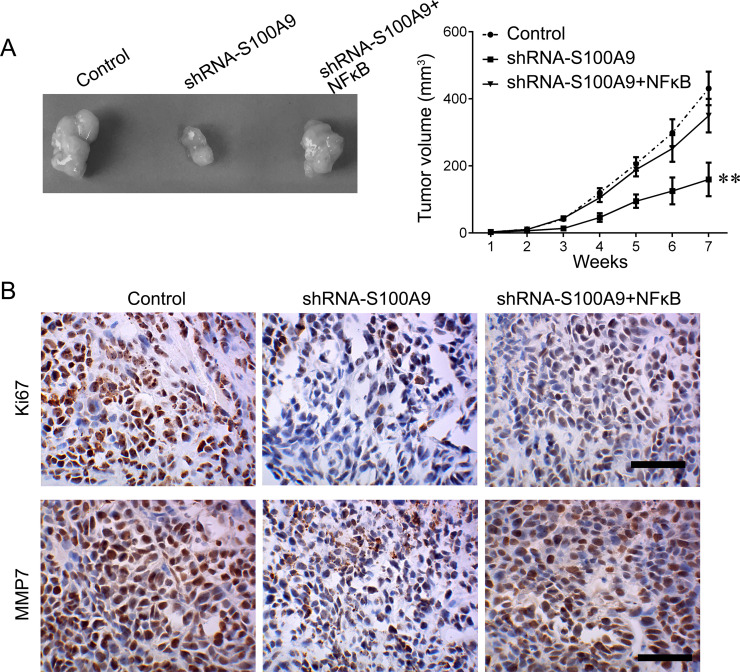
Activation of NF-κB signaling frequently facilitates tumor growth and metastasis. To further investigate the underlying mechanism by which S100A9 inhibits HPC cell proliferation and invasion, we analyzed the expression of key molecules of NF-κB signaling in Fadu cells that knocked down S100A9. We found that downregulation of S100A9 significantly reduced the expression of NF-κB, p-NF-κB, and Bcl-2, as well as the expression of MMP7 and MMP2 (Fig. 3A). We then cotransfected Fadu cells with shRNA-S100A9 and NF-κB-expressed plasmid. The results showed that upregulation of NF-κB reactivated the NF-κB signaling that was inactivated by shRNA-S100A9 transfection (Fig. 3B). Moreover, we tested whether the biological function of S100A9 was achieved via NF-κB signaling. Restoration of NF-κB expression sufficiently reversed the inhibitory effects on cell proliferation and invasion induced by downregulation of S100A9 (Fig. 3C and D). We further confirmed the role of S100A9 in vivo. Tumor growth was significantly suppressed by knockdown of S100A9 (Fig. 4A). This effect was reversed by induction of NF-κB. We also detected the expression of proliferative marker Ki-67 and the expression of proinvasive gene MMP7 by using immunohistochemical (IHC) staining. Their expression was notably downregulated after knockdown of S100A9. Once NF-κB expression was upregulated, the effects of S100A9 on Ki-67 MMP7 expression were attenuated (Fig. 4B). These findings demonstrated that the inhibitory effects of S100A9 knockdown on HPC progression are achieved through the NF-κB pathway.
Figure 3.
Restoration of NF-κB abolishes S100A9 knockdown-mediated inhibition of tumor growth in vitro and in vivo. (A) Fadu cells were transfected with shRNA-S100A9 for 48 h. Expression of NF-κB, p-NF-κB, Bcl-2, MMP7, and MMP2 proteins was measured by Western blot. (B) Fadu cells were transfected with shRNA-S100A9 alone or together with NF-κB-expressed plasmid for 48 h. Expression of NF-κB, p-NF-κB, Bcl-2, MMP7, and MMP2 proteins was measured by Western blot (**p < 0.01). (C) CCK-8 was used to measure the cell proliferation after the indicated treatment. (D) Transwell assay was used to measure the invasion ability of Fadu cells after the indicated treatment (*p < 0.05, **p < 0.01). Data are expressed as mean ± standard deviation.
Figure 4.
