Abstract
Aberrant expression of microRNA-92a (miR-92a) has been investigated in various cancers. However, the function and mechanism of miR-92a in acute myeloid leukemia (AML) remain to be elucidated. Our data showed that miR-92a was evidently downregulated and methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) was remarkably upregulated in AML cell lines HL-60 and THP-1. Dual luciferase reporter assay revealed that MTHFD2 was a direct target of miR-92a. Gain- and loss-of-function analysis demonstrated that MTHFD2 knockdown or miR-92a overexpression notably inhibited proliferation and promoted apoptosis of AML cell lines. Restoration of MTHFD2 expression reversed proliferation inhibition and apoptosis induction of AML cells triggered by miR-92a. Moreover, an implanted tumor model in mice indicated that miR-92a overexpression dramatically decreased tumor growth and MTHFD2 expression in vivo. Taken together, our results suggest that miR-92a inhibits proliferation and induces apoptosis by directly regulating MTHFD2 expression in AML. miR-92a may act as a tumor suppressor in AML, providing a promising therapeutic target for AML patients.
Key words: miR-92a, Methylenetetrahydrofolate dehydrogenase 2 (MTHFD2), Acute myeloid leukemia (AML), Proliferation, Apoptosis
INTRODUCTION
Acute myeloid leukemia (AML) is the most common malignant myeloid disorder, mainly occurring in elderly patients, and characterized by the accumulation of blast cell and the blockage of myeloid differentiation in the bone marrow1. Cytogenetic and molecular genetic abnormalities lead to the clonal expansion of early hematopoietic progenitor cells and obstruct the hematopoiesis of normal bone marrow. Subsequently, AML patients suffer from symptoms of anemia and thrombocytopenia2. Although many AML patients have a response to induction chemotherapy, the persistence of residual blast cells in the bone marrow leads to a high relapse rate3. Therefore, it is urgently needed to illuminate the pathogenesis and identify new therapeutic targets for AML.
Methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) is an enzyme encoded by the nuclear MTHFD2 gene on chromosome 2 in humans4, which can produce a dual-function mitochondrial enzyme with methylenetetrahydrofolate dehydrogenase and cyclohydrolase activities5. MTHFD2 is the most distinctively expressed metabolic enzyme between cancer cells and normal cells, including normal proliferating cells6. A previous study of mRNA profiles spanning 19 cancer types revealed that MTHFD2 mRNA and protein expression was elevated in many cancers, and MTHFD2 knockdown resulted in an obvious suppression of proliferation and cell death in diverse cancer cell types7. MTHFD2 was overexpressed in breast cancer, and MTHFD2 silencing by RNAi blocked proliferation, invasion, and migration in breast cancer cell lines8,9. Moreover, MTHFD2 suppression could impair growth and induce myeloid differentiation in AML cell lines and promote apoptosis in FLT3-ITD mutant AML6.
MicroRNAs (miRNAs) are small noncoding RNA molecules that usually lead to gene silencing at the posttranscriptional level through binding to complementary sequences in the 3′-untranslated region (3′-UTR) of their target mRNAs10. miRNAs have been used as diagnostic and prognosis biomarkers because of their function as oncogenes or tumor suppressors in cancer11. Ample evidence has demonstrated that various miRNAs are closely associated with AML, such as miR-181a, miR-193a, and miR-15512–14. miR-92a has been shown to play a crucial role in different cancers. For instance, miR-92a overexpression was related to tumor metastasis and poor prognosis in colorectal cancer15, and promoted cell proliferation and invasion of cervical cancer by targeting FBXW716. Downregulation of an miR-17-92a cluster induced autophagy through targeting autophagy-related gene ATG7 in prostate cancer cells17. Moreover, a previous study found that downregulation of miR-92a in plasma was identified as a novel biomarker for the detection of leukemia18. However, few documents have reported the role of miR-92a in the development of AML.
In the present study, we aimed to investigate the effect of miR-92a on AML cells and its underlying molecular mechanisms.
MATERIALS AND METHODS
Cell Culture
The human bone marrow stromal cell line HS-5 and human AML cell lines HL-60 and THP-1 were purchased from the American Tissue Culture Collection (ATCC; Manassas, VA, USA) and cultured at 37°C in humidified 5% CO2. All cells were grown in RPMI-1640 medium (Gibco, Grand Island, NY, USA) with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin added.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from the harvested HL-60, THP-1, and HS-5 cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed to cDNAs by M-MLV reverse transcriptase (Invitrogen). An Applied Biosystems 7500 Real-time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) and a SYBR Premix Ex Taq™ kit (Takara Bio, Otsu, Japan) were used to conduct the qRT-PCR. For miR-92a detection, a TaqMan microRNA assay (Ambion, Austin, TX, USA) was performed. The relative expression of miR-92a and MTHFD2 mRNA was calculated using the 2−ΔΔCt method with β-actin serving as the internal reference.
Western Blot Analysis
The BCA Protein Assay Kit (Pierce, Rockford, IL, USA) was used to detect the concentration of protein extracted from cultured cells or resected tumors. Equivalent protein samples were separated by 10% SDS-PAGE and electrotransferred to polyvinylidene fluoride (PVDF; Amersham Pharmacia, Little Chalfont, UK). The membrane was incubated with the primary antibodies for MTHFD2 (Cell Signaling Technology, Danvers, MA, USA), Ki-67 (Cell Signaling Technology), Bax (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Bcl-2 (Santa Cruz Biotechnology), CytC (Santa Cruz Biotechnology), or β-actin (Cell Signaling Technology), followed by horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology). The blots were detected using an enhanced chemiluminescence kit (ECL; Amersham Biosciences, Piscataway, NJ, USA) and visualized with the Image Quant software (Molecular Dynamics, Sunnyvale, CA, USA).
Cell Transfection
miR-92a mimics (miR-92a), miR-92a inhibitor (anti-miR-92a), siRNA targeting MTHFD2 (si-MTHFD2), and the scramble negative controls (miR-control and si-control) were synthesized by Dharmacon (Lafayette, CO, USA). pcDNA empty vector and pcDNA-MTHFD2 were obtained from Invitrogen. To overexpress or inhibit miR-92a, HL-60 and THP-1 cells were transfected with miR-92a mimics or anti-miR-92a using Lipofectamine 2000 (Invitrogen). To knock down or upregulate MTHFD2, si-MTHFD2 or pcDNA-MTHFD2 was transfected into HL-60 and THP-1 cells according to the Lipofectamine 2000 specification.
Flow Cytometry
Cell apoptosis was determined according to the manufacturer’s instructions included within the Annexin-V-FITC Apoptosis Detection Kit (Sigma-Aldrich, St. Louis, MO, USA). Briefly, cells were harvested, washed, and resuspended in PBS, followed by incubation with staining solution (5 μl of annexin V-FITC and 10 μl of PI) in the dark for 15 min at room temperature. Cells were then analyzed using FACSCalibur Flow Cytometer (BD Biosciences, San Jose, CA, USA) with FACSDiva software V6.1.3 (BD Biosciences).
Cell Viability Assays
HL-60 and THP-1 cells transfected with the miR-92a mimic, si-MTHFD2 or miR-92a mimic, and pcDNA-MTHFD2 were seeded into a 96-well plate at a density of 4 × 104 cells per well. After 24, 48, and 72 h of incubation, cell viability was determined by the cell counting kit-8 (CCK-8; Dojindo, Kumamoto, Japan) following the manufacturer’s protocol. An enzyme-linked immunosorbent assay reader (Bio-Rad Laboratories, Hercules, CA, USA) was used to measure the absorbance of each well at 450 nm.
Luciferase Reporter Assay
To determine whether MTHFD2 is a direct target of miR-92a, the luciferase reporter assay was performed. The 3′-UTR of MTHFD2 mRNA containing wild-type (WT) or mutant (MUT) binding sequence of miR-92a was amplified and inserted into the downstream of firefly luciferase gene in the pGL3 vector (Promega, Madison, WI, USA) to construct the pGL3-WT-MTHFD2 or pGL3-MUT-MTHFD2 reporter. HL-60 and THP-1 cells were seeded into 12-well plates at a density of 2 × 105. When cells reached 80% confluence, luciferase reporters were cotransfected with the miR-92a mimic or miR-control into HL-60 and THP-1 cells using Lipofectamine 2000. At 48 h after transfection, luciferase activity was measured with a Dual-Luciferase® Reporter Assay System (Promega) and normalized to Renilla luciferase activity.
Tumor Formation in Nude Mice
A total of 3 × 106 HL-60 cells with miR-92a or miR-control transfection were suspended in PBS and subcutaneously injected into the right flank of nude mice aged 4–6 weeks (n = 6 per group). A slide caliper was used to measure tumor length and width every 5 days in order to calculate tumor volume according to the formula: volume = ½ × length × width2. Thirty-five days later, tumors were harvested, and tumor weights were measured immediately after mice were sacrificed. qRT-PCR and Western blot were applied to test the expression of MTHFD2 in resected tumors. All animal procedures were performed with the approval of the Local Medical Experimental Animal Care Commission.
Statistical Analysis
SPSS 19.0 software (SPSS, Chicago, IL, USA) was utilized to perform the statistical analysis. All data were presented as means ± SD. Student’s t-test or one-way ANOVA was used to compare the difference between two or more groups. A value of p < 0.05 was set as the statistical significance.
RESULTS
Expression of miR-92a and MTHFD2 in AML Cell Lines
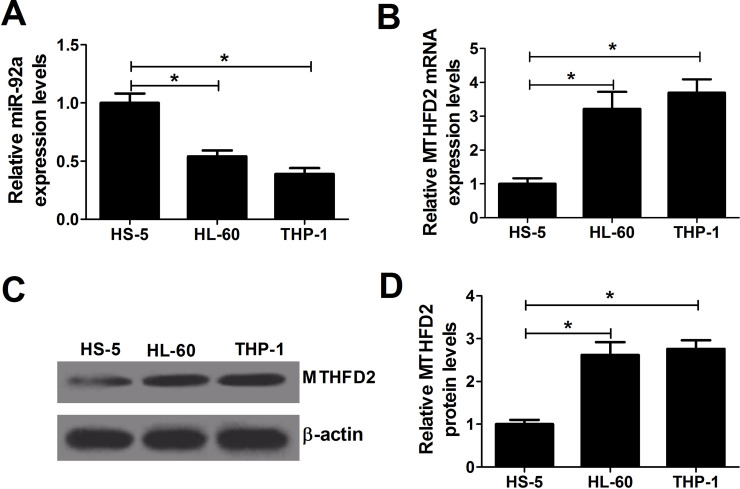
To explore the role of miR-92a in AML, qRT-PCR analysis was first performed to detect the expression of miR-92a in AML cell lines HL-60 and THP-1, and normal bone marrow stromal cell line HS-5. miR-92a expression was less in HL-60 and THP-1 cells than in HS-5 cells (Fig. 1A). MTHFD2 has been found to be upregulated in many cancers. Thus, the expression level of MTHFD2 was examined. The results revealed that MTHFD2 mRNA (Fig. 1B) and protein (Fig. 1C and D) were observably increased in HL-60 and THP-1 cells compared with those in HS-5 cells. These data hinted that miR-92a and MTHFD2 may be associated with the progression of AML.