Knockdown of S100A9 suppresses HPC growth in vivo. (A) The xenografted tumor obtained from nude mice (left) and the tumor volume measured every week (right) (**p < 0.01). (B) The expression of Ki-67 and MMP7 proteins was measured by IHC staining in tumor section. Scale bar: 100 μm. Data are expressed as mean ± standard deviation.
DISCUSSION
In this study, we identified the potential biomarker protein for HPC using 2D-DIGE proteomics analysis. We identified eight upregulated proteins, including FGB, PKM, PSMB5, FABP5, S100A9, ANXA2, LGALS7, and VIM, and four downregulated proteins, including KRT1, HSPB1, GLRX3, and PRDX4 (Table 1), in metastatic tissues, nonmetastatic tissues, advanced tumor tissues, and early tumor tissues. Among these, S100A9 expression was further confirmed in a large additional cohort. Our data showed that a higher S100A9 level was an independent prognostic factor for predicting the prognosis of HPC patients.
S100A9 is a calcium-binding protein, participating in the inflammatory process and development of various tumors12. Pei et al. found that NF-κB expression in diabetic hearts was increased, together with the elevation of S100A9 during doxorubicin-induced cardiac inflammation, revealing the S100A9-associated molecular signaling pathways that mediate doxorubicin-induced cardiotoxicity in diabetic hearts13. Recent studies have indicated that S100A9 may be a biomarker for various cancers. For example, the serum level of S100A9 in colorectal cancer patients was significantly increased and was positively correlated with the malignant features of colorectal cancer14. Transcript levels of S100A9 were significantly decreased in malignant oral squamous cell carcinoma. A close correlation between the expression levels of S100A9 and pathological characteristics was also observed, representing a promising marker for the evaluation of the risk potential of suspicious oral lesions in molecular pathology15. Zhou et al.’s finding indicated that S100A9 could serve as a reliable predictor of therapeutic response and independent prognostic factors of survival in advanced extranodal NK/T-cell lymphoma patients treated with pegaspargase/gemcitabine16. In breast cancer, strong expression and secretion of S100A9 were associated with the loss of estrogen receptor in breast cancer and were involved in the poor prognosis of Her2+/basal-like subtypes of breast cancer17,18. Here, for the first time, we have identified S100A9 as a potential biomarker that can be applied to the early diagnosis of HPC and have highlighted the utility of onco-proteogenomics for identifying protein markers that can be applied to disease-oriented translational medicine19.
In addition, knockdown of S100A9 expression that is upregulated in human HPC cell lines induced significant inhibition on cell growth and invasion ability. In line with our results in this study, the expression of S100A9 was found to be increased in human osteosarcoma issues and was positively correlated with clinical classification and survival rate. Downregulation of S100A9 inhibited osteosarcoma cellular proliferation, migration, and invasion in vitro and suppressed tumor formation in vivo with the reduction on the Ki-67 proliferation index20. Knockdown of S100A9 repressed the protein levels of phospho-ERK1/2 and phospho-p65 and prompted upregulation of p21 and p27, leading to inactivation of cyclin-dependent kinase 2 (CDK2) and CDK4, suggesting that downregulation of S100A9 inhibits osteosarcoma cell growth through inactivation of the MAPK and NF-κB signaling pathways20. S100A9 homodimers induced the expression of proinflammatory cytokines (IL-6, IL-8, and IL-1β) in a TLR4- and EGFR-dependent manner in human breast cancer cells in vitro18. It was reported that the release of S100A9 protein into the extracellular milieu enhanced oral cancer cell invasion, transendothelial monocyte migration, and angiogenic activity through the activation of NF-κB and STAT321. In addition, Wu et al. and Wilson et al. found a similar mechanism by which S100A9 promoted cell growth and invasion in hepatocellular carcinoma. They demonstrated that S100A9 bound to the receptor for advanced glycation end products (RAGE) and stimulated RAGE-dependent MAPK signaling cascades to promote cell growth and invasion in hepatocellular carcinoma22,23. Interestingly, Iotzova-Weiss et al. showed that RAGE and S100A9 were important factors driving normal and tumor keratinocyte proliferation. RAGE and S100A9 were transcriptionally upregulated in squamous cell carcinoma (SCC). The proliferation and migratory activities of SCC keratinocytes were induced by exposure to exogenous S100A9, which may be through inducing phosphorylation of p38 followed by activation of ERK1/224. Consistent with these findings, we found that downregulation of S100A9 significantly reduced the expression of NF-κB, phosphorylation of NF-κB and Bcl-2, as well as the expression of MMP7 and MMP2. We also found that restoration of NF-κB expression sufficiently reversed the inhibitory effects on cell proliferation and invasion induced by downregulation of S100A9 in vitro and in vivo, demonstrating that downregulation of S100A9 repressed the HPC cancer cells’ ability for growth and invasion through NF-κB signaling.