Figure 1.
Expression of microRNA-92a (miR-92a) and methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) in acute myeloid leukemia (AML) cell lines. (A, B) Quantitative real-time PCR (qRT-PCR) analysis was conducted to determine the expression of miR-92a and MTHFD2 in AML cell lines (HL-60 and THP-1) and normal bone marrow stromal cell HS-5. (C, D) Western blot analysis was performed to detect the protein level of MTHFD2 in HL-60, THP-1, and HS-5 cells. *p < 0.05 versus HS-5.
MTHFD2 Is a Direct Target of miR-92a
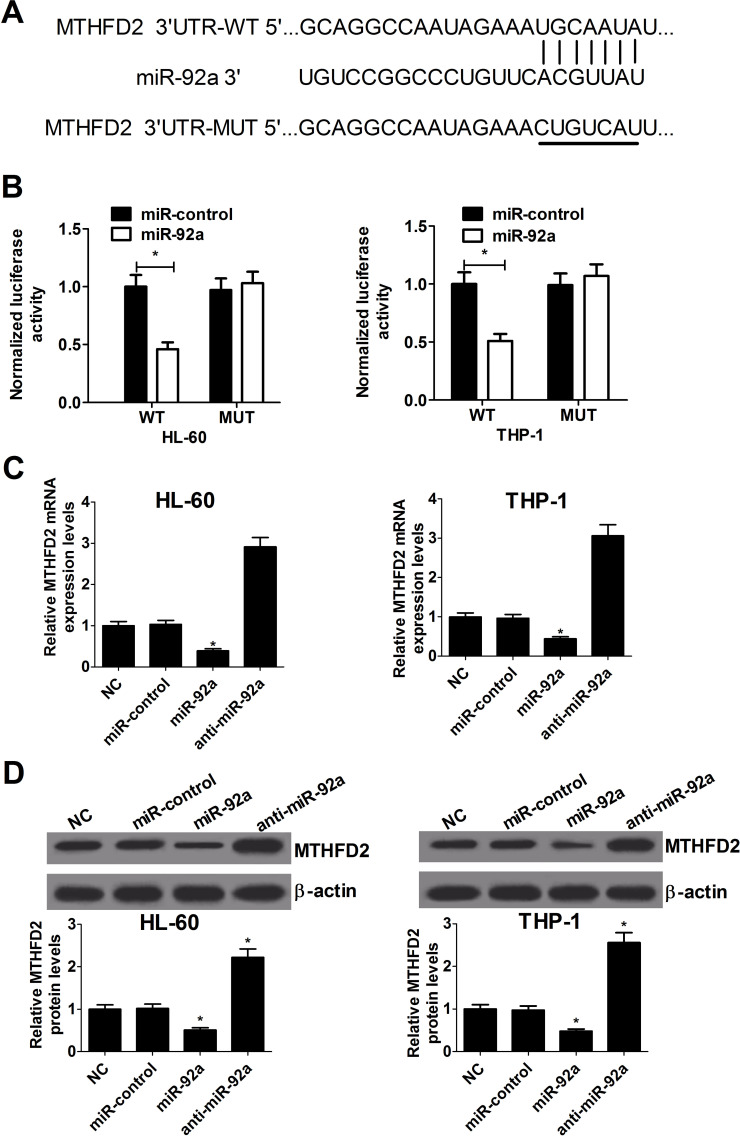
As is well known, miRNAs exert a role of suppressing the expression of their target genes. Considering the reverse expression level of miR-92a and MTHFD2 in AML cell lines, we suspected that MTHFD2 was targeted by miR-92a. Therefore, the online TargetScan database was used to predict potential target genes of miR-92a. The results showed that the 3′-UTR of MTHFD2 contained a putative binding sequence of miR-92a (Fig. 2A). To verify whether miR-92a could directly target MTHFD2, a dual luciferase reporter assay was conducted in AML cells. Cotransfection of pGL3-WT-MTHFD2 with miR-92a brought about an obvious decrease in luciferase activity compared with the control, but no significant difference was found in the luciferase activity of pGL3-MUT-MTHFD2 plasmid between the miR-92a mimic and the control group in HL-60 and THP-1 cells (Fig. 2B). To further confirm whether miR-92a could regulate MTHFD2 expression, HL-60 and THP-1 cells were transfected with miR-92a mimic or inhibitor. miR-92a overexpression resulted in a decrease in mRNA and protein levels of MTHFD2 in both HL-60 and THP-1 cells (Fig. 2C and D). Conversely, MTHFD2 mRNA and protein levels were notably elevated in anti-miR-92a-transfected HL-60 and THP-1 cells. Taken together, these results revealed that miR-92a directly targets MTHFD2 and negatively regulates the expression of MTHFD2 in AML cells.
Figure 2.
MTHFD2 is a direct target of miR-92a in AML cells. (A) The putative and mutated miR-92a binding sequences in the 3′-untranslated region (3′-UTR) of MTHFD2 were shown. (B) Luciferase reporter assay in HL-60 and THP-1 cells cotransfected with wild-type or mutant MTHFD2 3′-UTR and miR-92a mimics or miR-control. (C) qRT-PCR analysis of MTHFD2 mRNA expression in miR-92a mimic- or anti-miR-92a-transfected HL-60 and THP-1 cells. (D) Western blot analysis of the relative level of MTHFD2 protein in HL-60 and THP-1 cells transfected with miR-92a mimics or anti-miR-92a. *p < 0.05 versus miR-control.
MTHFD2 Knockdown Inhibits Proliferation and Induces Apoptosis of AML Cell Lines
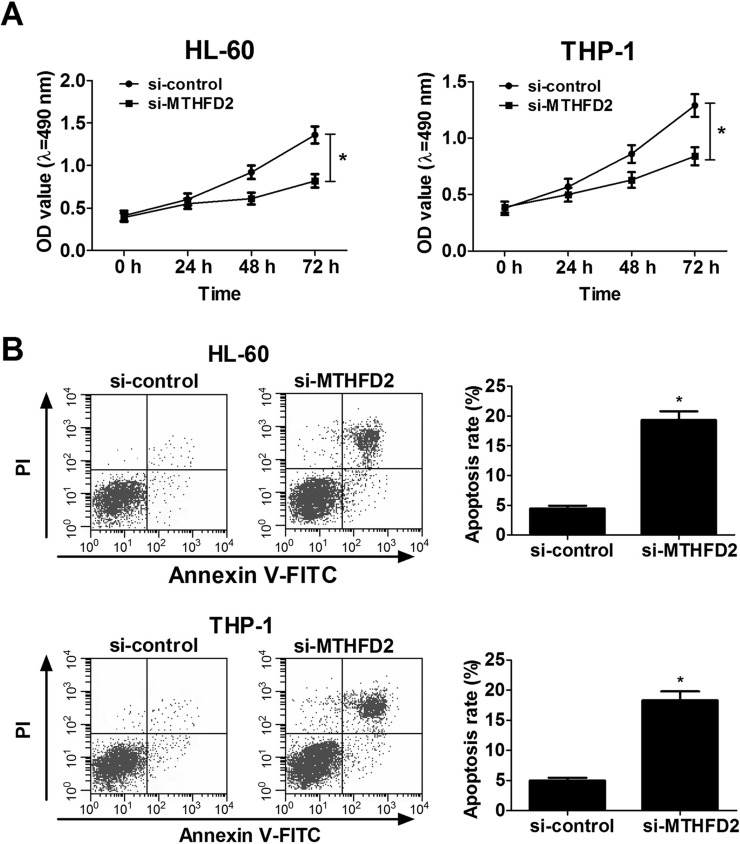
To confirm the role of MTHFD2 in AML, a loss-of-function assay was performed by transfecting si-MTHFD2 into HL-60 and THP-1 cells. The influence of MTHFD2 on the proliferation and apoptosis of AML cells was determined by the CCK-8 assay and flow cytometry analysis, respectively. The CCK-8 assay determined that MTHFD2 knockdown markedly suppressed cell proliferation of HL-60 and THP-1 cells (Fig. 3A). Flow cytometry analysis revealed that apoptosis rates were evidently enhanced in si-MTHFD2-transfected HL-60 and THP-1 cells (Fig. 3B). Collectively, these results demonstrated that MTHFD2 knockdown hindered proliferation and promoted apoptosis of AML cells.
Figure 3.
Downregulation of MTHFD2 by si-MTHFD2 knockdown decreases proliferation and increases apoptosis of AML cell lines. HL-60 and THP-1 cells were transfected with si-control or si-MTHFD2. (A) Cell viability of transfected HL-60 and THP-1 cells was determined by the cell counting kit-8 (CCK-8) assay at different time points (0, 24, 48, and 72 h). (B) Cell apoptosis was measured in transfected HL-60 and THP-1 cells at 48 h with annexin-V/PI double staining followed by flow cytometry analysis. *p < 0.05 versus si-control.