In conclusion, for the first time we have identified the oncogenic protein, S100A9, using 2D-DIGE proteomics analysis. We also demonstrated the oncogenic role of S100A9 in HPC. A high expression of S100A9 is associated with a poor prognosis for HPC patients. Downregulation of S100A9 significantly inhibited HPC cancer cells’ growth and migratory activity in vitro and in vivo through the inactivation of NF-κB signaling. Our study provides a potential novel diagnostic biomarker and therapeutic target for HPC treatment.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Nos. 81302355 and 81201740), the Natural Science Foundation of Hunan Province (No. 2013FJ4225), and the Xiangya Famous Doctors Foundation.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Newman JR, Connolly TM, Illing EA, Kilgore ML, Locher JL, Carroll WR. Survival trends in hypopharyngeal cancer: A population-based review. Laryngoscope 2015;125:624–9. [DOI] [PubMed] [Google Scholar]
- 2. Hall SF, Griffiths R. Did the addition of concomitant chemotherapy to radiotherapy improve outcomes in hypopharyngeal cancer? A population-based study. Curr Oncol. 2016;23:266–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan JY, Wei WI. Current management strategy of hypopharyngeal carcinoma. Auris Nasus Larynx 2013;40:2–6. [DOI] [PubMed] [Google Scholar]
- 4. Kalfert D, Pesta M, Kulda V, Topolcan O, Ryska A, Celakovsky P, Laco J, Ludvikova M. MicroRNA profile in site-specific head and neck squamous cell cancer. Anticancer Res. 2015;35:2455–63. [PubMed] [Google Scholar]
- 5. Szentkuti G, Danos K, Brauswetter D, Kiszner G, Krenacs T, Csako L, Repassy G, Tamas L. Correlations between prognosis and regional biomarker profiles in head and neck squamous cell carcinomas. Pathol Oncol Res. 2015;21:643–50. [DOI] [PubMed] [Google Scholar]
- 6. Zhou L, Cheng L, Tao L, Jia X, Lu Y, Liao P. Detection of hypopharyngeal squamous cell carcinoma using serum proteomics. Acta Otolaryngol. 2006;126:853–60. [DOI] [PubMed] [Google Scholar]
- 7. Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: Two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 2003;3:36–44. [DOI] [PubMed] [Google Scholar]
- 8. Colantonio DA, Chan DW. The clinical application of proteomics. Clin Chim Acta 2005;357:151–8. [DOI] [PubMed] [Google Scholar]
- 9. Tian WD, Li JZ, Hu SW, Peng XW, Li G, Liu X, Chen HH, Xu X, Li XP. Proteomic identification of alpha-2-HS-glycoprotein as a plasma biomarker of hypopharyngeal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:9021–31. [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu G, Cai G, Liu Y, Tan H, Yu C, Huang M, Wei M, Li S, Cui X, Huang D, Tian Y, Zhang X. Quantitative iTRAQ LC-MS/MS proteomics reveals transcription factor crosstalk and regulatory networks in hypopharyngeal squamous cell carcinoma. J Cancer 2014;5:525–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang J, Zhou M, Zhao R, Peng S, Luo Z, Li X, Cao L, Tang K, Ma J, Xiong W, Fan S, Schmitt DC, Tan M, Li X, Li G. Identification of candidate biomarkers for the early detection of nasopharyngeal carcinoma by quantitative proteomic analysis. J Proteomics 2014;109:162–75. [DOI] [PubMed] [Google Scholar]
- 12. Leanderson T, Liberg D, Ivars F. S100A9 as a pharmacological target molecule in inflammation and cancer. Endocr Metab Immune Disord Drug Targets 2015;15:97–104. [DOI] [PubMed] [Google Scholar]