Restoration of MTHFD2 Expression Relieves the Proliferation Inhibition Triggered by miR-92a Overexpression in AML Cells
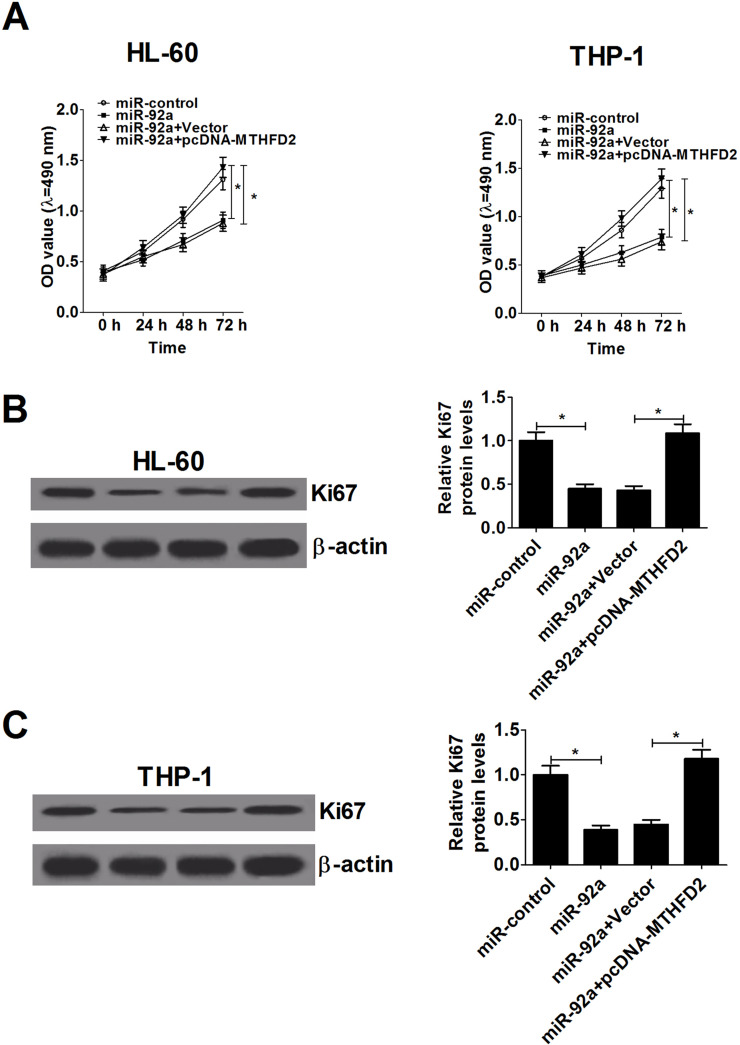
To further investigate the effect of miR-92a on the proliferation capability of AML cells and the possible mechanism, HL-60 and THP-1 cells were transfected with miR-92a mimics or cotransfected with miR-92a mimics and pcDNA-MTHFD2. The results showed that miR-92a upregulation dramatically inhibited proliferation in both HL-60 and THP-1 cells, whereas cotransfection of pcDNA-MTHFD2 abrogated this antiproliferation effect in the same cells (Fig. 4A). Moreover, proliferation-related protein Ki-67 was markedly decreased in miR-92a mimic-transfected HL-60 and THP-1 cells. However, MTHFD2 overexpression abolished the inhibitory effect of miR-92a on Ki-67 expression (Fig. 4B and C). To sum up, these results demonstrated that miR-92a overexpression impeded the proliferation of AML cells through suppressing MTHFD2.
Figure 4.
miR-92a suppresses proliferation of AML cells by targeting MTHFD2. HL-60 and THP-1 cells were transfected with either miR-92a mimics or in combination with pcDNA-MTHFD2. (A) CCK-8 analysis of cell viability at indicated time points (0, 24, 48, and 72 h) in transfected HL-60 and THP-1 cells. (B, C) Western blot analysis indicated the level of proliferation-related protein Ki-67 at 48 h in transfected HL-60 (B) and THP-1 cells (C). *p < 0.05 versus controls.
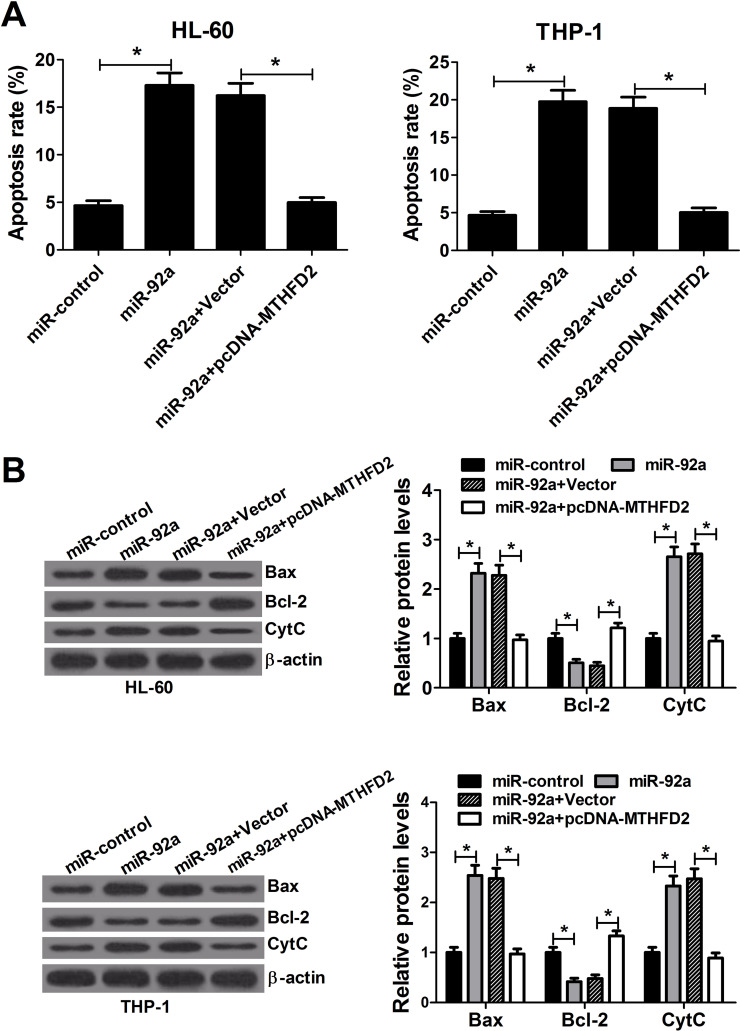
Ectopic Expression of MTHFD2 Reverses the Apoptosis Induction Elicited by miR-92a in AML Cells
To better understand the underlying mechanism of the growth inhibition caused by miR-92a, flow cytometry analysis was carried out to estimate the effect of miR-92a overexpression on apoptosis in AML cells. HL-60 and THP-1 cells were transfected with miR-92a mimics or in combination with pcDNA-MTHFD2. Overexpression of miR-92a resulted in a prominent augmentation of apoptosis in HL-60 and THP-1 cells, whereas enforced expression of MTHFD2 overturned this effect (Fig. 5A). Moreover, Western blot was performed to detect apoptosis-related protein. Notably, Bax and CytC were evidently increased, and Bcl-2 was distinctly decreased in miR-92a mimic-transfected HL-60 and THP-1 cells (Fig. 5B). In contrast, these effects were significantly rescued by the cotransfection of MTHFD2. Altogether, these data suggested that miR-92a overexpression promoted the apoptosis of AML cells by regulating MTHFD2.
Figure 5.
miR-92a induces apoptosis of AML cells by targeting MTHFD2. HL-60 and THP-1 cells were transfected with miR-92a mimics or cotransfected with miR-92a mimics and pcDNA-MTHFD2. (A) Flow cytometry analysis was performed to analyze the apoptosis rate of HL-60 and THP-1 cells at 48 h after transfection. (B) The level of apoptosis-related proteins Bax, Bcl-2, and CytC was evaluated in HL-60 and THP-1 cells by Western blot 48 h posttransfection. *p < 0.05 versus controls.
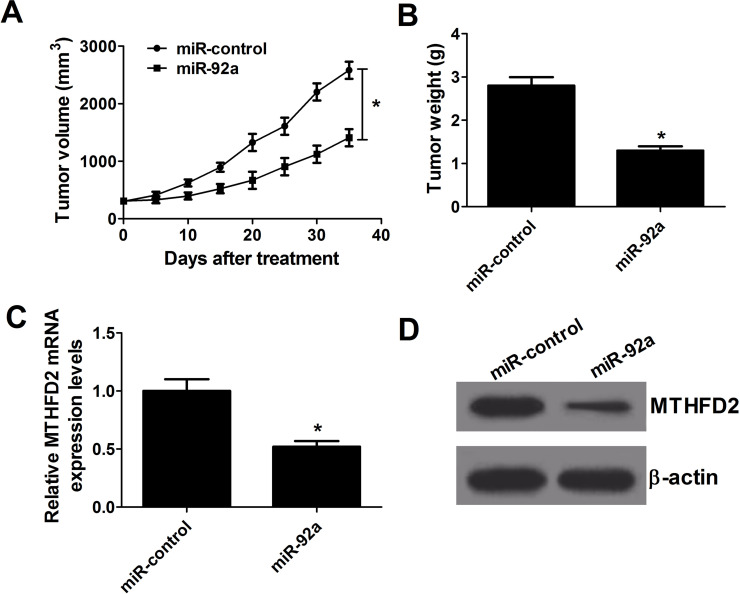
miR-92a Upregulation Inhibits AML Tumor Growth In Vivo
To validate the biological function of miR-92a in the tumor growth of an AML mouse model, HL-60 cells with miR-92a or miR-control transfection were injected subcutaneously into nude mice for tumor formation. The tumor volume was examined every 5 days. After 35 days, mice were sacrificed, and tumor weights were measured. A remarkable inhibition was observed in the growth of miR-92a-derived xenograft tumor compared with the control group (Fig. 6A). Likewise, mice implanted with miR-92a-transfected HL-60 cells produced tumors that weighed less compared to those in the control group (Fig. 6B). Furthermore, miR-92a overexpression led to an apparent decrease in MTHFD2 mRNA and protein expression in the excised tumors (Fig. 6C and D). Therefore, overexpression of miR-92a suppressed AML tumorigenesis in vivo.
Figure 6.
miR-92a overexpression reduced the growth of AML cells in vivo. A xenograft model was constructed by subcutaneous injection of HL-60 cells transfected with miR-92a or miR-control. (A) Tumor size was measured with a caliper every 5 days. (B) Tumors were isolated, and weights were assessed after mice were sacrificed at day 35. (C, D) The mRNA and protein level of MTHFD2 in removed tumors. *p < 0.05 versus miR-control.
DISCUSSION
In the present study, we found that miR-92a was evidently downregulated and MTHFD2 was remarkably upregulated in AML cells. Subsequent dual luciferase reporter analysis confirmed that MTHFD2 is a direct target of miR-92a. MTHFD2 knockdown or miR-92a overexpression inhibited proliferation and induced apoptosis of AML cells. Moreover, restoration of MTHFD2 expression reversed proliferation inhibition and apoptosis induction of AML cells triggered by miR-92a. Furthermore, it was revealed that miR-92a overexpression dramatically reduced AML tumor growth in vivo. All these findings demonstrated that miR-92a suppressed AML tumorigenesis in vitro and in vivo by inhibiting proliferation and inducing apoptosis through directly regulating MTHFD2 expression.