- 13. Pei XM, Tam BT, Sin TK, Wang FF, Yung BY, Chan LW, Wong CS, Ying M, Lai CW, Siu PM. S100A8 and S100A9 are associated with doxorubicin-induced cardiotoxicity in the heart of diabetic mice. Front Physiol. 2016;7:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shu P, Zhao L, Wagn J, Shen X, Zhang X, Shen S, Ma J, Li X. [Association between serum levels of S100A8/S100A9 and clinical features of colorectal cancer patients]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2016;41:553–59. [DOI] [PubMed] [Google Scholar]
- 15. Reckenbeil J, Kraus D, Probstmeier R, Allam JP, Novak N, Frentzen M, Martini M, Wenghoefer M, Winter J. Cellular distribution and gene expression pattern of Metastasin (S100A4), Calgranulin A (S100A8), and Calgranulin B (S100A9) in oral lesions as markers for molecular pathology. Cancer Invest. 2016;34:246–54. [DOI] [PubMed] [Google Scholar]
- 16. Zhou Z, Li Z, Sun Z, Zhang X, Lu L, Wang Y, Zhang M. S100A9 and ORM1 serve as predictors of therapeutic response and prognostic factors in advanced extranodal NK/T cell lymphoma patients treated with pegaspargase/gemcitabine. Sci Rep. 2016;6:23695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bao YI, Wang A, Mo J. S100A8/A9 is associated with estrogen receptor loss in breast cancer. Oncol Lett. 2016;11:1936–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bergenfelz C, Gaber A, Allaoui R, Mehmeti M, Jirstrom K, Leanderson T, Leandersson K. S100A9 expressed in ER(-)PgR(-) breast cancers induces inflammatory cytokines and is associated with an impaired overall survival. Br J Cancer 2015;113:1234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan NJ, Chen HM, Song W, Zhang ZY, Zhang MD, Feng LY, Gao CF. Macrophage mannose receptor 1 and S100A9 were identified as serum diagnostic biomarkers for colorectal cancer through a label-free quantitative proteomic analysis. Cancer Biomark. 2016;16:235–43. [DOI] [PubMed] [Google Scholar]
- 20. Cheng S, Zhang X, Huang N, Qiu Q, Jin Y, Jiang D. Down-regulation of S100A9 inhibits osteosarcoma cell growth through inactivating MAPK and NF-kappaB signaling pathways. BMC Cancer 2016;16:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fang WY, Chen YW, Hsiao JR, Liu CS, Kuo YZ, Wang YC, Chang KC, Tsai ST, Chang MZ, Lin SH, Wu LW. Elevated S100A9 expression in tumor stroma functions as an early recurrence marker for early-stage oral cancer patients through increased tumor cell invasion, angiogenesis, macrophage recruitment and interleukin-6 production. Oncotarget 2015;6:28401–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu R, Duan L, Cui F, Cao J, Xiang Y, Tang Y, Zhou L. S100A9 promotes human hepatocellular carcinoma cell growth and invasion through RAGE-mediated ERK1/2 and p38 MAPK pathways. Exp Cell Res. 2015;334:228–38. [DOI] [PubMed] [Google Scholar]
- 23. Wilson CL, Jurk D, Fullard N, Banks P, Page A, Luli S, Elsharkawy AM, Gieling RG, Chakraborty JB, Fox C, Richardson C, Callaghan K, Blair GE, Fox N, Lagnado A, Passos JF, Moore AJ, Smith GR, Tiniakos DG, Mann J, Oakley F, Mann DA. NFkappaB1 is a suppressor of neutrophil-driven hepatocellular carcinoma. Nat Commun. 2015;6:6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iotzova-Weiss G, Dziunycz PJ, Freiberger SN, Lauchli S, Hafner J, Vogl T, French LE, Hofbauer GF. S100A8/A9 stimulates keratinocyte proliferation in the development of squamous cell carcinoma of the skin via the receptor for advanced glycation-end products. PLoS One 2015;10:e120971. [DOI] [PMC free article] [PubMed] [Google Scholar]