Increased evidence has revealed that miR-92a is abnormally expressed and closely associated with the pathological process in various cancers. For instance, miR-92a was acknowledged to be upregulated in cervical cancer16, colorectal cancer19, and gastric cancer20. Previous studies suggested that locked nucleic acid-induced miR-92a deficiency could prevent proliferation and promote apoptosis in colorectal cancer21 and acute megakaryoblastic leukemia22. Moreover, cell viability was inhibited and apoptosis was induced by miR-92a blockage in gastric cancer23. However, downregulation of miR-92a expression was observed in other cancers, such as breast cancer24 and melanoma25. Additionally, miR-92a inhibited adhesion, invasion, and proliferation of ovarian cancer cells by inhibiting integrin α5 expression26, which is consistent with our results that miR-92a was downregulated in AML cells and inhibits proliferation and induces apoptosis of AML cells. The previous findings, together with our results, demonstrate that miR-92a may function as an oncogene or tumor suppressor in different cancers.
It is well known that miRNAs are involved in tumorigenesis through modulating the expression of their target genes. Previous studies revealed that miR-92a functioned as an oncogene or tumor suppressor in various cancers through suppressing its targets, including PHLPP227, p5728, BCL2L1129, FBXW716, MYCBP230, integrin α526, and CDH131. In the present study, TargetScan prediction demonstrated that MTHFD2 was a target of miR-92a. Subsequently, a luciferase reporter assay confirmed that miR-92a directly targeted the MTHFD2 3′-UTR in HL-60 and THP-1 cells. Finally, Western blot analysis indicated that miR-92a could inhibit MTHFD2 expression in the AML cell lines HL-60 and THP-1. All these results suggest that MTHFD2 is a direct target gene of miR-92a in AML cells.
MTHFD2, a mitochondrial enzyme, was illustrated to be decidedly upregulated in many cancers7. A previous study revealed that MTHFD2 promotes rapid cell growth in mice during embryonic development32. MTFHD2 had the potential to promote proliferation independent of its dehydrogenase activity and colocalized to DNA synthesis sites in the nucleus33. Additionally, it was illuminated that elevated expression of MTHFD2 was associated with the increased risk of bladder cancer34. The present study showed that MTHFD2 was upregulated, and MTHFD2 knockdown inhibited proliferation and induced apoptosis of AML cells. Moreover, forced expression of MTHFD2 abated miR-92a induced an antiproliferative and a pro-apoptotic effect in AML cells. In agreement with our findings, a previous study revealed that MTHFD2 was a functional target of miR-9, and MTHFD2 knockdown mimicked the antiproliferative and proapoptotic activity of miR-9 overexpression in breast cancer cells9.
In summary, miR-92a overexpression suppressed proliferation and induced apoptosis of AML cells through targeting MTHFD2. Our findings propose that miR-92a could be a promising therapeutic target for patients with AML. Because miRNAs can be regulated, one miRNA can control multiple targets, and one target can be controlled by many miRNAs, the regulatory network involving miRNAs is extremely complicated. Therefore, further efforts are still needed to explore the deep regulatory mechanism of miRNAs in AML.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Kumar CC. Genetic abnormalities and challenges in the treatment of acute myeloid leukemia. Genes Cancer 2011;2:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Estey E, Döhner H. Acute myeloid leukaemia. Lancet 2006;368:1894–907. [DOI] [PubMed] [Google Scholar]
- 3. Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–52. [DOI] [PubMed] [Google Scholar]
- 4. Kazuo M, Sumio S. Oligo-capping: A simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 1994;138:171–4. [DOI] [PubMed] [Google Scholar]
- 5. Xu X, Qiao M, Zhang Y, Jiang Y, Wei P, Yao J, Gu B, Wang Y, Lu J, Wang Z. Quantitative proteomics study of breast cancer cell lines isolated from a single patient: Discovery of TIMM17A as a marker for breast cancer. Proteomics 2010;10:1374–90. [DOI] [PubMed] [Google Scholar]
- 6. Pikman Y, Puissant A, Alexe G, Furman A, Frumm S, Ross L, Chen L, Fenouille N, Bassil CF, Lewis CA. Targeting MTHFD2 in acute myeloid leukemia. Blood 2015;126:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lehtinen L, Ketola K, Mäkelä R, Mpindi JP, Viitala M, Kallioniemi O, Iljin K. High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget 2013;4:48–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Selcuklu SD, Donoghue MT, Rehmet K, de Souza Gomes M, Fort A, Kovvuru P, Muniyappa MK, Kerin MJ, Enright AJ, Spillane C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012;287:29516–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 11. Croce C. Introduction to the role of microRNAs in cancer diagnosis, prognosis, and treatment. Cancer J. 2012;18:213–4. [DOI] [PubMed] [Google Scholar]
- 12. Huang X, Schwind S, Santhanam R, Eisfeld AK, Chiang C-l, Lankenau M, Yu B, Hoellerbauer P, Jin Y, Tarighat SS. Targeting the RAS/MAPK pathway with miR-181a in acute myeloid leukemia. Oncotarget 2016;7:59273–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xing CY, Hu XQ, Xie FY, Yu ZJ, Li HY, Wu JB, Tang LY, Gao SM. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589:1981–7. [DOI] [PubMed] [Google Scholar]
- 14. Gerloff D, Grundler R, Wurm A, Bräuer-Hartmann D, Katzerke C, Hartmann J, Madan V, Müller-Tidow C, Duyster J, Tenen D. NF-κB/STAT5/miR-155 network targets PU. 1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia 2015;29:535–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou T, Zhang G, Liu Z, Xia S, Tian H. Overexpression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Int J Colorectal Dis. 2013;28:19–24. [DOI] [PubMed] [Google Scholar]
- 16. Zhou C, Shen L, Mao L, Wang B, Li Y, Yu H. miR-92a is upregulated in cervical cancer and promotes cell proliferation and invasion by targeting FBXW7. Biochem Biophys Res Commun. 2015;458:63–9. [DOI] [PubMed] [Google Scholar]
- 17. Guo J, Mei Y, Li K, Huang X, Yang H. Downregulation of miR-17-92a cluster promotes autophagy induction in response to celastrol treatment in prostate cancer cells. Biochem Biophys Res Commun. 2016;478:804–10. [DOI] [PubMed] [Google Scholar]
- 18. Tanaka M, Oikawa K, Takanashi M, Kudo M, Ohyashiki J, Ohyashiki K, Kuroda M. Downregulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PloS One 2009;4:e5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ke TW, Wei PL, Yeh KT, Chen WTL, Cheng YW. MiR-92a promotes cell metastasis of colorectal cancer through PTEN-mediated PI3K/AKT pathway. Ann Surg Oncol. 2015;22:2649–55. [DOI] [PubMed] [Google Scholar]
- 20. Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer 2011;47:784–91. [DOI] [PubMed] [Google Scholar]
- 21. Ahmadi S, Sharifi M, Salehi R. Locked nucleic acid inhibits miR-92a-3p in human colorectal cancer, induces apoptosis and inhibits cell proliferation. Cancer Gene Ther. 2016;23:199–205. [DOI] [PubMed] [Google Scholar]
- 22. Sharifi M, Salehi R. Blockage of miR-92a-3p with locked nucleic acid induces apoptosis and prevents cell proliferation in human acute megakaryoblastic leukemia. Cancer Gene Ther. 2016;23:29–35. [DOI] [PubMed] [Google Scholar]
- 23. Zhou H, Zhang Y, Wei X, Zheng A, Dai D, Zhang H, Cao H. Downregulation of miR-92a inhibits cell proliferation and invasion but induces apoptosis of gastric cancer by regulating FBXW7. Int J Clin Exp Pathol. 2016;9:4283–91. [Google Scholar]
- 24. Nilsson S, Möller C, Jirström K, Lee A, Busch S, Lamb R, Landberg G. Downregulation of miR-92a is associated with aggressive breast cancer features and increased tumour macrophage infiltration. PLoS One 2012;7:e36051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA 2006;103:9136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohyagi-Hara C, Sawada K, Kamiura S, Tomita Y, Isobe A, Hashimoto K, Kinose Y, Mabuchi S, Hisamatsu T, Takahashi T. miR-92a inhibits peritoneal dissemination of ovarian cancer cells by inhibiting integrin α5 expression. Am J Pathol. 2013;182:1876–89. [DOI] [PubMed] [Google Scholar]
- 27. Rao E, Jiang C, Ji M, Huang X, Iqbal J, Lenz G, Wright G, Staudt L, Zhao Y, McKeithan T. The miRNA-17∼ 92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia 2012;26:1064–72. [DOI] [PubMed] [Google Scholar]
- 28. Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niu H, Wang K, Zhang A, Yang S, Song Z, Wang W, Qian C, Li X, Zhu Y, Wang Y. [Corrigendum] miR-92a is a critical regulator of the apoptosis pathway in glioblastoma with inverse expression of BCL2L11. Oncol Rep. 2014;32:1319. [DOI] [PubMed] [Google Scholar]
- 30. Venza M, Visalli M, Beninati C, Benfatto S, Teti D, Venza I. miR-92a-3p and MYCBP2 are involved in MS-275-induced and c-myc-mediated TRAIL-sensitivity in melanoma cells. Int Immunopharmacol. 2016;40:235–43. [DOI] [PubMed] [Google Scholar]
- 31. Chen Zl, Zhao Xh, Wang JW, Li BZ, Wang Z, Sun J, Tan FW, Ding DP, Xu XH, Zhou F. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J Biol Chem. 2011;286:10725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Di Pietro E, Wang XL, MacKenzie RE. The expression of mitochondrial methylenetetrahydrofolate dehydrogenase-cyclohydrolase supports a role in rapid cell growth. Biochim Biophys Acta 2004;1674:78–84. [DOI] [PubMed] [Google Scholar]
- 33. Sheppard NG, Jarl L, Mahadessian D, Strittmatter L, Schmidt A, Madhusudan N, Tegnér J, Lundberg EK, Asplund A, Jain M. The folate-coupled enzyme MTHFD2 is a nuclear protein and promotes cell proliferation. Sci Rep. 2015;5:15029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andrew AS, Gui J, Sanderson AC, Mason RA, Morlock EV, Schned AR, Kelsey KT, Marsit CJ, Moore JH, Karagas MR. Bladder cancer SNP panel predicts susceptibility and survival. Hum Genet. 2009;125:527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]